Abstract
The Escherichia coli β sliding clamp, which is encoded by the dnaN gene, is reported to interact with a variety of proteins involved in different aspects of DNA metabolism. Recent findings indicate that many of these partner proteins interact with a common surface on the β clamp, suggesting that competition between these partners for binding to the clamp might help to coordinate both the nature and order of the events that take place at a replication fork. The purpose of the experiments discussed in this report was to test a prediction of this model, namely, that a mutant β clamp protein impaired for interactions with the replicative DNA polymerase (polymerase III [Pol III]) would likewise have impaired interactions with other partner proteins and hence would display pleiotropic phenotypes. Results discussed herein indicate that the dnaN159-encoded mutant β clamp protein (β159) is impaired for interactions with the α catalytic subunit of Pol III. Moreover, the dnaN159 mutant strain displayed multiple replication and repair phenotypes, including sensitivity to UV light, an absolute dependence on the polymerase activity of Pol I for viability, enhanced Pol V-dependent mutagenesis, and altered induction of the global SOS response. Furthermore, epistasis analyses indicated that the UV sensitivity of the dnaN159 mutant was suppressed by (not epistatic with) inactivation of Pol IV (dinB gene product). Taken together, these findings suggest that in the dnaN159 mutant, DNA polymerase usage, and hence DNA replication, repair, and translesion synthesis, are altered. These findings are discussed in terms of a model to describe how the β clamp might help to coordinate protein traffic at the replication fork.
It has become increasingly apparent in recent years that DNA replication, recombination, and repair are intimately related to each other (reviewed in reference 7). For example, the Escherichia coli RecA protein is not only required for most homologous recombination but is also required for DNA replication under normal growth conditions (7, 28), as well as for the proper regulation of global cellular responses following DNA damage (13, 47), including replication of damaged DNA (13, 47). Moreover, the fact that the same DNA structure may serve as a substrate for both DNA replication and homologous recombination indicates that mechanisms must exist for coordinately regulating these processes in vivo (48). Consistent with the need for such regulation, processing of replication intermediates by recombination is proposed to serve as one cause of genome instability and is suspected to be a contributing factor to the development of certain cancers in humans (25). The fact that E. coli possesses at least five distinct DNA polymerases and that humans possess at least 16 DNA polymerases (reviewed in references 11, 12, and 48) further underscores the need organisms have for a higher-order regulatory system that acts to coordinate the actions of the cell's different DNA polymerases with those of its various repair and recombination proteins (45, 46).
As discussed below, findings from a variety of different groups have recently begun to suggest possible mechanisms for how organisms might coordinate the events at a replication fork. It has been known for some time now that the α catalytic subunit of the E. coli replicative DNA polymerase (polymerase III [Pol III]) is, by itself, a distributive enzyme (19). In order for Pol III to replicate the bacterial chromosome, it must become a highly processive enzyme. Pol III is composed of 10 distinct subunits (19, 29). The β subunit, also referred to as the β sliding clamp, is required for Pol III processivity (5, 29). β exists as a homodimer in solution. In the crystal, it forms a ring-shaped molecule with an opening in the middle sufficiently large to accommodate duplex DNA (20, 41). β is “loaded” onto DNA by another component of Pol III, the five-subunit γ clamp loader complex, which acts to open one of the two dimer interfaces (19). Once loaded, β interacts with the α catalytic subunit to tether it to the DNA, thereby conferring an amazing degree of processivity upon the polymerase (29). Consistent with the structure of the β clamp being ideally suited for its role in DNA replication, the processivity subunits of eukaryotic and bacterial phage T4 replicative polymerases, proliferating cell nuclear antigen (PCNA) and gp45, respectively, possess similar structures, despite their lack of amino acid sequence similarity (22, 31).
Although clamp proteins were first discovered because of their role in DNA replication, it soon became clear that they participate in other DNA metabolic events as well. In the bacterial phage T4 system, Tinker and colleagues (50) demonstrated the ability of the T4 gp45 clamp to interact with the modified RNA polymerase of the phage to activate transcription from late-gene promoters. Likewise, the human clamp protein PCNA is known to interact with a variety of proteins involved in DNA replication, repair, chromatin remodeling, and cell cycle progression (reviewed in reference 51). Consistent with these findings, it has recently been reported that the E. coli β clamp protein interacts with other proteins in addition to components of Pol III, including the other four E. coli DNA polymerases (3, 26, 27, 45, 46), MutS (27), DNA ligase (27), and the DnaA-like replication protein Hda (23). β is also proposed to play an important role in DNA damage checkpoint control (33, 36). Taken together, these findings suggest that clamp proteins function as mobile platforms on DNA to which a variety of partner proteins may attach, and as such, likely participate in processes over and above DNA replication (reviewed in reference 48).
Recently, Dalrymple et al. (8) described a consensus sequence (QL[S/D]LF), referred to as the eubacterial clamp-binding motif, that appears to mediate, at least in part, interactions of certain partner proteins with β (8). This motif is believed to interact with a hydrophobic pocket at the base of the C-terminal tail of the β clamp (see Fig. 1). A variety of recent reports have substantiated this proposal (4, 6, 16, 24, 26) and collectively suggest that partner proteins compete with each other for binding to the β clamp. Thus, although not yet demonstrated, it has become generally accepted that the β clamp plays an important role in coordinating protein traffic at the replication fork. The purpose of the experiments discussed in this report was to test the model that the β clamp helps to coordinate protein traffic at the replication fork. I hypothesized that if partners do in fact compete with each other for binding to the clamp, then a mutant β protein impaired for interactions with Pol III would likewise display impaired interactions with other partner proteins, and as a result, would display multiple replication and/or repair defects. As discussed below, I determined that the mutant β clamp protein encoded by the dnaN159 allele was impaired for interactions with the α catalytic subunit of Pol III. Moreover, this mutant displayed multiple replication and repair defects including (i) an enhanced SOS mutator phenotype, (ii) an absolute dependence upon the polymerase activity of DNA polymerase I for survival, (iii) altered induction of the global SOS response, and (iv) sensitivity to UV light. Further characterization of the dnaN159 mutant strain indicated that UV sensitivity was suppressed by the ΔdinB::kan allele. These results are discussed in terms of a model for how β might help to regulate protein traffic at the replication fork.
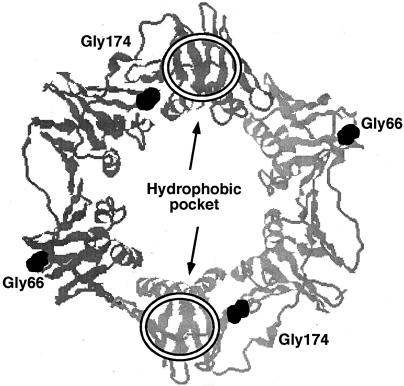
FIG. 1.
Structure of the E. coli β sliding clamp. The relative positions of the glycine-66 (Gly66) and glycine-174 (Gly174) residues, which are replaced by glutamic acid and alanine, respectively, in β159 are shown. The approximate location of the hydrophobic pocket with which the eubacterial clamp-binding motif has been shown to interact is indicated by the oval within each β protomer. This figure was preparing by using RasMac (http://mc2.cchem.berkeley.edu/Rasmol/) and the coordinates for the β clamp from the PDB (2POL).
MATERIALS AND METHODS
E. coli strains, plasmid DNAs, and bacteriological techniques.
E. coli strains were routinely grown in Luria-Bertani (LB) medium (30). When necessary, the following antibiotics were used at the indicated concentrations: ampicillin, 150 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 40 μg/ml; tetracycline, 12 μg/ml, and rifampin, 50 μg/ml. Bacterial transformation was performed using calcium chloride as described previously (43).
E. coli strains and plasmid DNAs used in this study are described in Table 1. E. coli DH5α was used as the host for cloning. All strains are derivatives of E. coli K-12 and were constructed using P1vir-mediated generalized transduction as described previously (30). In most cases, transduction of each allele was confirmed by measuring UV sensitivity and/or by diagnostic PCR. The dnaN allele used in this study was originally referred to as dnaN59 (39) but has since been renamed dnaN159 by the E. coli Genetic Stock Center (Yale University). Therefore, this allele is referred to as dnaN159 throughout this report. Transduction of dnaN159 was performed by selection for the closely linked tnaA300::Tn10 allele and was confirmed by verifying temperature sensitivity and by nucleotide sequencing the PCR-amplified dnaN allele (discussed below).
TABLE 1.
E. coli strains and plasmid DNAs used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Source or construction |
|---|---|---|
| E. coli strains | ||
| DH5α | endA1 hsdR17(rK− mK+) glnV44 thi-1 recA1 gyrA relA Δ(lacZYA-argF)U169 deoR (φ80dlacΔ[lacZ]M15) | Laboratory stock |
| KA473 | fhuA22 ompF627 relA1 thyA148 metB1 tnaA300::Tn10 | T. Katayama (18) |
| KA474 | Same as KA473 but with dnaN159b | T. Katayama (18) |
| RW118 | thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4(Oc) rpsL31 xyl-5 mtl-1 argE3(Oc) thi-1 sulA211 | R. Woodgate (15) |
| MS100 | Same as RW118 but with dnaN+tnaA300::Tn10 | P1(KA473) × RW118 |
| MS101 | Same as RW118 but with dnaN159 tnaA300::Tn10 | P1(KA474) × RW118 |
| RW434 | Same as RW118 but with lexA3(Ind−) | R. Woodgate (10) |
| RW542 | Same as RW118 but with lexA51(Def) | R. Woodgate (10) |
| MS102 | Same as RW434 but with dnaN+tnaA300::Tn10 | P1(KA473) × RW434 |
| MS103 | Same as RW434 but with dnaN159 tnaA300::Tn10 | P1(KA474) × RW434 |
| MS104 | Same as RW542 but with dnaN+tnaA300::Tn10 | P1(KA473) × RW542 |
| MS105 | Same as RW542 but with dnaN159 tnaA300::Tn10 | P1(KA474) × RW542 |
| CS5430 | ΔuvrB::Cmr | N. Goosen (32) |
| MS106 | Same as MS100 but with ΔuvrB::Cmr | P1(CS5430) × MS100 |
| MS107 | Same as MS101 but with ΔuvrB::Cmr | P1(CS5430) × MS101 |
| RW120 | Same as RW118 but with ΔumuDC595::cat | R. Woodgate (15) |
| MS108 | Same as RW120 but with dnaN+tnaA300::Tn10 | P1(KA473) × RW120 |
| MS109 | Same as RW120 but with dnaN159 tnaA300::Tn10 | P1(KA474) × RW120 |
| MG1655c | rph-1 | CGSCd |
| JW165 | lacZ53 thyA36 IN(rrnD-rrnE)1 rpsL151 rha-5 malB45 deoC2 | CGSC |
| JW164 | Same as JW165 but with polA1 | CGSC |
| CJ278 | Δ(gal-bio) thi-1 relA1 spoT1 ΔpolA::kan | C. Joyce (17) |
| CJ225 | CJ278 transformed with pJC100 (polA+) | C. Joyce (17) |
| CJ231 | CJ278 transformed with pCJ102 (polA1 [polA 5′ exo]) | C. Joyce (17) |
| CJ233 | CJ278 transformed with pCJ103 (polA Klenow) | C. Joyce (17) |
| RW520 | Same as RW118 but with ΔumuDC596::ermGT | R. Woodgate (56) |
| MS110 | Same as RW520 but with ΔuvrB::Cmr | P1(CS5430) × RW520 |
| MS111 | Same as MS110 but with dnaN+tnaA300::Tn10 | P1(KA473) × MS110 |
| MS112 | Same as MS110 but with dnaN159 tnaA300::Tn10 | P1(KA474) × MS110 |
| STL1336 | thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4(Oc) rfbD1 mgl-51 rpsI31 kdgK51 xyl-5 mtl-1 argE3(Oc) thi-1 Δ(araD-polB)::Ω | M. Goodman via J. Courcelle (37) |
| MS113 | Same as MS106 but with Δ(araD-polB)::Ω | P1(STL1336) × MS106 |
| MS114 | Same as MS107 but with Δ(araD-polB)::Ω | P1(STL1336) × MS107 |
| FC1237 | ara Δ(lac-proB) ΔdinB::kan | P. Foster via B. Strauss (40) |
| MS115 | Same as MS106 but with ΔdinB::kan | P1(FC1237) × MS106 |
| MS116 | Same as MS107 but with ΔdinB::kan | P1(FC1237) × MS107 |
| SY2 | ΔlacX74 rpsL araD139 Δ(ara-leu)7697 galU galK hsr hsm+sulA::lacZ′YA::kan | H. Ohmori (57) |
| MS117 | Same as SY2 but with dnaN+tnaA300::Tn10 | P1(KA473) × SY2 |
| MS118 | Same as SY2 but with dnaN159 tnaA300::Tn10 | P1(KA474) × SY2 |
| Plasmids | ||
| pWSK29 | Apr; pSC101 derivative | Laboratory stock (54) |
| pJD100 | pWSK29 bearing the dnaN+ gene | This work |
| pCJ100 | Cmr; F bearing the polA+ gene | C. Joyce (17) |
| pCJ102 | Cmr; pCJ100 bearing the polA1 (polA 5′ exo) gene | C. Joyce (17) |
| pCJ103 | Cmr; pCJ100 bearing the polA Klenow gene | C. Joyce (17) |
| pGBT9 | AprTRP1; yeast two-hybrid vector bearing the Gal4 DNA binding domain | Laboratory stock |
| pGBα542-1160 | AprTRP1; pGBT9 bearing α(542-1160) | This work |
| pGAD424 | AprLEU2; yeast two-hybrid vector bearing the Gal4 activation domain | Laboratory stock |
| pGADdnaN | AprLEU2; pGAD424 bearing dnaN+ | This work |
| pGADdnaN159 | AprLEU2; pGAD424 bearing dnaN159 | This work |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance.
The dnaN59 allele has been renamed dnaN159 by the E. coli Genetic Stock Center. Based on DNA sequence analysis, the dnaN159 allele contains two nucleotide alterations, resulting in G66E and G174A amino acid substitutions (see Materials and Methods).
The MG1655 strain used corresponds to the exact same isolate whose complete genome was sequenced by Blattner and colleagues (2).
CGSC, E. coli Genetic Stock Center.
pJD100 is a pWSK29 (54) derivative that contains the wild-type dnaN gene in an EcoRI-SalI fragment positioned downstream of the lac promoter. It was cloned in two steps using the EcoRI-XhoI fragment from pACYCdnaA (43) and the XhoI-SalI fragment from pJRC210 (42). In the absence of added isopropyl-β-d-thiogalactopyranoside (IPTG), pJD100 expresses β from the native dnaN promoters. During exponential growth, these promoters are weak (52). Consistent with this finding, Western blot analyses revealed that the steady-state level of β expressed from pJD100 in the absence of added IPTG was less than the physiological levels (data not shown). However, in the presence of 50 μM IPTG, the steady-state level of β expressed from pJD100 was near the physiological levels (data not shown). Therefore, 50 μM IPTG was included in the growth media in all experiments described in this report using this plasmid.
Nucleotide sequence analysis of the dnaN159 allele.
The dnaN159 allele was recently reported to contain two substitutions: a dG-to-dA mutation changing glycine-66 to glutamine and a dG-to-dC mutation changing glycine-174 to alanine (14). To confirm that all dnaN159-bearing strains shown in Table 1 possessed both mutations, the dnaN159 allele of each strain was PCR amplified using primer JK-28+2 (5′-CATTGCCAATGCCAACTTTACC-3′), which is located in the C-terminal end of the dnaA gene (which is upstream of dnaN), and primer RecF-reverse (5′-ACCTACCAGAAAGTTAAAGCCG-3′), which is located in the N-terminal end of the recF gene (which is downstream of dnaN). The resulting PCR fragment was purified (QIAGEN) and subjected to automated DNA sequence analysis (Roswell Park Cancer Institute Biopolymer Facility, Buffalo, N.Y.). In addition to confirming that each strain contained the correct dnaN159 sequence, it was discovered that the substitution at position 66 was actually GGA to GAA, resulting in a glycine-to-glutamic acid substitution, not a glycine-to-glutamine substitution as previously reported (14). Both the G66E (GGA→GAA) and G174A (GGC→GCC) substitutions were confirmed by sequencing the dnaN159 allele from HC123 obtained from the E. coli Genetic Stock Center (data not shown).
Saccharomyces cerevisiae two-hybrid analysis.
The dnaN+ and dnaN159 genes, as well as amino acid residues 542 to 1160 of the α catalytic subunit of Pol III, were each PCR amplified using the following primers, which contain either an EcoRI (upstream) or BamHI (downstream) restriction digestion site to facilitate their subsequent cloning into pGAD424 or pGBT9, respectively: β Y2H Top (5′-AAAGAATTCGGAGCCGGAATGAAATTTACCGTAGAACGTGAGC-3′), β Y2H Bottom (5′-AAGGATCCCGTCGACTTACAGTCTCATTGGCATGACAACATAAGC-3′), α 542 Y2H Top (5′-AAAGAATTCGGAGCCGGAATGTCTGAACCACGTTTCGTACACCTGCGG-3′), and α CT-end Y2H Bottom (5′-TTTAAGGATCCCGTCGACTTAGTCAAACTCCAGTTCCACCTGCTCCG-3′). After confirmation of the structures of the various plasmid constructs, both the pGBT9 and pGAD424 plasmids, as well as appropriate combinations of their different derivatives, were transformed into S. cerevisiae HF7c (Clontech) by the lithium acetate method (1) or by electroporation (Bio-Rad) as described elsewhere. Representative transformants were grown in liquid synthetic dropout medium lacking leucine and tryptophan (1) at 30°C. To test for interactions between β and α, cultures grown overnight were washed twice with sterile 0.8% saline and adjusted to an optical density at 600 nm (OD600) of ∼1.0, and 10-μl aliquots of each appropriate dilution (undiluted [10 μl], 10−1, and 10−2) were spotted onto solid synthetic dropout medium lacking leucine and tryptophan (control plate) or lacking leucine, tryptophan, and histidine (to score for protein-protein interactions), and the plates were incubated at 30°C for 2 or 3 days.
Measuring UV sensitivity.
Cultures grown overnight in M9 minimal medium (30) supplemented with glucose (0.2%), thiamine (1 μg/ml), and Casamino Acids (0.5%) were subcultured in the same medium at 30°C with shaking. After cultures reached mid-exponential phase (OD595 of ∼0.6), 6 ml of each culture was transferred to a 100-mm-diameter, sterile, glass petri dish for irradiation. Irradiation was performed using a 15-W germicidal bulb (254-nm wavelength; General Electric) set at a height such that it delivered UV light at a dose of 1 J/m2/s for uvrB+ strains or at a dose 0.3 J/m2/s for ΔuvrB::Cmr strains, as measured using a UV radiometer (VWR). Aliquots corresponding to the desired UV doses were removed from the shaking petri dish by hand, serially diluted in sterile 0.8% saline, and plated onto solid LB medium. Survival was calculated after incubation for 1 or 2 days at the appropriate temperature. Colonies were counted using the colony count feature of the Gel Doc 1000 (Bio-Rad). Survival at each UV dose was calculated after defining growth at the 0 J/m2 UV dose for each strain as 100% survival. Each UV killing curve was performed at least three different times.
β-Galactosidase assays.
Fresh cultures of E. coli MS117 (relevant genotype, dnaN+ sulA::lacZ′YA::kan) and MS118 (relevant genotype, dnaN159 sulA::lacZ′YA::kan) grown overnight were diluted 1:100 into M9 minimal medium supplemented with glucose (0.2%), thiamine (1 μg/ml), and Casamino Acids (0.5%) and grown at 30°C with shaking. When cultures reached mid-exponential phase (OD595 of ∼0.5), β-galactosidase activity was measured using the chloroform-sodium dodecyl sulfate method described by Miller (30). Each data point represents an average of three values.
RESULTS
The purpose of the experiments discussed in this report was to test the hypothesis that the β clamp plays a role in coordinating protein traffic at the replication fork by forcing partner proteins to compete with each other for binding to it. I hypothesized that if this model is correct and if partners bind to similar surfaces on β, then a mutant clamp protein with impaired interactions with Pol III would be similarly affected for interactions with other partner proteins, resulting in multiple replication and/or repair defects. The temperature-sensitive dnaN159 allele encodes a mutant form of the β clamp (referred to as β159 in this report) bearing two amino acid substitutions, a glycine-to-glutamic acid substitution at position 66 (G66E) and a glycine-to-alanine substitution at position 174 (G174A) (see Materials and Methods). Results discussed below indicate that the mutant β159 protein has impaired interactions with Pol III. Moreover, the results of this study demonstrate that the dnaN159 allele displays numerous mutant phenotypes, consistent with the idea that the β clamp plays important roles in replication, repair, and damage tolerance.
The mutant β159 clamp protein has impaired interactions with the replicative DNA polymerase.
Most, if not all, of the partner proteins that interact with β appear to do so, at least in part, through contact with variants of a pentapeptide sequence (QL[S/F]LF) that has been termed the eubacterial clamp-binding motif (8). This motif is believed to interact with a hydrophobic pocket at the base of the C-terminal tail of each β protomer. Although G174, which is present within a solvent-exposed loop of β, is not actually part of this hydrophobic pocket, it is very close to the pocket (Fig. 1). Moreover, the loop containing G174 has been suggested to form part of the surface on β that is contacted directly by two well-conserved leucine residues adjacent to the more rigidly defined five-residue eubacterial clamp-binding motif (6). On the basis of these observations, I hypothesized that the temperature sensitivity of the β159 mutant protein might be due, at least in part, to an impaired interaction of the mutant β159 protein with the α catalytic subunit of Pol III.
Interactions of wild-type β and β159 with the α catalytic subunit of Pol III were examined by yeast two-hybrid analysis. Since β has been reported to interact with elements of α confined to the region between residue 542 and its C terminus at position 1160 [α(542-1160)], I focused on determining whether β159 was impaired for interactions with this region. An interaction between wild-type β and α(542-1160) was observed, as expected (Fig. 2). However, β159 was severely impaired for interactions with α(542-1160), as indicated by its poor growth on medium lacking histidine.
FIG. 2.
Interactions of the wild-type β and the mutant β159 proteins with the α catalytic subunit of Pol III. Yeast two-hybrid analysis was performed as described in Materials and Methods using yeast strain HF7c (MATa ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3 gal4-542 gal80-538 LYS2::GAL1-HIS3 URA3::(GAL4 17-mers)3-CYC1-lacZ). Gal4-AD, Gal4 activation domain; Gal4-DBD, Gal4 DNA binding domain.
The dnaN159 mutant E. coli strain is sensitive to UV irradiation.
If the β clamp plays an important role in coordinating protein traffic at the replication fork, then the ability of one partner to compete with another for binding to the clamp must be determined, at least in part, by the relative affinities of the different partners for the clamp, as well as their respective concentrations within the cell. Thus, perturbation of one or more of these clamp-partner interactions could have a ripple effect, leading to a reduction in the overall efficiency of replication and repair. In light of my finding that β159 had impaired interactions with Pol III, I next asked whether this mutant had impaired interactions with any other partners that act in DNA repair or damage tolerance.
Sensitivity to UV light is a hallmark for a DNA repair or damage tolerance defect (13). Therefore, initial efforts were focused on determining whether the dnaN159 allele sensitized E. coli to UV light. UV sensitivity of a dnaN159 mutant strain was measured at both 30 and 37°C: UV sensitivity could not be measured at higher temperatures due to the thermolability of the dnaN159 strain. At 30°C, the UV sensitivity of the dnaN159 strain (MS101) was indistinguishable from that of the isogenic dnaN+ partner strain (MS100; data not shown). However, at the semipermissive temperature of 37°C, the dnaN159 strain was reproducibly approximately threefold more sensitive to UV light than the isogenic dnaN+ partner was (data not shown).
It seemed likely that the extent of UV sensitivity of the dnaN159 mutant might be exacerbated in the absence of functional nucleotide excision repair (NER) due to the increased dependence upon other cellular functions to repair or tolerate UV-induced DNA lesions, one or more of which is presumably impaired in the dnaN159 mutant strain. Therefore, to avoid the possibility that a repair or damage tolerance defect of the dnaN159 mutant might be masked by NER, the UV sensitivity of the dnaN159 mutant was measured in a uvrB::Cmr genetic background in which NER had been abolished. The dnaN159 ΔuvrB::Cmr strain (MS107) was ∼10-fold more UV sensitive than the isogenic dnaN+ partner strain (MS106) was at 30°C (Fig. 3A). Importantly, this UV sensitivity was suppressed by pJD100, which expresses near-physiological levels of wild-type β, but not the parent plasmid (pWSK29) lacking the dnaN gene (data not shown). This result confirmed that the UV sensitivity was due to the dnaN159 mutation. Moreover, the UV sensitivity of the dnaN159 ΔuvrB::Cmr mutant, but not the dnaN+ strain, was greater at 37°C, such that it was ∼30-fold more UV sensitive than the isogenic dnaN+ strain (Fig. 3B). Taken together, these results suggest that the mutant β159 protein has impaired interactions with one or more partners that act in DNA repair and/or damage tolerance in vivo and demonstrate that this defect(s) is largely compensated for by NER. On the basis of these results, additional experiments were designed to determine the mechanistic basis for the UV sensitivity of the dnaN159 mutant.
FIG. 3.
A dnaN159 mutant strain is sensitive to UV light. The UV sensitivities of strains MS106 (relevant genotype, dnaN+ ΔuvrB::Cmr) and MS107 (relevant genotype, dnaN159 ΔuvrB::Cmr) were measured at 30 °C (A) and 37°C (B) as described in Materials and Methods. Representative results are shown.
The DNA polymerase activity of Pol I is absolutely required in a dnaN159 mutant strain.
As part of an effort to elucidate the mechanistic basis for the UV sensitivity of the dnaN159 mutant, epistasis analysis was performed. For these experiments, a series of isogenic strains was constructed. Each pair in this series contained either the dnaN+ or dnaN159 allele, together with a mutation affecting a different partner protein known to interact with β in vitro. I hypothesized that if the UV sensitivity of the dnaN159 mutant strain were due to an impaired interaction of β159 with a particular partner protein, then isogenic dnaN159 and dnaN+ strains bearing a mutation affecting this partner would exhibit comparable sensitivities to UV. As discussed in more detail below, all of the desired strains could be constructed except for one: the combination of dnaN159 and ΔpolA was lethal.
Initially, an attempt was made to transduce the dnaN159 allele into the polA1 mutant strain JW164 using P1vir by selecting for tetracycline resistance conferred by the nearby tnaA300::Tn10 allele (dnaN and tnaA are ∼90% linked): the polA1 allele bears an amber mutation resulting in a truncated form of Pol I that retains the 5′→3′ exonuclease activity but completely lacks detectable polymerase and 3′→5′ exonuclease activities (21). However, I was unable to transduce the temperature-sensitive dnaN159 phenotype into the polA1 strain, despite the fact that the temperature sensitivity conferred by dnaN159 could be transduced into the polA+ strains MG1655 and JW165 (Table 2). Control transductions using a P1vir lysate propagated on the dnaN+ tnaA300::Tn10 strain KA473 to infect the polA1 mutant strain JW164 or the polA+ strains MG1655 and JW165 were efficient (Table 2).
TABLE 2.
A dnaN159 mutant strain requires the polymerase, but not the 5′ exonuclease activity of DNA polymerase I
| Straina |
polA allele located on:
|
No. of tnaA300::Tn10 transductants (Tcr CFU)b using:
|
||
|---|---|---|---|---|
| Chromosome | F factor | P1 vir (tnaA300::Tn10 dnaN+) | P1 vir (tnaA300::Tn10 dnaN159)c | |
| MG1655 | polA+ | None | 376 | 1,720 (18/20) |
| JW165 | polA+ | None | 12 | 307 (28/29) |
| JW164 | polA1 (polA 5′ exo) | None | 174 | 460 (0/12)d |
| CJ278 | ΔpolA::kan | None | 23 | 146 (0/60)d |
| CJ225 | ΔpolA::kan | polA+ | 2,744 | 6,456 (75/82) |
| CJ231 | ΔpolA::kan | polA1 (polA 5′ exo) | 760 | 318 (0/86) |
| CJ233 | ΔpolA::kan | polA Klenow | 2,106 | 9,744 (61/76) |
Strains are described in Table 1; JW165 and JW164 are isogenic with each other except at polA.
Tcr, tetracycline resistant.
Individual clones were picked and transferred (in duplicate) onto LB plates supplemented with tetracycline and incubated overnight at 30 or 42°C. Values in parentheses indicate the number of temperature-sensitive clones observed relative to the total number transferred.
Approximately 50% (10 of 22) of the JW164 dnaN159 transductants and approximately 20% (18 of 78) of the CJ278 dnaN159 transductants did not grow at either 30 or 42°C when transferred to LB plates.
The G174A substitution in dnaN159 destroys a HaeIII restriction site (34). Therefore, the possibility that the dnaN159 allele was transduced into strain JW164 but that the polA1 allele suppressed its temperature sensitivity was investigated by restriction analysis of PCR-amplified fragments corresponding to the middle region of the dnaN gene flanking the HaeIII site. PCR fragments corresponding to six independent tetracycline-resistant dnaN159 tnaA300::Tn10 transductants of the polA1 strain JW164 were digested with HaeIII. All six were susceptible to HaeIII digestion, confirming that none of the six contained the G174A substitution (data not shown). In contrast to these findings, ∼90% of the tetracycline-resistant dnaN159 tnaA300::Tn10 transductants of the isogenic polA+ strain JW165 tested were temperature sensitive (Table 2). Furthermore, PCR products corresponding to four independent JW165 (polA+) transductants from this same experiment were resistant to HaeIII digestion, confirming the presence of the G174A substitution (data not shown). Taken together, these findings indicate that the combination of dnaN159 and polA1 is synthetically lethal.
To determine whether the polymerase and/or 3′→5′ exonuclease activity(ies) of Pol I is required in the dnaN159 mutant, an attempt was made to transduce the dnaN159 allele into an independent set of isogenic strains differing only in their polA alleles. These strains, originally constructed by Joyce and Grindley (17), each bear the ΔpolA::kan allele on their chromosome. In addition, three of the four contain an F factor directing expression of polA+, the polA 5′ exonuclease fragment (i.e., polA1), or the polA Klenow fragment. As shown in Table 2, transduction experiments utilizing these strains reproduced results observed with strains JW164 and JW165: the dnaN159 allele could not be transduced into the strain bearing the ΔpolA::kan allele (CJ278) or into the ΔpolA::kan strain bearing the F-encoded polA1 allele (CJ231) but could be moved into the ΔpolA::kan strain bearing the F-encoded polA+ allele (CJ225). Interestingly, the dnaN159 allele could also be transduced into the ΔpolA::kan strain bearing the polA Klenow (CJ233) allele on the F factor (Table 2). These results indicate that the dnaN159 mutant requires the polymerase, but not the 5′→3′ exonuclease activity of Pol I. Further experiments are required to determine whether the dnaN159 mutant requires the 3′→5′ exonuclease activity of Pol I for viability.
The UV sensitivity of the dnaN159 mutant is suppressed by ΔdinB::kan and is exacerbated by ΔumuDC596::ermGT.
The fact that the polymerase activity of DNA Pol I is required for viability in the dnaN159 mutant suggests that an impaired β-Pol I interaction does not cause the UV sensitivity of the dnaN159 strain. Since interactions of β with DNA Pol II, Pol IV, and Pol V appear to be important for the proper function of these polymerases in vivo, the next efforts were focused on determining whether one of these gene products was responsible for the UV sensitivity of the dnaN159 mutant.
Deletion of polB, which encodes Pol II, had no discernible effect on UV sensitivity of the dnaN159 strain (data not shown). In striking contrast to this result with polB, deletion of dinB, which encodes DNA Pol IV, eliminated UV sensitivity of the dnaN159 ΔuvrB::Cmr strain (Fig. 4). Importantly, the UV sensitivities of the dnaN159 ΔuvrB::Cmr ΔdinB::kan and dnaN+ ΔuvrB::Cmr ΔdinB::kan mutants were each comparable to that of the dnaN+ ΔuvrB::Cmr (compare Fig. 2 and 3), indicating the following. (i) Deletion of dinB did not affect UV sensitivity of the dnaN+ ΔuvrB::Cmr strain. (ii) The UV sensitivity of the dnaN159 strain was suppressed by deletion of dinB, rather than being epistatic with ΔdinB::kan. Furthermore, the finding that the UV sensitivities of the isogenic dnaN+ and dnaN159 ΔdinB::kan strains were indistinguishable from each other suggests that Pol IV may compete effectively with components of Pol III for binding to the mutant β159 clamp protein in vivo, thereby affecting DNA replication and resulting in UV sensitivity.
FIG. 4.
The UV sensitivity of the dnaN159 mutant is epistatic with ΔdinB::kan. The UV sensitivities of strains MS115 (relevant genotype, dnaN+ ΔuvrB::Cmr ΔdinB::kan) and MS116 (relevant genotype, dnaN159 ΔuvrB::Cmr ΔdinB::kan) were measured at 30°C as described in Materials and Methods. Representative results are shown.
In contrast to the ΔdinB::kan strain, the UV sensitivity of the dnaN159 ΔuvrB::Cmr ΔumuDC596::ermGT strain was exacerbated by a factor of more than threefold compared to the UV sensitivity of the isogenic dnaN159 ΔuvrB::Cmr umuD+C+ strain (i.e., the dnaN159 ΔuvrB::Cmr strain was ∼30-fold more UV sensitive than the isogenic dnaN+ strain compared to the ∼10-fold sensitization observed in the umuD+C+ genetic background; compare Fig. 3 and 5). These results indicate that the UV sensitivity of the dnaN159 mutant was not due to a umuDC-dependent function and suggest that the umuDC gene products contribute in a significant way to survival in a dnaN159 mutant strain lacking functional NER.
FIG. 5.
The UV sensitivity of the dnaN159 mutant strain is exacerbated by ΔumuDC596::ermGT. The UV sensitivities of strains MS111 (relevant genotype, dnaN+ ΔumuDC596::ermGT ΔuvrB::Cmr) and MS112 (relevant genotype, dnaN159 ΔumuDC596::ermGT ΔuvrB::Cmr) were measured at 30°C as described in Materials and Methods. Representative results are shown.
Pol V-dependent SOS mutagenesis is enhanced in the dnaN159 mutant.
The enhanced UV sensitivity of the dnaN159 ΔuvrB::Cmr ΔumuD596::ermGT strain (Fig. 5) is presumably attributable to the loss of the polymerase activity of Pol V. As a test of whether Pol V plays a larger role in replication in the dnaN159 ΔuvrB::Cmr mutant (MS107) following UV irradiation, its proficiency in SOS mutagenesis was measured using an argE3(Oc)→Arg+ reversion assay (53). As shown in Fig. 6, the frequency of argE3(Oc)→Arg+ revertants was approximately 50% higher in the mutant than in the isogenic dnaN+ strain. Nearly identical results were obtained using isogenic strains bearing the ΔuvrC::Cmr allele in place of ΔuvrB::Cmr (data not shown). Furthermore, the frequencies of UV-induced His+ reversion and rifampin resistance were each approximately twofold higher in the dnaN159 mutant than in the isogenic dnaN+ strain (Fig. 6). In contrast, neither UV-induced Arg+ or His+ reversion nor rifampin resistance was observed using dnaN+ ΔuvrB::Cmr or dnaN159 ΔuvrB::Cmr strains lacking umuDC (data not shown). Taken together, these results suggest that Pol V plays a larger role in replicating damaged DNA in the dnaN159 mutant than in the isogenic dnaN+ strain.
FIG. 6.
Pol V-dependent SOS mutagenesis in a dnaN159 ΔuvrB::Cmr strain. Proficiency of strains MS106 (relevant genotype, dnaN+ ΔuvrB::Cmr) and MS107 (relevant genotype, dnaN159 ΔuvrB::Cmr) in Pol V-dependent SOS mutagenesis was measured using the argE3(Oc)→Arg+ (A) and hisG4(Oc)→His+ (B) reversion assays as described previously (53) or a rifampin resistance assay (C) as described previously (44). Strains were incubated at 30°C, and reversion and rifampin resistance were induced by irradiation with UV light (2 J/m2). The values are means ± standard errors of the means (error bars).
The global SOS response is chronically induced in the dnaN159 mutant.
Results discussed to this point suggest that replication forks are destabilized in the dnaN159 mutant, presumably due at least in part to an impaired interaction of the mutant β159 protein with Pol III, which in turn results in a chronic replication defect. If this were the case, induction of the global SOS response would be altered in the dnaN159 mutant strain. As a test of this model, a sulA::lacZ′YA::kan fusion was used to measure the kinetics of SOS induction in a pair of isogenic strains bearing either the dnaN+ or dnaN159 allele. Since sulA is SOS regulated, the extent to which the SOS response is induced (or not induced) in a population of cells is directly proportional to the level of β-galactosidase. In this analysis, strains were grown to mid-exponential phase at 30°C, and β-galactosidase activity was measured as a function of time both before and after UV irradiation.
As shown in Fig. 7, the basal level of SOS induction in the absence of DNA damage was approximately threefold higher in the dnaN159 mutant than in the isogenic dnaN+ strain. Moreover, the extent of SOS induction after UV irradiation was more robust (Fig. 7). Taken together, these findings indicate that induction of the global SOS response is altered in the dnaN159 mutant strain. However, it should be stressed that the precise extent of SOS induction in the dnaN159 mutant may be overestimated by this approach due to the fact that the steady-state level of the SOS-regulated sulA::lacZ′ gene product likely accumulates over time. Nonetheless, these results clearly indicate that the global SOS response is chronically induced at a low level in the dnaN159 strain, consistent with the idea that the mutant β159 protein exhibits impaired DNA replication. A similar level of chronic SOS induction (e.g., approximately three- to fivefold) has been reported for strains bearing mutations in other replication or repair genes, including uvrD, mutS, dam, as well as certain polA alleles (38).
FIG. 7.
Kinetics of induction of the global SOS response in the dnaN159 mutant. β-Galactosidase activity of strains MS117 (relevant genotype, dnaN+ sulA::lacZ′YA::kan) and MS118 (relevant genotype, dnaN159 sulA::lacZ′YA::kan) was assayed as described in Materials and Methods. Cultures were irradiated with UV (25 J/m2) 10 min after the initial β-galactosidase determination (0 min). The values are means ± standard errors of the means (error bars). Not all error bars are large enough to be visible beyond the symbols.
The temperature sensitivity of the dnaN159 mutant is exacerbated by lexA alleles that affect SOS induction.
In light of the fact that Pol IV and Pol V are SOS regulated, the finding that DNA polymerase usage appears to be altered in the dnaN159 mutant suggests that chronic SOS induction might be crucial for viability of the dnaN159 mutant. Alternatively, given the finding that inactivation of Pol IV (ΔdinB::kan) suppressed the UV sensitivity of the dnaN159 mutant, constitutive induction of the SOS response might perturb growth of the dnaN159 mutant. To distinguish between these two possibilities and determine whether the observed chronic, low-level induction of the SOS response influences viability of the dnaN159 mutant, the growth characteristics of isogenic strains bearing dnaN+ or dnaN159 together with either the lexA+, lexA51(Def), or lexA3(Ind−) allele were compared at different temperatures. The SOS response is constitutively expressed in mutants bearing the lexA51(Def) allele. In contrast, the lexA3(Ind−) gene product is refractory to RecA-mediated cleavage, and hence SOS cannot be induced in strains bearing the lexA3(Ind−) allele.
In most genetic backgrounds tested, the dnaN159 mutation confers thermolability at temperatures in excess of 37°C (14; data not shown). This thermolability was only modestly enhanced by the lexA3(Ind−) allele, as judged by the small size of the colonies at 37°C (Fig. 8). In striking contrast to the dnaN159 lexA3(Ind−) strain, the dnaN159 lexA51(Def) strain grew poorly at 30°C and was considerably more temperature sensitive than its isogenic dnaN159 lexA+ partner, displaying significantly reduced growth at both 35 and 37°C. Consistent with previous reports, all three dnaN+ derivatives grew well at all temperatures tested. Taken together, these findings indicate that a moderate level of chronic SOS induction is crucial for optimal growth of the dnaN159 mutant: either constitutive induction or, albeit to a lesser extent, the inability to induce the SOS response, is detrimental to the growth of the dnaN159 mutant.
FIG. 8.
Influence of different lexA alleles on the temperature sensitivity of the dnaN159 mutant. Exponential cultures of E. coli strains grown at 30°C in LB medium were serially diluted in sterile 0.8% saline as indicated and spotted onto solid LB medium. Plates were incubated at the indicated temperatures, and E. coli strains are described in Table 1. Although the lexA3(Ind−) dnaN159 mutant used in this study was largely proficient for growth at 37°C, Grompone et al. (14) reported that a similar strain displayed reduced survival (approximately 1,000-fold) at 37°C. I suggest that the different growth phenotypes observed for these two lexA3(Ind−) dnaN159 strains relates to their different pedigrees. Consistent with this notion, Michel and colleagues have observed significantly different growth phenotypes for the dnaE486 allele at 37°C, depending upon the genetic background of the strain (Bénédicte Michel, personal communication).
DISCUSSION
The purpose of the experiments discussed in this report was to test the model that the β clamp plays a biologically important role in coordinating protein traffic at the replication fork. The results of this study clearly demonstrate that the G66E and/or G174A substitutions of the mutant β159 clamp protein affect its interactions with the α catalytic subunit of Pol III, as well as interactions with a number of other partner proteins. As a result, the dnaN159 mutant strain displayed several striking phenotypes with respect to DNA replication and repair, consistent with the idea that the clamp acts as a scaffold to help coordinated replication and repair. These phenotypes include the following: (i) UV sensitivity which is dependent upon functional Pol IV, (ii) an enhanced Pol V-dependent mutation rate, (iii) chronic (and altered) induction of the global SOS response, and (iv) an absolute dependence upon the polymerase activity of Pol I for viability. Taken together, these findings indicate that DNA polymerase usage is altered in the dnaN159 mutant strain. Furthermore, these results provide direct evidence for a model in which the β clamp plays a biologically important role in coordinating protein traffic at the replication fork.
The finding that inactivation of dinB (Pol IV) suppressed the UV sensitivity of the dnaN159 ΔuvrB::Cmr strain suggests that Pol IV is able to compete effectively with Pol III for binding to the mutant β159 clamp to gain access to the replication fork and as a result confers UV sensitivity upon the dnaN159 mutant strain. On the basis of recent structural analyses of the β-Pol IV complex (4, 6), G66 is well removed from the surface of β that contacts Pol IV (Fig. 1). Although G174 is near the surface that contacts Pol IV, this residue does not interact directly with the polymerase. In contrast to these findings with Pol IV, the results discussed herein suggest that G174 (and/or G66E) is important for interaction with the α catalytic subunit of Pol III (Fig. 2). Thus, the G174A substitution (and possibly the G66E substitution as well) appears to differentially affect interactions of β with Pol III and Pol IV. This finding, suggesting that Pol III and Pol IV make unique contacts with β in addition to the hydrophobic pocket, is consistent with previous findings regarding interactions of β with the UmuD, UmuD′, and α catalytic subunit of Pol III (9).
In contrast to Pol IV, which impairs DNA replication and/or repair in the dnaN159 mutant after UV irradiation, Pol V plays an important role by enhancing survival of the dnaN159 mutant after UV irradiation (Fig. 5). The finding that Pol V-dependent mutagenesis is enhanced roughly twofold in the dnaN159 ΔuvrB::Cmr strain (Fig. 6) suggests that Pol V, like Pol IV, is able to effectively compete with Pol III for the β clamp, particularly after UV irradiation. However, unlike Pol IV, this competition is apparently beneficial to the dnaN159 mutant. The opposing consequences of the presumed competition of Pol IV and Pol V with Pol III in the dnaN159 mutant is presumably related to the fact that Pol V is considerably more proficient at replicating over UV-induced lesions, while Pol IV prefers to bypass other classes of DNA lesions (49).
DNA Pol I, like Pol V, plays a critically important role in the dnaN159 mutant strain. This conclusion is based on the fact that the dnaN159 allele confers synthetic lethality when combined with either the polA1 or ΔpolA::kan allele. Mutant E. coli strains deficient in the Pol I polymerase activity display a defect in the joining of Okazaki fragments (35). Interestingly, the growth defect of the polA mutant is largely suppressed by overexpression of the α subunit of Pol III (55). Taken together, these findings suggest that both Pol I and Pol III play critically important roles in Okazaki fragment synthesis and maturation. The impaired ability of the mutant β159 protein to interact with Pol III presumably results in an elevated number of single-stranded DNA gaps, particularly on the lagging strand, which in turn causes chronic, low-level induction of the global SOS response (Fig. 7). The requirement for the polymerase activity of Pol I in the dnaN159 mutant may relate to an increased role for Pol I in Okazaki fragment maturation and or repair synthesis (i.e., gap filling). Likewise, Pol V may also play an important role in Okazaki fragment synthesis and/or gap filling, thus explaining the higher UV-induced mutation frequency observed in the dnaN159 mutant. It is not yet known whether the mutant β159 protein has impaired interactions with Pol I or whether this interaction is even required for the proper function of Pol I in vivo. Finally, the dnaN159 ΔuvrB::Cmr Δ(araD-polB)::Ω strain was indistinguishable from the isogenic dnaN159 ΔuvrB::Cmr polB+ strain, suggesting that Pol II is unable to compete effectively with Pol III for the β clamp, at least under the conditions examined in this work. In conclusion, Pol I and Pol V appear to play more significant roles in the dnaN159 mutant, while Pol IV appears to impair growth after UV irradiation.
Duzen et al. previously demonstrated that UmuD, UmuD′, and the α catalytic subunit of Pol III each contact unique sites on β that are well removed from the hydrophobic pocket which interacts with the eubacterial clamp-binding motif (9). This fact, taken together with the findings of this study that DNA polymerase usage is altered in the dnaN159 mutant compared to the otherwise isogenic dnaN+ strain, suggests that these unique contacts are critically important for the temporal and spatial regulation of protein-protein interactions involving β and its different partners. Furthermore, the results of this study suggest that interactions involving these unique surfaces might play an important role in the process by which one partner gains access to a clamp that is on DNA but in association with a different partner. In this case, initial interaction of the incoming partner with β that is already in complex with a different partner might involve a unique surface of the clamp that is not recognized by the resident partner and is therefore accessible to the arriving protein. Subsequent interactions involving the clamp, two partner proteins, DNA, and possibly other proteins at the replication fork, presumably effect release of the first partner from the DNA and/or clamp, resulting in a partner switch. A key feature of this model is that by forcing partners to utilize a combination of unique and common contacts with the clamp, the cell may impose a hierarchy on the different partners such that only certain proteins are able to compete effectively with a particular β-partner-DNA complex for access to the replication fork. In other words, in order for one partner to effect a switch, the resident partner must not occlude the surface on β that is uniquely recognized by the incoming protein. This model would explain how β exchanges one partner for another despite the fact that most, if not all, of its partners interact with a common surface of the clamp (i.e., the hydrophobic pocket located at the base of the C-terminal tail of each β protomer, which interacts with the eubacterial clamp-binding motif) and would begin to explain how the cell might regulate the temporal ordering of the different interactions involving β to promote DNA transactions, such as Okazaki fragment maturation, which requires the repetitive sequential actions of Pol III, Pol I, and DNA ligase.
It was suggested previously that interactions of the different E. coli umuDC gene products with the β clamp constitute part of a higher-order regulatory system termed replication fork management that acts to control both the nature and order of the events that take place at the replication fork (45-48). Results discussed in this report provide further evidence in support of this model by suggesting a role for β in helping to effect switches between different DNA polymerases at the replication fork during replication and repair in vivo. Further genetic and biochemical characterization of the dnaN159 mutant, as well as other dnaN alleles, will better define the role of the β clamp in these switches.
Acknowledgments
I thank the anonymous reviewers for their comments. I also thank Mary Berlyn (E. coli Genetic Stock Center, Yale University), Justin Courcelle (Mississippi State), Nora Goosen (Leiden University), Cathy Joyce (Yale University), Tsutomu Katayama (Kyushu University), Charles McHenry (University of Colorado, Health Sciences Center), Haruo Ohmori (Kyoto University), Bernie Strauss (University of Chicago), and Roger Woodgate (NICHD/NIH) for generously providing E. coli strains; Jill Duzen for help with cloning plasmid constructs used in the yeast two-hybrid analysis; Steven Sandler (University of Massachusetts, Amherst) for helpful comments on an earlier version of this manuscript; and Kenneth Blumenthal (University at Buffalo) and members of my lab for comments and many helpful discussions.
This work was supported in part by Public Service Health grant GM066094; Howard Hughes Medical Institute Biomedical Research Support Program grant 53000261 to the School of Medicine and Biomedical Sciences, University at Buffalo, SUNY; and start-up funds from the School of Medicine and Biomedical Sciences, University at Buffalo, SUNY.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2001. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Blattner, F. R., G. R. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bonner, C. A., P. T. Stukenberg, M. Rajagopalan, R. Eritja, M. O'Donnell, K. McEntee, H. Echols, and M. F. Goodman. 1992. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J. Biol. Chem. 267:11431-11438. [PubMed] [Google Scholar]
- 4.Bunting, K. A., S. M. Roe, and L. H. Pearl. 2003. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 22:5883-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgers, P. M., A. Kornberg, and Y. Sakakibara. 1981. The dnaN gene codes for the beta subunit of DNA polymerase III holoenzyme of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:5391-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnouf, D. Y., V. Olieric, J. Wagner, S. Fujii, J. Reinbolt, R. P. Fuchs, and P. Dumas. 2004. Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J. Mol. Biol. 335:1187-1197. [DOI] [PubMed] [Google Scholar]
- 7.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 8.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duzen, J. M., G. C. Walker, and M. D. Sutton. 2004. Identification of specific amino acid residues in the E. coli beta processivity clamp involved in interactions with DNA polymerase III, UmuD and UmuD′. DNA Repair 3:301-312. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez De Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg, E. C., W. J. Feaver, and V. L. Gerlach. 2000. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc. Natl. Acad. Sci. USA 97:5681-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg, E. C., R. Wagner, and M. Radman. 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296:1627-1630. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 14.Grompone, G., M. Seigneur, S. D. Ehrlich, and B. Michel. 2002. Replication fork reversal in DNA polymerase III mutants of Escherichia coli: a role for the beta clamp. Mol. Microbiol. 44:1331-1339. [DOI] [PubMed] [Google Scholar]
- 15.Ho, C., O. I. Kulaeva, A. S. Levine, and R. Woodgate. 1993. A rapid method for cloning mutagenic DNA repair genes: isolation of umu-complementing genes from multidrug resistance plasmids R391, R446b, and R471a. J. Bacteriol. 175:5411-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeruzalmi, D., O. Yurieva, Y. Zhao, M. Young, J. Stewart, M. Hingorani, M. O'Donnell, and J. Kuriyan. 2001. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106:417-428. [PubMed] [Google Scholar]
- 17.Joyce, C. M., and N. D. Grindley. 1984. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J. Bacteriol. 158:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 19.Kelman, Z., and M. O'Donnell. 1995. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64:171-200. [DOI] [PubMed] [Google Scholar]
- 20.Kong, X. P., R. Onrust, M. O'Donnell, and J. Kuriyan. 1992. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69:425-437. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. Freeman, New York, N.Y.
- 22.Krishna, T. S., X. P. Kong, S. Gary, P. M. Burgers, and J. Kuriyan. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79:1233-1243. [DOI] [PubMed] [Google Scholar]
- 23.Kurz, M., B. Dalrymple, G. Wijffels, and K. Kongsuwan. 2004. Interaction of the sliding clamp beta-subunit and Hda, a DnaA-related protein. J. Bacteriol. 186:3508-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenne-Samuel, N., J. Wagner, H. Etienne, and R. P. Fuchs. 2002. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 3:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 26.Lopez de Saro, F. J., R. E. Georgescu, M. F. Goodman, and M. O'Donnell. 2003. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 22:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez de Saro, F. J., and M. O'Donnell. 2001. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl. Acad. Sci. USA 98:8376-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71:71-100. [DOI] [PubMed] [Google Scholar]
- 29.Maki, S., and A. Kornberg. 1988. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J. Biol. Chem. 263:6561-6569. [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Moarefi, I., D. Jeruzalmi, J. Turner, M. O'Donnell, and J. Kuriyan. 2000. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 296:1215-1223. [DOI] [PubMed] [Google Scholar]
- 32.Moolenaar, G. F., C. Moorman, and N. Goosen. 2000. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J. Bacteriol. 182:5706-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murli, S., T. Opperman, B. T. Smith, and G. C. Walker. 2000. A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth. J. Bacteriol. 182:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohmori, H., M. Kimura, T. Nagata, and Y. Sakakibara. 1984. Structural analysis of the dnaA and dnaN genes of Escherichia coli. Gene 28:159-170. [DOI] [PubMed] [Google Scholar]
- 35.Okazaki, R., M. Arisawa, and A. Sugino. 1971. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc. Natl. Acad. Sci. USA 68:2954-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opperman, T., S. Murli, B. T. Smith, and G. C. Walker. 1999. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 96:9218-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rangarajan, S., R. Woodgate, and M. F. Goodman. 1999. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc. Natl. Acad. Sci. USA 96:9224-9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.SaiSree, L., M. Reddy, and J. Gowrishankar. 2000. lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli. J. Bacteriol. 182:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakakibara, Y., and T. Mizukami. 1980. A temperature-sensitive Escherichia coli mutant defective in DNA replication: dnaN, a new gene adjacent to the dnaA gene. Mol. Gen. Genet. 178:541-553. [DOI] [PubMed] [Google Scholar]
- 40.Strauss, B. S., R. Roberts, L. Francis, and P. Pouryazdanparast. 2000. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J. Bacteriol. 182:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stukenberg, P. T., P. S. Studwell-Vaughan, and M. O'Donnell. 1991. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 266:11328-11334. [PubMed] [Google Scholar]
- 42.Sutton, M. D., M. F. Farrow, B. M. Burton, and G. C. Walker. 2001. Genetic interactions between the Escherichia coli umuDC gene products and the beta processivity clamp of the replicative DNA polymerase. J. Bacteriol. 183:2897-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutton, M. D., and J. M. Kaguni. 1995. Novel alleles of the Escherichia coli dnaA gene are defective in replication of pSC101 but not of oriC. J. Bacteriol. 177:6657-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutton, M. D., M. Kim, and G. C. Walker. 2001. Genetic and biochemical characterization of a novel umuD mutation: insights into a mechanism for UmuD self-cleavage. J. Bacteriol. 183:347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutton, M. D., I. Narumi, and G. C. Walker. 2002. Posttranslational modification of the umuD-encoded subunit of Escherichia coli DNA polymerase V regulates its interactions with the beta processivity clamp. Proc. Natl. Acad. Sci. USA 99:5307-5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutton, M. D., T. Opperman, and G. C. Walker. 1999. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD′ interact physically with the replicative DNA polymerase. Proc. Natl. Acad. Sci. USA 96:12373-12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 48.Sutton, M. D., and G. C. Walker. 2001. Managing DNA polymerases: coordinating DNA replication, DNA repair, and DNA recombination. Proc. Natl. Acad. Sci. USA 98:8342-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang, M., P. Pham, X. Shen, J.-S. Taylor, M. O'Donnell, M. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerase IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 50.Tinker, R. L., K. P. Williams, G. A. Kassavetis, and E. P. Geiduschek. 1994. Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell 77:225-237. [DOI] [PubMed] [Google Scholar]
- 51.Tsurimoto, T. 1999. PCNA binding proteins. Front. Biosci. 4:D849-D858. [DOI] [PubMed] [Google Scholar]
- 52.Villarroya, M., I. Perez-Roger, F. Macian, and M. E. Armengod. 1998. Stationary phase induction of dnaN and recF, two genes of Escherichia coli involved in DNA replication and repair. EMBO J. 17:1829-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker, G. C. 1977. Plasmid (pKM101)-mediated enhancement of repair and mutagenesis: dependence on chromosomal genes in Escherichia coli K-12. Mol. Gen. Genet. 152:93-103. [DOI] [PubMed] [Google Scholar]
- 54.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 55.Witkin, E. M., and V. Roegner-Maniscalco. 1992. Overproduction of DnaE protein (alpha subunit of DNA polymerase III) restores viability in a conditionally inviable Escherichia coli strain deficient in DNA polymerase I. J. Bacteriol. 174:4166-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodgate, R. 1992. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221-225. [DOI] [PubMed] [Google Scholar]
- 57.Yasuda, T., K. Morimatsu, T. Horii, T. Nagata, and H. Ohmori. 1998. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 17:3207-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]