Abstract
The mechanisms by which bacteria adopt and maintain individual shapes remain enigmatic. Outstanding questions include why cells are a certain size, length, and width; why they are uniform or irregular; and why some branch while others do not. Previously, we showed that Escherichia coli mutants lacking multiple penicillin binding proteins (PBPs) display extensive morphological diversity. Because defective sites in these cells exhibit the structural and functional characteristics of improperly localized poles, we investigated the connection between cell division and shape. Here we show that under semipermissive conditions the temperature-sensitive FtsZ84 protein produces branched and aberrant cells at a high frequency in mutants lacking PBP 5, and this phenotype is exacerbated by the loss of additional peptidoglycan endopeptidases. Surprisingly, certain ftsZ84 strains lyse at the nonpermissive temperature instead of filamenting, and inhibition of wild-type FtsZ forces some mutants into tightly wound spirillum-like morphologies. The results demonstrate that significant aspects of bacterial shape are dictated by a previously unrecognized relationship between the septation machinery and ostensibly minor peptidoglycan-modifying enzymes and that under certain circumstances improper FtsZ function can destroy the structural integrity of the cell.
Peptidoglycan does far more than serve as a protective dividing wall between the bacterial cell and its environment. For example, peptidoglycan-derived fragments act as virulence factors (7, 8), exacerbate the toxicity of gram-negative lipopolysaccharide (42), and trigger the innate immune system of organisms from fruit flies to humans (14, 15). Less appreciated is the role of peptidoglycan in imparting to bacteria specific shapes, a characteristic that contributes to survival by affecting growth and multiplication, differentiation, surface attachment, motility, and the ability to escape predatory organisms (13, 16, 18, 38, 39, 41, 46).
Although the protective function of the peptidoglycan exoskeleton has been studied for decades, the fundamental question of cell shape has become approachable only recently. On one hand, experiments suggest that peptidoglycan-synthesizing enzymes may be attached to internal filamentous scaffolds of MreB or Mbl proteins (9, 17, 22, 24), whose geometric distribution determines the cylindrical form of the bacterium. How these cytoskeletons assume their own forms, how they transduce this information to the cell wall, and what determines the original size and shape of the cylinder are open questions. On the other hand, the uniform cell shape of Escherichia coli also depends on the activities of a group of “nonessential” low-molecular-weight (LMW) penicillin binding proteins (PBPs) that make slight modifications to peptidoglycan structure (10, 30, 31). Shape defects in mutants lacking these proteins coincide with the presence of inappropriately placed, metabolically inert peptidoglycan that exhibits the characteristics of cell poles (12, 32). Because these inert patches distort the geometry of the sacculus, their spatial distribution may define the general shape of bacteria (44).
Normally, only peptidoglycan at the cell poles is inert to insertion or turnover (11). Therefore, because poles are created at septation, a defect originating during cell division might explain the propagation of misplaced polar material and the generation of different cell shapes. Because FtsZ is the central regulator of bacterial cell division (5, 23, 25), we hypothesized that this protein might also play a central role in shape determination (44). Three lines of evidence support such a role for FtsZ. First, FtsZ initiates and directs septum formation leading to synthesis of poles, and it is reasonable to assume that misplaced pole fragments in PBP mutants have the same origin. Second, FtsZ-dependent synthesis of inert rings of peptidoglycan occurs prior to septal invagination, suggesting that FtsZ initiates or localizes the synthesis of this unique polymer (11, 12). Third, such a role for FtsZ is supported by a retrospective analysis of mutations and circumstances that produce disparately shaped E. coli (44). In this work, we provide evidence for the idea that morphological variation emanates from an interaction between the FtsZ-dependent division pathway and the peptidoglycan modification pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions. The E. coli strains are described in Table 1. PS236 was a gift of J. Lutkenhaus and was used as the source of the ftsA12 allele (linked to leu::Tn10) for phage P1 transductions into other strains. WM1109 (MG1655 ftsZ84 leu::Tn10 Tetr) was a gift of W. Margolin. PBP genes were deleted by using phage P1 transduction to transfer genes marked with a kanamycin resistance cassette from a set of E. coli mutants (10) to E. coli MG1655 and WM1109. The resistance cassette was removed by RP4 resolvase so that multiple genes could be deleted from a single strain by repeated P1 transductions, as described previously (10, 21). Plasmid pPJ5 carries the wild-type dacA gene encoding PBP 5 (31), and pFAD38 (from J. Lutkenhaus) carries the sulA gene (2), both under the control of the arabinose promoter. Plasmid pAV2 was constructed by inserting a chloramphenicol resistance gene into the ampicillin resistance gene of pFAD38. The cat gene from pBAD18-Cm was amplified by PCR (primers 5′-CCGGAGCTGAATGAAGCC-3′ and 5′-CCTCCATCCAGTCTATTA-3′), and the linear product was blunt ended by treatment with Klenow fragment, phosphorylated by T4 polynucleotide kinase, and ligated into the FspI site of pFAD38 to yield pAV2.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Genotype or characteristics | FtsZ | PBP(s) deleted | Reference or source |
|---|---|---|---|---|
| Strains | ||||
| PS236 | W3110 ftsA12 leu::Tn10 | + | None | 34 |
| MG1655 | ilvG mutant rfb-50 rph-1 | + | None | W. Margolin |
| WM1109 | MG1655 ftsZ84(Ts) leu::Tn10 | ftsZ84 | None | W. Margolin |
| BMWM1-1 | WM1109 dacA::res | ftsZ84 | 5 | This study |
| AV5-2K | WM1109 dacB::Kan | ftsZ84 | 4 | This study |
| AV5-2 | WM1109 dacB::res | ftsZ84 | 4 | This study |
| AV6-1K | BMWM1-1 dacB::Kan | ftsZ84 | 4, 5 | This study |
| AV7-5K | WM1109 pbpG::Kan | ftsZ84 | 7 | This study |
| AV7-5 | WM1109 pbpG::res | ftsZ84 | 7 | This study |
| AV8-1K | BMWM1-1 pbpG::Kan | ftsZ84 | 5, 7 | This study |
| AV14-1K | MG1655 dacB::Kan | + | 4 | This study |
| AV14-1 | MG1655 dacB::res | + | 4 | This study |
| AV15-1K | MG1655 pbpG::Kan | + | 7 | This study |
| AV21-10 | MG1655 dacA::res | + | 5 | This study |
| AV22-1K | AV21-10 dacB::Kan | + | 4, 5 | This study |
| AV23-1K | AV21-10 pbpG::Kan | + | 5, 7 | This study |
| AV24-1K | AV5-2 pbpG::Kan | ftsZ84 | 4, 7 | This study |
| AV25-1K | AV7-5 dacC::Kan | ftsZ84 | 6, 7 | This study |
| AV26-1K | AV7-5 dacD::Kan | ftsZ84 | 7, DacD | This study |
| AV27-1K | AV14-1 pbpG::Kan | + | 4, 7 | This study |
| AV65-1 | AV23-1 ftsA12 leu::Tn10 (by transduction from PS236) | + | 5, 7 | This study |
| AV74-1K | AV21-10 dacC::Kan | + | 5, 6 | This study |
| AV75-1K | AV21-10 dacD::Kan | + | 5, DacD | This study |
| AV76-1K | MG1655 dacC::Kan | + | 6 | This study |
| AV77-1K | MG1655 dacD::Kan | + | DacD | This study |
| EC448 | MC4100 Δ(λattL-lom)::bla lacIqP208-ftsZ-gfp | + | None | 43 |
| AV40-1p | AV21-10 Δ(λattL-lom)::bla lacIqP208-ftsZ-gfp pAV2 | + | 5 | This study |
| AV42-2p | AV23-1K Δ(λattL-lom)::bla lacIqP208-ftsZ-gfp pAV2 | + | 5, 7 | This study |
| AV43-1p | AV27-1K Δ(λattL-lom)::bla lacIqP208-ftsZ-gfp pAV2 | + | 4, 7 | This study |
| AV44-1p | MG1655 Δ(λattL-lom)::bla lacIqP208-ftsZ-gfp pAV2 | + | None | This study |
| Plasmids | ||||
| pAV2 | SulA expression vector; Camr | This study | ||
| pFAD38 | SulA expression vector; Ampr | 2 | ||
| pPJ5 | PBP 5 expression vector; Camr | 31 | ||
| pCH189 | MinCD expression vector derived from pBAD33; Camr | P. de Boer | ||
| pJE44 | DicB expression vector derived from pBAD33; Camr | P. de Boer 19 |
Strains were maintained in Luria-Bertani (LB) broth or on LB agar plates at 30°C, supplemented when appropriate with ampicillin (50 μg/ml), chloramphenicol (20 μg/ml), or kanamycin (50 μg/ml), or with glucose (0.2%, wt/vol) to inhibit expression from the arabinose promoter. Overnight cultures were washed and diluted 1:50 into fresh LB medium containing 0, 0.25, 0.5, or 0.75% NaCl, supplemented with required antibiotics. Arabinose (0.2%, wt/vol, or 20 μM when inducing PBP 5 expression from pPJ5) was added to induce gene expression from the arabinose promoter. Growth was monitored by growing cultures in aerated flasks or by incubating them in a Bioscreen C Microbiology Reader (Labsystems, Helsinki, Finland). Chemicals were from Sigma Chemical Co. (St. Louis, Mo.).
Microscopic evaluation of morphological aberration and branching.
To compare the relative morphologies of different mutants, overnight cultures were diluted 1:100 into LB broth (0.5% salt) with appropriate antibiotics or arabinose. Samples were removed during mid-log phase (A600 = 0.3) and spotted onto 1.5% agar-coated glass slides, and the cells were allowed to settle and adhere for 5 min. Photographs were collected from random fields as described previously (31), and cell populations were evaluated visually for branching and morphological aberrations. Branches were defined as clearly elongated protrusions having a uniform width. All other bends, kinks, blebs, and imperfections were designated as “abnormalities.” For time-lapse microscopy, cells were incubated on a stage heated to 42°C, and individual cells and microcolonies were photographed at intervals.
Scanning electron microscopy (SEM).
Cells were grown to mid-log phase in LB medium plus 0.5% NaCl, collected and resuspended in phosphate-buffered saline, spotted onto glass coverslips coated with poly-l-lysine, fixed in Karnovsky's fixative (20), postfixed in 1% OsO4, and prepared as described previously (6). Cells were observed and photographed with a Hitachi 4700 field emission scanning electron microscope at original magnifications of 2,000 to 10,000 diameters.
RESULTS
Improper FtsZ84 function enhances cell branching in PBP mutants.
E. coli cells carrying the ftsZ84(Ts) allele are temperature sensitive when grown in LB medium containing low salt concentrations (≤0.5% NaCl) but grow normally in LB medium with high salt concentrations (1% NaCl), indicating that the activity of FtsZ84 can be manipulated by varying these conditions. Because ftsZ84 mutants exhibit a moderate increase in branching (1 to 2% of the population) (45), we reasoned that, if the septation machinery and the LMW PBPs cooperated to determine cell shape, then branching or abnormalities might be enhanced in ftsZ84 mutants lacking specific PBPs. Therefore, we deleted various PBPs from E. coli MG1655 ftsZ84 and observed the morphology of these strains at three temperatures (30, 37, and 42°C) and five salt concentrations (0, 0.25, 0.5, 0.75, and 1% NaCl), representing a range of FtsZ84 activities.
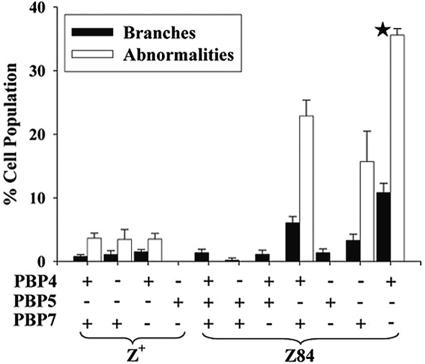
Deleting PBP 5 from an ftsZ84 mutant increased branching frequency by about sixfold at 37°C in LB medium with 0.5% NaCl (Fig. 1 and 2). Complementing this double mutant by expressing wild-type PBP 5 from pPJ5 in trans eliminated the deformities and branches (data not shown). No similarly significant aberrations occurred if PBP 5 or any other LMW PBPs were deleted from E. coli bearing the wild-type ftsZ gene (Fig. 2, columns 1 to 4, and data not shown). Also, no morphological effect was observed when PBP 1b, 4, 6, or 7 or DacD was deleted in the presence of the ftsZ84 allele, consistent with previous findings that among the PBPs the loss of PBP 5 influences cell shape most (30, 31) (Fig. 2, columns 6 and 7, and data not shown).
FIG. 1.
Branches and abnormalities in E. coli ftsZ84 ΔPBP 5. Cells grown at 30°C were washed, diluted into LB medium (0.5% NaCl), and incubated at 37°C until the culture reached mid-log phase. Arrowheads indicate some of the branches and morphological abnormalities. The image is a composite of cells from different fields. Bar, 10 μm.
FIG. 2.
Morphological abnormalities in PBP mutants expressing FtsZ or FtsZ84. E. coli strains were treated as described for Fig. 1. The star (★) indicates that the mutant was observed only at 30°C because the strain lysed at 37°C (see text).
Notwithstanding the principal role of PBP 5, branching frequency increased by an additional 80% in an ftsZ84 mutant from which both PBPs 5 and 7 were deleted (Fig. 2, column 11), in line with microscopic evidence that cell deformities increase when multiple LMW PBPs are deleted in concert with PBP 5 (30). On the other hand, an ftsZ84 mutant lacking PBPs 4 and 5 produced a ∼5-fold increase in cells with aberrant shapes, though branching was not enhanced significantly (Fig. 2, column 10). Thus, PBPs 4 and 7 must have different physiological responsibilities since deleting PBP 7 potentiated branching more than did the loss of PBP 4. As expected, an ftsZ84 strain lacking PBPs 4 and 7 was morphologically normal because PBP 5 was functional (Fig. 2, column 9).
Simple inactivation of FtsZ did not explain these unusual morphological problems. Filaments did not appear in any of the mutants grown at 30°C at any salt concentration or at 42°C in 1% NaCl (data not shown), conditions in which FtsZ84 function is sufficiently normal that wild-type division occurs. When incubated at 42°C in a low-salt medium, conditions in which FtsZ84 is completely inactive (1), cells formed uniformly smooth, unbranched filaments (data not shown). Also, when FtsZ84 was inhibited by expressing the SulA protein, the frequency of branching in filaments of an ftsZ84 ΔPBP 5 mutant dropped precipitously, from 52 to 6%. The number of branched filaments increased only in cultures incubated at an intermediate temperature (37°C) and NaCl concentration (0.5%) (Fig. 1 and 2). Thus, in the absence of PBP 5, morphological variation did not arise in cells expressing either functional or inactive FtsZ84. Instead, the phenotype depended on a partially functional FtsZ84 protein, and the results suggest that creating deformed and irregular cell shapes requires a specific FtsZ84-PBP combination.
FtsZ84-dependent lysis of specific PBP mutants.
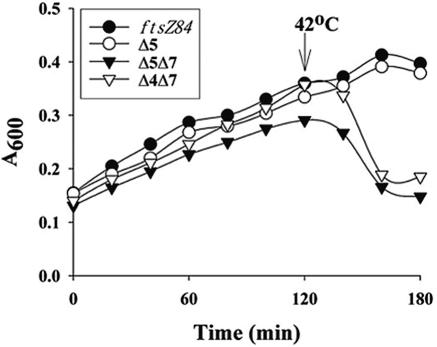
Although FtsZ is essential, cells with inactive FtsZ continue to metabolize and elongate at the nonpermissive temperature, their eventual death apparently a secondary consequence of the increased fragility of extraordinarily long filaments. Surprisingly, we discovered that in at least one genetic background FtsZ became essential for maintaining cellular integrity. When shifted to their nonpermissive temperature, mutants expressing FtsZ84 but missing either PBPs 5 and 7 or PBPs 4 and 7 did not filament but lysed instead (Fig. 3). When microcolonies on agar slides were shifted to the nonpermissive temperature, individual cells lost their rod shape, bloated slightly along their entire length, and burst (Fig. 4). This type of lysis was distinct from β-lactam-induced lysis, which is preceded by discrete bulges at the poles or septum. Once again, the phenotype depended on the impaired activity of FtsZ84: SulA inactivation of wild-type FtsZ did not induce lysis and inhibiting FtsZ84 with SulA reduced the rate of lysis by 88% (data not shown). These observations provide additional evidence that FtsZ interacts in a dramatic way with the cell's peptidoglycan-synthesizing apparatus to construct a functional cell wall, via a previously unknown pathway involving multiple LMW PBPs.
FIG. 3.
Lysis of PBP mutants expressing FtsZ84. Cells were treated as described for Fig. 1, and their growth was monitored in a Bioscreen Reader at 30°C and after shift to 42°C. Mutants lacking PBP 4, PBP 7, or PBPs 4 and 5 did not lyse (data not shown). Instead, these mutants grew as well as the ftsZ84 parental strain.
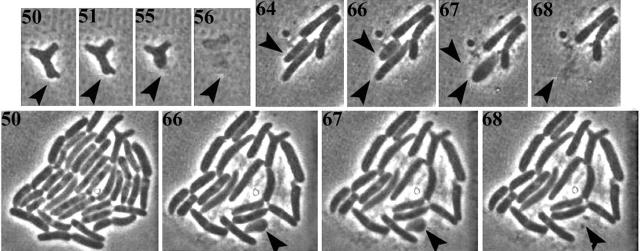
FIG. 4.
Time-lapse photomicroscopy of lysis of E. coli ftsZ84 ΔPBP 5 ΔPBP 7. Numbers indicate minutes elapsed after microcolonies on agar slides were shifted to 42°C. Arrowheads indicate individual cells that lyse over time. Comparing the large colony at 50 and at 68 min reveals that many cells lysed over that period.
Inhibition of wild-type FtsZ creates spiral cells in PBP mutants.
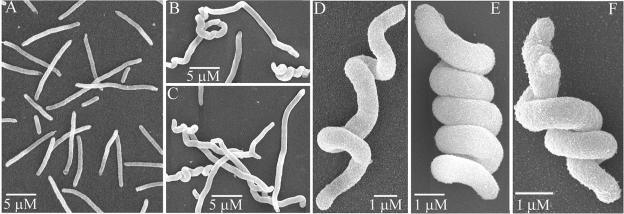
In control assays performed during the preceding experiments, we observed a revealing morphological oddity. When SulA was expressed in a ΔPBP 5 ΔPBP 7 double mutant expressing wild-type FtsZ, a third to half of the resulting cell filaments were twisted into spirals of varying pitch (Fig. 5B to F). Normal filaments without spirals were produced when SulA was expressed in a strain lacking PBPs 4 and 7 (Fig. 5A), indicating that the phenomenon depended on the loss of PBP 5. The phenotype included “tight” spirals of high helical pitch that evoked the image of tightly wound springs in which neighboring turns abutted one another (e.g., Fig. 5E and F) and “loose” spirals of low helical pitch in which successive turns were not in contact (e.g., Fig. 5D). Either arrangement could encompass the entire length of a filament (e.g., Fig. 5D and E) or be confined to a portion of only one end (e.g., Fig. 5B and C).
FIG. 5.
Spiral morphology induced by SulA-mediated inhibition of FtsZ in PBP mutants. Cells were grown overnight at 30°C in LB medium plus 0.2% glucose, washed and diluted into LB medium (0.5% NaCl) plus 0.2% arabinose to induce SulA expression, incubated at 30°C for 1 h and at 42°C for 1 h, and prepared for SEM. (A) E. coli ΔPBP 4 ΔPBP 7; (B to F) E. coli ΔPBP 5 ΔPBP 7.
To quantify these observations and see if other LMW PBPs participated in the spiral phenotype, we inhibited FtsZ in strains lacking one or two PBPs. In mutants lacking a single PBP, SulA inhibition of FtsZ produced spirals in the ΔPBP 5 strain but not in cells lacking PBP 4, 6, or 7 or DacD (Table 2, rows 1 to 5). In the ΔPBP 5 mutant, a small fraction of the cells were spiraled (2 to 3%), and these were of the loose variety. On the other hand, the frequency of spirals increased dramatically when PBP 4, 6, or 7 was deleted in concert with PBP 5. In each of these double mutants the numbers of cells with loose spirals increased to about 30% of the population, while about 3% of cells exhibited tight spirals (Table 2, rows 6 to 8). The ΔPBP 5 ΔDacD strain produced no more spiral cells than did the ΔPBP 5 single mutant (Table 2, row 9), and the strain lacking PBPs 4 and 7 produced no spirals of either type (Table 2, row 10; Fig. 5A). Thus, although the loss of PBP 5 was essential for the phenotype, the additional deletion of PBP 4, 6, or 7 enhanced the frequency and helical pitch of the resulting spirals.
TABLE 2.
Cell spirals in PBP mutants
| Strain namea | PBP(s) deleted | % of cell population with:
|
No. of cells counted | |
|---|---|---|---|---|
| Tight spirals | Loose spirals or twisted ends | |||
| 1, AV14-1K | 4 | —b | — | 400 |
| 2, AV21-10 | 5 | <0.5 | 2-3 | 528 |
| 3, AV76-1K | 6 | — | — | 300 |
| 4, AV15-1K | 7 | — | — | 400 |
| 5, AV77-1K | DacD | — | — | 430 |
| 6, AV22-1K | 5, 4 | 2.7 | 26 | 367 |
| 7, AV74-1K | 5, 6 | 3 | 30 | 432 |
| 8, AV23-1K | 5, 7 | 2.5 | 29 | 460 |
| 9, AV75-1K | 5, DacD | <0.5 | 2-3 | 350 |
| 10, AV27-1K | 4, 7 | — | — | 300 |
All strains carry the SulA expression plasmid, pFAD38.
The percentage of cells expressing the trait is less than 0.1%.
It was possible, though unlikely, that cell spiraling depended on the presence of an underlying helical FtsZ polymer. To test this, we examined the disposition of FtsZ-GFP in mutants expressing SulA. In the presence of SulA, FtsZ-green fluorescent protein fluorescence was distributed uniformly throughout the cytoplasm instead of in rings or helices (data not shown), suggesting that an altered FtsZ helical fiber did not create the spirals. A second possibility was that inhibition of septation itself was the mechanism driving spiral formation. This was also unlikely because, as mentioned, inactivating FtsZ84 at high temperature produced nonspiraled filaments. However, to be certain, we moved the temperature-sensitive ftsA12 allele from strain PS236 into the mutant AV23-1, which lacks PBPs 5 and 7. Inhibiting septation by inactivating FtsA12 at 42°C in this mutant (AV65-1) produced smooth nonspiraled filaments (data not shown). We also filamented strain AV23-1 by inactivating the essential septation protein PBP 3 with the PBP 3-specific antibiotic aztreonam. In these filaments, the ends of a very few cells (<1%) exhibited loose spirals comprising a single turn (Fig. 6). Thus, the extensive spiral phenotype did not arise by inhibiting septation per se but required the specific contribution of an FtsZ-dependent reaction.
FIG. 6.
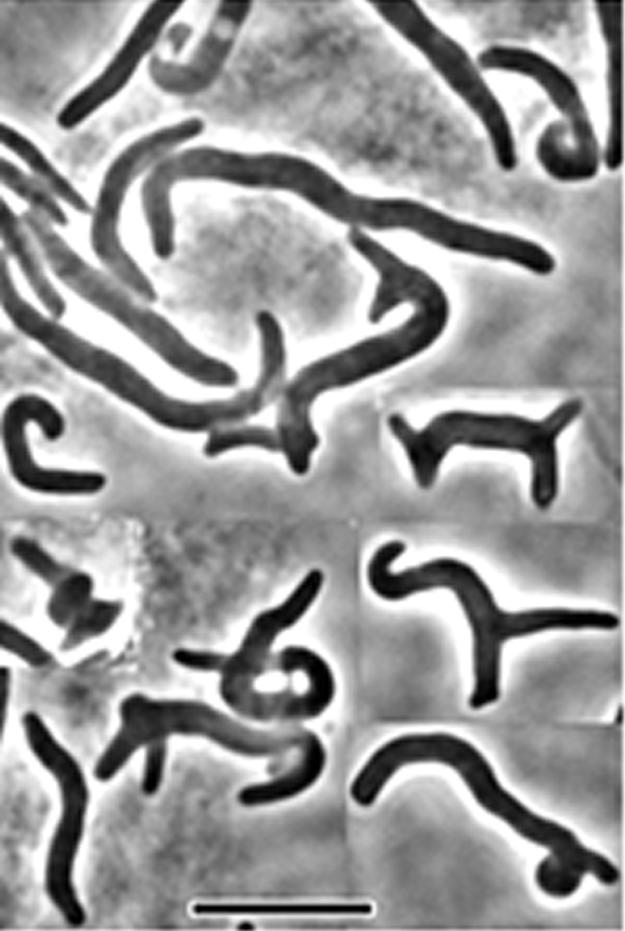
Morphology induced in filaments formed by inhibition of PBP 3. E. coli MG1655 ΔdacA ΔpbpG (ΔPBP 5 ΔPBP 7) was grown at 30°C in LB medium (0.5% NaCl). Cell division was inhibited by the addition of aztreonam (5 μg/ml), and the culture was incubated for an additional 60 min at 42°C before observation with phase microscopy. The figure is a composite of different fields and overrepresents the number of slightly spiraled cells in the population.
A third possibility was that spiral cells were created by a previously unknown characteristic or function of SulA. To determine if this might be true, we inhibited FtsZ with MinC, by overexpressing MinCD from plasmid pCH189 or by inducing MinC by expressing DicB from plasmid pJE44 (19, 33). If spiraling required a specific function of SulA, then spiral cells should not arise in these experiments. However, no matter how MinC was induced, spiral cells accumulated in a mutant lacking PBPs 5 and 7 (data not shown). When MinCD was overexpressed, 23% of the resulting cells displayed loose spirals and 4% had tight spirals; in the presence of DicB, 30% of the cells displayed loose spirals and 4.5% had tight spirals. These numbers were very close to and sometimes a little higher than the percentages of spirals induced by SulA (Table 2). The results make it unlikely that a special trait of SulA or MinC is the proximal cause of spiraling, though that possibility cannot be ruled out completely.
Finally, we noted that, out of several hundred unambiguous spirals observed by SEM, all were left-handed (Fig. 5 and data not shown), suggesting that the phenomenon was not accidental but reflected a heretofore-unknown relationship between FtsZ and the LMW PBPs.
DISCUSSION
The processes driving bacterial cell division versus those that regulate morphology have usually been treated as separate, independent pathways, although their activities must of necessity be interrelated. In contrast, the results presented here establish that FtsZ significantly influences gross cell shape in addition to its classic function of determining the site and progression of septation. The mechanism by which FtsZ mediates these morphological effects is surprising in that it entails an interaction with certain LMW PBPs or their products, which are otherwise dispensable for bacterial viability (10). Specifically, FtsZ and PBP 5 work together to generate the normally uniform, unbranched shape of E. coli, with PBPs 4, 6, and 7 playing accessory roles. Furthermore, under some circumstances, cell integrity itself depends on an undefined FtsZ-PBP relationship which, when disturbed, triggers cell dissolution.
There are previous, indirect indications of a possible connection between cell division and the activity of the LMW PBPs. For example, increased expression of PBP 5 forces E. coli to grow as spheres instead of rods (26) and restores normal division to cells carrying a temperature-sensitive version of the FtsI (PBP 3) protein (4). Because PBP 5 is a dd-carboxypeptidase, the results imply that reducing the number of pentapeptide side chains in peptidoglycan impedes the ability of the cell to elongate normally, perhaps by reducing the amount of substrate available for elongation driven by PBP 2 in favor of septation driven by PBP 3 (26). This “precursor balance” idea is at present no more than a hypothesis and considers only apportioning substrates between the transpeptidation enzymes PBPs 2 and 3. Certainly, no involvement of FtsZ was envisioned, so the phenotypes reported here (branching, shape abnormalities, and lysis) may reflect different aspects of substrate availability.
Recently, a more direct indication of the relationship between LMW PBPs and cell division was discovered by Morlot et al., who reported that the PBP 5-like dd-carboxypeptidase encoded by the dacA gene influences the course of cell division in Streptococcus pneumoniae (27). In an impressive technical display, Morlot et al. observed that in a dacA mutant the septation-specific high-molecular-weight PBP 2x formed a ring separate from and at least partially independent of the septal FtsZ ring (27). In the absence of this dd-carboxypeptidase the high-molecular-weight PBP ring wanders freely about the interior of the cell and may initiate peptidoglycan synthesis at inappropriate locations, thereby explaining the shape defects previously associated with this mutant (37). The morphological aberrations that we report here for E. coli might derive from a similar mechanism. In direct contrast to the proposal of Begg et al. (4), the evidence of Morlot et al. favors an interpretation in which the septation machinery prefers peptidoglycan composed of pentapeptides (27).
One unanswered question is whether the FtsZ and PBP 5 proteins interact directly or indirectly through downstream effectors or substrates. In Bacillus subtilis, the homologue of the E. coli PBP 5 dd-carboxypeptidase (itself named PBP 5) localizes strongly to the septum, though it also forms punctate spots along the lateral cell wall (36). The finding leaves open the possibility that the two proteins might interact directly, but it could just as well mean that PBP 5 is suitably positioned to control the level of a substrate required by an FtsZ-dependent reaction. Another possibility is that the interactions between these proteins differ in the two organisms. Unlike the situation in E. coli, deleting LMW PBPs from B. subtilis has no impact on vegetative cell morphology (35).
A particularly surprising aspect of our present findings for E. coli is that the morphological and lytic effects depend not on the inactivation of FtsZ but on the presence of a poorly functional FtsZ variant. Both branching and lysis occurred only when cells carrying the FtsZ84 protein were incubated in conditions in which the protein was expected to function imperfectly. In these semipermissible conditions, the altered activity of FtsZ84 may impair an interaction with PBP 5 or its peptidoglycan products or may lead it to perform its functions with greater promiscuity. Alternately, the observed morphological oddities could be created if a subset of inactive FtsZ84 molecules damages the function of a residual amount of active protein. In any event, inhibiting the nonessential LMW PBPs and partially inhibiting the activity of FtsZ could represent a therapeutic strategy different from that now pursued by targeting only the “essential” high-molecular-weight PBPs.
An even more unexpected and intriguing observation is that for the first time, as far as we know, E. coli was induced to adopt a morphology analogous to that of the spirillum-shaped bacteria. The fact that certain PBP mutants adopt remarkably spiral shapes when FtsZ is inhibited suggests yet another aspect of the interaction between LMW PBPs and the septation machinery. The mechanism underlying spiral shape is known in only two prokaryotes: periplasmic flagella create the helical cells of Borrelia burgdorferi and other spirochetes (28), and the curved cells of Caulobacter crescentus depend on the helical disposition of a recently identified cytoplasmic protein named crescentin (3). Neither of these mechanisms appears to exist for E. coli. In fact, one reason that the spiral phenotype is so intriguing is that SulA inhibits FtsZ polymerization (29, 40), seemingly ruling out a direct role for FtsZ in controlling the helical shape. One potential explanation is that FtsZ exhibits a new trait visible only in cells lacking certain PBPs. For example, spiral shape might be imposed on cells by the helical arrangement of a small number of residual FtsZ polymers (though this seems unlikely). Another possibility is that the spiral cells are generated by the emergence of a new internal polymer that directs cell shape or that the spirals reflect previously invisible characteristics of the mechanics of peptidoglycan deposition in the absence of certain PBPs and FtsZ.
Any model for the mechanism that creates bacterial shape must now take into account the role of PBP 5 (30, 31), the involvement of peptidoglycan endopeptidases, the generation and mislocalization of patches of inert peptidoglycan (12, 32), the participation of MreB (9, 17, 22, 24), and the influence of FtsZ. So far, the data can be accommodated in a generalized morphological pathway in which FtsZ or an FtsZ-dependent process acts on the cell wall to synthesize metabolically inert peptidoglycan (iPG), whose spatial distribution would dictate the overall geometry of the growing cell (44). Peptidoglycan-modifying enzymes would then moderate iPG synthesis by reducing the level of substrate (PBP 5) or by degrading extraneous iPG (PBPs 4 and 7) (44). In this scenario, FtsZ84 might drive morphological diversity by directing the synthesis of iPG in extraneous amounts or at inappropriate locations, perhaps by interfering with the proper polymerization, localization, or activity of Z rings. In any case, understanding the processes underlying these new relationships should shed light on the genesis, structure, and topological regulation of prokaryotic morphology in general.
Acknowledgments
We thank Edward Carlson and Donna Laturnus for performing the SEM.
This work was supported by grant 9982157 from the National Science Foundation and grant GM61019 from the National Institutes of Health.
REFERENCES
- 1.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 179:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausmees, N., J. R. Kuhn, and C. Jacobs-Wagner. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705-713. [DOI] [PubMed] [Google Scholar]
- 4.Begg, K. J., A. Takasuga, D. H. Edwards, S. J. Dewar, B. G. Spratt, H. Adachi, T. Ohta, H. Matsuzawa, and W. D. Donachie. 1990. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J. Bacteriol. 172:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 6.Carlson, E., J. Audette, L. Klevey, H. Nguyen, and P. Epstein. 1997. Ultrastructural and functional analyses of nephropathy in calmodulin-induced diabetic transgenic mice. Anat. Rec. 247:9-19. [DOI] [PubMed] [Google Scholar]
- 7.Cloud, K. A., and J. P. Dillard. 2002. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect. Immun. 70:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cookson, B. T., A. N. Tyler, and W. E. Goldman. 1989. Primary structure of the peptidoglycan-derived tracheal cytotoxin of Bordetella pertussis. Biochemistry 28:1744-1749. [DOI] [PubMed] [Google Scholar]
- 9.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria. Two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 10.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pedro, M. A., J. C. Quintela, J.-V. Höltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Pedro, M. A., K. D. Young, J.-V. Höltje, and H. Schwarz. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dusenbery, D. B. 1998. Fitness landscapes for effects of shape on chemotaxis and other behaviors of bacteria. J. Bacteriol. 180:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dziarski, R. 2003. Recognition of bacterial peptidoglycan by the innate immune system. Cell. Mol. Life Sci. 60:1793-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dziarski, R., K. A. Platt, E. Gelius, H. Steiner, and D. Gupta. 2003. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood 102:689-697. [DOI] [PubMed] [Google Scholar]
- 16.Faulkner, G., and R. A. Garduño. 2002. Ultrastructural analysis of differentiation in Legionella pneumophila. J. Bacteriol. 184:7025-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall mophogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 18.Hay, N. A., D. J. Tipper, D. Gygi, and C. Hughes. 1999. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J. Bacteriol. 181:2008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of DMinC/MinD and DMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karnovsky, M. J. 1965. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27:137-138A. [Google Scholar]
- 21.Kristensen, C. S., L. Eberl, J. M. Sanchez-Romero, M. Givskov, S. Molin, and V. de Lorenzo. 1995. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J. Bacteriol. 177:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroos, L., and J. R. Maddock. 2003. Prokaryotic development: emerging insights. J. Bacteriol. 185:1128-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 24.Margolin, W. 2003. Bacterial shape: growing off this mortal coil. Curr. Biol. 13:R705-R707. [DOI] [PubMed] [Google Scholar]
- 25.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 26.Markiewicz, Z., J. K. Broome-Smith, U. Schwarz, and B. G. Spratt. 1982. Spherical E. coli due to elevated levels of d-alanine carboxypeptidase. Nature 297:702-704. [DOI] [PubMed] [Google Scholar]
- 27.Morlot, C., M. Noirclerc-Savoye, A. Zapun, O. Dideberg, and T. Vernet. 2004. The d,d-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 51:1641-1648. [DOI] [PubMed] [Google Scholar]
- 28.Motaleb, M. A., L. Corum, J. L. Bono, A. F. Elias, P. Rosa, D. S. Samuels, and N. W. Charon. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA 97:10899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee, A., C. N. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson, D. E., and K. D. Young. 2001. Contributions of PBP 5 and dd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, D. E., and K. D. Young. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsen, T., A. S. Ghosh, M. B. Goldberg, and K. D. Young. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popham, D. L., M. E. Gilmore, and P. Setlow. 1999. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J. Bacteriol. 181:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheffers, D. J., L. J. Jones, and J. Errington. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51:749-764. [DOI] [PubMed] [Google Scholar]
- 37.Schuster, C., B. Dobrinski, and R. Hakenbeck. 1990. Unusual septum formation in Streptococcus pneumoniae mutants with an alteration in the d,d-carboxypeptidase penicillin-binding protein 3. J. Bacteriol. 172:6499-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shigematsu, M., A. Umeda, S. Fujimoto, and K. Amako. 1998. Spirochaete-like swimming mode of Campylobacter jejuni in a viscous environment. J. Med. Microbiol. 47:521-526. [DOI] [PubMed] [Google Scholar]
- 39.Tóth, L., and K. Kato. 1997. Size-selective grazing of bacteria by Bosmina longirostris—an image-analysis study. J. Plankton Res. 19:1477-1493. [Google Scholar]
- 40.Trusca, D., S. Scott, C. Thompson, and D. Bramhill. 1998. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J. Bacteriol. 180:3946-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Loosdrecht, M. C., W. Norde, and A. J. Zehnder. 1990. Physical chemical description of bacterial adhesion. J. Biomater. Appl. 5:91-106. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J. E., P. F. Jorgensen, E. A. Ellingsen, M. Almiof, C. Thiemermann, S. J. Foster, A. O. Aasen, and R. Solberg. 2001. Peptidoglycan primes for LPS-induced release of proinflammatory cytokines in whole human blood. Shock 16:178-182. [DOI] [PubMed] [Google Scholar]
- 43.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49:571-580. [DOI] [PubMed] [Google Scholar]
- 45.Yu, X. C., and W. Margolin. 2000. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J. Bacteriol. 182:6203-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, J., K. Jäger, T. Black, K. Zarka, O. Koksharova, and C. P. Wolk. 2001. HcwA, an autolysin, is required for heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 183:6841-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]