Abstract
The holoenzyme of adenosylcobalamin-dependent ethanolamine ammonia lyase undergoes suicidal inactivation during catalysis as well as inactivation in the absence of substrate. The inactivation involves the irreversible cleavage of the Co-C bond of the coenzyme. We found that the inactivated holoenzyme undergoes rapid and continuous reactivation in the presence of ATP, Mg2+, and free adenosylcobalamin in permeabilized cells (in situ), homogenate, and cell extracts of Escherichia coli. The reactivation was observed in the permeabilized E. coli cells carrying a plasmid containing the E. coli eut operon as well. From coexpression experiments, it was demonstrated that the eutA gene, adjacent to the 5′ end of ethanolamine ammonia lyase genes (eutBC), is essential for reactivation. It encodes a polypeptide consisting of 467 amino acid residues with predicted molecular weight of 49,599. No evidence was obtained that shows the presence of the auxiliary protein(s) potentiating the reactivation or associating with EutA. It was demonstrated with purified recombinant EutA that both the suicidally inactivated and O2-inactivated holoethanolamine ammonia lyase underwent rapid reactivation in vitro by EutA in the presence of adenosylcobalamin, ATP, and Mg2+. The inactive enzyme-cyanocobalamin complex was also activated in situ and in vitro by EutA under the same conditions. Thus, it was concluded that EutA is the only component of the reactivating factor for ethanolamine ammonia lyase and that reactivation and activation occur through the exchange of modified coenzyme for free intact adenosylcobalamin.
Certain enzymes utilize the high reactivity of free radicals to catalyze the reactions that are difficult to catalyze by ionic mechanisms (6). Since the radicals involved in the catalysis are highly reactive, radical enzymes have a general tendency to undergo suicide inactivation during catalysis or inactivation by oxygen (40). Therefore, protection of the radical intermediates against undesired side reactions or reactivation of the inactivated enzymes is of essential importance for radical-catalyzed enzymatic reactions.
Adenosylcobalamin (AdoCbl)-dependent enzymes are known to catalyze reactions including carbon skeleton rearrangements, heteroatom eliminations, and intramolecular amino group migrations by radical mechanisms (5, 13). An adenosyl radical formed by homolytic cleavage of the Co-C bond of enzyme-bound AdoCbl is directly involved in catalysis. The holoenzymes of diol dehydratase and glycerol dehydratase undergo suicide inactivation by glycerol during catalysis (3, 29, 30, 44) or O2 inactivation in the absence of substrate (25, 30, 38, 47). This phenomenon is of special interest because glycerol is a physiological substrate for these enzymes (1, 17, 18, 41, 42). This apparent inconsistency was solved by previous findings that the glycerol-inactivated enzymes in permeabilized cells (in situ) of Klebsiella oxytoca and Klebsiella pneumoniae undergo rapid reactivation in the presence of ATP, Mg2+ (or Mn2+), and AdoCbl (20, 46). The genes encoding the proteins (reactivating factors) involved in the reactivation of diol dehydratase (27) and glycerol dehydratase (39) were identified in previous studies and named ddrAB (pduGH) and gdrAB (dhaB4/dhaF and orf2b/dhaG), respectively. The gene products have been characterized, and the mechanism of reactivation of inactivated holoenzymes by the corresponding reactivating factors has also been proposed (21, 28, 36, 43). The ADP-bound form of reactivating factor induces conformational change of the enzyme through tight complex formation with it, resulting in release of the enzyme-bound modified coenzyme. Finally, the complex dissociates in the presence of ATP, forming apoenzyme that can be reconstituted into active holoenzyme with intact AdoCbl. We proposed that these reactivating factors are a new type of molecular chaperones that are involved in the reactivation of already-folded, inactivated enzymes (21, 28, 43).
The next question is whether such reactivating factors are general for AdoCbl-dependent enzymes. To answer this question, a reactivating factor for ethanolamine ammonia lyase (EC 4.3.1.7) (EAL) has been sought. EAL catalyzes AdoCbl-dependent conversion of ethanolamine to acetaldehyde (11). The enzyme was first discovered by Bradbeer in a choline-fermenting Clostridium sp. (10) and then shown to be present in many bacteria in which exogenous vitamin B12 is required for growth on ethanolamine. They include Escherichia (35), Klebsiella (35), Salmonella (12), and Bacillus (48). It was shown with Escherichia coli that the enzyme is induced when the bacterium is grown in the presence of both ethanolamine and vitamin B12 (7, 8). The enzyme is involved in anaerobic fermentation (10) or catabolism (12, 35) of this substrate or the utilization of this substrate as a nitrogen source in aerobic growth (12, 35). The holoenzyme of EAL (holoEAL) undergoes suicidal inactivation by ethanolamine during catalysis and inactivation in the absence of substrate (22). In either case, the irreversible cleavage of the Co-C bond of enzyme-bound AdoCbl takes place, and the modified coenzyme is kept tightly bound, resulting in the irreversible inactivation of the enzyme. Such suicide inactivation seemed enigmatic, because ethanolamine is an essential substrate for the growth of these bacteria when it is used as the sole source of carbon, energy, or nitrogen. The inactivation might be reversed if the reactivating factor for EAL was present and removed the damaged cofactor from the enzyme like diol dehydratase-reactivating factor (DdrAB) and glycerol dehydratase-reactivating factor (GdrAB).
It had been reported that the eut operon of Salmonella enterica serovar Typhimurium encodes proteins essential for the cobalamin-dependent utilization of ethanolamine (31). The eut operon consists of 17 genes (23), including the genes encoding large and small subunits of EAL (eutB and eutC, respectively) (15). Although the functions of several gene products have been postulated based on genetic analyses and homology searches for deduced amino acid sequences (23, 32, 33, 37), these functions have not been wholly established. Furthermore, the functions of several other gene products remained unclear. From genetic evidence that eutA mutants are sensitive to inhibition by cyanocobalamin (CN-Cbl) (32), it was suggested that one possible function for the product of eutA was as a reactivating factor of EAL (23). Fragmentary similarity between EutA and DarA or GarA also suggests this possibility (40).
In this paper, we report the ATP-dependent reactivation of the inactivated holoEAL of E. coli and identification of an EAL-reactivating factor. Gene organization of the E. coli eut operon (Fig. 1) is very similar to that of S. enterica serovar Typhimurium LT2 (23, 49). We identified eutA, the gene adjacent to the 5′ terminal of the EAL genes (eutBC) in the E. coli eut operon, as the gene encoding an EAL-reactivating factor by coexpression, followed by the in situ assay of reactivating activity. Direct evidence is also presented here which shows that purified EutA alone functions in vitro as a reactivating factor for EAL.
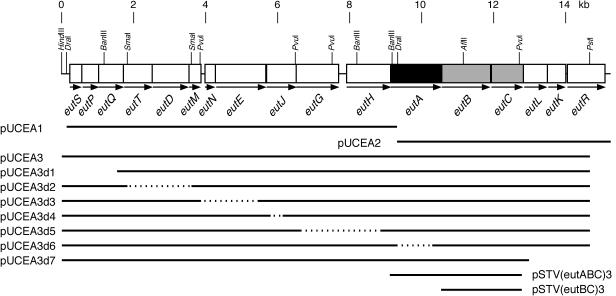
FIG. 1.
Gene organization of the E. coli eut operon and insert DNA of plasmids used in this study. The map is drawn to scale. Genes are indicated by boxes. The DNA regions carried by mutant plasmids are shown with thick bars below the map. Dotted lines indicate deletions. The eutB and eutC genes encode the α- and β-subunits of EAL, respectively. The restriction enzymes used for the construction of deletion mutants are HindIII/BsmI for pUCEA3d1, SmaI for pUCEA3d2, AatII/NheI for pUCEA3d3, SalI/EcoRV for pUCEA3d4, RsrII/ScaI for pUCEA3d5, DraI/SacII for pUCEA3d6, and AatII/PstI for pUCEA3d7.
MATERIALS AND METHODS
Materials.
Crystalline AdoCbl was a gift from Eisai Co., Ltd. (Tokyo, Japan). CN-Cbl was obtained from Glaxo Research Laboratories (Greenford, United Kingdom). All other chemicals and the enzymes used for construction of plasmids were commercial products of the highest grade available and were used without further purification.
Bacterial strains, phage, plasmids, and culture conditions.
For expression of the chromosomal eut operon, E. coli JM109 was aerobically grown at 30°C in a synthetic medium containing glycerol, ethanolamine, and CN-Cbl, as described by Scarlett and Turner (35). Cells were harvested at a late logarithmic phase.
The genes in the eut operon were isolated from λ phage DNA of clone 421 (clone name 4E10) of the Kohara E. coli genomic λ phage library (24) (kindly provided by Hirotada Mori, Research and Education Center for Genetic Information, Nara Institute of Science and Technology, Nara, Japan). This clone contains a 17.9-kb chromosomal DNA insert covering all of the genes of the eut operon. pUC119, pSTV28, and pSTV29 were used as vectors. E. coli JM109 and SCS110 (Stratagene) were used as hosts for the preparation of plasmids. Transformation of E. coli was carried out by the electroporation method of Dower et al. (14). Recombinant strains carrying plasmids were grown aerobically at 30°C in Luria-Bertani medium containing 50 μg of ampicillin/ml (for strains carrying plasmids derived from pUC119) or 50 μg of chloramphenicol/ml (for strains carrying plasmids derived from pSTV28 and pSTV29). For inducible expression of cloned genes, isopropyl-1-thio-β-d-galactopyranoside was added to a concentration of 1 mM when the culture reached an optical density at 600 nm of approximately 1.0. Cells were harvested at about 2 h after the induction was initiated.
Preparation of permeabilized cells.
Permeabilized cells were prepared by treatment with 1% (vol/vol) toluene as described previously (20), except that the treatment was performed on a small scale with 1.5-ml microtubes.
Preparation of homogenates and cell extracts.
Cells were suspended in 50 mM potassium phosphate buffer (pH 8.0) containing 10 mM ethanolamine (for cell extracts) or containing both 10 mM ethanolamine and 1% Brij 35 (for homogenates) and disrupted by sonication for 3 min at 240 W with a Kaijo (Tokyo, Japan) TA-5287 ultrasonicator. The resulting homogenate was centrifuged for 30 min at 20,600 × g, and the supernatant was used as cell extract.
EAL and protein assays.
The recombinant apoenzyme of EAL (apoEAL) of E. coli purified to homogeneity from E. coli JM109 harboring the expression plasmid pUSI2ENd (EAL) (K. Akita and T. Toraya, unpublished data) was used throughout this study. All the enzyme assays were carried out in darkness to avoid photolysis of AdoCbl. The amount of acetaldehyde formed from ethanolamine by the EAL reaction was determined by the 3-methyl-2-benzothiazolinone hydrazone method, as described previously for the assay of diol dehydratase (45). One unit of EAL is defined as the amount of enzyme activity that catalyzes the formation of 1 μmol of acetaldehyde per min at 37°C. The protein concentration of purified EutA was determined by measuring the absorbance at 280 nm. The molar absorption coefficient at 280 nm (ɛM,280) calculated by the method of Gill and von Hippel (19) for EutA from its deduced amino acid composition was 27,910 M−1 cm−1. Based on the predicted molecular weight, ɛ1%,280 was calculated to be 5.63 for EutA.
Assay of EAL-reactivating factor.
The reactivating factor assay was also carried out in darkness. In situ reactivation of the inactivated holoenzyme and in situ activation of the enzyme-CN-Cbl complex were assayed with toluene-treated (permeabilized) cells (20) by incubation at 37°C for appropriate time periods in the presence of 10 mM ethanolamine, 15 μM AdoCbl, 3 mM ATP, and 3 mM MgCl2 in 30 mM potassium phosphate buffer (pH 8.0). The reaction was terminated by the addition of potassium citrate buffer (pH 3.6) to a final concentration of 50 mM, and the amount of acetaldehyde formed was determined as described above.
In vitro reactivation of inactivated holoEAL and in vitro activation of the EAL-CN-Cbl complex were assayed by incubation with an appropriate amount of EutA, 20 mM ATP, 20 mM MgCl2, 10 μM AdoCbl, and 25 mM potassium phosphate buffer (pH 8.0), in a total volume of 50 μl. After the reaction mixtures were incubated at 37°C for appropriate time periods, potassium citrate buffer (pH 3.6) was added to a final concentration of 50 mM to terminate the reaction. The amount of acetaldehyde formed was determined after appropriate dilution.
DNA manipulations.
Standard recombinant DNA techniques were performed as described by Sambrook et al. (34). Restriction endonucleases and the enzymes for construction of plasmids were used according to the manufacturer's instructions. Cloning of PCR products into pCR2.1 was carried out with the Original TA cloning kit (Invitrogen).
Constructions of plasmids.
The gene organization of the E. coli K-12 eut operon is illustrated in Fig. 1 (23, 49). The 9.2-kb DraI and the 7.4-kb DraI-SmaI fragments from λ phage DNA prepared from clone 421 of the Kohara E. coli genomic λ phage library (24) were ligated to an SmaI digest of pUC119 to construct pUCEA1 and pUCEA2, respectively. The 14.7-kb HindIII-PstI fragment from the same phage DNA was inserted between the HindIII-PstI sites of pUC119 to construct pUCEA3. In these plasmids, cloned genes were under the control of the lac promoter. A series of deletion mutants of pUCEA3 (pUCEA3d1 to pUCEA3d7) were constructed by ligation of several DNA fragments from pUCEA1 and pUCEA3 formed by digestion with the appropriate restriction endonucleases listed in the legend to Fig. 1. The 3′-terminal region of eutC with the NdeI and BanIII sites was amplified by PCR with Taq DNA polymerase, 5′ primer (TCCATATGTGAGTCATCGATACGCCGTTTACTCGCCGCGTATGG), and 3′ primer (ACAGATCTTATCGGGTCATGTTGATGCCGGATG) (restriction sites added are indicated by underlining, and the sequence complementary to the termination codon is indicated by italics) and cloned into pCR2.1 to construct pCR(eutCc). The DNA fragment containing the 5′ terminus of eutA was prepared by hybridization of two synthetic oligo-DNAs (TATGAACACTCGCCAGCTATTGAGCGTCGGTAT, where the initiation codon is shown in italics, and CGATACCGACGCTCAATAGCTGGCGAGTGTTCA) and ligated to the 4.1-kb NdeI-BanIII fragment from pCR(eutCc) to produce pCR(eutAn/Cc). Plasmid pSTV28 was subjected to BanIII digestion and Klenow fragment treatment and then digested by SacI. The resulting 2.8-kb fragment and the 430-bp SacI-PvuII fragment from pCR(eutAn/Cc) were ligated to construct pSTV(eutAn/Cc). The 3.5-kb BanIII-PvuI fragment from pUCEA3d2 was ligated to the 3.2-kb BanIII-PvuI fragment from pSTV(eutAn/Cc) to construct pSTV(eutABC). The 3.7-kb NdeI-BglII fragment from pSTV(eutABC) and the 4.4-kb NdeI-BglII fragment from pCXV(DD) (27) were ligated to construct pCXV(eutABC). To produce pCXV(eutBC), the 610-bp NdeI-AflII fragment from pUSI2ENd(EAL) (the expression plasmid for E. coli EAL genes; Akita and Toraya, unpublished), the 1.7-kb AflII-BglII fragment from pSTV(eutABC), and the 4.4-kb NdeI-BglII fragment from pCXV(DD) were ligated. The 3.7-kb and 2.3-kb BamHI-BglII fragments from pCXV(eutABC) and pCXV(eutBC) were inserted into the BamHI site of pSTV29 to produce pSTV(eutABC)2 and pSTV(eutBC)2, respectively. To eliminate in-frame gene fusions between the vector's lacZα and the 5′ ends of cloned genes in these plasmids, pSTV(eutABC)2 and pSTV(eutBC)2 were digested with BamHI, followed by Klenow fragment treatment and self ligation. The resulting plasmids were designated pSTV(eutABC)3 and pSTV(eutBC)3, respectively. The 1.2-kb NdeI-SacII fragment from pCXV(eutABC) and the 0.3-kb SacII-BsrDI fragment from pUCEA2R (containing the same region of the eut operon as pUCEA2, but in the opposite direction) were cloned between the NdeI and EcoRI sites of pUSI2ENd(EAL) (the cohesive ends formed by BsrDI and EcoRI digestion had been blunted by treating with T4 DNA polymerase and Klenow fragment, respectively, prior to ligation) to construct pUSI2ENd(eutA).
Purification of recombinant EutA protein.
Recombinant EutA protein was purified to homogeneity from extracts of E. coli JM109 harboring expression plasmid pUSI2ENd(eutA) that was aerobically grown at 37°C in Luria-Bertani medium containing ampicillin (50 μg/ml). Isopropyl-1-thio-β-d-galactopyranoside was added to a concentration of 1 mM for induction, and cells were harvested in the late logarithmic phase. Purification was carried out at 0 to 4°C. Throughout the purification steps, purity of the protein in each fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Wet cells (16.5 g) were suspended in 40 ml of 50 mM potassium phosphate buffer (pH 8.0) containing 2 mM EDTA and 2 mM phenylmethanesulfonyl fluoride and disrupted by sonication for 10 min at 240 W. After centrifugation at 23,700 × g for 30 min, the supernatant was collected. The precipitate was washed with 40 ml of the same buffer, and the washing buffer was combined with the supernatant (cell extract). Ninety-percent-saturated ammonium sulfate solution in 50 mM potassium phosphate buffer (pH 8.0) containing 2 mM EDTA was added to the cell extract to 35% saturation. After centrifugation, the precipitate was dissolved in 60 ml of 50 mM potassium phosphate buffer (pH 8.0) containing 2 mM EDTA and 2 mM phenylmethanesulfonyl fluoride and dialyzed for 1 day against 4 liters of 10 mM potassium phosphate buffer (pH 8.0) containing 0.5 mM EDTA and then for additional 1 day against 2 liters of 5 mM potassium phosphate buffer (pH 8.0) containing 0.5 mM EDTA.
The dialysate was applied to a DEAE-cellulose column (bed volume, 200 ml) that had been equilibrated with 10 mM potassium phosphate buffer (pH 8.0) containing 0.5 mM EDTA. The column was washed successively with 1 liter of the same buffer and 1 liter of 10 mM potassium phosphate buffer (pH 8.0) containing 0.5 mM EDTA and 100 mM KCl and then developed with 1 liter of 10 mM potassium phosphate buffer (pH 8.0) containing 0.5 mM EDTA and 150 mM KCl. The EutA-containing fractions were pooled and concentrated to 100 ml by ultrafiltration through a Diaflo PM-10 membrane (Millipore). The concentrate was dialyzed for 1 day against 4 liters of 7 mM potassium phosphate buffer (pH 8.0) and concentrated again to 25 ml. The concentrate obtained was dialyzed for 1 day against 2 liters of 7 mM potassium phosphate buffer (pH 8.0).
The dialysate was applied to a hydroxyapatite column (bed volume, 100 ml) which had been equilibrated with 7 mM potassium phosphate buffer (pH 8.0). The column was developed with 340 ml of the same buffer. The EutA-containing fractions were pooled and concentrated to about 3.5 ml by ultrafiltration through a Diaflo PM-10 membrane and Centriplus YM-10 (Millipore). EutA was finally purified by gel filtration on a Sephadex G-200 column (bed volume, 180 ml) in 10 mM potassium phosphate buffer (pH 8.0), concentrated by ultrafiltration, and stored at −80°C until use.
RESULTS
Induction of EAL in E. coli, K. oxytoca, and K. pneumoniae.
It was reported that E. coli and S. enterica serovar Typhimurium LT2 produce EAL when grown in the presence of both ethanolamine and vitamin B12 (7, 8, 31). When E. coli JM109 was grown aerobically in the synthetic medium containing glycerol as a carbon source, ethanolamine as a sole nitrogen source, and CN-Cbl (40 ng/ml), as described by Scarlett and Turner (35), induction of enzyme activity was certainly observed upon the addition of AdoCbl. The induced level of EAL activity (0.077 U/mg of protein in cell extract compared with the uninduced level of <0.002 U/mg) was roughly in the same range as activity reported for E. coli NCIMB 8114 and Klebsiella pneumoniae subsp. pneumoniae (formerly Klebsiella aerogenes) NCIMB 8267 (0.10 to 0.23 U/mg of protein in cell extract) (35) and S. enterica serovar Typhimurium LT2 (0.045 U/mg) (31). EAL activity was induced in K. oxytoca ATCC 8724 and K. pneumoniae ATCC 25955 as well, when they were grown under the same conditions (data not shown).
In situ reactivation of inactivated holoEAL of E. coli.
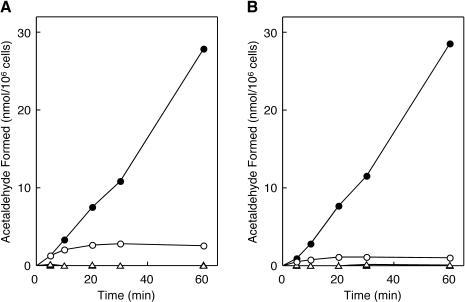
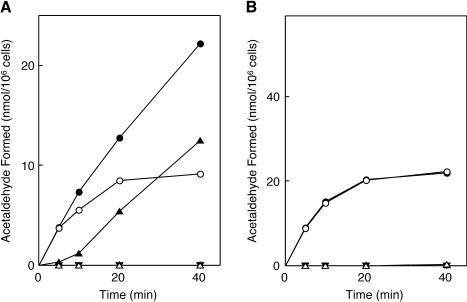
To examine whether the inactivated holoEAL is reactivated, the time course of the EAL reaction was determined with toluene-treated (permeabilized) cells of induced E. coli JM109 (the so-called in situ system) (20). As shown in Fig. 2A, the deamination of ethanolamine by permeabilized E. coli JM109 cells with added AdoCbl was accompanied by concomitant inactivation and ceased almost completely within 20 min. However, when ATP and Mg2+ were supplemented in the reaction mixture in addition to AdoCbl, deamination of ethanolamine proceeded apparently without significant inactivation, even 60 min after the reaction was initiated. Meanwhile, enzyme activity with added AdoCbl in permeabilized E. coli cells grown in the presence of 2 μg of CN-Cbl/ml was about 40% of that in cells grown in the presence of 40 ng of CN-Cbl/ml (Fig. 2B). However, almost the same level of enzyme activity was attained upon the addition of ATP and Mg2+ together with AdoCbl as was observed with cells grown on 40 ng of CN-Cbl/ml. These results suggested that about 60% of the enzyme in the cells grown in 2 μg of CN-Cbl/ml had been converted to the inactive enzyme-CN-Cbl complex or the inactivated holoenzyme under the growth conditions and that the inactive enzyme-cobalamin complexes underwent ATP-dependent reactivation or activation. It was amazing that the presence of a 50-fold excess of CN-Cbl in a growing culture resulted in only 60% reduction of enzyme activity. This fact suggests in vivo lowering of EAL affinity for CN-Cbl by a reactivating factor in the presence of ATP and Mg2+, as in the case of diol and glycerol dehydratases (28, 43). Despite the presence of the inactivated holoenzyme or inactive enzyme-CN-Cbl complex, enzyme activity was not increased at all in the presence of ATP and Mg2+ without AdoCbl, indicating that free AdoCbl is also absolutely required for the (re)activation. The absolute requirement of free AdoCbl and lack of requirement of a reducing agent for (re)activation eliminated the involvement of an adenosylating enzyme [ATP:cob(I)alamin adenosyltransferase; BtuR] and suggested the exchange of modified coenzyme or CN-Cbl for AdoCbl in the (re)activation process. The presence of a similar reactivation system was observed in the permeabilized cells of K. oxytoca ATCC 8724 and K. pneumoniae ATCC 25955 when cultivated under the conditions identical to those for E. coli (data not shown).
FIG. 2.
Time courses of in situ deamination of ethanolamine by permeabilized E. coli cells that were grown in medium containing ethanolamine and CN-Cbl. Toluene-treated cells of E. coli JM109 (7.2 × 106 cells) grown in the medium containing CN-Cbl at concentrations of 40 ng/ml (A) and 2 μg/ml (B) were incubated at 37°C for the indicated times in the following mixtures: 30 mM potassium phosphate buffer (pH 8.0), 50 mM KCl, 10 mM ethanolamine, with (circles) or without (triangles) 15 μM AdoCbl, in the presence (solid symbols) or absence (open symbols) of 3 mM ATP plus 3 mM MgCl2, in a total volume of 1.0 ml. The amount of acetaldehyde formed was determined as described in the text.
In vitro reactivation of inactivated holoEAL of E. coli.
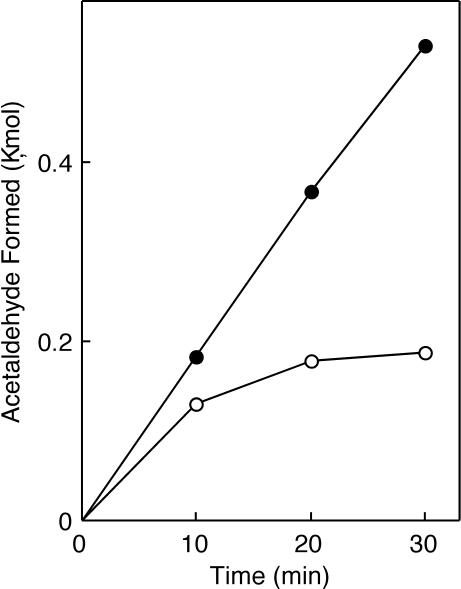
In vitro reactivation of ethanolamine-inactivated holoEAL in the presence of ATP, Mg2+, and AdoCbl was also examined. One of the typical results with homogenates is shown in Fig. 3. The reactivation was observed with not only homogenates but also with cell extracts (data not shown) of E. coli JM109 grown in the synthetic glycerol-ethanolamine medium containing CN-Cbl. The efficiency of reactivation was dependent in a nonlinear manner on the protein concentration of cell extracts (data not shown). To simplify the interpretation of the data, EAL reactivation was examined in situ in the following studies.
FIG. 3.
Time courses of ethanolamine deamination by homogenates of E. coli cells that were grown in the medium containing ethanolamine and CN-Cbl. Homogenates of E. coli JM109 grown in the medium containing 40 ng of CN-Cbl/ml were incubated at 37°C for the indicated time in the following mixtures: 30 mM potassium phosphate buffer (pH 8.0), 50 mM KCl, 10 mM ethanolamine, 15 μM AdoCbl, in the presence (solid circles) or absence (open circles) of ATP plus MgCl2 (3 mM each), in a total volume of 0.5 ml. The amount of acetaldehyde formed was determined as described in the text.
In situ reactivation of inactivated holoEAL overexpressed in recombinant E. coli.
We assumed that a certain protein factor was responsible for the reactivation of inactivated holoEAL. If this is the case, the gene(s) encoding the EAL-reactivating factor might be present in the vicinity of the EAL genes (eutBC). As shown in Fig. 1, there is a specific region in E. coli genomic DNA that is highly homologous to the S. enterica serovar Typhimurium LT2 eut operon (23, 49). The composition and the order of genes in this region are identical to those of the S. enterica serovar Typhimurium LT2 eut operon. Identities of the amino acid sequences of the products of eut genes between E. coli and S. enterica serovar Typhimurium LT2 are 79 to 98%, suggesting that this homologous region is essential for the cobalamin-dependent utilization of ethanolamine in E. coli as well. Actually, high EAL activity in recombinant E. coli cells overexpressing eutB and eutC of E. coli has been demonstrated (Akita and Toraya, unpublished). Therefore, we searched for the gene(s) encoding the hypothetical EAL-reactivating factor in the E. coli eut operon. We cloned DNA segments containing several parts of the E. coli eut operon into plasmid pUC119 (Fig. 1). The cloned genes of the operon can be expressed with the lac promoter on the vector, and thus their expression does not require ethanolamine and cobalamin. Therefore, the cloned genes can be expressed without induction of the chromosomal eut operon. This enabled us to study reactivation without disturbance by the chromosomal gene-derived products. The plasmid pUCEA3 carries a 14.7-kb DNA segment that covers most of the eut operon except the region encoding the C-terminal 146 amino acid residues of EutR (Fig. 1). The eutR gene encodes a positive regulator for the eut operon in S. enterica serovar Typhimurium LT2 (23). In contrast, pUCEA2 contains less than half of the downstream region (∼5.8 kb) of the eut operon and lacks the region encoding the N-terminal 63 amino acid residues of EutA. We examined the permeabilized cells of recombinant E. coli carrying pUCEA2 and pUCEA3 for reactivating factor activity. One of the typical results is shown in Fig. 4. The deamination of ethanolamine by permeabilized cells of both recombinant E. coli strains with added AdoCbl was accompanied by concomitant inactivation and ceased almost completely within 20 min in the absence of ATP and Mg2+. However, when ATP and Mg2+ were supplemented in the reaction mixture in addition to AdoCbl, an initial, rapid phase of ethanolamine deamination was followed by a slower but almost constant rate of the deamination reaction in the permeabilized cells of E. coli carrying pUCEA3 (Fig. 4A) but not in the permeabilized cells of E. coli carrying plasmid pUCEA2 (Fig. 4B). Therefore, it is strongly suggested that a certain protein factor(s) encoded by the gene(s) in the upstream region of eut operon that is lacking in pUCEA2 is essential for the reactivation of inactivated holoEAL.
FIG. 4.
Time courses of in situ deamination of ethanolamine by recombinant E. coli. Permeabilized cells of E. coli carrying pUCEA3 (6.6 × 106 cells) (A) or pUCEA2 (1.1 × 106 cells) (B) were incubated at 37°C for the indicated times in the following mixtures: 30 mM potassium phosphate buffer (pH 8.0), 50 mM KCl, 10 mM ethanolamine, 15 μM AdoCbl, in the presence (solid circles) or absence (open circles) of ATP plus MgCl2 (3 mM each), in a total volume of 1.0 ml. The amount of acetaldehyde formed was determined as described in the text.
Identification of the gene(s) encoding an essential component of the reactivating factor.
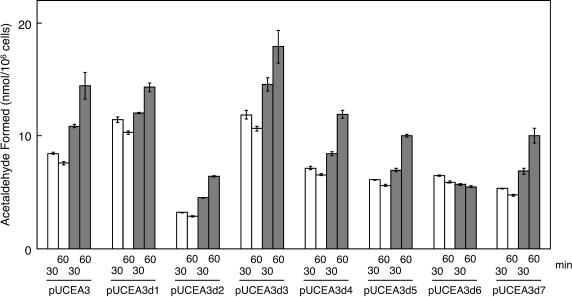
To identify the gene(s) encoding an essential component of the EAL-reactivating factor, we constructed a series of deletion mutant plasmids of pUCEA3 (Fig. 1). To measure the capability of reactivation with permeabilized E. coli cells carrying each plasmid (Fig. 5), the amounts of acetaldehyde formed at 60 min of incubation in the presence and absence of ATP and Mg2+ were compared to that at 30 min in the absence of ATP and Mg2+ (essentially, all the holoenzyme present in the reaction mixture had been inactivated by this time). There were obvious differences among strains in the level of enzyme activity itself. In the presence of ATP and Mg2+, permeabilized cells of E. coli strains harboring either plasmid pUCEA3d1, pUCEA3d2, pUCEA3d3, pUCEA3d4, pUCEA3d5, or pUCEA3d7 showed obvious reactivation of the inactivated holoEAL, as did the strain carrying pUCEA3, although the extent of reactivation varied among strains. In contrast, reactivation did not occur at all with the permeabilized E. coli cells harboring pUCEA3d6. pUCEA3d6 lacked only eutA, compared with pUCEA3 (Fig. 1). From these results, it can be concluded that EutA, the gene product of eutA, is an EAL-reactivating factor itself or at least its essential component.
FIG. 5.
Identification of the gene(s) encoding the protein(s) essential for reactivation. Permeabilized cells of recombinant E. coli strains harboring various plasmids (1.4 × 107 to 4.7 × 107 cells) were incubated at 37°C for 30 and 60 min in the following mixtures: 30 mM potassium phosphate buffer (pH 8.0), 50 mM KCl, 10 mM ethanolamine, 15 μM AdoCbl, in the presence (grey bars) or absence (open bars) of ATP plus MgCl2 (3 mM each), in a total volume of 1.0 ml. The amount of acetaldehyde formed was determined as described in the text. Error bars, average ± SD for the results of three experiments.
The search for an auxiliary protein(s).
Since reactivating factors for diol dehydratase and glycerol dehydratase consist of two kinds of subunits (21, 43), there might be an auxiliary protein(s) for the EAL-reactivating factor. By coexpression experiments, we searched for a gene(s) in the eut operon with a product(s) that potentiates the EAL-reactivating activity of EutA. However, no evidence was obtained that suggested the presence of such a gene(s) in the eut operon of E. coli. It was therefore strongly suggested that EutA is the only component of the EAL-reactivating factor.
In situ activation of the enzyme-CN-Cbl complex in recombinant E. coli.
Inactivation of holoEAL during catalysis involves the irreversible cleavage of the Co-C bond of enzyme-bound AdoCbl and is brought about by tight binding of the modified coenzyme. Therefore, the inactive enzyme-CN-Cbl complex can be considered a model of the inactivated holoenzyme. We examined whether the permeabilized cells of E. coli carrying pSTV(eutABC)3 and pSTV(eutBC)3 are capable of activation of the enzyme-CN-Cbl complex. In situ formation of inactive complex with CN-Cbl was almost completed within a few minutes of preincubation in both strains (data not shown). When ATP and Mg2+ were supplemented together with AdoCbl in the preincubated reaction mixture containing the enzyme-CN-Cbl complex, about 75% of the activity was restored in the permeabilized E. coli cells carrying pSTV(eutABC)3 (Fig. 6A), but not at all in the permeabilized E. coli cells carrying pSTV(eutBC)3 (Fig. 6B). Such activation did not occur at all in the absence of ATP and Mg2+ or even in the presence of ATP and Mg2+ without added AdoCbl. Prolonged preincubation with CN-Cbl led to a sudden decrease in capability of activation, probably due to the lability of the EAL-reactivating factor (data not shown). Activation of the enzyme-CN-Cbl complex by ATP, Mg2+, and AdoCbl was observed in the cell extracts of induced E. coli JM109 and recombinant E. coli carrying pUCEA3 or pSTV(eutABC)3 as well (data not shown).
FIG. 6.
Time courses of in situ activation of the enzyme-CN-Cbl complex in recombinant E. coli. Permeabilized cells of E. coli carrying pSTV(eutABC)3 (A) or pSTV(eutBC)3 (B) (each, 1.6 × 108 cells) were incubated at 37°C for 1 min with (triangles and inverted triangles) or without (circles) 7.5 μM CN-Cbl in 40 mM potassium phosphate buffer (pH 8.0) containing 50 mM KCl and 5 mM ethanolamine in a total volume of 6.0 ml. The 0.2-ml aliquots were drawn from the mixtures and added to the following mixtures: 30 mM potassium phosphate buffer (pH 8.0), 60 mM KCl, 10 mM ethanolamine, and 15 μM AdoCbl, in the presence (solid symbols) or absence (open symbols) of ATP plus MgCl2 (3 mM each), to a final volume of 1.0 ml. In one control experiment, ATP and MgCl2 were added but AdoCbl was not added (solid inverted triangles). The reaction mixtures were incubated at 37°C for the indicated times, and the acetaldehyde formed was determined as described in the text.
High-level expression and purification of recombinant EutA.
To obtain direct evidence for the function of EutA as an EAL-reactivating factor, the EutA protein overexpressed in E. coli JM109 was purified to homogeneity. EutA migrated as a single band with an Mr of 50,000 upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis, in good agreement with the predicted molecular weight of EutA (49,599). The NH2-terminal six-amino-acid sequence determined by Edman sequencing was MNTRQL. This coincided with that deduced from the nucleotide sequence of the eutA gene. EutA migrated as a very broad band upon PAGE under nondenaturing conditions.
In vitro reactivation of the ethanolamine-inactivated holoEAL by purified EutA.
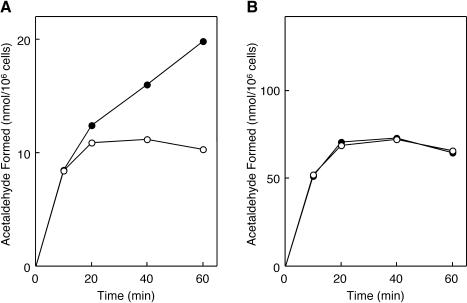
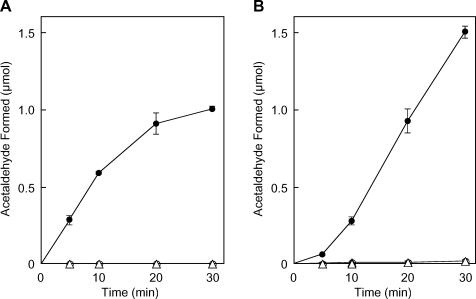
The purified recombinant EutA was examined in vitro for activity as a reactivating factor for EAL. When the holoEAL suicidally inactivated during deamination of ethanolamine was incubated with EutA in the presence of AdoCbl, ATP, and Mg2+, rapid reactivation of EAL took place (Fig. 7A). No restoration of EAL activity was observed without EutA or with EutA but in the absence of ATP and Mg2+. By comparing the activity of reactivated enzyme with that of the noninactivated control, it is evident that 46% of original EAL activity was restored under the conditions. HoloEAL inactivated by O2 in the absence of substrate also underwent rapid reactivation by the factor in the presence of AdoCbl, ATP, and Mg2+ (data not shown). Thus, it can be concluded that EutA alone actually functions as a reactivating factor for both the suicidally inactivated and O2-inactivated holoEAL in vitro.
FIG. 7.
Time courses of in vitro reactivation of suicidally inactivated holoEAL (A) and in vitro activation of the EAL-CN-Cbl complex (B) by purified EutA. Suicidally inactivated holoEAL was formed by incubation of purified apoEAL (0.11 U; 5.9 μg) at 37°C for 45 min with 20 μM AdoCbl in 40 mM potassium phosphate buffer (pH 8.0) containing 0.4 M ethanolamine and 0.4% Brij 35 in a total volume of 25 μl. The EAL-CN-Cbl complex was formed by incubation of apoEAL (0.11 U; 5.9 μg) at 37°C for 15 min with 4 μM CN-Cbl in 40 mM potassium phosphate buffer (pH 8.0) containing 0.4 M ethanolamine and 0.4% Brij 35 in a total volume of 25 μl. In vitro reactivation and activation were assayed as described in the text with (circles) or without (triangles) 33 μg of EutA in the presence (solid symbols) and absence (open symbols) of ATP plus MgCl2 (20 mM each). AdoCbl was added to a final concentration of 10 μM. Noninactivated EAL was treated similarly to controls to estimate the extent of (re)activation. Error bars, average ± SD for the results of three experiments.
In vitro activation of the EAL-CN-Cbl complex by purified EutA.
The results shown in Fig. 7B indicate that the inactive complex of EAL with CN-Cbl, an adenine-lacking cobalamin, also underwent rapid activation in vitro by EutA in the presence of free AdoCbl, ATP and Mg2+. Again, no activation occurred without EutA or with EutA but in the absence of ATP and Mg2+. By comparison with the no-CN-Cbl control, it is evident that approximately 71% of the inactive EAL-CN-Cbl complex was activated by EutA under the experimental conditions. By increasing the EutA/EAL ratio, restoration of EAL activity to up to 95% of original levels was observed (data not shown).
DISCUSSION
In this paper, we report for the first time that inactivated holoEAL in permeabilized cells, homogenates, and cell extracts of E. coli undergoes reactivation by a certain protein(s). The eutA gene in the eut operon was identified as the gene encoding this protein. It was also shown here that the purified recombinant EutA protein alone reactivated the inactivated holoEAL and activated the EAL-CN-Cbl complex in the presence of AdoCbl, ATP, and Mg2+ to approximately 46 and 71% of the original activity, respectively. Thus, it was concluded that EutA is the only component of an EAL-reactivating factor. Roof and Roth (32) demonstrated by a genetic study with S. enterica serovar Typhimurium LT2 that eutA mutants defective in ethanolamine utilization under aerobic conditions with added CN-Cbl are partially rescued by exogenously supplied AdoCbl. This finding suggested that one possible function of EutA was as a DdrAB-like reactivating factor for EAL (23), which is consistent with our conclusions. The eutA gene exists immediately upstream of the EAL genes (eutBC) in the eut operon and encodes a polypeptide consisting of 467 amino acid residues (Fig. 8A) with a predicted molecular weight of 49,599. The deduced amino acid sequence of the EutA protein was compared with those of other proteins in databases with the BLAST program (2), but EutA did not show significant overall homology to other proteins. As pointed out by Kofoid et al. (23), E. coli EutA showed local homologies to EutJ in both E. coli and S. enterica serovar Typhimurium LT2 and to E. coli DnaK and the other Hsp70 family molecular chaperones. The capability of in situ reactivation of holoEAL was not affected by the deletion of eutJ (Fig. 5). Moreover, eutJ could not replace eutA in the eutA mutant at all. Previous research first discovered reactivating factors for diol and glycerol dehydratases (27, 39) and reported that there are fragmentary sequence similarities between large subunits of diol and glycerol dehydratases-reactivating factors (DdrA and GdrA) and Hsp70 family molecular chaperones (21, 28). Furthermore, these regions were found to be conserved in EutA as well (Fig. 8B), although there is no overall similarity between them except that between DdrA and GdrA. These three conserved regions constitute part of the ADP-binding site of human Hsp70 (9, 16). Therefore, it is strongly suggested that the reactivating factors and Hsp70 family molecular chaperones evolved from a common ancestor protein and share the ATP/ADP-switching mechanism (40).
FIG. 8.
Deduced amino acid sequence of E. coli EutA (A) and fragmentary sequence homology between reactivating factors and Hsp70 family molecular chaperones (B). Positions that show fragmentary homology in panel B are underlined in panel A. Identical and similar amino acid residues are indicated by asterisks and dots, respectively, in panel B. Eco, E. coli; Kox, K. oxytoca; Kpn, K. pneumoniae; Cco, Clostridium cochlearium; hum, human.
Inactivation of EAL during catalysis involves irreversible cleavage of the Co-C bond of AdoCbl. The modified coenzyme remains tightly bound to the enzyme. Thus, the reactivation of inactivated holoEAL must require the readenosylation of enzyme-bound modified coenzyme or the exchange of modified coenzyme for intact coenzyme. If the former was the case, ATP and a reducing agent would be necessary. If the latter was the case, AdoCbl would be necessary. The absolute requirements of AdoCbl and ATP but not of a reducing agent for reactivation strongly suggest that the reactivation of inactivated holoEAL by the EAL-reactivating factor takes place by the ATP-dependent cobalamin-exchange mechanism. The same requirements for the activation of inactive enzyme-CN-Cbl complex also suggest this mechanism of (re)activation. Such reactivation by the ATP-dependent exchange of enzyme-bound modified coenzyme for intact AdoCbl has already been demonstrated to occur in the reactivation of inactivated holodiol dehydratase (28, 43) and hologlycerol dehydratase (21, 36) by their reactivating factors. In these enzymes, the ADP-bound forms of reactivating factors induce conformational change of the enzymes through complex formation, resulting in the release of enzyme-bound modified coenzyme (21, 28). Upon exchange of ADP for ATP at the nucleotide-binding site, the reactivating factors are converted into the low-affinity form for the dehydratases; thus, the complexes with dehydratases dissociate into the reactivating factors and the free apoenzymes that are reconstitutable to the active holoenzymes (21, 28). The reactivating factor does not become a final constituent of the enzyme system. Since the reactivating factors have extremely low but distinct levels of ATPase activity (21, 28, 36), the ATP-bound, low-affinity form of the factors is back converted into the ADP-bound, high-affinity form. That is, the factors' affinity for the enzymes is controlled by the bound nucleotides. These features of action mechanisms of diol and glycerol dehydratase reactivating factors are quite analogous to those of representative molecular chaperones, such as GroEL and DnaK. Therefore, we concluded that the reactivating factors for diol and glycerol dehydratases are a new type of molecular chaperones that participate in the reactivation of the already-folded but inactivated enzymes. Recently, the crystal structure of the nucleotide-free form of GdrAB of K. pneumoniae was reported by Liao et al. (26). The crystal structure of DdrAB of K. oxytoca is very similar to that of GdrAB (40).
All the data presented here strongly suggest that the reactivation of inactivated holoEAL occurs similarly, although the detailed mechanism of the EAL-reactivating factor has not yet been elucidated. Baker et al. (4) reported that AdoCbl-dependent β-lysine mutase undergoes concomitant inactivation during catalysis that accompanies irreversible cleavage of the Co-C bond of AdoCbl. They noted that this inactivation is prevented by the addition of ATP and a sulfhydryl protein named E2. However, it might be possible that this phenomenon is not due to the prevention of inactivation but to the reactivation of the inactivated holoenzyme and that E2 is a kind of reactivating factor for this enzyme. It was reported that there is a glmL (mutL) gene between the glmS (mutS) and glmE (mutE) genes for clostridial glutamate mutase (50). The function of its product is not known at present. Since the deduced amino acid sequence of GlmL (MutL) shows fragmentary similarity to DdrA, GdrA, and EutA (Fig. 8B), it might be a reactivating factor for glutamate mutase or its major component. Therefore, such reactivating factors are not special but rather general for the AdoCbl-dependent enzymes. This seems reasonable, because AdoCbl-dependent enzymes catalyze reactions by radical mechanism and tend to undergo suicide inactivation during catalysis or inactivation by oxygen in the absence of substrate (40).
Acknowledgments
This work was supported in part by Grant-in-Aid for Scientific Research ([B] 13480195 and Priority Areas 753) to T.T. from the Ministry of Education, Science, Sports, and Culture, Japan.
We thank Hirotada Mori for providing clone 421 of the Kohara E. coli genomic λ phage library. We also thank Kumiko Ito, Ruriko Ogawa, and Yukiko Kurimoto for assistance in manuscript preparation.
REFERENCES
- 1.Abeles, R. H., A. M. Brownstein, and C. H. Randles. 1960. β-Hydroxypropionaldehyde, an intermediate in the formation of 1,3-propanediol by Aerobacter aerogenes. Biochim. Biophys. Acta 41:530-531. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachovchin, W. W., R. G. Eagar, Jr., K. W. Moore, and J. H. Richards. 1977. Mechanism of action of adenosylcobalamin: glycerol and other substrate analogues as substrates and inactivators for propanediol dehydratase-—kinetics, stereospecificity, and mechanism. Biochemistry 16:1082-1092. [DOI] [PubMed] [Google Scholar]
- 4.Baker, J. J., C. van der Drift, and T. C. Stadtman. 1973. Purification and properties of β-lysine mutase, a pyridoxal phosphate and B12 coenzyme dependent enzyme. Biochemistry 12:1054-1063. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee, R. (ed.). 1999. Chemistry and biochemistry of B12. John Wiley & Sons, New York, N.Y.
- 6.Banerjee, R. (ed.). 2003. Chemical reviews, vol. 103. Radical enzymology. American Chemical Society, Washington, D.C.
- 7.Blackwell, C. M., F. A. Scarlett, and J. M. Turner. 1977. Microbial metabolism of amino alcohols. Control of formation and stability of partially purified ethanolamine ammonia-lyase in Escherichia coli. J. Gen. Microbiol. 98:133-139. [DOI] [PubMed] [Google Scholar]
- 8.Blackwell, C. M., and J. M. Turner. 1978. Microbial metabolism of amino alcohols. Formation of coenzyme B12-dependent ethanolamine ammonia-lyase and its concerted induction in Escherichia coli. Biochem. J. 176:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradbeer, C. 1965. The clostridial fermentations of choline and ethanolamine. I. Preparation and properties of cell-free extracts. J. Biol. Chem. 240:4669-4674. [PubMed] [Google Scholar]
- 11.Bradbeer, C. 1965. The clostridial fermentations of choline and ethanolamine. II. Requirement for a cobamide coenzyme by an ethanolamine deaminase. J. Biol. Chem. 240:4675-4681. [PubMed] [Google Scholar]
- 12.Chang, G. W., and J. T. Chang. 1975. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature 254:150-151. [DOI] [PubMed] [Google Scholar]
- 13.Dolphin, D. (ed.). 1982. B12, vol. 2. John Wiley & Sons, New York, N.Y.
- 14.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faust, L. R. P., J. A. Connor, D. M. Roof, J. A. Hoch, and B. M. Babior. 1990. Cloning, sequencing, and expression of the genes encoding the adenosylcobalamin-dependent ethanolamine ammonia-lyase of Salmonella typhimurium. J. Biol. Chem. 265:12462-12466. [PubMed] [Google Scholar]
- 16.Flaherty, K. M., C. DeLuca-Flaherty, and D. B. McKay. 1990. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346:623-628. [DOI] [PubMed] [Google Scholar]
- 17.Forage, R. G., and M. A. Foster. 1982. Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. J. Bacteriol. 149:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forage, R. G., and E. C. C. Lin. 1982. DHA system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J. Bacteriol. 151:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 20.Honda, S., T. Toraya, and S. Fukui. 1980. In situ reactivation of glycerol-inactivated coenzyme B12-dependent enzymes, glycerol dehydratase and diol dehydratase. J. Bacteriol. 143:1458-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajiura, H., K. Mori, T. Tobimatsu, and T. Toraya. 2001. Characterization and mechanism of action of a reactivating factor for adenosylcobalamin-dependent glycerol dehydratase. J. Biol. Chem. 276:36514-36519. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, B. H., and E. R. Stadtman. 1968. Ethanolamine deaminase, a cobamide coenzyme-dependent enzyme. II. Physical and chemical properties and interaction with cobamides and ethanolamine. J. Biol. Chem. 243:1794-1803. [PubMed] [Google Scholar]
- 23.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 25.Lee, H. A., Jr., and R. H. Abeles. 1963. Purification and properties of dioldehydrase, an enzyme requiring a cobamide coenzyme. J. Biol. Chem. 238:2367-2373. [PubMed] [Google Scholar]
- 26.Liao, D.-I., L. Reiss, I. Turner, Jr., and G. Dotson. 2003. Structure of glycerol dehydratase reactivase: a new type of molecular chaperone. Structure 11:109-119. [DOI] [PubMed] [Google Scholar]
- 27.Mori, K., T. Tobimatsu, T. Hara, and T. Toraya. 1997. Characterization, sequencing, and expression of the genes encoding a reactivating factor for glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. J. Biol. Chem. 272:32034-32041. [DOI] [PubMed] [Google Scholar]
- 28.Mori, K., and T. Toraya. 1999. Mechanism of reactivation of coenzyme B12-dependent diol dehydratase by a molecular chaperone-like reactivating factor. Biochemistry 38:13170-13178. [DOI] [PubMed] [Google Scholar]
- 29.Pawelkiewicz, J., and B. Zagalak. 1965. Enzymic conversion of glycerol into β-hydroxypropionaldehyde in a cell-free extract from Aerobacter aerogenes. Acta Biochim. Pol. 12:207-218. [PubMed] [Google Scholar]
- 30.Poznanskaya, A. A., M. I. Yakusheva, and V. A. Yakovlev. 1977. Study on the mechanism of action of adenosylcobalamin-dependent glycerol dehydratase from Aerobacter aerogenes. II. The inactivation kinetics of glycerol dehydratase complexes with adenosylcobalamin and its analogs. Biochim. Biophys. Acta 484:236-243. [DOI] [PubMed] [Google Scholar]
- 31.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roof, D. M., and J. R. Roth. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171:3316-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roof, D. M., and J. R. Roth. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimunium. J. Bacteriol. 174:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Scarlett, F. A., and J. M. Turner. 1976. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J. Gen. Microbiol. 95:173-176. [DOI] [PubMed] [Google Scholar]
- 36.Seifert, C., S. Bowien, G. Gottschalk, and R. Daniel. 2001. Identification and expression of the genes and purification and characterization of the gene products involved in reactivation of coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. Eur. J. Biochem. 268:2369-2378. [DOI] [PubMed] [Google Scholar]
- 37.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stroinski, A., J. Pawelkiewicz, and B. C. Johnson. 1974. Allosteric interactions in glycerol dehydratase. Purification of enzyme and effects of positive and negative cooperativity for glycerol. Arch. Biochem. Biophys. 162:321-330. [DOI] [PubMed] [Google Scholar]
- 39.Tobimatsu, T., H. Kajiura, M. Yunoki, M. Azuma, and T. Toraya. 1999. Identification and expression of the genes encoding a reactivating factor for adenosylcobalamin-dependent glycerol dehydratase. J. Bacteriol. 181:4110-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toraya, T. 2003. Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem. Rev. 103:2095-2127. [DOI] [PubMed] [Google Scholar]
- 41.Toraya, T., S. Honda S. Kuno, and S. Fukui. 1978. Coenzyme B12-dependent diol dehydratase: regulation of apoenzyme synthesis in Klebsiella pneumoniae (Aerobacter aerogenes) ATCC 8724. J. Bacteriol. 135:726-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toraya, T., S. Kuno, and S. Fukui. 1980. Distribution of coenzyme B12-dependent diol dehydratase and glycerol dehydratase in selected genera of Enterobacteriaceae and Propionibacteriaceae. J. Bacteriol. 141:1439-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toraya, T., and K. Mori. 1999. A reactivating factor for coenzyme B12-dependent diol dehydratase. J. Biol. Chem. 274:3372-3377. [DOI] [PubMed] [Google Scholar]
- 44.Toraya, T., T. Shirakashi, T. Kosuga, and S. Fukui. 1976. Substrate specificity of coenzyme B12-dependent diol dehydrase: glycerol as both a good substrate and a potent inactivator. Biochem. Biophys. Res. Commun. 69:475-480. [DOI] [PubMed] [Google Scholar]
- 45.Toraya, T., K. Ushio, S. Fukui, and H. P. C. Hogenkamp. 1977. Studies on the mechanism of the adenosylcobalamin-dependent diol dehydrase reaction by the use of analogs of the coenzyme. J. Biol. Chem. 252:963-970. [PubMed] [Google Scholar]
- 46.Ushio, K., S. Honda, T. Toraya, and S. Fukui. 1982. The mechanism of in situ reactivation of glycerol-inactivated coenzyme B12-dependent enzymes, glycerol dehydratase and diol dehydratase. J. Nutr. Sci. Vitaminol. 28:225-236. [DOI] [PubMed] [Google Scholar]
- 47.Wagner, O. W., H. A. Lee, Jr., P. A. Frey, and R. H. Abeles. 1966. Studies on the mechanism of action of cobamide coenzymes. Chemical properties of the enzyme-coenzyme complex. J. Biol. Chem. 241:1751-1762. [PubMed] [Google Scholar]
- 48.Wolf, J. B., and R. N. Brey. 1986. Isolation and genetic characterizations of Bacillus megaterium cobalamin biosynthesis-deficient mutants. J. Bacteriol. 166:51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto, Y., H. Aiba, T. Baba, K. Hayashi, T. Inada, K. Isono, T. Itoh, S. Kimura, M. Kitagawa, K. Makino, T. Miki, N. Mitsuhashi, K. Mizobuchi, H. Mori, S. Nakade, Y. Nakamura, H. Nashimoto, T. Oshima, S. Oyama, N. Saito, G. Sampei, Y. Satoh, S. Sivasundaram, H. Tagami, H. Takahashi, J. Takeda, K. Takemoto, K. Uehara, C. Wada, S. Yamagata, and T. Horiuchi. 1997. Construction of a contiguous 874-kb sequence of the Escherichia coli K-12 genome corresponding to 50.0-68.8 min on the linkage map and analysis of its sequence features. DNA Res. 4:91-113. [DOI] [PubMed] [Google Scholar]
- 50.Zelder, O., B. Beatrix, U. Leutbecher, and W. Buckel. 1994. Characterization of the coenzyme-B12-dependent glutamate mutase from Clostridium cochlearium produced in Escherichia coli. Eur. J. Biochem. 226:577-585. [DOI] [PubMed] [Google Scholar]