Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that causes chronic lung infections in cystic fibrosis (CF) patients. One characteristic of P. aeruginosa CF isolates is the overproduction of the exopolysaccharide alginate, controlled by AlgR. Transcriptional profiling analyses comparing mucoid P. aeruginosa strains to their isogenic algR deletion strains showed that the transcription of cyanide-synthesizing genes (hcnAB) was ∼3-fold lower in the algR mutants. S1 nuclease protection assays corroborated these findings, indicating that AlgR activates hcnA transcription in mucoid P. aeruginosa. Quantification of hydrogen cyanide (HCN) production from laboratory isolates revealed that mucoid laboratory strains made sevenfold more HCN than their nonmucoid parental strains. In addition, comparison of laboratory and clinically derived nonmucoid strains revealed that HCN was fivefold higher in the nonmucoid CF isolates. Moreover, the average amount of cyanide produced by mucoid clinical isolates was 4.7 ± 0.85 μmol of HCN/mg of protein versus 2.4 ± 0.40 μmol of HCN/mg of protein for nonmucoid strains from a survey conducted with 41 P. aeruginosa CF isolates from 24 patients. Our data indicate that (i) mucoid P. aeruginosa regardless of their origin (laboratory or clinically derived) produce more cyanide than their nonmucoid counterparts, (ii) AlgR regulates HCN production in P. aeruginosa, and (iii) P. aeruginosa CF isolates are more hypercyanogenic than nonmucoid laboratory strains. Taken together, cyanide production may be a relevant virulence factor in CF lung disease, the production of which is regulated, in part, by AlgR.
Pseudomonas aeruginosa is an opportunistic pulmonary pathogen of patients with cystic fibrosis (CF), where it is the major cause of morbidity and mortality. P. aeruginosa is able to persist and exacerbate damage in the lungs that ultimately results in respiratory failure. One unique feature of P. aeruginosa CF isolates is overproduction of the exopolysaccharide alginate that phenotypically results in a mucoid colony morphology (23). The first committed step for alginate production is transcriptional activation of the algD gene (12), an event that requires the AlgR regulator and the alternative sigma factor AlgU (AlgT) (11, 14, 26, 34, 35, 38, 39, 46). Mucoidy is also associated with the chronic phase of CF airway disease where the bacteria acquire increased resistance to various antibiotics and phagocytic cells. Furthermore, increasing evidence suggests that P. aeruginosa may be in a microaerophilic or anaerobic microenvironment trapped within biofilms in the thick mucus lining the airways of CF patients (48, 55, 59). Under such conditions, the organisms are able to produce alginate and maintain mucoidy (25, 48, 56, 59).
It has long been recognized that P. aeruginosa generates poisonous cyanide as a secondary metabolite (7). Hydrogen cyanide (HCN) is produced from glycine (7, 54) in a poorly understood oxidative reaction catalyzed by HCN synthase (8, 9, 53) whose expression requires the las/rhl tandem of the intercellular signaling process known as quorum sensing (43, 44). HCN is not produced when the organism is grown under strict anaerobic conditions when supplied nitrate as a terminal electron acceptor (7) but rather occurs optimally at low oxygen tensions (∼5%) during the transition from exponential to stationary phase when bacteria are at high cell densities and fully capable of quorum sensing (6, 10, 43, 44). An early report (22) describing the detection of HCN in P. aeruginosa-infected wounds from burn patients gave cause to believe that the cyanogenic properties of the organism may be a contributing factor in its pathogenicity. Further support for this hypothesis stems from recent studies demonstrating that a mutant defective in hcnC had a strongly reduced ability to kill the nematode Caenorhabditis elegans in an experimental infection model (18). In addition, recent transcriptional profiling analyses revealed that mucoid P. aeruginosa actively transcribes hcnA, encoding HCN synthase (16), further suggesting that cyanide production may be an important virulence factor.
The present study extends the recent work (29) examining the molecular basis underlying the ability of AlgR to control virulence in P. aeruginosa. Through the use of Affymetrix GeneChip technology, we previously identified many potential genes under AlgR control, one of which was hcnA (30). Here, we show that (i) mucoid P. aeruginosa strains produce copious amounts of HCN, (ii) AlgR regulates this process, and (iii) HCN production in clinical CF isolates is significantly elevated over that of laboratory strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The P. aeruginosa strains used in this study are listed in Table 1. P. aeruginosa strains PAO6857 (mucB::Tcr), PAO568 (mucA2), PAR568 (mucA2 ΔalgR), and PAR6857 (mucB::Tcr ΔalgR) were grown in modified Pseudomonas isolation media (20 g of peptone/liter, 7 mM MgCl2, 59 mM K2SO4, 2% glycerol, pH 7.0) under microaerophilic conditions for S1 nuclease protection assays. Overnight cultures were diluted 1:100 in 500-ml aeration flasks capped with rubber stoppers and then placed at 37°C with slow shaking at 100 rpm for microaerophilic growth conditions. These cultures were grown to an optical density at 600 nm (OD600) of 0.4. The plasmid used for algR complementation was pCMR7 (algR+) (38). Routine overnight cultures were grown in Luria-Bertani broth at 37°C in a rotary shaker incubator. Carbenicillin (300 μg/ml) was supplemented as needed for plasmid maintenance and genetic manipulations.
TABLE 1.
Strains used in this study
| P. aeruginosa | Genotype | Phenotypea | Reference or source |
|---|---|---|---|
| PAO1 | Wild type | NM | 27 |
| PAO381 | leu | NM | 17 |
| PDO300 | mucA2 | M | 32 |
| PDR300 | mucA2 ΔalgR | NM | This study |
| PAO6857 | mucB::Tcr | M | 47 |
| PAR6857 | mucB::Tcr ΔalgR | NM | This study |
| PA0568 | mucA2 | M | 17 |
| PAR568 | mucA2 ΔalgR | NM | This study |
| FRD1 | mucA | M | 21 |
| FRD1R | mucA algR::Smr | NM | 31 |
| TUMC92 | Clinical isolate | NM | This study |
| TUMC-92R | ΔalgR | NM | This study |
| TUMC197 | Clinical isolate | NM | This study |
| TUMC-197R | ΔalgR | NM | This study |
| Escherichia coli DH-5α | Invitrogen |
M, mucoid; NM, nonmucoid.
DNA manipulations.
The algR gene was deleted from strains as previously described (30). Briefly, an algR deletion plasmid (pRKO442 [ΔalgR]) was introduced into clinical and laboratory P. aeruginosa strains by triparental conjugation (28), and double recombinants were obtained by selection of carbenicillin-sensitive and sucrose-resistant colonies (15). Deletion of algR was confirmed by PCR with oligonucleotide primers ArgHF (5′-ATATATGAGCTCGGACCTGTCCGACCTGTTCC-3′) and HemCR (5′-ATATATGAGCTCGGCTGGCGTAGGTGTTCGAG-3′) and Southern blot (data not shown) and Western blot analysis with anti-AlgR (see Fig. 2A and 4B) (13).
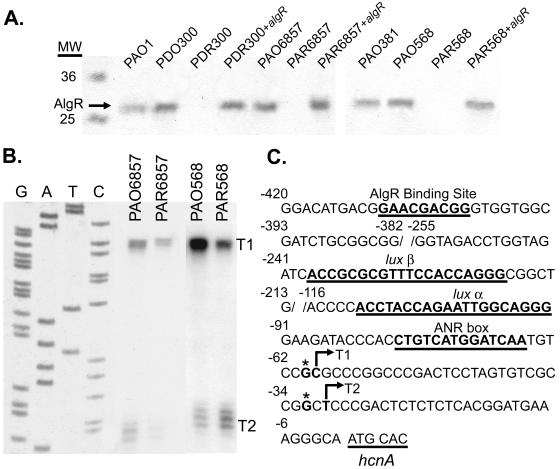
FIG. 2.
AlgR controls the expression of hcnA in mucoid P. aeruginosa. S1 nuclease protection assay of the hcnA promoter region. A. Western blot analysis of PAO1, PDO300 (mucA2), PDR300 (mucA2 ΔalgR), PAO6857 (mucB::Tcr), and PAR6857 (mucB::Tcr ΔalgR) using anti-AlgR 3H-9 confirmed the deletion of the 29-kDa transcriptional regulator, AlgR, in strains PAR6857, PDR300, and PAR568. MW, molecular weight. B. Mapped transcriptional start sites T1 and T2 of the mucoid strains PAO6857 and PAO568 and their algR deletion strains PAR6857 and PAR568. C. Schematic of the hcnA promoter sequence. T1 and T2, mapped transcriptional start sites (indicated with arrows); ANR box, ANR binding site; lux α and lux β, LasR and RhlR binding sites.
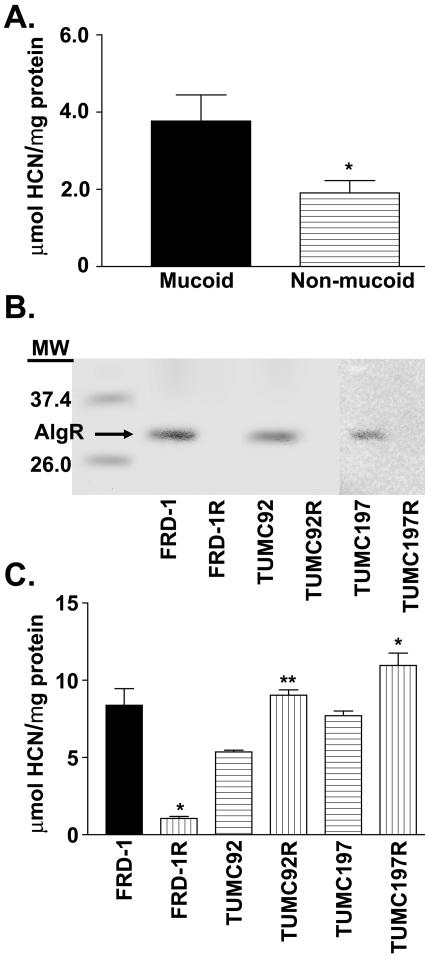
FIG. 4.
Mucoid clinical CF isolates produce more HCN than nonmucoid isolates. A. Mean quantification of HCN produced in 4 h from 20 mucoid and 21 nonmucoid CF clinical isolates. Mucoid isolates produce approximately twofold more HCN than nonmucoid isolates. *, P < 0.05, as determined by unpaired t test. B. Western blot analysis of clinical isolates FRD1, TUMC92, and TUMC197 and their algR deletion strains FRD1R (algR::Smr), TUMC-92R (ΔalgR), and TUMC-197R (ΔalgR) using anti-AlgR. C. Quantification of HCN produced in 4 h from clinical isolates TUMC197 and TUMC92 and their algR deletion strains, TUMC-197R and TUMC-92R, and from the mucoid CF isolate FRD1 and its nonmucoid algR strain FRD1R. Solid bar, mucoid strain; horizontally lined bars, nonmucoid strains; vertically lined bars, algR deletion strains. *, P < 0.05, as determined by unpaired t test. **, P < 0.01, as determined by unpaired t test.
Western blot analysis of AlgR.
P. aeruginosa strains were grown aerobically in Luria-Bertani broth at 37°C overnight. The bacteria were collected by centrifugation and resuspended in 50 mM Tris-HCl (pH 8.0)-150 mM NaCl and lysed by sonication. Total protein concentrations were quantified by the Bradford protein assay (Bio-Rad). Cell extracts (30 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 5 to 20% polyacrylamide gradient gels and transferred to a polyvinylidene difluoride membrane (GE Osmonics). The membranes were probed using a 1:2,000 dilution of anti-AlgR mouse monoclonal antibody (13) followed by a 1:2,000 dilution of horseradish peroxidase-conjugated goat anti-mouse monoclonal antibody and the signal was detected using the Opti-4CN substrate kit (Bio-Rad).
Qualitative gaseous HCN production assay of P. aeruginosa strains.
An initial screen of P. aeruginosa CF isolates was performed by testing qualitatively for gaseous HCN production using a modified method of Castric and Castric (5). Whatman filter paper strips were soaked with 5 mg each of copper(II) ethyl acetoacetate and 4,4′-methylenebis-(N,N-dimethylaniline) dissolved in 2.5 ml of chloroform and allowed to air dry. The strips were placed in the lids of L agar plates streaked with P. aeruginosa CF clinical isolates and incubated inverted overnight at 37°C. Relative HCN production was indicated by a change in the color of the strips from white to blue. As a control, L agar plates with no bacteria and the qualitative assay strip were incubated in the same incubator with CF isolates on different plates at the same time.
Quantitative HCN production assay of P. aeruginosa strains.
Twelve nonmucoid and mucoid clinical isolates from the same patient were quantified for HCN production. In addition, 17 isolates from 12 other patients were randomly selected for HCN production. A significantly modified protocol of Gallagher and Manoil was used to measure HCN (18). Laboratory and clinical P. aeruginosa isolates were grown on 8.5-cm-diameter plates of Pseudomonas isolation agar for 16 to 24 h at 37°C in triplicate and then enclosed without the lid in sealed chambers containing 1 ml of 4 M NaOH. After a 4-h incubation at 30°C, the NaOH was diluted to 0.09 M with H2O. Next, the samples were further diluted in 0.09 M NaOH to bring the cyanide concentration to within the linear detection range (0 to 15 μM potassium cyanide [KCN]). Cyanide levels were quantified by comparison with KCN standards dissolved in 0.09 M NaOH as follows. Samples (210 μl) were mixed with 700 μl of a 1:1 mixture of 0.1 M o-dinitrobenzene in ethylene-glycol monomethyl ether and 0.2 M p-nitrobenzaldehyde in ethylene-glycol monmethyl ether. After a 30-min incubation at 22°C, the OD578 was measured as previously described (18). Total protein was determined for each strain by collecting bacteria grown on an agar plate and resuspending the cells in 5 ml of 0.85% NaCl. After centrifugation (10,500 × g), the cells were lysed and protein precipitated in 5% trichloroacetic acid (37). Protein pellets were resuspended in 1 ml of 50 mM KH2PO4 and the total amount of protein was determined by Bradford assay (Bio-Rad). Cyanide production is expressed as the ratio between the total amount (micromoles) of HCN and the total amount (milligrams) of bacterial protein recovered from cells grown on a petri dish.
S1 nuclease protection assays.
RNA for S1 nuclease protection assays was isolated from PAO6857 (mucB::Tcr), PAR6857 (PAO6857 ΔalgR), PAO568 (mucA2), and PAR568 (PAO568 ΔalgR) grown to stationary phase using CsCl as previously described (35). An S1 nuclease protection assay was performed as previously described using 100 μg of RNA with the following modifications (35). The 519-bp region of the hcnA promoter ranging from −495 to +24 (numbering relative to translational start site) was cloned into M13mp18. Single-stranded phages were isolated and used as the template for the uniformly labeled ([α-32P]dCTP; NEN DuPont) single-stranded DNA probe. The probe was digested using BglI and purified on a 5% polyacrylamide gel. The probe was hybridized to 100 μg of RNA from the above strains and treated with S1 nuclease, and the products were separated on a sequencing gel adjacent to a sequencing ladder generated using the same oligonucleotide, hcnAprimext (5′-GTGTTGACGACGTTCAAGAAGGTGCAT-3′), as the probe.
AlgR gel mobility shift assay.
Binding of AlgR to the hcnA promoter region was examined using native AlgR, purified as previously described (38). A 99-bp DNA fragment containing the hcnA promoter (−495 to −396 in relation to the translational start site) was excised from pCRhcnA (hcnA promoter in pCR2.1) by EcoRI and end-labeled with [γ-32P]ATP (6,000 Ci/mmol; NEN Dupont) using T4 polynucleotide kinase (Invitrogen, Carlsbad, Calif.). The probe was purified by passing through a G-25 Sephadex microspin column (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). Binding reactions were carried out as described previously with some modifications (38). Briefly, the probe was mixed with 100 ng of AlgR containing 25 mM Tris-HCl (pH 8.0), 0.5 mM dithiothreitol, 20 mM KCl, 0.5 mM EDTA, 5% glycerol, 10 μg of salmon sperm DNA, and an additional 0.25 μg of pUC12 per ml as nonspecific competitor DNA. Competition assays were performed by the addition of unlabeled pCRhcnA, to determine the specificity of AlgR. After incubation for 10 min at room temperature, the samples were separated by electrophoresis on a 5% native polyacrylamide gel with Sharp's buffer (6.7 mM Tris-HCl [pH 8.0], 3.3 mM sodium acetate, 1.0 mM EDTA) used as a running buffer for approximately 1.5 h at 30 mA. Subsequently, the gel was dried and bands were visualized by autoradiography.
RESULTS
Cyanide production is elevated in mucoid P. aeruginosa.
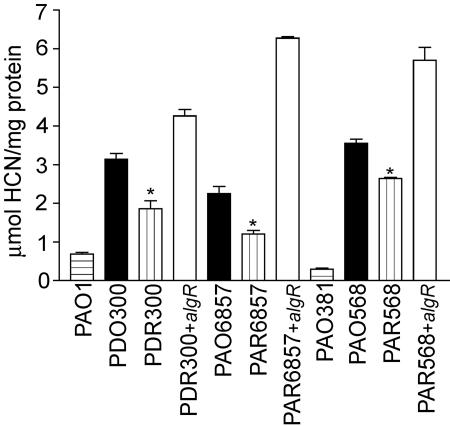
Analysis of the transcriptional profile comparing mucA2 and mucB::Tcr mucoid P. aeruginosa strains to their isogenic algR deletion strains showed that hcnA transcription was ∼3-fold lower in the algR mutant strains (unpublished data). These results are consistent with those of Firoved and Deretic, who reported that hcnA transcription is sixfold greater in the mucA22 mucoid strain PAO578I than in its isogenic algU mutant (16). Since AlgR is required for alginate production (11) and controlled by AlgU (35), we postulated that AlgR may be activating hcnA transcription in mucoid P. aeruginosa bacteria. Therefore, cyanide was measured directly in mucoid and nonmucoid laboratory strains of PAO1 and PAO381. The nonmucoid laboratory strains PAO1 and PAO381 produced an average of 0.49 μmol of cyanide/mg of protein. In contrast, the mucA mucoid derivatives of these strains, PDO300 and PAO568, respectively, each produced approximately seven times the amount of cyanide (Fig. 1). In addition, PAO6857 (mucB::Tcr) produced 3.3-fold more cyanide than PAO1 (Fig. 1). Taken together, these results indicate that mucoid derivatives of P. aeruginosa produce more cyanide than their nonmucoid parental strains.
FIG. 1.
Cyanide production is controlled by AlgR and elevated in laboratory mucoid P. aeruginosa. Quantification of HCN (micromoles per milligram of protein) produced in 4 h from nonmucoid parent strains PAO1 and PAO381 (horizontally lined bars), mucoid derivative strains PAO568, PAO6857, and PDO300 (solid bars), subsequent isogenic algR deletion strains PAR568, PAR6857, and PDR300 (vertically lined bars), and complemented algR deletion strains PDR300+algR, PAR6857+algR, and PAR568+algR (open bars). All of the parent mucoid strains produced elevated levels of HCN compared to their relative algR deletion counterparts. *, P of <0.05 when compared to the parent strain, as determined by unpaired t test.
Mucoid P. aeruginosa requires AlgR for complete hcnA expression and cyanide production.
To determine if AlgR plays a role in a mucA2 derivative of P. aeruginosa, algR was deleted from P. aeruginosa strains PAO568 and PDO300 (Fig. 2A). An S1 nuclease protection assay performed on the hcnA promoter from PAO568 and its algR deletion strain PAR568 revealed that transcription from hcnA promoter T1 was elevated in PAO568 compared to the same promoter in PAR568 (Fig. 2B). HCN production was then quantified in the mucoid mucA2 strains PDO300 and PAO568 and their respective algR deletion strains. This comparison revealed that the mucoid mucA parental strain produced an average of 1.5-fold more cyanide than the algR mutants (Fig. 1). Moreover, overexpression of algR in trans in the algR deletion strains restored cyanide production (Fig. 1).
To determine if AlgR affected hcnA transcription in a mucB mucoid P. aeruginosa strain, algR was deleted in PAO6857 (mucB::Tcr) to generate the strain PAR6857 (mucB::Tcr ΔalgR) (Fig. 2A). An S1 nuclease protection analysis was performed on the hcnA promoter by using total RNA from these strains grown microaerophilically and revealed AlgR activated the T1 and T2 promoters of hcnA (Fig. 2B and C). Moreover HCN determinations revealed that PAO6857 produced 1.9-fold more HCN per milligram of protein than its algR deletion strain PAR6857 (Fig. 1). This result is consistent with the transcriptional profile analysis of the same strain and its algR deletion mutant that indicated hcnA is activated threefold by AlgR. Taken together, these results suggest that AlgR activates cyanide production by ∼2-fold in mucoid laboratory strains of P. aeruginosa, irrespective of the nature of the mutation (mucA, mucB) conferring the mucoid phenotype.
AlgR binds to the hcnA promoter region.
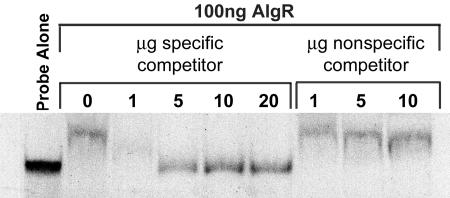
Since hcnA transcription is algR dependent and there is an AlgR binding site within the hcnA promoter (−410 to −402; GAACgACGG, where the lowercase “g ” represents a departure from the reverse and complement of the algD/algC consensus AlgR binding sequence [30]), we tested the ability of purified AlgR to bind the hcnA promoter region in an in vitro gel mobility shift assay. AlgR caused a shift in mobility compared to the probe alone, indicating that AlgR is capable of binding to the hcnA promoter region (Fig. 3). The addition of nonradioactive hcnA (specific competitor) reduced the amount of probe shifted by AlgR in a dose-dependent manner, indicating specificity of AlgR for the hcnA promoter region. Additionally, nonradioactive pUC12 was added as a nonspecific competitor to ensure that competition was not due to nonspecific AlgR DNA binding. These results provide in vitro evidence that AlgR binds specifically to the hcnA promoter DNA.
FIG. 3.
AlgR binds specifically to the hcnA promoter. Competitive gel mobility shift assay of AlgR on the hcnA promoter region. The probe-alone lane contains only labeled hcnA promoter (99 bp, −495 to −396 in relation to the translational start site) from plasmid pCRhcnA. All other lanes contain the same amount of labeled hcnA promoter plus 100 ng of purified AlgR. For the specific competitor, the given micrograms of undigested pCRhcnA was added to the DNA/AlgR mix. For the nonspecific competitor, the given micrograms of undigested pUC12 was added to the DNA/AlgR mix.
Clinical P. aeruginosa CF isolates produce elevated cyanide levels.
To determine the prevalence of cyanide production among P. aeruginosa CF isolates, we performed a qualitative HCN production assay on 167 clinical P. aeruginosa isolates from 103 CF patients. This assay showed that 74% of all isolates examined produced HCN (data not shown). Furthermore, 83% of CF patients from the Tulane University Medical Center were found to carry at least one positive P. aeruginosa HCN-producing strain. To confirm these results, the amount of cyanide produced by 41 separate clinical CF P. aeruginosa isolates from 24 CF patients was quantified. The average amount of cyanide produced by all 41 strains was 3.5 μmol of cyanide/mg of protein, indicating that the majority of CF patients surveyed harbored P. aeruginosa strains that constitutively produced HCN when tested in vitro. When HCN production by both mucoid and nonmucoid isolates from 12 patients was compared, the amount of cyanide detected averaged 4.4 ± 1.1 and 2.5 ± 0.5 μmol of HCN/mg of protein, respectively. We then examined eight mucoid strains from additional patients for whom there were no nonmucoid isolates and found an average amount of 5.2 ± 1.2 μmol of HCN/mg of protein produced. We also examined nine nonmucoid strains from patients that did not carry mucoid isolates and determined that these strains produced an average of 1.36 ± 0.6 μmol of HCN/mg of protein. Overall, the amount of cyanide produced by mucoid P. aeruginosa CF isolates was approximately twice as high as that of nonmucoid CF isolates (Fig. 4A). This is in agreement with the results we obtained with the mucoid laboratory strains of P. aeruginosa.
AlgR activates cyanide production in mucoid CF clinical isolates and represses HCN production in nonmucoid CF isolates.
In order to assess the role of AlgR in clinical CF isolates on HCN production, algR was deleted from the well-characterized mucoid CF isolate FRD1 (Fig. 4B). The amount of cyanide produced by the mucoid CF isolate FRD1 was compared with its isogenic algR mutant which revealed that FRD1 algR produced only 1.05 μmol of HCN/mg of protein, an amount eightfold lower than that for FRD1 (Fig. 4C). Interestingly, among CF clinical isolates, there were nonmucoid isolates that produced copious amounts of HCN (for example, TUMC92, 5.5 μmol of HCN/mg of protein, and TUMC197, 6.25 μmol of HCN/mg of protein) despite their nonmucoid phenotype. Thus, to ascertain the relevance of AlgR in regulating HCN production in nonmucoid clinical CF isolates, isogenic algR mutants were constructed for the nonmucoid hypercyanogenic clinical isolates TUMC92 and TUMC197 (Fig. 4B) and cyanide production was measured. We observed a threefold increase in cyanide production in the clinical algR mutant TUMC-197R and a 1.8-fold increase in cyanide production from the algR clinical isolate TUMC-92R (Fig. 4C). These results are consistent with our observation that AlgR repressed HCN production in the wild-type nonmucoid P. aeruginosa strain PAO1 (30). These results provide strong evidence that AlgR activates cyanide production in mucoid CF clinical isolates and represses its production in nonmucoid isolates.
DISCUSSION
The effects of HCN have been examined on many cell types (2, 24, 41). In these studies, HCN exposure resulted in neuronal necrosis (41) and inhibition of metalloenzymes including cytochrome c oxidase (49). One report of KCN exposure to the immortalized epithelial lung A549 cells resulted in double-stranded DNA breaks below 0.5 Mbp, indicating that endogenous nuclease activity was induced in a dose-dependent manner after KCN exposures (50). These data suggest that A549 cells exposed to >1 mM KCN may undergo necrosis. Cell death via necrosis results in increased inflammation and infiltration of polymorphonuclear leukocytes to the site of infection. Excess inflammation due to migration of polymorphonuclear leukocytes and lung necrosis are pathological hallmarks of CF airway infection (1).
In this study, we quantified the amount of HCN produced by mucoid and nonmucoid laboratory and CF isolates of P. aeruginosa and showed that AlgR, a transcriptional regulator of alginate biosynthesis (11, 38-40) and twitching motility (51, 52), plays a central role in controlling HCN production. Although HCN-producing P. aeruginosa strains have been isolated from burn patients (22), the only evidence that HCN may be a virulence factor in P. aeruginosa stems from an experimental infection model utilizing C. elegans (18). Since at physiological pH and ambient temperatures and above, cyanide exists predominantly as volatile HCN gas, it is possible to envision that HCN generated by P. aeruginosa CF strains could diffuse into the environment through exhaled breath with no deleterious affects. However, a recent report (19) describing the detection of cyanide in CF patient sputum could be interpreted as indicating that cyanide, in the form of either gaseous HCN or soluble cyanide (CN−), remains in the thick mucus layer harboring P. aeruginosa biofilms, thus being potentially available for diffusion into surrounding lung epithelial cells.
The conditions in P. aeruginosa-infected lung tissue appear to be optimal for cyanide production given that the thick CF mucus (48, 55, 59) provides an ideal environment for the growth of cells to high cell densities as a biofilm under microaerophilic conditions, processes controlled in part by quorum sensing (43, 44). Interestingly, there are two reported promoters for the hcnA gene, T1, controlled by quorum-sensing regulators alone, and T2, which appears to rely on a synergistic action of LasR, RhlR, and ANR (Fig. 2C) (43). Five regulatory proteins have been identified for the hcnA promoter: GacA (45), ANR (45, 62), LasR, RhlR (43), and RsmA (42). The global regulator GacA positively controls HCN synthesis as well as other secondary metabolites and exoenzymes (45). P. aeruginosa gacA or anr mutants produce very little HCN (45, 62). One positive regulator of anaerobic respiration, ANR, is required for anaerobic growth of P. aeruginosa (58). LasR and RhlR are quorum-sensing regulators required for transcription of the hcnA promoter (43). RsmA (regulator of secondary metabolites) functions as a pleiotropic posttranscriptional regulator of HCN synthesis directly and also indirectly by negatively regulating the amounts of quorum sensing N-acylhomoserine lactones (42). Our data provide strong evidence that AlgR also regulates hcnA transcription by affecting both T1 and T2 promoters, indicating that AlgR and LasR and/or RhlR and ANR coordinately regulate these promoters. Moreover, we have discovered through transcriptional profiling experiments that there are other genes (hemN, PA1557, and arcD) of the AlgR regulon that are coregulated by AlgR and ANR (30). Further studies are needed to elucidate the mechanism by which these transcriptional regulators may potentially interact.
Under anaerobic growth conditions, P. aeruginosa is able to produce alginate and maintain its production (25, 48, 56, 59). Our survey of both laboratory and CF isolates demonstrated that mucoid P. aeruginosa produced more HCN than nonmucoid strains. These findings are consistent with the fact that environmental conditions favoring cyanogenesis by P. aeruginosa also favor alginate production (25, 48, 55, 59). Alginate is considered to be a major virulence factor in the CF lung (23) with the first committed step of biosynthesis being the transcription of algD, which is activated in mucoid P. aeruginosa (12). Transcription of algD requires AlgR as well as the alternative sigma factor AlgU (AlgT) (11, 14, 26, 34, 35, 38, 39, 46). AlgR is currently known as a transcriptional activator of the algD (11, 38-40) and algC (61) promoters and is required for type IV pilus function (51, 52). The switch from the nonmucoid to the mucoid phenotype in P. aeruginosa is known to involve mutations in mucA or mucB, resulting in activation of AlgU (AlgT) (20, 33, 34, 36, 57) and subsequently AlgR (35).
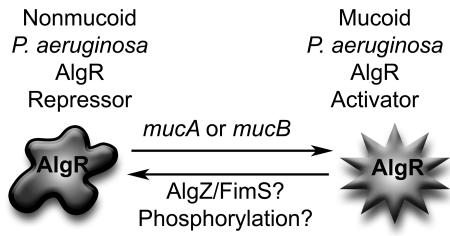
Recently, it was shown that AlgR is capable of acting as a repressor of transcription on the hcnA promoter and HCN production in the nonmucoid strain PAO1 (30). Results from our present study indicate that AlgR is an activator of hcnA transcription and HCN production in both mucoid laboratory and CF clinical isolates. Taken together, these results strongly suggest that AlgR is capable of switching from a repressor in the nonmucoid phenotype to an activator in mucoid P. aeruginosa (Fig. 5). This interpretation is consistent with results demonstrating that the switch from nonmucoid to mucoid phenotypes involving mutations in mucA or mucB results in activation of AlgU (AlgT) (20, 33, 34, 36, 57) and also AlgR (35). The resultant activation of AlgR may be due to increased levels or posttranslational modifications; however, our data do not discern between these two possible mechanisms for AlgR activation of cyanide production. There is evidence that phosphorylation of AlgR is required for twitching motility (52), yet the kinase for AlgR has not been identified. The protein involved in the ability of AlgR to switch from a repressor to an activator of twitching motility may be AlgZ/FimS, as an insertional inactivation of algZ/fimS results in the loss of twitching motility (50). We have preliminary evidence indicating that AlgZ/FimS is also involved in activation of AlgR in mucoid P. aeruginosa, consistent with the findings on AlgZ/FimS in regards to the twitching motility phenotype.
FIG. 5.
Schematic representing possible mechanisms for AlgR activation. Transcriptional profiling experiments have shown that AlgR is capable of repressing transcription of hcnA and other genes (30). We have shown that AlgR activates hcnA transcription and HCN production in mucoid P. aeruginosa. Mutations in mucA (4, 34), mucB (33), or mucD (3) result in release of the alternative sigma factor AlgU, resulting in increased transcription of algR (35). AlgZ/FimS has been shown to be a repressor of alginate production (60) and an activator of twitching motility (51, 52). However phosphorylation does not play a role in AlgR activation of the alginate system (31) but is necessary for activation of twitching motility (52). These results imply that there may be more than one way to activate AlgR in P. aeruginosa. The hcnA promoter is the only known example where AlgR acts as a repressor in the nonmucoid state and as an activator in the mucoid phenotype. Rounded star, repressor AlgR; pointed star, activated AlgR.
Because cyanide could have extremely potent detrimental affects on respiratory epithelial cells, novel strategies for the future treatment of CF airway disease may involve inhibitors of both the quorum sensing and AlgR regulatory cascades, both of which are required for cyanide biosynthesis in P. aeruginosa.
Acknowledgments
This work was supported by LEQSF (1999-02) RD-A-42 and HEF (2000-05)-06 grants from the State of Louisiana Board of Regents and by National Institutes of Health (NIH) grant RO1-AI050812-01A2 (to M.J.S.), National Science Foundation grant MCB 012680 (to D.A.K.), NIH grant R01-40541, and the Cystic Fibrosis Foundation (D.J.H.). The cost of the P. aeruginosa GeneChips was defrayed in part by subsidy grant no. 024 from Cystic Fibrosis Foundation Therapeutics, Inc., to M.J.S.
Microarray equipment and technical support were supplied by the GeneChip Bioinformatics core at the Louisiana State University Health Sciences Center.
REFERENCES
- 1.Boat, T. F., M. J. Welsh, and A. L. Beaudet. 1989. Cystic fibrosis, p. 2649-2680. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic basis of inherited disease, 6th ed., vol. II. McGraw-Hill, New York, N.Y. [Google Scholar]
- 2.Borron, S. W., and F. J. Baud. 1996. Acute cyanide poisoning: clinical spectrum, diagnosis and treatment. Arch. Toxicol. Ind. Hyg. 47:307-322. [PubMed] [Google Scholar]
- 3.Boucher, J. C., J. M. Martinez-Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, J. C., H. Yu, M. H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castric, K. F., and P. A. Castric. 1983. Method for detection of cyanogenic bacteria. Appl. Environ. Microbiol. 45:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castric, P. A. 1983. Hydrogen cyanide production by Pseudomonas aeruginosa at reduced oxygen levels. Can. J. Microbiol. 29:1344-1349. [DOI] [PubMed] [Google Scholar]
- 7.Castric, P. A. 1975. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21:613-618. [DOI] [PubMed] [Google Scholar]
- 8.Castric, P. A. 1994. Influence of oxygen on the Pseudomonas aeruginosa hydrogen cyanide synthase. Curr. Microbiol. 29:19-21. [Google Scholar]
- 9.Castric, P. A. 1981. The metabolism of hydrogen cyanide by bacteria, p. 233-261. In B. Vennesland, E. E. Conn, C. J. Knowles, J. Westley, and F. Wissing (ed.), Cyanide in biology. Academic Press Ltd., London, United Kingdom.
- 10.Castric, P. A., R. F. Ebert, and K. F. Castric. 1979. The relationship between growth phase and cyanogenesis in Pseudomonas aeruginosa. Curr. Microbiol. 2:287-292. [Google Scholar]
- 11.Deretic, V., R. Dikshit, W. M. Konyecsni, A. M. Chakrabarty, and T. K. Misra. 1989. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J. Bacteriol. 171:1278-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic, V., J. F. Gill, and A. M. Chakrabarty. 1987. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J. Bacteriol. 169:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deretic, V., N. S. Hibler, and S. C. Holt. 1992. Immunocytochemical analysis of AlgP (HP1), a histonelike element participating in control of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 174:824-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeVries, C. A., and D. E. Ohman. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnesberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firoved, A. M., and V. Deretic. 2003. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J. Bacteriol. 185:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fyfe, J. A. M., and J. R. W. Govan. 1980. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J. Gen. Microbiol. 119:443-450. [DOI] [PubMed] [Google Scholar]
- 18.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 Kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallagher, L. A., C. Manoil, and J. L. Burns. 2003. Presented at Pseudomonas 2003, Quebec City, Canada.
- 20.Goldberg, J. B., W. L. Gorman, J. Flynn, and D. E. Ohman. 1993. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J. Bacteriol. 175:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg, J. B., and D. E. Ohman. 1984. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J. Bacteriol. 158:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldfarb, W. B., and H. Margraf. 1967. Cyanide production by Pseudomonas aeruginosa. Ann. Surg. 165:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govan, J. R. W., and V. Deretic. 1995. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greer, J. J., and E. Jo. 1995. Effects of cyanide on neural mechanism controlling breathing in neonatal rat in vivo. Neurotoxicology 16:211-215. [PubMed] [Google Scholar]
- 25.Hassett, D. J. 1996. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J. Bacteriol. 178:7322-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hershberger, C. D., R. W. Ye, M. R. Parsek, Z.-D. Xie, and A. M. Chakrabarty. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative σ factor (σE). Proc. Natl. Acad. Sci. USA 92:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holloway, B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572-581. [DOI] [PubMed] [Google Scholar]
- 28.Konyecsni, W. M., and V. Deretic. 1988. Broad-host-range plasmid and M13 bacteriophage-derived vectors for promoter analysis in Escherichia coli and Pseudomonas aeruginosa. Gene 74:375-386. [DOI] [PubMed] [Google Scholar]
- 29.Lizewski, S. E., D. S. Lundberg, and M. J. Schurr. 2002. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 70:6083-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lizewski, S. E., J. R. Schurr, D. W. Jackson, A. Frisk, A. J. Carterson, and M. J. Schurr. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 186:5672-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, S., U. Selvaraj, D. E. Ohman, R. Quarless, D. J. Hassett, and D. J. Wozniak. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malhotra, S., L. A. Silo-Suh, K. Mathee, and D. E. Ohman. 2000. Proteome analysis of the effect of mucoid conversion on global protein expression in Pseudomonas aeruginosa strain PAO1 shows induction of the disulfide bond isomerase, DsbA. J. Bacteriol. 182:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, D. W., M. J. Schurr, M. H. Mudd, and V. Deretic. 1993. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol. Microbiol. 9:497-506. [DOI] [PubMed] [Google Scholar]
- 34.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. W. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, D. W., M. J. Schurr, H. Yu, and V. Deretic. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to stress response. J. Bacteriol. 176:6688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathee, K., C. J. McPherson, and D. E. Ohman. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J. Bacteriol. 179:3711-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.May, T. B., and A. M. Chakrabarty. 1994. Isolation and assay of Pseudomonas aeruginosa alginate. Methods Enzymol. 235:295-304. [DOI] [PubMed] [Google Scholar]
- 38.Mohr, C. D., N. S. Hibler, and V. Deretic. 1991. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J. Bacteriol. 173:5136-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohr, C. D., J. H. J. Leveau, D. P. Krieg, N. S. Hibler, and V. Deretic. 1992. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 174:6624-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohr, C. D., D. W. Martin, W. M. Konyecsni, J. R. Govan, S. Lory, and V. Deretic. 1990. Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 172:6576-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niquet, J., R. A. Baldwin, S. G. Allen, D. G. Fujikawa, and C. G. Wasterlain. 2003. Hypoxic neuronal necrosis: protein synthesis-independent activation of a cell death program. Proc. Natl. Acad. Sci. USA 100:2825-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pessi, G., and D. Haas. 2001. Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200:73-78. [DOI] [PubMed] [Google Scholar]
- 43.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 46.Schurr, M. J., D. W. Martin, M. H. Mudd, N. S. Hibler, J. C. Boucher, and V. Deretic. 1993. The algD promoter: regulation of alginate production by Pseudomonas aeruginosa in cystic fibrosis. Cell. Mol. Biol. Res. 39:371-376. [PubMed] [Google Scholar]
- 47.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the σE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 49.Solomonson, L. P. 1981. Cyanide as a metabolic inhibitor, p. 11-28. In C. J. Knowles, J. Westley, J. Wissing, and F. Wissing (ed.), Cyanide in biology. Academic Press, London, United Kingdom.
- 50.Vock, E. H., W. K. Lutz, P. Hormes, H. D. Hoffmann, and S. Vamvakas. 1998. Discrimination between genotoxicity and cytotoxicity in the induction of DNA double-strand breaks in cells treated with etoposide, melphalan, cisplatin, potassium cyanide, Triton X-100, and gamma-irradiation. Mutat. Res. 413:83-94. [DOI] [PubMed] [Google Scholar]
- 51.Whitchurch, C. B., R. A. Alm, and J. S. Mattick. 1996. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 93:9839-9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitchurch, C. B., T. E. Erova, J. A. Emery, J. L. Sargent, J. M. Harris, A. B. Semmler, M. D. Young, J. S. Mattick, and D. J. Wozniak. 2002. Phosphorylation of the Pseudomonas aeruginosa response regulator AlgR is essential for type IV fimbria-mediated twitching motility. J. Bacteriol. 184:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wissing, F. 1983. Anaerobic column chromatography in the presence of detergents and its application to bacterial HCN-producing enzymes. J. Microbiol. Methods 1:31-39. [Google Scholar]
- 54.Wissing, F. 1974. Cyanide formation from oxidation of glycine by a Pseudomonas species. J. Bacteriol. 117:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyckoff, T. J., B. Thomas, D. J. Hassett, and D. J. Wozniak. 2002. Static growth of mucoid Pseudomonas aeruginosa selects for non-mucoid variants that have acquired flagellum-dependent motility. Microbiology 148:3423-3430. [DOI] [PubMed] [Google Scholar]
- 57.Xie, Z. D., C. D. Hershberger, S. Shankar, R. W. Ye, and A. M. Chakrabarty. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J. Bacteriol. 178:4990-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye, R. W., D. Haas, J. O. Ka, V. Krishnapillai, A. Zimmermann, C. Baird, and J. M. Tiedje. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 177:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]
- 60.Yu, H., M. Mudd, J. C. Boucher, M. J. Schurr, and V. Deretic. 1997. Identification of the algZ gene upstream of the response regulator AlgR and its participation in control of alginate production in Pseudomonas aeruginosa. J. Bacteriol. 179:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zielinski, N. A., A. M. Chakrabarty, and A. Berry. 1991. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266:9754-9763. [PubMed] [Google Scholar]
- 62.Zimmermann, A., C. Reimmann, M. Galimand, and D. Haas. 1991. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5:1483-1490. [DOI] [PubMed] [Google Scholar]