Abstract
Intracellular glucose in Escherichia coli cells imported by phosphoenolpyruvate-dependent phosphotransferase system-independent uptake is phosphorylated by glucokinase by using ATP to yield glucose-6-phosphate. Glucokinases (EC 2.7.1.2) are functionally distinct from hexokinases (EC 2.7.1.1) with respect to their narrow specificity for glucose as a substrate. While structural information is available for ADP-dependent glucokinases from Archaea, no structural information exists for the large sequence family of eubacterial ATP-dependent glucokinases. Here we report the first structure determination of a microbial ATP-dependent glucokinase, that from E. coli O157:H7. The crystal structure of E. coli glucokinase has been determined to a 2.3-Å resolution (apo form) and refined to final Rwork/Rfree factors of 0.200/0.271 and to 2.2-Å resolution (glucose complex) with final Rwork/Rfree factors of 0.193/0.265. E. coli GlK is a homodimer of 321 amino acid residues. Each monomer folds into two domains, a small α/β domain (residues 2 to 110 and 301 to 321) and a larger α+β domain (residues 111 to 300). The active site is situated in a deep cleft between the two domains. E. coli GlK is structurally similar to Saccharomyces cerevisiae hexokinase and human brain hexokinase I but is distinct from the ADP-dependent GlKs. Bound glucose forms hydrogen bonds with the residues Asn99, Asp100, Glu157, His160, and Glu187, all of which, except His160, are structurally conserved in human hexokinase 1. Glucose binding results in a closure of the small domains, with a maximal Cα shift of ∼10 Å. A catalytic mechanism is proposed that is consistent with Asp100 functioning as the general base, abstracting a proton from the O6 hydroxyl of glucose, followed by nucleophilic attack at the γ-phosphoryl group of ATP, yielding glucose-6-phosphate as the product.
Growth of Escherichia coli by using various fermentable sugars as carbon sources, including glucose, maltose, galactose, and sucrose, primarily involves the phosphoenolpyruvate-dependent phosphotransferase system (PTS) (reviewed in reference 54). However, a secondary, PTS-independent system for utilization of glucose also exists, consisting of glucose uptake by galactose permease (GalP; galactose proton symporter), followed by phosphorylation by glucokinase (GlK; EC 2.7.1.2) to yield the metabolic intermediate glucose-6-phosphate. Although glk mutant strains of E. coli (43) and Bacillus subtilis (63) are not visibly physiologically impaired, this enzyme retains the important function of phosphorylating any free intracellular glucose. Free cytoplasmic glucose may arise from di-saccharide hydrolysis, for example, the cleavage of trehalose phosphate in Bacillus subtilis (25), or from metabolism of maltose or isomaltose (61). Indeed, studies of a PTS− E. coli strain have shown that a growth rate approximately 89% that of wild-type cells can be obtained by overexpression of GalP alone, suggesting that glucose transport, not GlK-dependent phosphorylation, is limiting growth (28). There is considerable industrial interest in enhancing the ability of E. coli to transport and phosphorylate glucose in a PTS-independent manner due to the ability of these strains to direct more carbon flux to aromatic synthesis pathways (20, 21, 28).
Microbial glucokinases can be divided into three families based on sequence comparisons. Group I (protein families database [PFAM] accession number PF04587) (7) consists of ATP- and ADP-dependent glucokinases (EC 2.7.1.147) from archaea (reviewed in reference 60) and have also been recently identified in eukaryotes (56). This group also includes a novel, bifunctional ADP-dependent GlK/PfK enzyme from Methanococcus jannaschii (59). Group I is the only group for which crystal structures have been determined to date. Group II glucokinases (PFAM accession numbers PF02685 and COG0837) are ATP-dependent glucokinases that do not have the classical repressor open reading frame kinase (ROK) sequence motif (69) and consist of over 50 full and partial protein sequences, including E. coli GlK (43). The overwhelming number of these sequences (49 of 52) are from bacteria, including both cyanobacteria (8 sequences) and proteobacteria (41 sequences). Group III consists of ATP-dependent glucokinases from both archaea (24) and bacteria (23, 63, 64) that possess the ROK sequence signature (PFAM accession number PF00480) and have a conserved CXCGX(2)GCXE motif (conserved Cys residues are highlighted) (42). Mutagenesis of any of these Cys residues to Ala in Bacillus subtilis GlK results in an inactive enzyme, suggesting their functional importance (42). The ATP/polyphosphate glucokinase from Mycobacterium tuberculosis (30) and glucomannokinase from Anthrobacter sp. strain KM (45) as well as the strictly polyphosphate-dependent GlK from Microlunatus phosphovorus (66) are also members of this group.
Enzymes that transfer a phosphoryl group to the 6 hydroxyl group of a hexose include both hexokinases (EC 2.7.1.1), having broad sugar specificity, and glucokinases (EC 2.7.1.2), more specific for glucose (reviewed in reference 72). In many cases, these enzymes have been somewhat arbitrarily classified as one or the other, owing to incomplete experimental data on their sugar specificity. Glucokinases that utilize ATP as a phosphoryl donor may, in addition, use other nucleoside triphosphates (37, 58) or polyphosphate (reviewed in reference 53) as substrates or both ATP and polyphosphate (30, 52). Recently, a strictly polyphosphate-dependent glucokinase (EC 2.7.1.63) has been purified from Microlunatus phosphovorus (66). GlK from E. coli has been cloned, purified, and studied kinetically (43). It is a cytoplasmic enzyme having 321 residues and a monomeric mass of 35 kDa. This enzyme shows much greater activity with glucose than with either mannose or galactose and shows no activity with fructose, thereby defining it as a glucokinase (43).
Several crystal structures of hexokinases have been determined, including hexokinase A(PI) (10) and B(PII) (4, 38), both from Saccharomyces cerevisiae, rat type I and Schistosoma mansoni hexokinase (46), human brain type I (1, 2, 3, 57), and, recently, human type IV (glucokinase) (36). Only three microbial ADP-dependent GlK structures are available, all from sequence group I. These include the enzyme from Thermococcus litoralis bound to ADP (33), Pyrococcus horikoshii GlK (70), and Pyrococcus furiosus GlK bound to glucose and AMP (34). However, no structures of microbial glucokinases from either group II or group III are currently known.
We have determined the first structure of a member of the group II microbial glucokinase family, that from E. coli O157:H7 (ecGlK). This structure reveals a dimeric enzyme that has a similar fold to human and yeast hexokinases, indicative of a common ancestral enzyme, although the sequence identity is low (16 to 18%). Key residues responsible for glucose and nucleotide binding and catalysis are conserved, both in sequence as short motifs and in structure. The structure of ecGlK is distinct from that of the ADP-dependent GlKs from Archaea. Glucose binding results in domain closure, as found in both archaeal GlKs and hexokinases.
MATERIALS AND METHODS
Cloning, expression, and purification.
The gene for ecGlK was amplified from E. coli O157:H7 genomic DNA (50) obtained from the American Type Culture Collection by using primers from Integrated DNA Technologies (Coralville, Iowa) and Pfu DNA polymerase (Stratagene, La Jolla, Calif.). The amplicon was cloned into a pET15 vector derivative in frame with an N-terminal, noncleavable His6 tag by using a BamHI/EcoRI cloning strategy and was transformed into E. coli BL21(DE3) for expression. For protein production, a 1-liter culture of LeMaster medium (27) containing ampicillin at a concentration of 100 μg/liter was inoculated with a 100-ml overnight culture and grown for 2 h at 37°C. isopropyl-β-d-thiogalactopyranoside (Sigma) was added at a final concentration of 0.1 mM, and the culture continued for 6 h. Cells were harvested by centrifugation (4,000 × g at 4°C for 25 min) and stored at −20°C.
For purification, the cell pellet was resuspended in 30 ml of lysis buffer (50 mM Tris-Cl [pH 7.5], 400 mM NaCl, 10 mM β-mercaptoethanol, 5% (wt/vol) glycerol, 1× BugBuster cell lysis detergent (Novagen), 300 U of bezonase nuclease (Novagen), 1.5 mg of lysozyme (Sigma), and 1 tablet of complete EDTA-free protease inhibitor cocktail (Roche Molecular Biologicals). This lysate was applied to a 2-ml bed volume of DEAE-Sepharose (Amersham) packed in an Econo column (Bio-Rad) and equilibrated with the same buffer. Following incubation, the mixture was poured into an Econo column, and the flowthrough was collected. This was applied to a 4-ml bed volume of Ni-NTA resin (Qiagen) preequilibrated in the same buffer. Following washing, first in buffer with 1 M NaCl, followed by buffer with 0.3 M NaCl, proteins were eluted by using 25 ml of 200 mM imidazole, pH 8. Protein fractions were checked for purity by sodium dodecyl sulfate and native polyacrylamide gel electrophoresis; pure fractions were concentrated, and buffer was exchanged into 20 mM Tris (pH 8), 0.2 M NaCl, 5% glycerol, and 10 mM dithiothreitol by ultrafiltration (Centriprep, Millipore). Approximately 8 mg of pure GlK protein was obtained per liter of culture. Protein concentration was determined by the method of Bradford (14).
DLS.
Dynamic light scattering (DLS) was performed by using a DynaPro MSPRII molecular sizing instrument (Proterion Corp., Piscataway, N.J.) and analyzed by using Dynamics V6 software. A volume of 20 μl of protein (6.3 mg/ml) in buffer (20 mM Tris-Cl [pH 8], 0.2 M NaCl, 5% glycerol, 10 mM dithiothreitol) was analyzed in a 96-well plate at room temperature.
Crystallization.
Crystals of apo-ecGlK were obtained by the hanging drop vapor diffusion method in drops containing 2 μl of SeMet-labeled protein (6.8 mg/ml) and 4 μl of reservoir solution [1.7 M (NH4)2SO4, 0.1 M Tris-Cl (pH 8.5)] suspended over 1 ml of reservoir solution. The crystals belong to the space group P43212 with the cell dimensions a = b = 81.5 and c = 234.7 Å; the crystals contain two molecules in the asymmetric unit.
Crystals of the ecGlK-glc complex were obtained by the hanging drop vapor diffusion method in drops containing 2 μl of SeMet-labeled protein (6.8 mg/ml) and 4 μl of reservoir solution (18.5 to 20%, wt/vol) (PEG6000; 0.1 M Tris-Cl buffer [pH 8.5], 0.2 M MgCl2) with the addition of 2 to 3 mM ADP and 2 mM glucose suspended over 1 ml of reservoir solution. The crystals belong to the space group P21 with the following cell dimensions: a = 78.5, b = 53.6, and c = 91.1 Å; β = 113.0°. The crystals contain two molecules in the asymmetric unit.
Data collection, phasing, and refinement.
Prior to data collection the crystals were immersed for 10 s in a cryoprotectant solution containing either 2 M (NH4)2SO4, 0.1 M Tris-Cl (pH 8.5), 3 M sodium formate (for apo-ecGlK) or 23% (wt/vol) PEG6000, 0.1 M Tris-Cl buffer (pH 8.5), 0.2 M MgCl2, 20% (wt/vol) glycerol (for ecGlK-glc), mounted in a nylon loop and flash-cooled in a cold stream of N2 gas at 100 K. Data were collected at the beamlines X8C and X25 of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, by using a quantum 4 charge-coupled device detector (X8C) or Q-315 detector (X25) and were processed with either HKL2000 (49) or d*TREK (51).
The structure of ecGlK was determined by using a three-wavelength multiwavelength anomalous diffraction experiment at the Se K edge from SeMet-labeled apo-protein (Table 1). All 10 expected Se sites were identified by using the program SOLVE (68). Density modification and model building were performed by using RESOLVE (67), resulting in a model containing 73% (472 of 642) main chain and 67% (3,284 of 4,911) total atoms. Further model building was performed by using O (35), alternating with cycles of refinement by using the program Refmac5 (47). The model has been refined to a final R factor of 0.200 and Rfree of 0.271 at a 2.3-Å resolution with no σ-cutoff. The model contains two molecules in the asymmetric unit and includes residues 2 to 321 in each monomer and 387 water molecules.
TABLE 1.
Data collection and refinement statistics
| Type of value | P43212 | P21 | |||
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å<952A>) | 0.980178 | 0.979962 | 0.964711 | 0.97868 | 1.10000 |
| Cell | |||||
| a (Å<952A>) | 81.40 | 81.37 | 81.26 | 81.47 | 78.42 |
| b (Å<952A>) | 81.40 | 81.37 | 81.26 | 81.47 | 53.54 |
| c (Å<952A>) | 234.54 | 234.49 | 234.16 | 234.71 | 90.90 |
| β (deg) | 90 | 90 | 90 | 90 | 113.0 |
| Resolution (Å<952A>) | 50-2.58 | 50-2.58 | 50-2.58 | 50-2.30 | 50-2.20 |
| Last shell (Å<952A>) | 2.67-2.58 | 2.67-2.58 | 2.67-2.58 | 2.38-2.30 | 2.28-2.20 |
| Rsym | 0.079 (0.220) | 0.071 (0.198) | 0.073 (0.237) | 0.046 (0.185) | 0.051 (0.171) |
| Completeness (%) | 94.7 (99.9) | 92.4 (96.0) | 94.6 (100) | 96.3 (84.9) | 89.5 (63.4) |
| I/σ(I) | 14.4 (13.9) | 14.9 (6.5) | 13.4 (11.2) | 23.4 (9.3) | 12.1 (3.4) |
| No. of reflections | 250,724 | 114,817 | 198,266 | 220,456 | 167,451 |
| No. of unique reflections | 24,444 | 22,045 | 24,279 | 34,804 | 31,835 |
| Wilson B-factor | 42.0 | 46.1 | |||
| Refinement | |||||
| R/Rfree | 0.200/0.271 | 0.193/0.265 | |||
| No. of non-H protein atoms, chain A (B) | 2,451 (2,457) | 2,452 (2,470) | |||
| No. of water molecules | 387 | 348 | |||
| B-factor (Å<952A>2), chain A (B) | |||||
| Main chain atoms | 29.2 (41.6) | 44.4 (48.8) | |||
| Side chain atoms | 31.8 (44.1) | 47.4 (51.6) | |||
| Water molecules | 40.0 | 51.9 | |||
| Glucose molecules | 37.7 (40.4) | ||||
| rmsd bond length (Å<952A>) | 0.019 | 0.015 | |||
| rmsd bond angle (°) | 1.75 | 1.65 | |||
| Ramachandran plot (% of residues in region) | |||||
| Most favored | 89.3 | 89.5 | |||
| Disallowed | 0.0 | 0.2 | |||
The structure of the ecGlK-glc complex was solved by molecular replacement by using the program MOLREP (71) from the CCP4 suite (73) with apo-ecGlK used as the starting model. Refinement was performed by using the program REFMAC5 (47), giving a final R factor of 0.193 and Rfree of 0.265 at a 2.2-Å resolution with no σ-cutoff. The model contains two molecules in the asymmetric unit and includes residues 3 to 321 (monomer A) and 2 to 321 (monomer B), one molecule of glucose bound to each monomer, and 465 water molecules. Data collection and refinement statistics are summarized in Table 1. Both models have good geometry without outliers, as shown by the program PROCHECK (39).
Coordinates.
Coordinates of ecGlK have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) (11) with accession codes 1Q18 (apo form) and 1SZ2 (glucose complex).
RESULTS AND DISCUSSION
Structure of the GlK monomer.
The structure of apo-ecGlK from the P43212 crystal form was determined by a three-wavelength multiwavelength anomalous diffraction experiment from SeMet-labeled protein (26) and refined to an R factor of 0.200 (Rfree = 0.271) at a 2.3-Å resolution. This model contains two molecules in the asymmetric unit and includes residues 2 to 321 in each monomer. Data collection and refinement statistics are presented in Table 1.
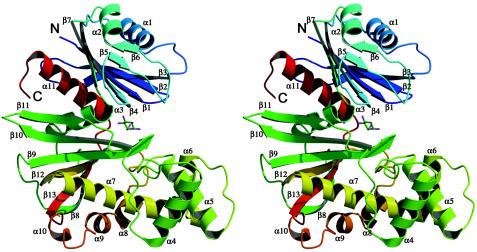
Each monomer of ecGlK consists of a small α/β domain made of two noncontiguous segments (residues 2 to 110 and 300 to 321) and a larger α+β domain (residues 111 to 299) (Fig. 1). A total of 13 β-strands and 11 α-helices make up the monomer and are labeled consecutively from the N to C terminus, as shown in Fig. 1. The small domain consists of a single, central, five-stranded mixed β-sheet (β3-β2-β1-β4-β7), with β2 antiparallel to the rest. This β-sheet is flanked on one face by a pair of α-helices (α1 and α2) and a β-hairpin (β5-β6) and on the opposite face by a pair of α-helices (α3 and α11). This five-turn-long α-helix (α11, residues 301 to 321) is not contiguous with the rest of the domain and comes from the C-terminal end of the monomer. It forms part of the interface between the large and small domains.
FIG. 1.
Ribbon model of the ecGlK monomer. Model uses rainbow colors from the N terminus (blue) to the C terminus (red). β-strands and α-helices are numbered sequentially from the N to C terminus. This and subsequent figures were prepared with either PyMOL (18; http://www.pymol.org) or Molscript (19) and Raster3D (41).
The large domain contains a mixed, six-stranded β-sheet (β8-β13-β12-β9-β10-β11) with β8 and β10 antiparallel to the rest. One face of this sheet is adjacent to a cluster of seven α-helices (α4 to α10), while the other face is directed toward the small domain. A longer central helix, α7, forms the core of the α-helix cluster. The interface between the large and small domains forms the active site cleft ∼28 Å wide and ∼20 Å deep. A single helix, α3 (residues 100 to 109), connects the two domains.
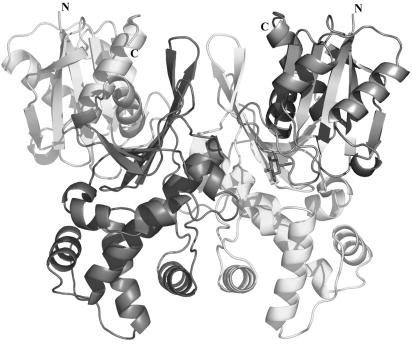
ecGlK dimer structure.
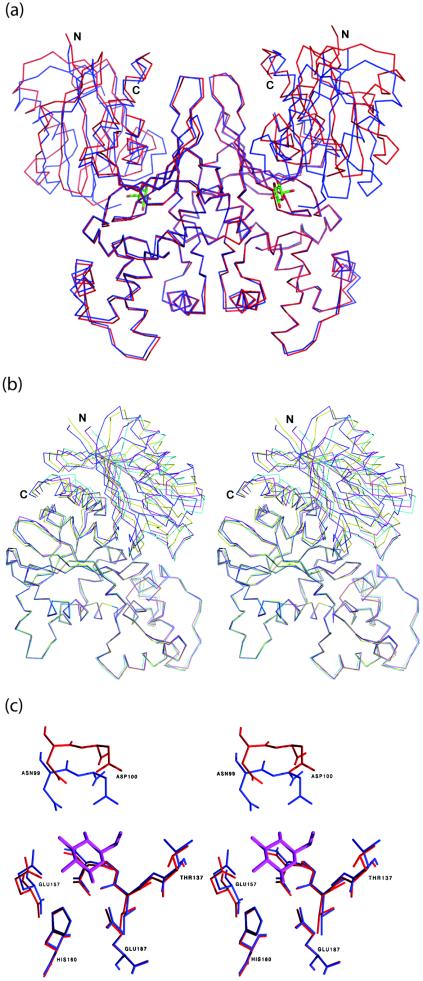
Analysis of purified ecGlK by DLS suggested it to be a dimer in solution. Crystallographic analysis of ecGlK revealed a dimer within the asymmetric unit. The association of ecGlK monomers to form the dimer structure (Fig. 2) occurs through interactions between the large domains of each monomer such that both active site clefts are solvent accessible. Secondary structure elements contributing to the dimer interface include helix α4 and adjacent loops, the C-terminal tip of helix α7, strand β10 and the loop connecting this to strand β11 (Fig. 2). The total buried surface area upon dimer formation is ∼3,060 Å2 for both monomers, equivalent to ∼10% of the accessible surface area of each monomer. Generally, ATP-dependent GlKs of bacterial or archaeal origin are dimeric enzymes, although the GlK from the archaeon Aeropyrum pernix is monomeric (24, 58), as is human glucokinase (36).
FIG. 2.
Ribbon model of the ecGlK dimer. The small domain is depicted in light gray, and the large domain is in dark gray for one monomer (left); the corresponding domains of the second monomer (right) are shown in dark and light gray, respectively. The two glucose molecules bound to the dimer are shown in stick representation.
Many hydrogen bonds and van der Waals contacts are formed between the two monomers (chains A and B) of ecGlK. Hydrogen bonds are formed between the side chains of Arg150(A) and Asp148(B), as well as Asp162(A) and Lys284(B). There are also backbone H-bonds between Leu250(A) and Glu157(B), as well as numerous water-mediated hydrogen bonds. Contacts between the two monomers are also provided through stacking interactions between the side chains of Phe287 of one monomer and His160 of the other. As described in the following, Glu157 and His160 are also part of the glucose-binding site.
Comparison with yeast and human hexokinases.
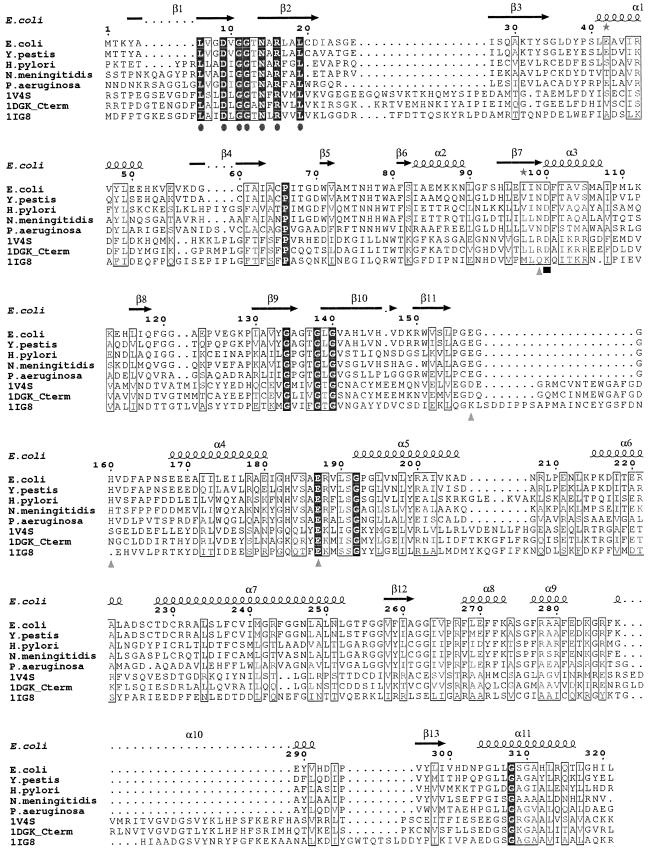
A recent exhaustive analysis of over 17,000 sequences of kinases and their relationship to structure (16) classified ecGlK (COG0837) within the RNase H-like kinase group, with representative structures from hexokinase (4), glycerol kinase (32), and acetate kinase (15). These kinases are also members of the sugar kinase-heat shock protein 70-actin superfamily (12, 31) and are classified within the actin-like ATPase superfamily within SCOP (5). An alignment of selected GlK protein sequences is shown in Fig. 3.
FIG.3.
Sequence alignment of representative members of group II glucokinases (PFAM PF02685) as well as human glucokinase (PDB 1V4S) (36), human brain hexokinase I (PDB 1DGK) (3), and hexokinase PII (PDB 1IG8) (38) by means of the ClustalW program (17). Identical residues are shaded. Residues associated with glucose binding (gray triangles) and the catalytic Asp (black rectangle) are indicated. The conserved β-strand-loop-β-strand motif associated with ATP-binding (Leu6-Leu19) with identical residues indicated by dark gray ovals is highlighted. The secondary structure elements (α-helices and β-strands) of ecGlK are depicted above the sequence alignment. This figure was prepared by using ESPript (22).
A search for similar structures by using the DALI server (29; http://www.ebi.ac.uk/dali/) found that the most similar structures were human brain hexokinase I (PDB code 1QHA) (57) and hexokinase B (also known as hexokinase PII) from S. cerevisiae (PDB 2YHX) (4). Yeast hexokinase PII is somewhat larger than ecGlK, comprising 486 residues (38), while human hexokinase type I is much larger, consisting of two ∼50-kDa chains (72). Each chain contains two globular units, an N-terminal regulatory domain (residues 1 to 474) and a C-terminal catalytic domain (residues 475 to 917) separated by a long α-helical linker (1). Each domain of human hexokinase I is structurally similar to the yeast hexokinase monomer (1) and to monomeric human glucokinase (36). Superposition of ecGlK with the human brain hexokinase I catalytic domain gave a root mean square deviation (rmsd) of 1.66 Å for 157 Cα atoms and 1.88 Å for 188 Cα atoms for human glucokinase, while a similar superposition between ecGlK and yeast hexokinase PII gave an rmsd of 1.77 Å for 149 Cα atoms. A redetermination of the yeast hexokinase PII structure reported by Anderson et al. (4) with the correct amino acid sequence (PDB 1IG8) (38) reveals a very similar fold for the two yeast PII hexokinase structures.
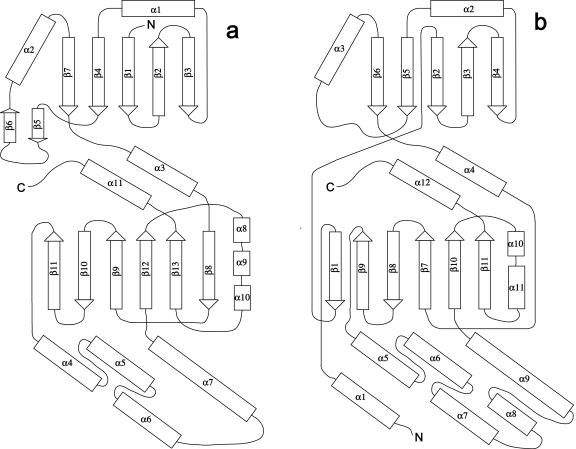
A superposition of ecGlK and yeast and human hexokinases is shown in Fig. 4. Structural similarities are most pronounced in the core regions of the structures. This structural similarity exists despite low (∼16 to 18%) sequence identity between ecGlK and the two hexokinases. Bacterial glucokinases, of which ecGlK is a member, along with yeast and human hexokinases had previously been identified as members of the “hexokinase family,” sharing several short sequence motifs and predicted to have similar folds (13). Comparison of the structures of ecGlK and human hexokinase I shows that the fold of the small domain is very similar, except for the absence of the β-hairpin (β5-β6) in human hexokinase I (Fig. 5). There are greater differences between the large domains (Fig. 5). The core mixed β-sheet (β13-β12-β9-β10-β11) is preserved in both hexokinase and ecGlK, although there is one extra β-strand (β8) at the C-terminal side of the sheet in ecGlK and one extra β-strand (β1) which comes from the N-terminal segment at the opposite side of the sheet in hexokinase (Fig. 5). Absent from ecGlK are specific structural features of eukaryotic hexokinases, including an N-terminal mitochondrial membrane-targeting sequence (46), and distinct, specific binding sites for the allosteric inhibitor glucose-6-phosphate (1) or nucleotide related to dissociation of hexokinase from the membrane (57). Evidently, yeast and human hexokinases as well as ecGlK evolved from a common ancestor, retaining similar overall structures while diverging in sequence.
FIG. 4.
Structure superposition of ecGlK (red), S. cerevisiae hexokinase PII (PDB 1IG8, green) (38), and the catalytic domain (residues 475 to 917) of human brain hexokinase I (PDB 1QHA, blue) (57). The dotted line delineates the small domain (top) from the large domain (bottom).
FIG. 5.
Comparison of fold topologies of ecGlK monomer (a) and catalytic domain (residues 475 to 917) (b) of human brain hexokinase I (PDB 1IG8).
A comparison of the structures of ecGlK and the ADP-dependent GlKs revealed no significant structural similarity between these two groups of glucokinases. Both enzymes consist of a small and a large domain, with the active site cleft between the domains. The folds of both small and large domains differ in ecGlK and the ADP-dependent GlKs. This result is consistent with the idea that ADP-dependent GlKs adopt a ribokinase-like fold (16, 33).
In rat hexokinase (46) and human hexokinase I (1), the dimer is formed through the association of N- and C-terminal domains from the two respective chains, yielding a head-to-tail arrangement. Dimerization is not essential for human hexokinase I in vitro, as monomeric enzyme retains activity (72). A comparison of the dimerization interfaces of ecGlK with the interface of human hexokinase I (PDB 1QHA) reveals that opposite faces of the respective monomers are associated with the dimer interface in these two enzymes, although the monomers themselves are structurally similar (Fig. 4). Similarly, the region of ecGlK involved in dimerization is not structurally conserved in the S. cerevisiae hexokinase PII (PDB 2YHX) structure (4). The monomer-dimer equilibrium of yeast PII hexokinase is influenced by pH, ionic strength, glucose concentration, and phosphorylation at Ser14 (8). Whether yeast hexokinase PII functions as a monomer or dimer in vivo is unclear.
A further difference between ecGlK and the ADP glucokinases is at the level of quaternary structure. The ADP-dependent GlK from P. furiosus does appear to be a dimer both in solution and in the crystal, with a disulfide bond between the side chains of Cys94 (34). The nearly identical enzyme activity of the C94S mutant GlK, which does not dimerize, compared with that of the native enzyme, as well as the lack of sequence conservation of this Cys residue in other GlKs suggests that the covalently linked dimer is not the physiologically relevant structure of this enzyme (34). The two ADP-dependent GlKs structurally characterized from P. horikoshii (70) and T. litoralis (33) are monomeric enzymes.
Glucose-binding site.
Crystals of apo-ecGlK soaked in reservoir solution containing low concentrations of glucose immediately cracked and dissolved, prohibiting structure determination. Cocrystallization experiments of ecGlK in the presence of glucose yielded a new crystal form in space group P21. The structure of the ecGlK-glc complex was determined by molecular replacement by using the native structure as the search model and was refined to an R factor of 0.193 (Rfree = 0.265) at a resolution of 2.2 Å.
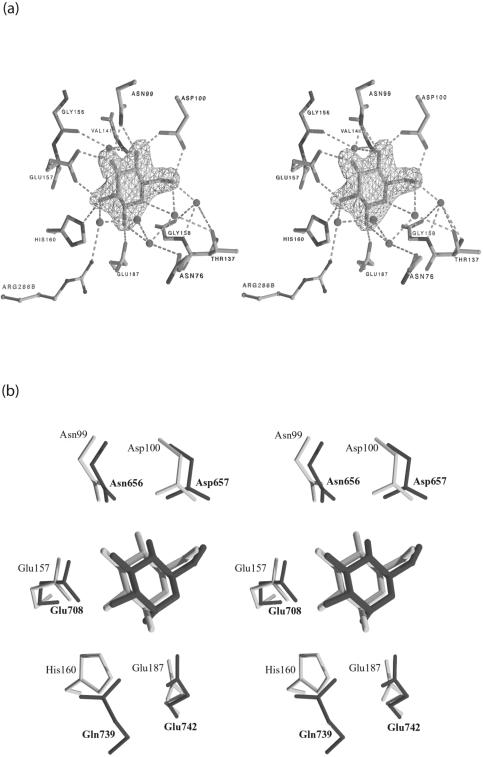
Inspection of the initial Fo-Fc difference map in the active site region revealed the presence of density corresponding to bound glucose in both ecGlK monomers (Fig. 6a). The bound glucose molecule is in a chair conformation and adopts the β-anomeric configuration. Both glucose molecules are well ordered with low B-factors, indicating good occupancy for their respective binding sites.
FIG. 6.
(a) Fo-Fc omit map of the ecGlK active site region, showing electron density for glucose. Glucose and water molecules were omitted prior to refinement. This map is contoured at the level of 3σ. Hydrogen bonds between glucose and ecGlK active site residues and waters are shown as dashed lines. (b) Structural superposition of the active site regions of ecGlK-glc (light gray) and human brain hexokinase I (PDB 1DGK; dark gray) depicted in stereo. The superposition was generated by using the atoms of the glucose molecule and residues corresponding to Asn99, Asp100, Glu157, and Glu187 of ecGlK.
Each glucose molecule participates in an extensive hydrogen bonding network within the active site pocket (Table 2 and Fig. 6a). All of the interacting residues are highly conserved within the related sequences of group II GlKs (PFAM accession number P46880) (Fig. 3). The residues Asn99, Asp100, Glu157, and Glu187 are also conserved both in sequence (Fig. 3) and structurally (Fig. 6b) in human hexokinase I. In human hexokinase I (PDB 1DGK), structurally equivalent residues to those of ecGlK (in parentheses) are Glu708 (Glu157), Gln739 (His160), Glu742 (Glu187), Asn656 (Asn99), and Asp657 (Asp100). Site-specific mutations of Asp657, Glu708, and Glu742 of human hexokinase I have been previously shown to abolish activity in vitro (6). The Glu708Ala and Glu742Ala mutations reduced the KM for glucose by 50- and 14-fold, respectively (6). A water-mediated hydrogen bond between the O6 atom of glucose and the amide N of Gly138 is also present in human hexokinase I.
TABLE 2.
Summary of direct and water-mediated hydrogen bonds between ecGlK and glucosea
| Atom 1 | Atom 2 | Distance (Å) |
|---|---|---|
| Glucose O1 | Glu187 OE2 | 2.6 |
| Glucose O1 | Wat117 | 2.9 |
| Wat117 | Asn76 OD1 | 3.0 |
| Glucose O2 | Glu157 OE1 | 2.5 |
| Glucose O2 | His160 NE2 | 3.0 |
| Glucose O2 | Wat319 | 2.7 |
| Wat319 | Arg286 NH2 (B) | 2.9 |
| Glucose O3 | Glu157 OE2 | 2.8 |
| Glucose O3 | Asn99 ND2 | 2.8 |
| Glucose O4 | Asp100 OD1 | 2.5 |
| Glucose O4 | Wat87 | 2.8 |
| Wat87 | Val141 N | 3.1 |
| Wat87 | Gly156 O | 2.9 |
| Glucose O6 | Asp100 OD2 | 2.6 |
| Glucose O6 | Wat77 | 2.7 |
| Wat77 | Gly138 N | 2.9 |
| Wat77 | Thr137 OG1 | 3.0 |
Wat, water.
Intrinsic flexibility of ecGlK.
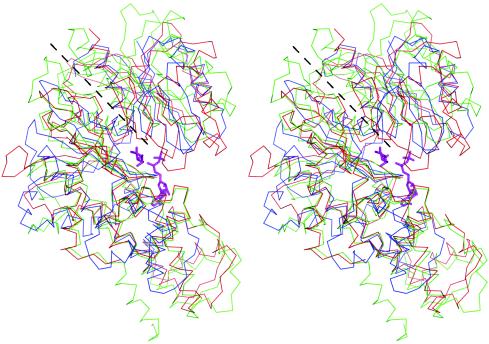
The cracking and dissolving of apo-ecGlK crystals when soaked in the presence of glucose was suggestive of ligand-induced conformational changes in the enzyme, analogous to those found initially in yeast hexokinase (9, 65) and subsequently in human hexokinase I (2) and ADP-dependent GlKs (34). Superposition of yeast hexokinase and its complex with glucose revealed closing of the domains relative to one another, effectively burying the substrate (65). A similar finding has been observed with P. furiosus GlK in the presence of bound glucose (34), where comparison of this structure with the related apo-GlK from T. litoralis showed a maximal shift in Cα positions of 12 Å at the tip of the small domain.
The dimer interfaces for ecGlK and ecGlK-glc are very similar but not identical. In ecGlK-glc, the H bond between Asp148OD1(A)-Arg150NH1(B) is not maintained. Dimers of ecGlK and ecGlK-glc were superimposed by using Cα atoms of the large domains (residues 120 to 300) of both molecules of the dimer, giving an rmsd of 0.42 Å for 372 Cα atoms. While the large domains are fixed through their interactions in the dimer, the small domains in both monomers are rotated by ∼15°, resulting in a maximum Cα displacement of ∼10 Å (Fig. 7a). As a consequence of this movement, the active site cleft becomes more closed.
FIG.7.
Domain-domain movements of ecGlK. (a) Superposition of dimers of ecGlK (red) and ecGlK-glc (blue). The superposition is based on the large domains of each model, showing the large relative movement of the small domains. Glucose is shown in stick representation. (b) Superposition of the large domains of ecGlK and ecGlK-glc with both monomers within the asymmetric unit in both crystal forms, showing the intrinsic conformational flexibility of ecGlK. Shown are the two monomers of ecGlK (monomer A, blue; monomer B, yellow) and the two monomers of ecGlK-glc (monomer A, cyan; monomer B, magenta). (c) Close-up of superposition of monomer A of ecGlK (red) and ecGlK-glc (blue) showing the glucose-binding site. Glucose is shown in stick representation.
Comparison of the overall structures of the two monomers of the apo-ecGlK dimer reveals that a few loops within the small domain have different conformations in the two monomers. Superposition of the two apo-ecGlK monomers by using only the large domains reveals the intrinsic conformational flexibility of this enzyme (Fig. 7b), as has been previously observed with yeast hexokinase in solution studies (55). The maximal Cα displacement for residues in the small domain in this superposition is for Thr78, displaced by 7 Å, part of the loop which closes the glucose-binding site. Superposition of the two monomers gave an rmsd of 1.3 Å for 320 Cα atoms, while superposition of Cα atoms of the large domain alone gave an rmsd of 0.38 Å for 186 Cα atoms. In ecGlK-glc, glucose binding stabilizes these flexible loops, resulting in their adopting the same conformation in both monomers. These observations imply that while glucose binding stabilizes the closed form, domain-domain movements occur in apo-ecGlK independently of glucose binding and reflect the intrinsic flexibility of the enzyme.
The only structural change observed within the small domains themselves upon glucose binding is a movement of the loop consisting of residues 73 to 79. Between large and small domains, a number of hydrogen bonds are broken as a result of domain closure including those between Glu315NE2 and Trp131 O, Glu315OE1 and Trp151 N, and Asn303OD1 and Arg16NH1 as well as Thr32OG1 and Arg16NE. New hydrogen bonds formed after domain closure include those between Asn303OD and Arg16NE and between Thr32OG and Arg16NH1. Several van der Waals contacts are also broken and reformed as a consequence of domain movement. Of those residues involved in glucose binding, only Asn99 and Asp100 undergo significant movement in comparison to apo-ecGlK and glucose-bound ecGlK (Fig. 7c).
Putative ATP-binding site.
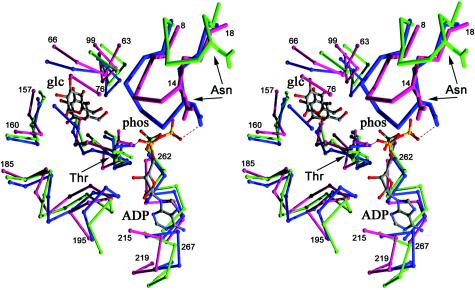
Although we could not obtain a complex between ecGlK and ADP/Mg2+, either in the presence or absence of glucose, a comparison of ecGlK with the structures of mutant human hexokinase I bound to ADP-glucose (3) or yeast hexokinase PII bound to sulfate (38) offers insights into the likely ATP-binding site of ecGlK. The ADP-binding site of a quadruple mutant of hexokinase I has been determined (PDB 1DGK) (3). Indeed, the nucleotide-binding site is structurally conserved in all members of this superfamily, as predicted by Bork et al. (12).
First, the positions of the sulfate anion of apo-yeast hexokinase PII and Pα of ADP from the human hexokinase binary complex superimpose, with a 0.65-Å distance between the P and S atoms, respectively. This position has been identified as a high-affinity anion binding site in a number of hexokinase structures (10, 46). In the case of human hexokinase I, the α-phosphoryl group makes hydrogen bonds with Thr680OG1 and Thr680N as well as with Thr863N. In yeast hexokinase PII, Ser419OG1, the structural equivalent of Thr863, forms a hydrogen bond with an O atom of the sulfate anion. Other hydrogen bonds with the sulfate are formed with Ser419N, Thr234OG1, and Thr234N. This last residue is the yeast equivalent of Thr680 of human hexokinase. Superposition of these models with ecGlK reveals that Thr137 of ecGlK is structurally equivalent to Thr234 of yeast hexokinase and Thr680 of human hexokinase. In addition, Thr137N could participate in a hydrogen bond with the α-phosphoryl group, analogous to that of Thr234N and Thr680N of yeast and human hexokinases, respectively.
As with other kinases, a metal ion, such as Mg2+ or Mn2+, is expected to be an essential component of the catalytic machinery (40). No cocrystal structure of hexokinase or glucokinase with bound Mg2+ has yet been reported. The side chains of Thr680 (Thr137 of ecGlK), Asp532 (Asp9 of ecGlK) or Asp861 (HK1; PDB 1DGK) appear proximal enough to the phosphoryl groups of the nucleotide binding site that they could participate in Mg2+ binding. Both Thr137 and especially Asp9 are highly conserved in the sequences of group II GlKs (Fig. 3). A combination of modeling (3) and electron paramagnetic resonance studies in solution (48) suggest that Mn2+ or Mg2+ may only form water-mediated interactions with the enzyme. Consistent with the importance of Asp532, the mutations Asp532Lys and Asp532Glu have been shown to decrease kcat of human hexokinase I by 1,000- and 200-fold, respectively (74). A hydrated Mg2+ binding site has also been suggested for the P. furiosus ADP-dependent glucokinase (34).
In the human hexokinase I complex, key residues interacting with ADP include Thr680OG1 (Pα), Thr683OG1 (Pβ and O5′ of ribose), and Asn537ND2 (Pβ). Asn537 is part of a loop, conserved in sequence between ecGlK (Asn14) and yeast hexokinase (Asn91), and is associated with nucleotide binding. A Thr680Val mutant of hexokinase I showed a decrease in kcat of ∼2,000-fold, while the Thr680Ser mutant only decreased ∼2.5-fold, showing the importance of this hydrogen bond in catalysis (74).
A conserved sequence and structural motif consisting of two β-strands connected by a loop within the small domain (residues Leu6-Leu19 of ecGlK) (Fig. 3) has the sequence L-(A/V)-X-D-X-G-G-T-N-X-R-X-X-L (conserved Asp and Gly residues are in boldface) and is proximal to the ATP phosphoryl group binding site. In particular, the residues equivalent to Leu6, Asp9, Gly11, Gly12, Asn14, Arg16, and Leu19 are completely conserved in the sequence alignment of ecGlK, human hexokinase, and yeast hexokinase as well as human glucokinase (Fig. 3). A similar motif [X2-D-(I/L/V)-G-G-(S/T-X3); conserved Asp and Gly residues are in boldface] is conserved as well in group III (ROK) glucokinases (30, 45) and is equivalent to a portion of the phosphate 1 motif identified originally by Bork et al. (13). The two conserved Gly residues contribute to formation of the loop and could form main chain hydrogen bonds with the ATP phosphates. Indeed, the mutation Gly534Ala of hexokinase I (Gly11 of ecGlK) results in a decrease of kcat by 4,000-fold (75). In the human hexokinase I-ADP-glucose complex, this loop has adopted the most closed conformation and makes direct hydrogen bonds between the phosphate O atom bridging the α and β phosphoryl groups and Ala536N, as well as between a Pβ O atom and Asn537N (Fig. 8). Importantly, in the human hexokinase I binary complex, Thr536 has been mutated to Ala, yet kcat/KM for neither glucose nor ATP is significantly perturbed (3). This argues that the backbone conformation of this loop is important for nucleotide binding, rather than the presence of the Thr536 side chain specifically, although modeling suggests that Thr536OG can evidently form a hydrogen bond with the Pα-Pβ bridging O (data not shown). Asn537ND2 also forms a hydrogen bond with an O atom of Pβ. We suggest that similar interactions would be found in yeast hexokinase and ecGlK in the presence of bound nucleotide and glucose. It is evident that a secondary conformational change of this conserved strand-loop-strand motif must occur upon nucleotide binding, resulting in further domain closure, in addition to that which occurs upon glucose binding (Fig. 8).
FIG. 8.
Potential ATP-binding site of ecGlK. Superposition of the ADP-binding site of the catalytic domain (residues 475 to 917) of mutant human hexokinase bound to ADP and glucose (PDB 1DGK; blue) (3), yeast PII hexokinase bound to sulfate (PDB 1IG8; green) (38), and ecGlK (magenta). Hydrogen bonds between Thr and Asn of human hexokinase and ADP are shown. The conserved sequence motif near the ATP-binding site [L-(A/V)-X-D-X-G-G-T-N-X-R-X-X-L] is shown as thicker lines.
Catalytic mechanism.
The expected chemical mechanism of catalysis for ecGlK, analogous to that of other kinases, is SN2 nucleophilic attack of the O6 atom of glucose on the electropositive P atom of the γ-phosphoryl group of ATP. Initial abstraction of the proton from the CH2OH group of O6 is presumably performed by Asp100 acting as a general base. Asp100OD2 is positioned 2.7 Å from O6 and is well oriented to fulfill this role (Fig. 6a). The Asp100 side chain position is anchored by hydrogen bonds to O4 of glucose and Asn99OD1, a highly conserved residue (Fig. 3). This mechanism is consistent with the complete conservation of Asp100 in group II glucokinase sequences and in human hexokinase (Asp657 of human hexokinase I) and glucokinase. Of the residues involved in glucose binding, the mutant Asp657Ala showed the largest effect on activity, resulting in a reduction in kcat of 100-fold relative to wild-type enzyme (6).
In the ADP-dependent glucokinases, the residue Asp451 of T. litoralis GlK is predicted to function as a general base. This residue interacts with the O6 atom of glucose and when mutated to Ala shows a specific activity of <0.001% compared to wild-type enzyme (34). A structurally equivalent residue, Asp440 of P. furiosus GlK, is predicted to function as a general base in this enzyme (34).
In human hexokinase I, a second residue, Lys621, is also within hydrogen bonding distance of Glc O6 and has been suggested as a possible catalytic residue (46). However, there is no structural equivalent of the Lys621 residue in ecGlK. Modeling of ATP bound to ecGlK, based on the superposition with ADP-bound human hexokinase I, positions the O6 atom of glucose to within a suitable distance of the γ-phosphoryl group for in-line nucleophilic attack. The distance between O6 of glucose and the Pα atom of mutant human hexokinase I (PDB 1DGK) is 5.6 Å. To accomplish the correct orientation of the γ-phosphoryl group, the ATP β- and γ-phosphoryl groups would need to adopt an extended conformation. No specific residue required to function as a general acid, responsible for protonating ADP as the leaving group, has been identified in hexokinase, although the possibility that it arises from a water molecule coordinated to Mg2+ has been suggested (3).
The kinetic mechanism of several glucokinases has been investigated and found to have a preferred order of substrate addition and product release. In the ATP-dependent glucokinases from Zymomonas mobilis (62), Propionibacterium shermanii, (37) and rat liver hexokinase IV (glucokinase, 44), the preferred order of substrate addition is glucose (or 2-deoxyglucose) followed by ATP or Mg2+. This kinetic mechanism in consistent with the ecGlK crystal structures, in that glucose binding stabilizes a closed form of ecGlK that can bind ATP, in turn resulting in a small but important conformational change necessary to form a catalytically competent form of the enzyme.
Acknowledgments
We thank Leon Flaks (NSLS; beamline X8C) and Michael Becker (NSLS; beamline X25) for assistance in data collection, Stephane Raymond for maintenance of the computing environment, and Frederic Ouellet and J. Sivaraman for assistance in protein purification and crystallization.
This research was supported in part by the Canadian Institutes of Health Research grant 200103GSP-90094-GMX-CFAA-19924 to M.C.
REFERENCES
- 1.Aleshin, A. E., C. Zeng, G. P. Bourenkov, H. D. Bartunik, H. J. Fromm, and R. B. Honzatko. 1998. The mechanism of regulation of hexokinase: new insights from the crystal structure of recombinant human brain hexokinase complexed with glucose and glucose-6-phosphate. Structure 6:39-50. [DOI] [PubMed] [Google Scholar]
- 2.Aleshin, A. E., C. Zeng, H. D. Bartunik, H. J. Fromm, and R. B. Honzatko. 1998. Regulation of hexokinase I: crystal structure of recombinant human brain hexokinase complexed with glucose and phosphate. J. Mol. Biol. 282:345-357. [DOI] [PubMed] [Google Scholar]
- 3.Aleshin, A. E., C.Kirby, X. Liu, G. P. Bourenkov, H. D. Bartunik, H. J. Fromm, and R. B. Honzatko. 2000. Crystal structure of mutant monomeric hexokinase I reveals multiple ADP binding sites and conformational changes relevant to allosteric regulation. J. Mol. Biol. 296:1001-1015. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, C. M., R. E. Stenkamp, and T. A. Steitz. 1978. Sequencing a protein by X-ray crystallography. II. Refinement of yeast hexokinase B co-ordinates and sequence at 2.1 Å resolution. J. Mol. Biol. 123:15-33. [DOI] [PubMed] [Google Scholar]
- 5.Andreeva, A., D. Howorth, S. E. Brenner, T. J. Hubbard, C. Chothia, and A. G. Murzin. 2004. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 32:D226-D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arora, K. K., C. R. Filburn, and P. L. Pedersen. 1991. Glucose phosphorylation. Site-directed mutations which impair the catalytic function of hexokinase. J. Biol. Chem. 266:5359-5362. [PubMed] [Google Scholar]
- 7.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behlke, J., K., Heidrich, M. Naumann, E-C. Müeller, A. Otto, R. Reuter, and T. Kriegel. 1998. Hexokinase 2 from Saccharomyces cerevisiae: regulation of oligomeric structure by in vitro phosphorylation at serine-14. Biochemistry 37:11989-11995. [DOI] [PubMed] [Google Scholar]
- 9.Bennett, W. S., Jr., and T. A. Steitz. 1978. Glucose-induced conformational change in yeast hexokinase. Proc. Natl. Acad. Sci. USA 75:4848-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett, W. S., Jr., and T. A. Steitz. 1980. Structure of a complex between yeast hexokinase A and glucose. I. Structure determination and refinement at 3.5 Å resolution. J. Mol. Biol. 140:183-209. [DOI] [PubMed] [Google Scholar]
- 11.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cyle proteins, sugar kinases, actin and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bork, P., C. Sander, and A. Valencia. 1993. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase and galactokinase families of sugar kinases. Protein Sci. 2:31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 15.Buss, K. A., D. R. Cooper, C. Ingram-Smith, J. G. Ferry, D. A. Sanders, and M. S. Hasson. 2001. Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol. 183:680-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheek, S., H. Zhang, and N. V. Grishin. 2002. Sequence and structure classification of kinases. J. Mol. Biol. 320:855-881. [DOI] [PubMed] [Google Scholar]
- 17.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delano, W.L. 2002. The PyMOL molecular graphics system on the World Wide Web (http://www.pymol.org).
- 19.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:132-134. [DOI] [PubMed] [Google Scholar]
- 20.Flores, N., J. Xiao, A. Berry, F. Bolivar, and F. Valle. 1996. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat. Biotechnol. 14:620-623. [DOI] [PubMed] [Google Scholar]
- 21.Flores, S., G. Gosset, N. Flores, A. A. de Graaf, and F. Bolivar. 2002. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by 13C labeling and NMR spectroscopy. Metab. Eng. 4:124-137. [DOI] [PubMed] [Google Scholar]
- 22.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, T., and P. Schönheit. 2003. ATP-dependent glucokinase from the hyperthermophilic bacterium Thermotoga maritima represents an extremely thermophilic ROK glucokinase with high substrate specificity. FEMS Microbiol. Lett. 226:405-411. [DOI] [PubMed] [Google Scholar]
- 24.Hansen, T., B. Reichstein, R. Schmid, and P. Schönheit. 2002. The first archaeal ATP-dependent glucokinase from the hyperthermophilic crenarchaeon Aeropyrum pernix, represents a monomeric, extremely thermophilic ROK glucokinase with broad hexose specificity. J. Bacteriol. 184:5955-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helfert, C., S. Gotsche, and M. K. Dahl. 1995. Cleavage of trehalose-phosphate in Bacillus subtilis is catalyzed by α-phospho-α-(1,1)glucosidase encoded by the treA gene. Mol. Microbiol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 26.Hendrickson, W. A. 1991. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254:51-58. [DOI] [PubMed] [Google Scholar]
- 27.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez-Montalvo, V., A. Martinez, G. Hernandez-Chavez, F. Bolivar, F. Valle, and G. Gosset. 2003. Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotech. Bioeng. 83:687-694. [DOI] [PubMed] [Google Scholar]
- 29.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh, P-C., B. C. Shenoy, D. Samols, and N. F. B. Phillips. 1996. Cloning, expression and characterization of polyphosphate glucokinase from Mycobacterium tuberculosis. J. Biol. Chem. 271:4909-4915. [DOI] [PubMed] [Google Scholar]
- 31.Hurley, J. H. 1996. The sugar kinase/heat shock protein 70/actin superfamily: Implications of conserved structure for mechanism. Annu. Rev. Biomol. Struct. 25:137-162. [DOI] [PubMed] [Google Scholar]
- 32.Hurley, J. H., H. R. Faber, D. Worthylake, N. D. Meadow, and S. Roseman. 1993. Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science 259:673-677. [PubMed] [Google Scholar]
- 33.Ito, S., S. Fushinobu, I. Yoshioka, S. Koga, H. Matsuzawa, and T. Wakagi. 2001. Structural basis for the ADP-specificity of a novel glucokinase from a hyperthermophilic archaeon. Structure (Cambridge) 9:205-214. [DOI] [PubMed] [Google Scholar]
- 34.Ito, S., S. Fushinobu, J.-J. Jeong, I. Yoshioka, S. Koga, H. Shoun, and T. Wakagi. 2003. Crystal structure of an ADP-dependent glucokinase from Pyrococcus furiosus: implications for a sugar-induced conformational change in ADP-dependent kinase. J. Mol. Biol. 331:871-883. [DOI] [PubMed] [Google Scholar]
- 35.Jones, T. A., J.-Y. Zou, S. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:100-119. [DOI] [PubMed] [Google Scholar]
- 36.Kamata, K., M. Mitsuya, T. Nishimura, J. Eiki, and Y. Nagata. 2004. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure (Cambridge) 12:429-438. [DOI] [PubMed] [Google Scholar]
- 37.Kowalczyk, T. H., P. J. Horn, W.-H. Pan, and N. F. B. Phillips. 1996. Initial rate and equilibrium isotope exchange studies on the ATP-dependent activity of polyphosphate glucokinase from Propionibacterium shermanii. Biochemistry 35:6777-6785. [DOI] [PubMed] [Google Scholar]
- 38.Kuser, P. R., S. Krauchenco, O. A. C. Antunes, and I. Polikarpov. 2000. The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J. Biol. Chem. 275:20814-20821. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski, R. A., M. W. McArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:282-291. [Google Scholar]
- 40.Matte, A., L. W. Tari, and L. T. J. Delbaere. 1998. How do kinases transfer phosphoryl groups? Structure 6:413-419. [DOI] [PubMed] [Google Scholar]
- 41.Merritt, E. A., and M. E. P. Murphy. 1994. Raster3D version 2.0-a program for photorealistic molecular graphics. Acta Crystallogr. Sect. D 50:869-873. [DOI] [PubMed] [Google Scholar]
- 42.Mesak, L. R., F. M. Mesak, and M. K. Dahl. 2004. Bacillus subtilis GlcK activity requires cysteines within a motif that discriminates microbial glucokinases into two lineages. BMC Microbiol. 4:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, D., C. Schneider-Fresenius, R. Horlacher, R. Peist, and W. Boos. 1997. Molecular characterization of glucokinase from Escherichia coli K-12. J. Bacteriol. 179:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monasterio, O., and M. L. Cardenas. 2003. Kinetic studies of rat liver hexokinase D (′glucokinase') in non-co-operative conditions show an ordered mechanism with MgADP as the last product to be released. Biochem. J. 371:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukai, T., S. Kawai, H. Matsukawa, Y. Matuo, and K. Murata. 2003. Characterization and molecular cloning of a novel enzyme, inorganic polyphosphate/ATP-glucomannokinase, of Anthrobacter sp. strain KM. Appl. Environ. Microbiol. 69:3849-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulichak, A. M., J. E. Wilson, K. Padmanabhan, and R. M. Garavito. 1998. The structure of mammalian hexokinase-1. Nat. Struct. Biol. 7:555-560. [DOI] [PubMed] [Google Scholar]
- 47.Murshudov, G. N., A. A. Vagin, A. Lebedev, K. S. Wilson, and E. J. Dodson. 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. Sect. D 55:247-255. [DOI] [PubMed] [Google Scholar]
- 48.Olsen, L. R., and G. H. Reed. 1993. The structure of the MnII-ADP-nitrate-lyxose complex at the active site of hexokinase. Arch. Biochem. Biophys. 251:97-103. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 50.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 51.Pflugrath, J. W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. Sect. D 55:1718-1725. [DOI] [PubMed] [Google Scholar]
- 52.Phillips, N. F. B., P. J. Horn, and H. G. Wood. 1993. The polyphosphate- and ATP-dependent glucokinase from Propionibacterium shermanii: Both activities are catalyzed by the same protein. Arch. Biochem. Biophys. 300:309-319. [DOI] [PubMed] [Google Scholar]
- 53.Phillips, N. F. B., P. C. Hsieh, and T. H. Kowalczyk. 1999. Polyphosphate glucokinase. Prog. Mol. Subcell. Biol. 23:101-125. [DOI] [PubMed] [Google Scholar]
- 54.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1996. Phoephoenolpyruvate: carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt and R. Curtiss (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington D.C.
- 55.Reid, C., and R. P. Rand. 1997. Probing protein hydration and conformational states in solution. Biophys. J. 72:1022-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ronimus, R. S., and H. W. Morgan. 2004. Cloning and biochemical characterization of a novel mouse ADP-dependent glucokinase. Biochem. Biophys. Res. Commun. 315:652-658. [DOI] [PubMed] [Google Scholar]
- 57.Rosano, C., E. Sabini, M. Rizzi, D. Deriu, G. Murshudov, M. Bianchi, G. Serafini, M. Magnani, and M. Bolognesi. 1999. Binding of non-catalytic ATP to human hexokinase I highlights the structural components for enzyme-membrane association control. Structure Fold Des. 7:1427-1437. [DOI] [PubMed] [Google Scholar]
- 58.Sakuraba, H., Y. Mitani, S. Goda, Y. Kawarabayasi, and T. Ohshima. 2003. Cloning, expression and characterization of the first archaeal ATP-dependent glucokinase from aerobic hyperthermophilic archaeon Aeropyrum pernix. J. Biochem. 133:219-224. [DOI] [PubMed] [Google Scholar]
- 59.Sakuraba, H., I. Yoshioka, S. Koga, M. Takahashi, Y. Kitahama, T. Satomura, R. Kawakami, and T. Ohshima. 2002. ADP-dependent glucokinase/phosphofructokinase, a novel bifunctional enzyme from the hyperthermophilic archaeon Methanococcus jannaschii. J. Biol. Chem. 277:12495-12498. [DOI] [PubMed] [Google Scholar]
- 60.Sakuraba, H., S. Goda, and T. Ohshima. 2004. Unique sugar metabolism and novel enzymes of hyperhermophilic archaea. Chem. Rec. 3:281-287. [DOI] [PubMed] [Google Scholar]
- 61.Schönert, S., T. Buder, and M. K. Dahl. 1998. Identification and enzymatic characterization of the maltose-inducible α-glucosidase MalL (sucrase-isomaltase-maltase) of Bacillus subtilis. J. Bacteriol. 180:2574-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scopes, R. K., and D. R. Bannon. 1995. Kinetic analysis of the activation of Zymomonas mobilis glucokinase by phosphate. Biochim. Biophys. Acta 1249:173-179. [DOI] [PubMed] [Google Scholar]
- 63.Skarlatos, P., and M. K. Dahl. 1998. The glucose kinase of Bacillus subtilis. J. Bacteriol. 180:3222-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Späth, C., Kraus, A., and W. Hillen. 1997. Contribution of glucose kinase to glucose repression of xylose utilization in Bacillus megaterium. J. Bacteriol. 179:7603-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steitz, T. A., M. Shoham, and W. S. Bennett, Jr. 1981. Structural dynamics of yeast hexokinase during catalysis. Phil. Trans. R. Soc. Lond. 293:43-52. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka, S., S.-O. Lee, K. Hamaoka, J. Kato, N. Takiguchi, K. Nakamura, H. Ohtake, and A. Kuroda. 2003. Strictly polyphosphate-dependent glucokinase in a polyphosphate accumulating bacterium, Microlunatus phosphovorus. J. Bacteriol. 185:5654-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. Sect. D 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Titgemeyer, F., J. Reizer, A. Reizer, and M. H. Saier, Jr. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349-2354. [DOI] [PubMed] [Google Scholar]
- 70.Tsuge, H., H. Sakuraba, T. Kobe, A. Kujime, N. Katunuma, and T. Ohshima. 2002. Crystal structure of the ADP-dependent glucokinase from Pyrococcus horikoshii at 2.0-Å resolution: a large conformational change in ADP-dependent glucokinase. Protein Sci. 11:2456-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vagin, A. A., and M. N. Isupov. 2001. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. Sect. D 57:1451-1456. [DOI] [PubMed] [Google Scholar]
- 72.Wilson, J. E. 1995. Hexokinases. Rev. Physiol. Biochem. Pharmacol. 126:65-198. [DOI] [PubMed] [Google Scholar]
- 73.Winn, M. D., A. W. Ashton, P. J. Briggs, C. C. Ballard, P. Patel. 2002. Ongoing developments in CCP4 for high-throughput structure determination. Acta Crystallogr. Sect. D 58:1929-1936. [DOI] [PubMed] [Google Scholar]
- 74.Zeng, C., A. E. Aleshin, J. B. Hardie, R. W. Harrison, and H. J. Fromm. 1996. ATP-binding site of human hexokinase as studied by molecular modeling and site-directed mutagenesis. Biochemistry 35:13157-13164. [DOI] [PubMed] [Google Scholar]
- 75.Zeng, C., A. E. Aleshin, C. Guanjin, R. B. Honzatko, and H. J. Fromm. 1998. The roles of glycine residues in the ATP-binding site of human brain hexokinase. J. Biol. Chem. 273:700-704. [DOI] [PubMed] [Google Scholar]