Abstract
A previous bioinformatics-based search for small RNAs in Escherichia coli identified a novel RNA named IS183. The gene encoding this small RNA is located between and on the opposite strand of genes encoding two transcriptional regulators of the acid response, gadX (yhiX) and gadW (yhiW). Given that IS183 is encoded in the gad gene cluster and because of its role in regulating acid response genes reported here, this RNA has been renamed GadY. We show that GadY exists in three forms, a long form consisting of 105 nucleotides and two processed forms, consisting of 90 and 59 nucleotides. The expression of this small RNA is highly induced during stationary phase in a manner that is dependent on the alternative sigma factor σS. Overexpression of the three GadY RNA forms resulted in increased levels of the mRNA encoding the GadX transcriptional activator, which in turn caused increased levels of the GadA and GadB glutamate decarboxylases. A promoter mutation which abolished gadY expression resulted in a reduction in the amount of gadX mRNA during stationary phase. The gadY gene was shown to overlap the 3′ end of the gadX gene, and this overlap region was found to be necessary for the GadY-dependent accumulation of gadX mRNA. We suggest that during stationary phase, GadY forms base pairs with the 3′-untranslated region of the gadX mRNA and confers increased stability, allowing for gadX mRNA accumulation and the increased expression of downstream acid resistance genes.
One of the main Escherichia coli defenses against extreme acid is based on the induction of two glutamate decarboxylase enzymes, GadA and GadB (5). These two enzymes use glutamate as a substrate to catalyze the production of γ-aminobutyric acid, a process that consumes an intracellular proton (10). The pool of glutamate is believed to be renewed by the GadC protein, which is a putative amino acid antiporter that exchanges γ-aminobutyric acid for external glutamate. The expression of gadA, gadB, gadC, and other genes of the glutamate-dependent acid response has been found to be extensively regulated. GadE, a LuxR family regulator, is essential for inducing gad genes in E. coli (12). Additionally, a poorly understood interplay between two opposing AraC-like regulators (GadX and GadW) contributes significantly to the regulation of expression of the two decarboxylase enzymes (13, 14). GadX transcriptionally activates GadA and GadB expression, and some studies suggest that GadW acts as a repressor to oppose this action (24, 27). Additional reported regulators of the gad system include the stationary-phase sigma factor σs, the cyclic AMP receptor protein CRP, and the nucleoid protein H-NS (13, 27).
Recent studies have led to the identification of a large number of small regulatory RNAs in E. coli (3, 6, 28). Many of these small RNAs still have uncharacterized functions. Among the small RNAs that have been studied, most bind to the Hfq protein and function by base pairing with target mRNAs (reviewed in reference 26). The Hfq binding small RNAs characterized thus far have been shown to function by three different mechanisms. The DsrA and RprA RNAs activate translation of the alternative sigma factor σS (15, 16). These small RNAs form base pairs with the 5′ leader sequence of the rpoS mRNA and inhibit the formation of a secondary structure that normally occludes the ribosome binding site (16, 17). The OxyS RNA is representative of a group of small RNAs that repress the translation of target mRNAs. The OxyS RNA forms base pairs with the fhlA mRNA across the ribosome binding site, thus blocking ribosome binding and translation (1, 2). A third mechanism used by small RNAs to control gene expression is the destabilization of mRNAs. The RyhB RNA forms base pairs with specific mRNAs encoding proteins involved in iron metabolism and targets them for degradation by RNase E (18, 19).
Here we show that a novel small RNA identified as IS183 in a screen for conserved promoter and terminator sequences controls the expression of genes involved in the glutamate-dependent acid response. This regulatory effect is mediated by positive regulation of the gadX mRNA, which encodes a transcriptional activator of the acid response system. Due to its location within the gad gene cluster and its role in regulating acid response genes, IS183 has been renamed GadY. The gadY gene sequence overlaps the 3′ end of the gadX mRNA encoded on the opposite strand. Since these sequences were found to be necessary for the GadY-dependent accumulation of gadX mRNA, we suggest that the base pairing between GadY and gadX in the overlapping region results in stabilization of the gadX mRNA transcript. This is the first example of a regulatory RNA that positively regulates the accumulation of its mRNA target.
MATERIALS AND METHODS
Plasmids and bacterial strains.
Standard molecular biology procedures were used for the isolation of genomic DNAs and plasmids, for restriction digests, for molecular cloning, and for transformation by electroporation or heat shock. Platinum Taq DNA polymerase (Invitrogen, Carlsbad, Calif.) was routinely used to amplify DNA fragments. The sequences of all fragments generated by PCR and all mutations generated by site-directed mutagenesis were confirmed by sequencing. All plasmids and bacterial strains used for this study are listed in Table 1. The sequences of all primers used for this study are given at the following web site: http://dir2.nichd.nih.gov/nichd/cbmb/segr/segrPublications.html.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild type | Lab collection |
| MC4100 | Wild type | Lab collection |
| GSO109 | MG1655 gadY−10 mutation | This study |
| GSO110 | MG1655 ΔgadX::kan Kanr | This study |
| GSO111 | MG1655 ΔgadY | This study |
| GSO81 | MC4100 hfq-1::Ω (Cmr) | 29 |
| GSO108 | MG1655 rpoS::Tn10 Tetr | This study |
| Plasmids | ||
| pACYC184 | Cloning vector (Cmr Tetr) | New England Biolabs |
| pCR2.1 TOPO | Cloning vector (Ampr Kanr) | Invitrogen |
| pKK177-3 | Expression vector (Ampr) | Lab collection |
| pRI | pKK177-3 with an EcoRI site at transcription start site (Ampr) | This study |
| pRI-GadY | pRI carrying gadY (Ampr) | This study |
| pRI-YdaG | pRI carrying gadY in antisense orientation (Ampr) | This study |
| placZ-gadXgadY−10 mutant | pACYC184 carrying PlacZ fused to gadXgadY−10 mutant (Cmr) | This study |
| placZ-gadXΔgadY | pACYC184 carrying PlacZ fused to gadXΔgadY (Cmr) | This study |
MG1655 was used as the E. coli parent strain for all strains unless otherwise designated. The rpoS::Tn10 (from GS015 [11]) and ΔgadX::kan (from EK426 [14]) alleles were moved into MG1655 by P1 transduction (25) to generate GSO108 and GSO110, respectively. Deletion of the gadY gene was carried out by the method reported by Datsenko and Wanner (9), using plasmid pKD4 (kanamycin resistant) and primers JK114 and JK115, to create strain GSO111. Disruption of the gadY promoter was carried out by the method reported by Court et al. (8), using the primer GadY-promoter, to create strain GSO109.
To allow for the overexpression of RNAs without additional sequence being added onto the 5′ end in the cloning process, we introduced an EcoRI site adjacent to the transcription initiation site of pKK177-3 by site-directed mutagenesis using primers JK17 and JK18 and a Quick Change mutagenesis kit (Stratagene, La Jolla, Calif.), generating plasmid pRI. For the overexpression of gadY, the appropriate sequences were amplified from MG1655 genomic DNA by the use of primers JK116 and JK118. For the overexpression of the antisense sequence of gadY, the appropriate sequences were amplified from MG1655 genomic DNA by the use of primers JK119 and JK120. The PCR fragments were then digested with EcoRI and HindIII and cloned into the corresponding sites of pRI to create plasmids pRI-GadY and pRI-YdaG. For the construction of plasmid placZ-gadXgadY −10 mutant, the lac promoter was PCR amplified from MG1655 genomic DNA by the use of primers lacZ-S2 and lacZ-A1. The gadX gene was PCR amplified from strain GSO109 genomic DNA by the use of primers GadX-S5 and GadX-A2. A lac promoter-gadX gene hybrid was then created by a PCR with primers lacZ-S3 and GadX-DN-Nest3 and was cloned into the BamHI and HindIII restriction sites of plasmid pACYC184. For the construction of plasmid placZ-gadXΔgadY, the lac promoter was PCR amplified from MG1655 genomic DNA by the use of primers lacZ-S2 and lacZ-A1. The gadX gene lacking its 3′ untranslated region (3′ UTR) was PCR amplified from GSO111 genomic DNA by the use of primers GadX-S5 and GadX-A2. A hybrid DNA containing the lac promoter and the gadX gene lacking its 3′ UTR was then created by a PCR with primers lacZ-S3 and GadX-DN-Nest3 and was cloned into the BamHI and HindIII restriction sites of plasmid pACYC184.
Growth conditions.
The strains were grown at 37°C in Luria-Bertani (LB) medium. Ampicillin (100 μg/ml), tetracycline (10 μg/ml), kanamycin (30 μg/ml), or chloramphenicol (30 μg/ml) was added when appropriate. Cells were harvested at the indicated times after 1:100 dilutions of overnight cultures. The typical optical densities at 600 nm for 1:5 dilutions of cells grown for 1, 2, 4, and 6 h were 0.06, 0.2, 0.7, and 0.9, respectively.
Northern analysis.
Total RNAs were isolated from cells at various stages of growth, as specified in the figure legends, by using Trizol reagent (Invitrogen) according to the manufacturer's protocols. For detection of the GadY small RNA, total RNAs (5 μg) were fractionated in 8% polyacrylamide-8 M urea gels and transferred to Zeta Probe GT membranes (Bio-Rad Laboratories, Hercules, Calif.). The JK138 oligonucleotide probe was 5′ end labeled with 32P by the use of T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.). The membranes were hybridized in Ultrahyb Oligo buffer (Ambion, Austin, Tex.) at 45°C and washed with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% sodium dodecyl sulfate (SDS). For the detection of mRNA species, total RNAs were fractionated in a 1% agarose-0.05 M MOPS-1 mM EDTA-3.3% formaldehyde gel and transferred to Zeta Probe GT membranes (Bio-Rad). The membranes were hybridized in Ultrahyb Oligo buffer (Ambion) at 45°C and then washed with 0.2× SSC plus 0.1% SDS. GadX mRNA was detected with the oligonucleotide probe GadX-A1. gadA mRNA was detected with the oligonucleotide probe GadAB-A1.
Primer extension analysis.
RNA samples (5 μg) were incubated with 0.5 pmol of a 5′-end-labeled primer specific for GadY (IS183-A4) for 5 min at 95°C and then cooled to 42°C. After the addition of deoxynucleoside triphosphates (a 1 mM concentration of each) and avian myeloblastosis virus reverse transcriptase (10 U; Life Sciences Inc., St. Petersburg, Fla.), the reactions were incubated for 1 h at 42°C. The cDNA products were fractionated in 8% polyacrylamide-urea gels.
3′ RACE analysis.
We performed 3′ rapid amplification of cDNA ends (RACE) assays as described previously (3). RNAs were ligated to the RNA adapter E1. Primer extension was carried out with primer E4. Amplification of the GadY cDNA was performed with Taq DNA polymerase and primers E4 and JK108. Amplification of the gadX cDNA was performed with Taq DNA polymerase, primer E4, and either primer GadX-S1 or primer GadX-S2. Amplified cDNA fragments were cloned into pCR2.1 TOPO and then sequenced.
Immunoprecipitation.
Cell extracts were prepared from cultures of strain MC4100 or GS081 that had grown for 16 h. Immunoprecipitations were performed as described previously (30), with 20 μl of Hfq antiserum or preimmune serum, 24 mg of protein A-Sepharose (Amersham Biosciences), and 200 μl of cell extract.
Protein identification.
Overnight cultures were diluted 1:100 and grown for 2 h. Aliquots were centrifuged, resuspended in 150 μl of a solution containing 4 mM Tris (pH 8.0), 30 mM KCl, 0.2 mM MgCl2, 1 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride, and sonicated for 10 s. Laemmli buffer (40 μl) was added, the samples were boiled at 95°C for 5 min, and 10 μl of each sample was analyzed by SDS-10% polyacrylamide gel electrophoresis. Bands of interest were excised and digested in the gel with trypsin (Roche, Indianapolis, Ind.) (1 μg/protein band) as described at the web site http://www.abrf.org/ResearchGroups/ProteinIdentification/EPosters/pirgprotocol.html. Peptide masses were determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF), and proteins were identified with Protein Prospector software (http://prospector.ucsf.edu/).
RESULTS
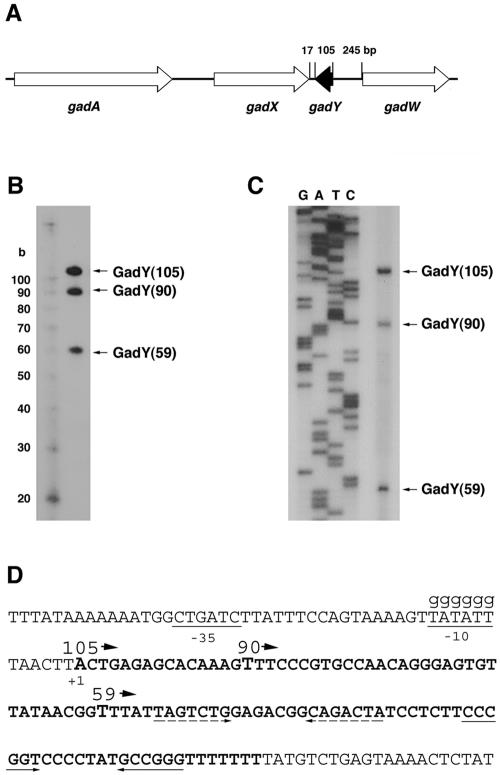
A 105-nucleotide small RNA encoded between gadX (yhiX) and gadW (yhiW).
A small RNA named IS183 was previously identified in a genome-wide computational screen for novel small RNA genes in E. coli (6). The gene was predicted based on the presence of conserved σ70 −35 and −10 promoter sequences followed by a potential Rho-independent transcription terminator. Two RNAs, of approximately 100 and 90 nucleotides, were reported to be detected by Northern analysis (6). This small RNA is encoded in an intergenic region between two open reading frames, gadX (yhiX) and gadW (yhiW), which code for transcriptional regulators of the glutamate-dependent acid response in E. coli (Fig. 1A) (14, 27). Due to the location of the IS183 gene in the middle of the gad gene cluster and the IS183 role in regulating acid response genes (see below), this small RNA has been renamed GadY. A sequence analysis showed that GadY is only present in strains that also encode gadX and gadW. Thus, the small RNA gene can be found in all sequenced strains of E. coli as well as Shigella, but not in other closely related organisms such as Salmonella and Klebsiella.
FIG. 1.
GadY exists as three forms. (A) Map of the gadAXYW acid response gene cluster. The gadY gene is shaded in black. (B) Northern blot detection of GadY. The total RNA (5 μg) from MG1655 cells grown to stationary phase (6 h) in LB medium was separated in an 8% polyacrylamide gel and then transferred to a nylon membrane. GadY RNA was detected with an oligonucleotide (JK138) that was complementary to GadY. Samples were run alongside Decade RNA markers (Ambion) with the indicated sizes. (C) Primer extension analysis of GadY RNA. Primer extension assays were performed with RNA isolated from MG1655 cells grown to stationary phase (4 h) in LB medium and with oligonucleotide IS183-A4, which is complementary to the 3′ end of GadY. (D) Sequence of the gadY gene in E. coli K-12. The −10 and −35 promoter sequences are underlined, and bold letters denote the gadY coding sequence. A promoter mutant (GSO109) was constructed in which the −10 sequence was changed to six guanine nucleotides, as indicated by lowercase letters. The 5′ ends of the three forms of the GadY RNA are displayed in larger letters and are labeled 105, 90, and 59, representing the sizes of these RNAs. The inverted repeat corresponding to the stem-loop of the transcription terminator is indicated by solid arrows. A putative single-stranded Hfq binding site (UUUAU) is adjacent to the putative stem-loop indicated by dashed arrows.
We first sought to characterize the GadY RNA in terms of its size. To confirm the initial identification of this RNA, we performed a Northern analysis, using an oligonucleotide probe capable of hybridizing 30 nucleotides upstream of the predicted Rho-independent terminator. Three species of the GadY small RNA, of approximately 100, 90, and 60 nucleotides, were detected in this analysis (Fig. 1B). We determined the boundaries of each of these RNA species by carrying out primer extension and 3′ RACE assays. Primer extension analysis identified three alternative 5′ ends of GadY, which are located at nucleotides 3662494, 3662509, and 3662540 of the E. coli genome sequence (Fig. 1C). 3′ RACE identified a single 3′ nucleotide located at nucleotide 3662598 of the E. coli genome sequence, which constitutes the 3′ end of the previously predicted Rho-independent terminator (data not shown). Thus, the three GadY RNA species are 105, 90, and 59 nucleotides in length and from here on will be referred to as GadY(105), GadY(90), and GadY(59) (Fig. 1C). The predicted σ70 −10 promoter sequence (TATATT) is located immediately upstream of the start of GadY(105) (Fig. 1D). GadY(90) and GadY(59) are believed to be processed forms of the full-length 105-nucleotide transcript due to the fact that the expression of all three forms was eliminated when we mutated the promoter of the full-length RNA (see below). The sequence at the 5′ end of both GadY(90) and GadY(59) is UUU, and both 5′ ends are adjacent to predicted stem-loop structures, but it is not known what features of the RNA direct processing.
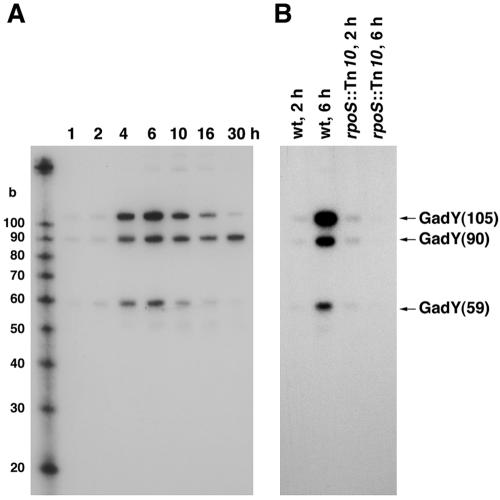
rpoS-dependent expression of GadY.
The expression profile of GadY was examined under different growth conditions. An overnight culture of wild-type E. coli cells was diluted 1:100 into fresh LB medium and incubated aerobically at 37°C. The total cellular RNA was harvested at various time points after inoculation, and Northern analysis was performed on equal amounts of RNA to determine the GadY RNA levels at each point during growth. The GadY RNA levels remained low during the first 2 h of growth, after which the amount of the small RNA increased dramatically. GadY RNA levels peaked after 6 h of growth and then slowly declined (Fig. 2A). The sharp increase in GadY expression corresponded with the entry of the E. coli cells into stationary phase. Interestingly, we observed a differential regulation of the three forms of the GadY small RNA. After 10 h of growth, the levels of the GadY(105) and GadY(59) forms of the RNA were substantially reduced while the levels of the GadY(90) RNA remained high (Fig. 2A). We also examined GadY levels in LB medium under low-pH conditions, but we did not detect any changes due to growth in acid (data not shown).
FIG. 2.
GadY expression is rpoS dependent. (A) GadY expression at different stages of growth. The total RNA was harvested at the indicated time points from MG1655 cells grown in LB medium. Samples (5 μg) were run alongside Decade RNA markers with the indicated sizes. (B) Level of GadY RNAs in wild-type and rpoS mutant strains. Total RNAs were harvested from MG1655 wild-type and rpoS::Tn10 mutant strains after 2 and 6 h of growth. For both panels, total RNAs were separated, transferred, and probed as described in the legend to Fig. 1B.
To test whether the increase in expression 6 h after dilution was dependent on the stationary-phase sigma factor σS, we examined GadY RNA levels in an rpoS mutant strain which lacks σS. Northern analysis detected very little GadY from wild-type or rpoS mutant cells grown to exponential phase (2 h). As expected, GadY RNA levels were high in the wild-type cells grown to stationary phase (6 h). This increase in GadY expression was abolished in an rpoS mutant, indicating that GadY expression is dependent on σS (Fig. 2B).
Hfq binding to GadY.
A growing number of small RNAs characterized from E. coli affect gene expression through the formation of base pair interactions with mRNAs, thereby exerting regulatory effects on the targets. All of the small RNAs that form base pairs with mRNAs require the activity of the RNA-binding protein Hfq. An earlier study that used microarrays to identify small RNAs that bind to Hfq suggested that GadY may coimmunoprecipitate with Hfq (30). For confirmation of this finding, total RNAs isolated from wild-type and hfq mutant cells, as well as RNAs that immunoprecipitated with control and Hfq-specific antisera, were specifically probed for GadY (Fig. 3). All three forms of GadY coimmunoprecipitated with Hfq, while no GadY was detected in the preimmune control precipitation. We also detected a fourth RNA band in the immunoprecipitation with an Hfq-specific antiserum. We suggest that this was a GadY degradation product that was enriched during the immunoprecipitation procedure, as it was never detected in total cellular RNA extracts. The Hfq protein has been shown to preferentially bind single-stranded AU-rich RNA sequences adjacent to stem-loop structures (29). Based on this observation, we propose that the Hfq binding site is the UUUAU sequence which is postulated to be a single-stranded region adjacent to a stem-loop and is present in all three forms of the RNA (Fig. 1D). The Northern analysis also showed that the strain harboring the hfq mutation contains no detectable GadY transcript (Fig. 3). We suggest that Hfq stabilizes the GadY RNA, similar to what is observed for other small RNAs. In part, the absence of GadY also may be due to the decreased σS expression observed in hfq mutant strains. Together, these results show that GadY is bound by Hfq and suggest that the small RNA acts by base pairing with target mRNAs.
FIG. 3.
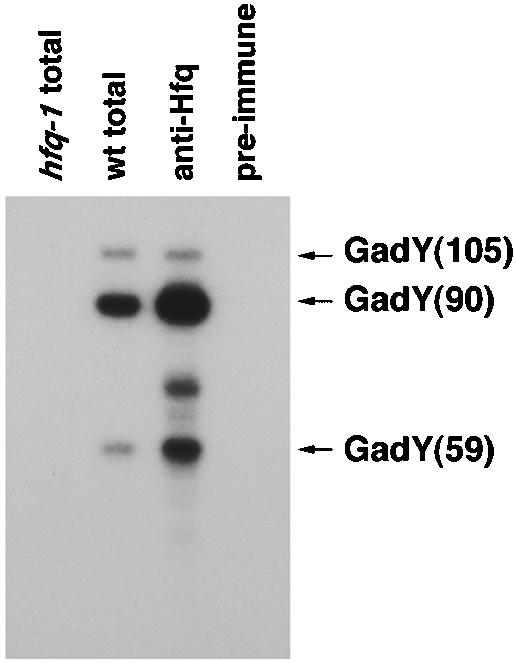
Hfq binding to GadY. Cell extracts were prepared from MC4100 and hfq-1 mutant cells grown for 16 h in LB medium. Immunoprecipitations were then carried out with the MC4100 extracts, using an Hfq antiserum or preimmune serum, and were compared to total RNAs isolated from 1/10 extract equivalents of the wild-type and hfq-1 mutant cells. The RNAs were separated, transferred, and probed as described in the legend to Fig. 1B. As observed in Fig. 2A, GadY(90) was much more abundant than GadY(105) or GadY(59) after 16 h. In addition, the GadY(105) or GadY(59) RNA species may have decreased stability under the conditions used to isolate the RNAs for this experiment.
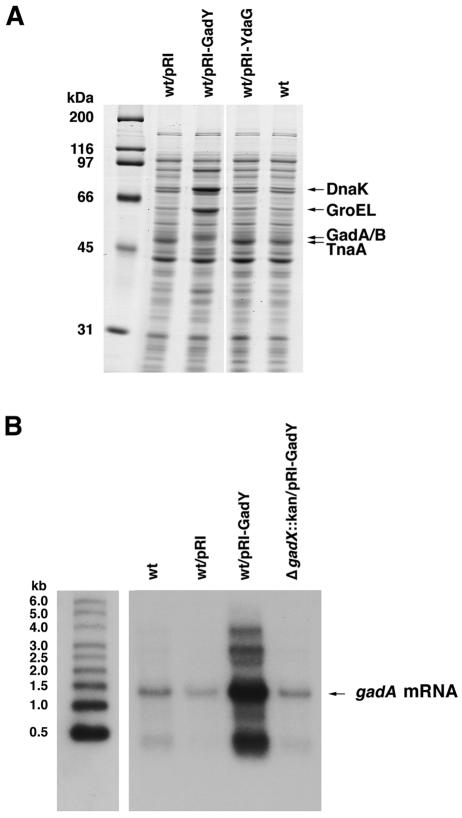
Effects of GadY overexpression.
Most of the characterized Hfq binding small RNAs affect the expression of target genes at a posttranscriptional level. Thus, we performed protein gel analysis to look for proteins whose expression was affected by GadY. A strain was constructed that contained the gadY gene cloned behind the strong tac promoter on a multicopy plasmid (pRI-GadY). All three forms of the GadY RNA were expressed at high levels from this plasmid (data not shown). In addition, a control plasmid was constructed with the gadY gene cloned behind the tac promoter in the antisense orientation (pRI-YadG). The protein profile of the GadY overexpression strain was compared with that of wild-type E. coli and the vector and antisense control strains. Protein bands whose intensities were either increased or decreased for the overexpressing strain were excised from the gel, digested with trypsin, and analyzed by MALDI-TOF mass spectroscopy. Based on this analysis, multiple proteins, including DnaK and GroEL, were highly induced, while the TnaA protein was reduced in its expression upon GadY overproduction (Fig. 4A). The peptide masses for one band with increased expression corresponded to the GadA and GadB proteins, which differ by only five amino acids and were indistinguishable in our analysis. The GadA and GadB isozymes are decarboxylase enzymes that convert glutamate to γ-aminobutyric acid, a reaction that consumes an intracellular proton (10). This GadA- and GadB-mediated reaction is one of at least three mechanisms used by E. coli to survive in a low-pH environment (5). Given the close proximity of the gadY and gadA genes on the chromosome, we chose gadA for further study (Fig. 1A).
FIG. 4.
Effect of GadY overproduction on GadA and GadB levels. (A) Protein expression patterns in cells overexpressing GadY. Equal amounts of total protein from MG1655 cells carrying vector pRI, pRI-GadY, or pRI-YdaG and grown in LB medium for 2 h were separated in SDS-10% polyacrylamide gels. The indicated bands were identified by in-gel trypsin digestion and MALDI-TOF mass spectrometry. (B) gadA mRNA levels in cells overexpressing GadY. Total RNAs (5 μg each) from MG1655, MG1655/pRI, MG1655/pRI-GadY, and MG1655 ΔgadX::kan/pRI-GadY grown in LB medium for 4 h were separated in a 1% agarose-0.05 M MOPS-1 mM EDTA-3.3% formaldehyde gel and then transferred to a nylon membrane. gadA mRNA was detected with an oligonucleotide (GadAB-A1) that was complementary to gadA. Samples were run alongside Millenium RNA size markers (Ambion), which were probed separately. The gadA mRNA migrated in the gel at the expected size of 1.4 kb. The two longer transcripts observed with GadY overproduction may correspond to gadAX and gadBC transcripts, and the shorter transcript is likely a degradation product.
We next sought to determine if gadA mRNA levels also increased in the presence of GadY, so we performed a Northern analysis to examine gadA mRNA expression in the wild-type strain overexpressing the small RNA. We also examined gadA mRNA levels in a gadX mutant strain overexpressing GadY since GadX is a known transcriptional activator of the gadA gene. When GadY was overexpressed in a wild-type strain, the levels of the 1.4-kb gadA mRNA increased 10-fold above the levels of the wild type alone (Fig. 4B). We also detected two larger RNA species and one smaller RNA species with GadY overproduction. Based on their respective sizes, the larger transcripts may be polycistronic gadAX and gadBC messages, while the smaller transcript is likely a degradation product. The positive regulatory effect of the GadY small RNA on gadA mRNA levels was lost when GadY was overexpressed in a gadX mutant strain, indicating that gadA induction by the GadY small RNA is dependent on gadX.
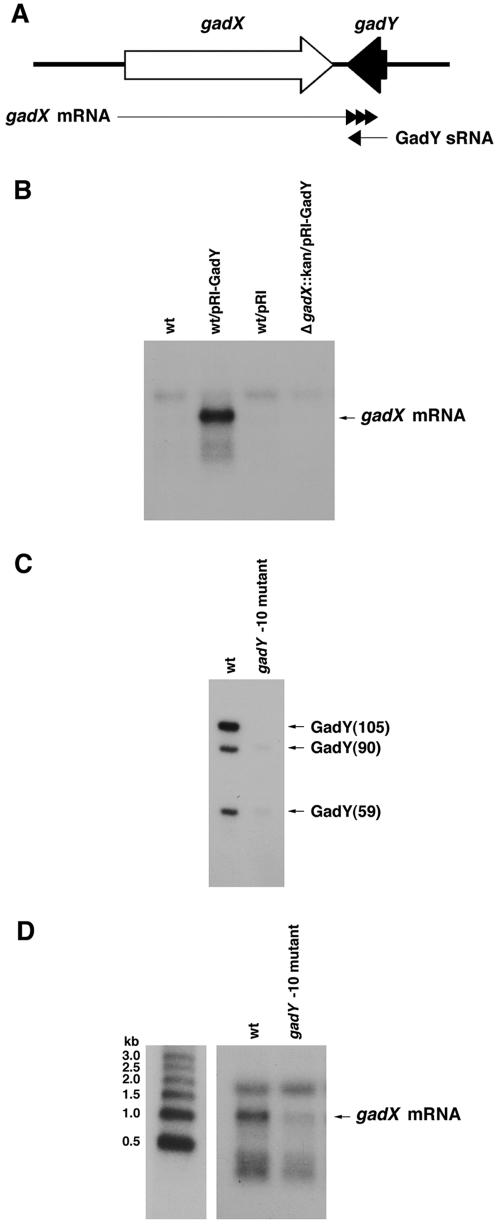
Possible base pairing between GadY and gadX mRNA.
Given that the gadX gene was necessary for gadA induction, we examined the gadX gene to identify a region that could form base pairs with GadY. No complementary sequences could be identified within the open reading frame of gadX. However, a closer examination of gene organization revealed that the 3′ end of the gadY gene and the stop codon of gadX are separated by only 17 nucleotides and that gadX does not display a recognizable Rho-independent terminator. 3′ RACE was performed to identify the 3′ end of the gadX mRNA. This analysis showed that the population of gadX mRNA species has heterogeneous 3′ ends (given at http://dir2.nichd.nih.gov/nichd/cbmb/segr/segrPublications.html). The majority of the mRNA species identified by RACE contained a 3′ UTR that overlapped the gadY gene by >30 nucleotides. The longest of these mRNA species contained a 3′ UTR that overlapped the gadY gene by 67 nucleotides (Fig. 5A).
FIG. 5.
Effects of increased and decreased GadY expression on gadX mRNA levels. (A) Map of overlap between GadY small RNA and gadX 3′ UTR. (B) gadX mRNA levels in cells overexpressing GadY. Total RNAs (5 μg each) from wild-type MG1655, MG1655/pRI-GadY, MG1655/pRI, and MG1655 ΔgadX::kan/pRI-GadY(105) grown in LB medium for 4 h were separated, transferred, and probed as described in the legend to Fig. 4B, except that the gadX mRNA was detected with an oligonucleotide (GadX-A1) that was complementary to gadX. (C) GadY RNA levels in gadY −10 promoter mutant strain. Total RNAs (5 μg each) from MG1655 and the gadY −10 mutant strain (GSO109) grown in LB medium for 6 h were separated, transferred, and probed as described in the legend to Fig. 1B. (D) gadX mRNA levels in cells with decreased GadY expression. Total RNAs (5 μg each) from MG1655 and the gadY −10 mutant grown in LB medium for 6 h were separated, transferred, and probed as described in the legend to Fig. 4B. The samples were run alongside Millenium RNA size markers, which were probed separately. The gadX mRNA migrated at the expected size of approximately 1 kb. The Northern blot in panel D was overexposed compared to that in panel B to allow detection of the gadX transcript from MG1655. The nonspecific hybridization to the 16S rRNA detected with the long exposure of panel D served as a loading control.
GadY positively controls gadX mRNA expression.
The finding that the 3′ UTR on the gadX mRNA could potentially form base pairs with the GadY small RNA led us to test whether the expression of gadX mRNA was affected by GadY expression. The GadY-overexpressing plasmid was introduced into wild-type and gadX mutant backgrounds. The cells were grown to stationary phase, total RNAs were harvested, and Northern analysis was performed to detect the gadX mRNA. The GadY-overexpressing strain displayed a dramatic accumulation of gadX mRNA to >20-fold higher levels than those observed in wild-type cells (Fig. 5B). This strong gadX induction would account for the increased expression of gadA upon GadY overexpression. Given that the region of potential GadY-gadX base pairing is at the 3′ ends of both of the transcripts, it is perhaps not surprising that even the overexpression of only the shortest form of GadY, GadY(59), resulted in high levels of gadX mRNA accumulation (data not shown).
We also tested whether gadX mRNA levels are affected in a gadY mutant strain. To accomplish this without disrupting the gadX gene, we constructed a gadY mutant strain in which expression of the small RNA was abolished. Mutagenesis was performed to replace the −10 promoter sequence of gadY (TATATT) with six guanine nucleotides that should no longer be recognized by the RNA polymerase holoenzyme (Fig. 1D). Northern analysis demonstrated that the expression of all three GadY RNAs was severely reduced in this −10 promoter mutant strain (Fig. 5C). This strain was then used to examine gadX mRNA levels in the absence of GadY. Wild-type and gadY −10 mutant cells were harvested during stationary phase, when GadY RNA levels were shown to be maximal. Total RNAs were isolated and tested for gadX mRNA levels by Northern analysis. These experiments revealed that gadX mRNA levels were reduced 4.5-fold in the gadY −10 promoter mutant strain (Fig. 5D). The GadY overexpression and mutant phenotypes indicated that GadY acts in a positive regulatory fashion upon gadX.
3′ UTR of gadX mRNA required for GadY-mediated regulation.
The transcriptional regulation of genes involved in the glutamate-dependent acid response is complex, with extensive cross activation and repression occurring between the different regulatory genes. To uncouple gadX transcription from this regulatory circuit, we placed two different forms of the gadX gene under control of the lac promoter in the plasmid pACYC184 (Fig. 6A). One plasmid contained the gadX gene under control of the E. coli lac promoter. This plasmid also contained the gadY gene with the previously described −10 promoter mutation, so GadY was not expressed from this construct (Fig. 6A). A second plasmid was constructed similarly with the gadX gene under control of the lac promoter, but in this instance the entire gadY gene was deleted and replaced with an 85-nucleotide scar (Fig. 6A). This deletion eliminated the region of overlap between the gadY gene and the gadX 3′UTR. These two plasmids were introduced into a strain carrying a chromosomal gadX deletion, and the GadY overexpression plasmid, which carries a compatible replicon, was transformed into each of these strains. The ability of GadY to control the expression of these two forms of gadX mRNA was tested by Northern analysis. In the strain containing gadX with a wild-type 3′ UTR, the gadX mRNA was induced threefold in the presence of GadY expressed from the second plasmid (Fig. 6B). In contrast, the gadX gene with the disrupted 3′ UTR was no longer induced by the GadY small RNA (Fig. 6B). These results indicate that sequences in the 3′ UTR of the gadX mRNA are necessary for GadY-dependent increases in the gadX mRNA levels and suggest that the base pairing of GadY and the 3′ UTR of the gadX mRNA increases its stability.
FIG. 6.
Effect of deleting gadX 3′ UTR. (A) Maps of placZ-gadXgadY −10 and placZ-gadXΔgadY constructs. (B) gadX mRNA levels from lacZ promoter fusion constructs in cells overexpressing GadY. Total RNAs (5 μg) from MG1655/placZ-gadXgadY −10 mutant pRI, MG1655/placZ-gadXgadY −10 mutant pRI-GadY, MG1655/placZ-gadXΔgadY pRI, and MG1655/placZ-gadXΔgadY pRI-GadY grown in LB medium for 4 h were separated, transferred, and probed as described in the legend to Fig. 4B. The reason for the lower gadX mRNA levels in MG1655/placZ-gadXΔgadY carrying pRI-GadY than in the same background strain carrying pRI is unknown.
DISCUSSION
GadY is unique in several ways with respect to other bacterial small RNAs. GadY is the first example of a bacterial small RNA that can form base pairs with the 3′ end of its target mRNA. Almost all of the characterized Hfq binding small RNAs modulate translation by forming base pairs with sequences spanning and adjacent to the ribosome binding site at the 5′ end of the target mRNA. In general, little is known about the 3′ UTRs of bacterial mRNAs and the type of regulatory signals they contain. More has been learned about regulation at the 3′ UTRs of eukaryotic transcripts, and several microRNAs in eukaryotic organisms have been shown to form base pairs within the 3′ UTRs of their target mRNAs (reviewed in reference 23). We suggest that the formation of base pairs between small RNAs and 3′ UTRs will likely be found for other RNAs. It is worth noting that the gadY-gadX gene arrangement couples the GadY small RNA to its target. Any mutations that occur within the pairing region will create a compensatory change on the opposite strand, with no net loss in complementarity between the small RNA and its mRNA target.
The GadY RNA is also the first E. coli small RNA that positively affects the accumulation of its target mRNA. So far, positive regulation has proved to be rare among RNA regulators. Only two small RNAs, DsrA and RprA of E. coli, have been found to positively regulate gene expression, and both of these function by enhancing translation of the rpoS target mRNA. Currently, no eukaryotic microRNAs are known to positively regulate gene expression, although it is conceivable that microRNAs with functions similar to GadY will be found.
The regulation of the genes involved in the glutamate-dependent acid response is complex and poorly understood. Control of this system has been shown to differ depending on the culture conditions (i.e., complex medium or minimal medium), and it involves multiple regulators (13). Under the conditions we used for our studies, specifically growth in LB broth at pH 7.0, gadX mRNA is expressed during stationary phase in a manner that is dependent on the secondary sigma factor σS (27). We have shown that GadY transcription is also dependent on σS. It was appealing to suggest that the gadX dependence on σS was mediated by GadY. We tested this possibility by overexpressing GadY in an rpoS mutant and found that the gadX mRNA did not accumulate in the rpoS mutant strain that overexpressed GadY (data not shown). This suggests that gadX mRNA accumulation during stationary phase is dependent on σS for transcription initiation as well as on stabilization via the GadY RNA.
We have not ruled out the possibility that GadY regulates multiple targets, some of which are encoded at separate locations of the chromosome. Protein gel analysis indicated that the expression of at least four proteins is directly or indirectly affected by this small RNA. Intriguingly, the TnaA protein, which displays GadY-dependent repression, has been implicated in protecting cells against alkaline stress (4). GadY targets could be regulated by all three forms of the RNA or could require one specific form of the GadY small RNA. Our GadY expression data suggest that the processing or stability of the three GadY small RNAs is different, as indicated by the fact that the 90-nucleotide form of the RNA persisted in late stationary phase while the full-length and 59-nucleotide forms became undetectable. If different forms of the small RNA are required for the regulation of individual target genes, it is possible that the targets are differentially regulated relative to each other.
GadY was shown to efficiently bind Hfq, an RNA binding protein required for the function of all small RNAs that form base pairs with target mRNAs. Although the mechanism of Hfq function is currently not fully understood, in vitro data for the OxyS and Spot42 small RNAs demonstrated that Hfq stimulates base pairing with the target mRNA (21, 22, 29). Current models of Hfq function indicate that the protein may help to unfold regions of base pairing, increase local concentrations of the small RNA and its target, or both. These proposed activities are believed to be essential because base pairing between most trans-encoded (encoded at a different location of the chromosome) small RNAs and their target mRNAs is short and noncontiguous. The GadY RNA is cis-encoded (encoded on the opposite strand from the target), so by definition there is 100% complementarity between the small RNA and its target. The question arises regarding whether Hfq is required for GadY base pairing with the gadX mRNA. It is possible that because of the extensive nature of the base pairing, Hfq is not needed to promote these interactions. This question is difficult to address, however, since GadY RNA levels were severely reduced in the Hfq mutant. Even if Hfq is not required for GadY base pairing with the gadX mRNA, it may play a role in promoting base pairing with an unknown trans-encoded target.
GadY represents a novel mechanism by which a small RNA can positively affect the accumulation of its target, in this case the gadX mRNA. We suggest that base pairing between the GadY small RNA and the gadX mRNA protects the mRNA from degradation by an RNase. This mechanism would be the opposite to the one used by the RyhB small RNA to regulate iron storage proteins. RyhB functions by base pairing with specific mRNAs and rendering the duplex RNA sensitive to RNase E degradation (18). Base pairing between GadY and the gadX mRNA could impede the binding or activity of an RNase that functions at the 3′ end of the gadX mRNA. E. coli encodes two exoribonucleases, polynucleotidephosphorylase and RNase II, whose activities can be blocked by secondary structures that create double-stranded RNA (7, 20). Due to the fact that gadX does not have an identifiable Rho-independent terminator comprised of a stem-loop, the unstructured 3′ end of the gadX mRNA could be particularly sensitive to this type of RNase activity. The base pairing of GadY with the 3′ UTR of gadX mRNA would create a double-stranded RNA duplex that could block the activity of such an RNase. Future experiments will be aimed at elucidating the mechanism of GadY protection of the gadX mRNA as well as determining whether other small RNAs act similarly to stabilize their target mRNAs.
Acknowledgments
We are grateful to Z. Ma and J. Foster for providing many reagents and sharing information. We thank S. Gottesman and members of our laboratory for helpful discussions, advice on techniques, and comments on the manuscript.
REFERENCES
- 1.Altuvia, S., A. Zhang, L. Argaman, A. Tiwari, and G. Storz. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 17:6069-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argaman, L., and S. Altuvia. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300:1101-1112. [DOI] [PubMed] [Google Scholar]
- 3.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 4.Bordi, C., L. Theraulaz, V. Mejean, and C. Jourlin-Castelli. 2003. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 48:211-223. [DOI] [PubMed] [Google Scholar]
- 5.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, S., E. A. Lesnik, T. A. Hall, R. Sampath, R. H. Griffey, D. J. Ecker, and L. B. Blyn. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems 65:157-177. [DOI] [PubMed] [Google Scholar]
- 7.Coburn, G. A., and G. A. Mackie. 1996. Overexpression, purification, and properties of Escherichia coli ribonuclease II. J. Biol. Chem. 271:1048-1053. [DOI] [PubMed] [Google Scholar]
- 8.Court, D. L., S. Swaminathan, D. Yu, H. Wilson, T. Baker, M. Bubunenko, J. Sawitzke, and S. K. Sharan. 2003. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene 315:63-69. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homola, A. D., and E. E. Dekker. 1967. Decarboxylation of gamma-hydroxyglutamate by glutamate decarboxylase of Escherichia coli (ATCC 11246). Biochemistry 6:2626-2634. [DOI] [PubMed] [Google Scholar]
- 11.Loewen, P. C., and B. L. Triggs. 1984. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J. Bacteriol. 160:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 13.Ma, Z., H. Richard, and J. W. Foster. 2003. pH-dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J. Bacteriol. 185:6852-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 16.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 18.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaren, R. S., S. F. Newbury, G. S. Dance, H. C. Causton, and C. F. Higgins. 1991. mRNA degradation by processive 3′-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J. Mol. Biol. 221:81-95. [PubMed] [Google Scholar]
- 21.Møller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 22.Møller, T., T. Franch, C. Udesen, K. Gerdes, and P. Valentin-Hansen. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16:1696-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson, P., M. Kiriakidou, A. Sharma, E. Maniataki, and Z. Mourelatos. 2003. The microRNA world: small is mighty. Trends Biochem. Sci. 28:534-540. [DOI] [PubMed] [Google Scholar]
- 24.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 25.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 27.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, A., K. M. Wassarman, C. Rosenow, B. C. Tjaden, G. Storz, and S. Gottesman. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50:1111-1124. [DOI] [PubMed] [Google Scholar]