Abstract
A large number of bacteria regulate chaperone gene expression during heat shock by the HrcA-CIRCE system, in which the DNA element called CIRCE serves as binding site for the repressor protein HrcA under nonstress conditions. In Caulobacter crescentus, the groESL operon presents a dual type of control. Heat shock induction is controlled by a σ32-dependent promoter and the HrcA-CIRCE system plays a role in regulation of groESL expression under physiological temperatures. To study the activity of HrcA in vitro, we purified a histidine-tagged version of the protein, and specific binding to the CIRCE element was obtained by gel shift assays. The amount of retarded DNA increased significantly in the presence of GroES/GroEL, suggesting that the GroE chaperonin machine modulates HrcA activity. Further evidence of this modulation was obtained using lacZ transcription fusions with the groESL regulatory region in C. crescentus cells, producing different amounts of GroES/GroEL. In addition, we identified the putative DNA-binding domain of HrcA through extensive protein sequence comparison and constructed various HrcA mutant proteins containing single amino acid substitutions in or near this region. In vitro and in vivo experiments with these mutated proteins indicated several amino acids important for repressor activity.
Caulobacter crescentus, a member of the α-subdivision of proteobacteria, responds to heat shock by elevating the levels of synthesis of over 20 polypeptides (16). Several heat shock-inducible genes in C. crescentus have been characterized, including dnaKJ (15), groESL (1), lon (37), hrcA/grpE (30), and ftsH (11), and they all are positively regulated by the alternative sigma factor of RNA polymerase σ32. The rpoH gene encoding σ32 in C. crescentus has also been characterized, and one of its promoters was shown to be σ32 dependent, indicating an autogenous control of rpoH transcription (28, 38). Furthermore, the levels of σ32 were shown to increase transiently during heat shock, and the increased transcription of rpoH seems to account for the induction of σ32 levels (28, 38).
Recent studies have shown that, as described for Escherichia coli (for a review, see reference 39), the heat shock protein (HSP) DnaK is a negative modulator of the heat shock response in C. crescentus, which acts by inhibiting σ32 activity and stimulating its degradation (7). However, despite the strong effect of DnaK levels on the induction phase of the response, downregulation of HSP synthesis is not affected by changes in the amount of this chaperone. Competition between σ32 and σ73, the major sigma factor in C. crescentus which was shown to be heat shock inducible, has been proposed as the most important factor controlling the shutoff of HSP synthesis during the recovery phase (7). Another important negative modulator of the heat shock response, the repressor protein HrcA in C. crescentus, has also been described (30). This protein, which is absent in E. coli, has been found to occur in a growing number of diverse bacterial species and primarily controls the expression of the major heat shock genes, the groESL and dnaKJ operons, by binding to the CIRCE (controlling inverted repeat of chaperone expression) element (18, 26). The HrcA-CIRCE system was shown to control groESL operon expression in C. crescentus, the groE genes presenting a dual type of control. Besides being heat shock inducible, groESL expression is cell cycle regulated during growth at normal temperatures, the regulatory region of the C. crescentus operon containing both a σ32 promoter and a CIRCE element (1, 5, 30).
With Bacillus subtilis, experimental evidence indicates that HrcA is maintained in an active conformation that is able to bind CIRCE through the GroE chaperonin system (25, 29). According to the model proposed, when increased formation of nonnative proteins occurs under heat shock conditions, the GroE system would be sequestered by these proteins and would no longer be available to activate HrcA, leading to derepression of heat shock genes controlled by the HrcA-CIRCE system.
In C. crescentus, however, mutations in CIRCE and hrcA produce loss of temporal control of the groE operon and increased levels of GroEL in cells growing at normal temperatures, with no effect on heat shock induction (5, 30). Thus, the HrcA-CIRCE system plays a role in the control of groE expression under nonstress conditions in C. crescentus, compared to heat shock induction reported for other bacteria.
In this report, we investigated the binding activity of C. crescentus HrcA to CIRCE in vitro, as well as a possible role of GroE chaperonin system in HrcA activity both in vitro and in vivo. Furthermore, a search for the HrcA DNA-binding domain was conducted by extensive protein sequence comparison. Based on the putative DNA-binding domain, several mutant HrcA proteins were obtained by site-directed mutagenesis, which were then analyzed for their activity both in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The C. crescentus strains NA1000, LS2293, SG300, and SG400 (Table 1) were grown at 30°C in PYE complex medium (27) or M2 minimal-salt medium (8) suplemented with 0.2% glucose (M2G) or 0.1% glucose and 0.1% xylose (M2GX). E. coli cells were cultured in Luria-Bertani medium at 37°C (31). The following E. coli strains were used: strain TG-1 was used for phage propagation and cloning, strain CJ236 was used to generate uracil-rich single-stranded DNA for site-directed mutagenesis, strain S17-1 was used as the donor strain in conjugations with C. crescentus, and strain DH5α was used to overexpress the recombinant HrcA proteins.
TABLE 1.
Bacterial strains
| Strain | Description | Reference |
|---|---|---|
| C. crescentus | ||
| NA1000 | Synchronizable CB15 derivative | 10 |
| LS2293 | NA1000 with hrcA disruption | 30 |
| SG300 | groESL under xylose-inducible promoter; conditional mutant | 7 |
| SG400 | dnaKJ under xylose-inducible promoter; conditional mutant | 7 |
| E. coli | ||
| TG-1 | supE hsdΔ5 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 13 |
| S17-1 | 294::RP4-2(Tc::Mu)(Km::Tn7) | 34 |
| DH5α | supE44 lacU169 (80 lacZ M15)hsdR17 recA1 endA11 gyrA96 thi-1 relA1 | 17 |
| CJ236 | dut ung thi relA pCJ195 | 21 |
Site-directed mutagenesis.
Site-directed mutagenesis was performed by the method of Kunkel et al. (21), using single-stranded DNA derived from C. crescentus hrcA cloned in M13mp19 as a template. The synthetic oligonucleotides Pro39Ala (AGACCGGTGAGGCCGTCGGTTCACG), Ser42Ala (AGCCCGTCGGTGCGCGCACCATTTC), Ser56Ala (GTCGCCGGCCGCGATCCGCAATACG), Arg58Ala (GGCCTCGATCGCCAATACGATGCAG), Pro81Ala (CCGGTCGTATGGCGACCCATGCGGG), Arg87Ala (CATGCGGGCCTGGCGATGTTCGTCG), and Glu301Ala (CTATATCGGGGCCGCCACACGTCTC), were used as primers to replace the corresponding bases (underlined) in the hrcA wild-type gene. The DNA fragments containing the mutations were ligated into pMR20, generating the plasmids pPro39Ala, pSer42Ala, pSer56Ala, pArg58Ala, pPro81Ala, pArg87Ala, and pGlu301Ala (Table 2), which were introduced by conjugation into C. crescentus LS2293 and NA1000 strains. The hrcA coding region, either the wild type or carrying substitutions corresponding to Pro81Ala and Arg87Ala, was cloned in the pProEx-1 expression vector, giving rise to pRB60, pMS3, and pMS4, respectively (Table 2).
TABLE 2.
Plasmids
| Plasmid | Description | Reference or source |
|---|---|---|
| placZ290 | lacZ transcription fusion vector, Tcr, IncP1; oriT | 14 |
| pAR31 | rpoH promoter region in plac290 | 28 |
| pMA11 | groESL promoter region in placZ290 | 1 |
| pRB19 | groESL promoter region with mutated CIRCE element in placZ290 | 5 |
| pMA100 | clpB promoter region in placZ290 | 7 |
| pDell | dnaKJ promoter region in placZ290 | 2 |
| pAS22 | Vector for expression of genes in C. crescentus from the Pxy/X promoter, Cmr; oriT | U. Jenal |
| pRB51 | pAS22 carrying groEL coding region | 7 |
| pMR20 | IncP1, oriT, Tcr | 30 |
| pRR312-3 | pMR20 carrying wild-type hrcA | 30 |
| pPro39Ala | pMR20 carrying hrcA gene with mutation corresponding to a substitution from proline at position 39 to alanine | This work |
| pSer42Ala | pMR20 carrying hrcA gene with mutation corresponding to a substitution from serine at position 42 to alanine | This work |
| pSer56Ala | pMR20 carrying hrcA gene with mutation corresponding to a substitution from serine at position 56 to alanine | This work |
| pArg58Ala | pMR20 carrying hrcA gene with mutation corresponding to a substitution from arginine at position 58 to alanine | This work |
| pPro81Ala | pMR20 carrying hrcA gene with mutation corresponding to a substitution from proline at position 81 to alanine | This work |
| pArg87Ala | pMR20 carrying hrcA gene with mutation corresponding to a substitution from arginine at position 87 to alanine | This work |
| pGlu301Ala | pMR20 carrying hrcA gene with mutation corresponding to a substitution from glutamate at position 301 to alanine | This work |
| pProEx-1 | His-tag expression vector | Gibco-BRL |
| pRB60 | pProEx-1 carrying the hrcA coding region | This work |
| pMS3 | pProEx-1 carrying the hrcA coding region with alanine replacing proline 81 | This work |
| pMS4 | pProEx-1 carrying hrcA coding region with alanine replacing arginine 87 | This work |
Overproduction and purification of recombinant HrcA.
Several conditions were tested to obtain recombinant HrcA in the soluble form; the best condition is described below. One liter of Luria-Bertani medium containing 200 μg of ampicillin/ml was inoculated with 10 ml of an overnight culture of E. coli DH5α carrying pRB60, pMS3, or pMS4 and incubated with vigorous aeration at 30°C until the optical density at 600 nm (OD600) was 0.5, when isopropyl-thiogalactopyranoside was added to 0.6 mM. After 3 h, the cells were pelleted, washed, and resuspended with 2 ml of buffer A (50 mM Tris-HCl [pH 8], 0.5 mM EDTA, 50 mM NaCl, 5% glycerol, and 150 μg of phenylmethylsulfonyl fluoride/ml). Cells were lysed by sonication, and cell debris was removed by centrifugation at 20,000 × g for 30 min. Under these conditions, most of the recombinant HrcA (wild-type and mutant proteins) was found in the soluble fraction. This fraction was then incubated with 2% deoxycholic acid for 10 min at 4°C, dialyzed against buffer I (10 mM Tris-HCl [pH 8], 100 mM NaCl, 10 mM imidazole) for 6 h and concentrated with a Centricon 30 filter device (Amicon). The resulting protein fraction was incubated for 2 h at 4°C with Ni-nitrilotriacetic acid agarose resin (QIAGEN) equilibrated in buffer I. The His6-HrcA protein was eluted with 5 ml of buffer I containing 100 and 150 mM imidazole, and the eluate was dialyzed for 2 h against buffer A. The concentration of purified His6-HrcA protein was determined with the bicinchoninic acid kit (Bio-Rad).
Gel shift assays.
Gel shift assays were used to study the binding of the HrcA protein to the CIRCE region, as previously described (6). The probe MA8 used for these assays was a 150-bp EcoRI-HindIII fragment from pMA18 (1) containing the C. crescentus groESL regulatory region. This DNA fragment was isolated from a polyacrylamide gel and end labeled with [γ-32P]ATP according to standard protocols (31). Labeled fragment (105 cpm) was mixed with 5 μg of purified His6-HrcA protein in DNA-binding buffer (10 mM Tris-HCl [pH 7.8], 10 mM MgCl2, 100 mM NaCl, 2 mM dithiothreitol, 1% glycerol, and 1 μg of salmon testis DNA) in a final volume of 30 μl. When noted, the E. coli protein GroEL (3 μM) and/or GroES (1 μM), kindly provided by A. C. R. da Silva (Universidade de São Paulo), was added to His6-HrcA, and the mixture was preincubated for 10 min at 30°C in DNA-binding buffer before the binding reaction was started by the addition of the probe. ATP and AMP-PCP were added to the binding reaction mixture to a final concentration of 30 μM when needed. The DNA-protein mixtures were incubated for 20 min at 30°C and loaded onto 8% nondenaturing polyacrylamide gel in 0.5× TBE buffer (4 mM Tris [pH 8], 4 mM boric acid, 1 mM EDTA). Gels were run in 0.5× TBE buffer at 4°C at 20 mA, dried under a vacuum, and subjected to autoradiography.
Binding assays with immobilized HrcA.
To investigate interactions between HrcA and GroEL, the binding of GroEL to immobilized His6-HrcA was analyzed according to a previously described method (9). Total protein extract obtained from E. coli overexpressing recombinant His6-HrcA was loaded onto an Ni-nitrilotriacetic acid agarose column. Following several washes with lysis buffer (50 mM NaH2PO4 [pH 8.0] containing 300 mM NaCl and 10 mM imidazole), a crude extract prepared from C. crescentus strain LS2293 (an hrcA null mutant, which presents higher GroEL levels) or purified GroEL was applied to the column. The column was washed once with lysis buffer, and then the bound proteins were eluted with the same buffer containing 100 mM imidazole. The eluate was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with anti-GroEL antiserum. As a control, the same experiment was carried out without loading the column with His6-HrcA. The binding reactions were performed at 4°C. The crude extract of LS2293 was prepared from cells obtained from 50 ml of an exponential-growth-phase culture. Cells were collected by centrifugation, resuspended in 5 ml of lysis buffer, and disrupted by sonication, and the insoluble debris was removed by centrifugation at 13,000 × g for 15 min at 4°C.
Immunoblots.
Aliquots of C. crescentus cells were collected, and total protein extracts were prepared as previously described (1). The protein extracts were resolved through SDS-PAGE, the proteins were transferred to nitrocellulose membranes, and the membranes were incubated with the appropriated antibodies, as previously described (5). The anti-HrcA antiserum was obtained from a rabbit immunized with a fusion protein overexpressed in E. coli, corresponding to about 30 kDa of the carboxy-terminal portion of C. crescentus HrcA containing an N-terminal histidine tag from the expression vector pProEx-1 (Gibco-BRL).
Heat shock promoter activity.
The lacZ transcriptional fusions pDel1, pMA11, pRB19, and pMA100 (Table 2) were introduced into C. crescentus strains NA1000 pRB51 and NA1000 pAS22, and pMA11 and pRB19 were introduced into strains SG300 and SG400. Overnight cultures of the resulting strains were diluted in 10 ml of PYE medium in triplicate and grown to an OD600 of 0.5. When noted, xylose was added to a final concentration of 0.1%. β-Galactosidase activity was measured according to Miller (24), with 50 to 100 μl of each culture. Assays were carried out in duplicate in at least three independent experiments.
RESULTS AND DISCUSSION
Specific binding of recombinant HrcA to the CIRCE element.
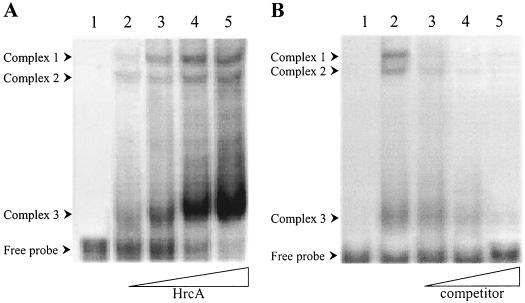
To obtain in vitro evidence of a physical interaction between CIRCE and HrcA, gel shift assays were performed using an amino-terminally histidine-tagged version of C. crescentus HrcA (His6-HrcA) and a 150-bp DNA probe (MA8) containing the promoter region and the CIRCE element of C. crescentus groESL operon. As shown in Fig. 1A, gel shift experiments with increasing amounts of His6-HrcA showed protein-dependent formation of three retarded bands. All three DNA-protein complexes were shown to be specific, since addition of increasing amounts of unlabeled probe as a specific competitor prevented the formation of all three complexes (Fig. 1B).
FIG. 1.
HrcA binds specifically to the groESL regulatory region. (A) Gel shift assay with DNA probe MA8 and increasing concentrations of C. crescentus HrcA. Lane 1, probe alone; lanes 2 to 5, 2.5, 5, 10, and 20 μg of purified HrcA. (B) Specific competition with unlabeled DNA MA8 in the gel shift assay. Lane 1, probe alone; lane 2, 5 μg of HrcA; lanes 3 to 5, 5 μg of HrcA plus 0.1 μM (lane 3), 0.5 μM (lane 4), or 1 μM (lane 5) of unlabeled probe. HrcA and the competitor DNA, when noted, were added to the binding buffer and incubated for 10 min at 30°C, and then the radiolabeled MA8 probe was added. The binding reaction was carried out at 30°C for 20 min. The complexes were resolved by SDS-PAGE as described in Material and Methods.
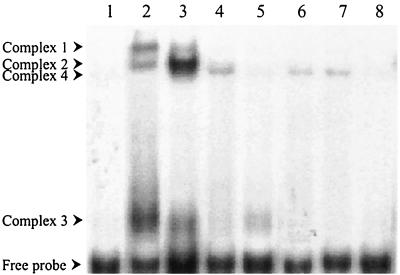
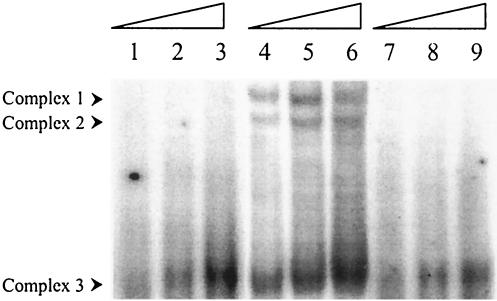
To investigate if C. crescentus HrcA DNA-binding activity could be chaperonin dependent, preincubation of His6-HrcA in binding buffer containing purified E. coli GroES and GroEL was carried out, and the DNA-binding assay was started by addition of the probe (MA8). As shown in Fig. 2, lane 3, when HrcA was preincubated with GroES/GroEL there was a significant increase in the amount of complex 2, with a concomitant decrease in the amounts of complexes 1 and 3. This corroborates the hypothesis proposed by Mogk et al. (25) that HrcA-binding activity is controlled by the availability of GroES/GroEL, which would stabilize HrcA in its active state, and suggests that complex 2 is the main complex, where HrcA is closer to its native conformation. In addition, a novel weak complex, which was named complex 4, appeared when the binding assay was carried out in the presence of GroES/GroEL. All four complexes are specific, since the amount of each complex decreased dramatically in the presence of the unlabeled probe (Fig. 2, lane 5).
FIG. 2.
Effect of GroES, GroEL, and ATP on HrcA-CIRCE binding. A gel shift assay was carried out with the MA8 probe alone (lane 1); with HrcA (lane 2); with HrcA, GroES, and GroEL (lane 3); with HrcA, GroES, GroEL, and ATP (lane 4); with HrcA, GroES, GroEL, and a specific competitor (lane 5); with GroEL (lane 6); with GroEL and ATP (lane 7); and with GroEL and a specific competitor (lane 8). The binding assay was carried out essentially as described in the legend to Fig. 1. The components of the different reaction mixtures include 5 μg of HrcA, 1 μg of GroES, 1 μg of GroEL, 1 mM ATP, and 0.5 μM competitor DNA.
Interestingly, when ATP was added to the binding reaction mixture containing HrcA, GroES and GroEL, the amount of complexes 1, 2, and 3 decreased drastically, whereas complex 4 was maintained (Fig. 2, lane 4). Hydrolysis of ATP seems to be important for this process, since the presence of a nonhydrolyzable ATP analog (AMP-PCP) had no effect in the formation of any of the four complexes (data not shown). The formation of complex 4 is intriguing, since an apparently identical complex was formed when the MA8 probe was incubated with GroEL alone or GroEL plus ATP (Fig. 2, lanes 6 and 7). This complex is also specific, because the addition of unlabeled probe prevented its formation (Fig. 2, lane 8).
Altered levels of GroES/GroEL affect groESL promoter activity in vivo.
To investigate if changes in GroES/GroEL levels could affect HrcA repressor activity in vivo, we introduced lacZ transcription fusions carrying the groESL promoter region either with a wild-type (pMA11) or a mutated (pRB19) CIRCE element into C. crescentus strains overexpressing or not overexpressing GroES/GroEL. The overexpressing strain NA1000 pRB51 carries a multicopy plasmid in which the groESL genes are under the control of the PxylX promoter (7). Thus, in the presence of xylose the levels of GroES/GroEL are two to threefold higher than in those in the control strain NA1000 pAS22, which carries only the chromosomal copy of the groESL genes (7). As the results shown in Table 3 indicate, β-galactosidase activity in C. crescentus cells carrying transcription fusion pMA11 was 30% lower in cells overexpressing GroES/GroEL (NA1000 pRB51) than in cells presenting normal levels of these chaperones (NA1000 pAS22). On the other hand, in cells carrying the transcription fusion pRB19, which contains a mutated CIRCE element, overexpression of GroES/GroEL had no effect on β-galactosidase activity levels. Since HrcA does not bind this mutated CIRCE element, at least not with the same affinity as the wild-type element (5, 30), no effect of excess GroES/GroEL was expected, as this experiment confirmed.
TABLE 3.
Effect of GroES/GroEL overexpression on transcription directed by heat shock gene promoters
| Transcription fusion | Promoter sourcea | β-Galactosidase activityb
|
||
|---|---|---|---|---|
| NA1000 pRB51 | NA1000 pAS22 | Ratioc | ||
| pMA11 | groE | 1,540 ± 110 | 2,200 ± 120 | 0.70 |
| pRB19 | groEmutCIRCE | 2,950 ± 200 | 3,100 ± 240 | 0.95 |
| pDe11 | dnaK | 1,410 ± 190 | 1,430 ± 160 | 0.99 |
| pMA100 | clpB | 2,790 ± 220 | 2,770 ± 230 | 1.00 |
The promoters indicated were transcriptionally fused to the lacZ gene in the plasmid placZ290, and the resulting constructs were established in strains NA1000 pRB51 (overexpression of GroES GroEL) and NA1000 pAS22 (normal levels of GroES GroEL).
Promoter activity was assayed by measuring β-galactosidase activity in cells grown to mid-log phase at 30°C in PYE with 0.1% xylose, as described in Materials and Methods. The results are reported in Miller units, and each represent the average of triplicate assays.
β-Galactosidase activity ratio was determined by dividing the activity measured with NA1000 pRB51 cells by the activity in NA1000 pAS22 cells.
As controls, lacZ transcription fusions with two other heat shock promoters which do not contain a CIRCE element, the dnaK promoter region (pDel1) and clpB promoter region (pMA100), were introduced into strains NA1000 pRB51 and NA1000 pAS22. No differences in β-galactosidase activity with both transcription fusions were observed, whether the cells overexpressed GroES/GroEL or not. Since groE, clpB, and dnaK promoters are all controlled by σ32, these results indicate that the effect of high GroES/GroEL levels in the groE promoter was due to the presence of CIRCE and not to the σ32 promoter. These results are in agreement with a positive role of GroES/GroEL on HrcA repressor activity, similar to the role that has been ascribed to B. subtilis (25, 29).
To further investigate the possible role of GroES/GroEL in the control of HrcA activity, we introduced transcription fusions pMA11 and pRB19 into two other C. crescentus strains, SG300 and SG400, in which the expression groESL and dnaKJ operons, respectively, are dependent on the presence of xylose (7). Thus, when xylose was removed from the growth medium, the levels of GroES/GroEL in SG300 cells decreased, being virtually absent after 5 h without xylose. Similarly, the levels of DnaK/DnaJ decreased drastically in SG400 cells growing in the absence of xylose. In addition, the decrease in GroES/GroEL levels in SG300 was accompanied by a concomitant increase in DnaK/DnaJ levels; conversely, the decrease in DnaK/DnaJ levels in SG400 was accompanied by an increase in GroES/GroEL levels (7). In both cases, there was an increase in σ32 levels, although the increase was greater in SG400 cells than in SG300 cells.
As shown in Table 4, when SG300 cells were grown in the absence of xylose and when the levels of GroES/GroEL decreased, β-galactosidase activity increased in cells carrying either transcription fusion pMA11 or pRB19, that is, independently of the presence or absence of a normal CIRCE element. However, when SG400 cells were grown in the absence of xylose and when DnaK/DnaJ levels decreased, with a concomitant rise in GroES/GroEL levels (7), β-galactosidase activity increased only in cells carrying transcription fusion pRB19, which contains a mutated CIRCE element. In cells carrying transcription fusion pMA11, which has a wild-type CIRCE element, no increase in β-galactosidase activity was observed. It should be noted that SG400 cells growing in the absence of xylose present higher levels of σ32 than SG300 cells under the same conditions; hence, β-galactosidase activity in SG400 pRB19 reached higher levels than in SG300 pRB19 cells. The fact that we observed differences in β-galactosidase activity levels when we compared cells carrying either pMA11 (wild-type CIRCE element) or pRB19 (mutated CIRCE element), but only in cells not depleted of GroES/GroEL (SG400 cells), was in accordance with the hypothesis that these chaperones are necessary for HrcA activity.
TABLE 4.
Effect of depletion GroES/GroEL or DnaK/DnaJ on transcription directed by groE and groEmutCIRCE promoter regions
| Transcription fusion | Promoter sourcea | Activity ratiob
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SG300
|
SG400
|
||||||||
| 0 h | 4 h | 6 h | 8 h | 0 h | 4 h | 6 h | 8 h | ||
| pMA11 | groE | 1.0 | 1.1 | 1.4 | 1.7 | 1.0 | 1.0 | 0.9 | 1.0 |
| pRB19 | groEmutCIRCE | 1.0 | 1.0 | 1.3 | 1.8 | 0.9 | 1.7 | 2.8 | 2.9 |
The promoters indicated were transcriptionally fused to the lacZ gene in the plasmid placZ290, and each construct was conjugated into Caulobacter strains SG300 and SG400.
Promoter activity was assayed by measuring β-galactosidase activity in cells of the indicated strains, grown to mid-log phase at 30°C in the presence of 0.1% xylose. Bacteria were washed and resuspended in M2G or M2GX and incubated at 30°C for the indicated times. The activity ratio was determined dividing the β-galactosidase activity measured in cells growing in M2G by the activity measured in cells growing in M2GX, at the indicated times after resuspension of the cells. The values shown represent the averages of four independent experiments.
Interaction of GroEL and HrcA in vitro.
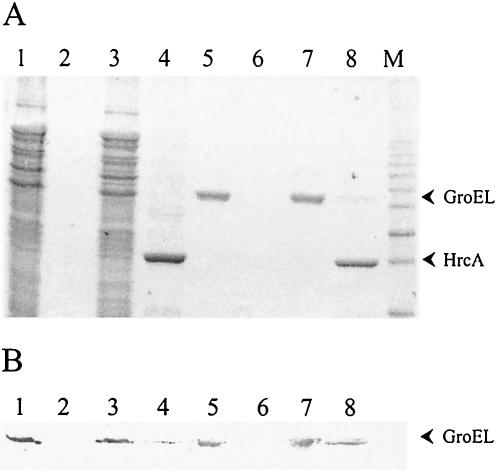
To demonstrate a physical interaction between GroEL and HrcA, the recombinant protein His6-HrcA was immobilized in a Ni-agarose column, and the C. crescentus total protein extract was applied to the column. After being washed, bound proteins were eluted with 100 mM imidazole buffer. Both His6-HrcA and GroEL were detected in the eluate, as determined by SDS-PAGE and immunoblot experiments (Fig. 3). No GroEL could be detected in the eluate of the control column, which was not loaded with HrcA. Similar results were obtained when purified E. coli GroEL was applied to the column instead of a total protein extract, confirming the direct interaction between HrcA and GroEL. Similar results were obtained with B. subtilis (29).
FIG. 3.
HrcA and GroEL interact in vitro. Fractions from a Ni-agarose control column (lanes 1, 2, 5, and 6) or from a Ni-agarose column loaded with His6-HrcA (lanes 3, 4, 7, and 8) to which was applied C. crescentus crude extract (lanes 1 to 4) or purified E. coli GroEL (lanes 5 to 8) by SDS-PAGE. The presence of HrcA and GroEL in the flowthrough (lanes 1, 3, 5, and 7) or eluate samples obtained with 100 mM imidazole buffer (lanes 2, 4, 6, and 8) was visualized by Coomassie blue staining of the gel (A) or by immunoblotting with anti-GroEL antibody (B). M, molecular weight marker.
Searching for HrcA DNA-binding domain by protein sequence comparison.
The observation that the CIRCE element is one of the most conserved regulatory sequences in bacteria (22, 32) suggests that the DNA-binding domain of HrcA proteins should present reasonable conservation in predicted structure and amino acid sequence. Thus, to identify this domain we used the programs MEME (Multiple Maximization Expectation for Motif Elicitation) (3) and MAST (Motif Alignment and Search Tool) (4) to analyze a set of 97 HrcA protein sequences taken from the National Center for Biotechnology Information database. We found that the most-conserved motif in the HrcA family is located in the N-terminal region of the protein and encompasses 37 amino acid residues, with the consensus sequence SsATIRNeMadLEelGllekpHtSsGRvPdkGyRyy, where the residues shown in uppercase letters are conserved in at least 70% of the HrcA proteins analyzed, and the remaining residues (in lowercase letters) are found in no less than 20% of the sequences. In C. crescentus HrcA, this region extends from residues 53 to 89 and was named conserved region 1 (Fig. 4A). The second-most-conserved region of HrcA is also located near the N terminus of the protein and comprises 29 amino acids; its consensus sequence is teRqlXILraIvedYiXtgePVGSktLse (the amino acid residues are shown as in the sequence above, except that X stands for residues conserved in <20% of the sequences). This motif extends from amino acids 19 to 47 in the Caulobacter HrcA protein and was named conserved region 2 (Fig. 4A).
FIG. 4.
Amino acid sequence comparison and predicted structure alignment of HrcA from different bacteria. (A) N-terminal portion of the ClustalW alignment of HrcA from different bacterial species with DeoR and LexA from E. coli. The two most-conserved regions in HrcA are in boxes. The sequences of the stabilization helix and recognition helix of the HTH motifs are indicated by dark shading (structurally determined for LexA and putative for HrcA and DeoR). The amino acids in the putative recognition helix are shown in boldface. The light-shaded region shows the position of the β-sheet in the winged helix domain of LexA (12) and its possible location in the HrcA proteins. The amino acids replaced in C. crescentus HrcA (this work) are shown in italics and underlined. (B) Portion of alignments with winged helix domains (PDB entries are shown between parentheses) obtained with the FUGUE system and with the ClustalW alignment of 97 HrcA sequences as input. The sequences of the putative HTH motifs are shaded, except in the Fur protein for clarity; helical regions common to all the structures are indicated by the letter H under the alignment. The fourth amino acid of each helix is shown in boldface. In the proteins with known structures, the region of the β-sheet of the winged helix domain is indicated by light shading, and the amino acids in lowercase are accessible to the solvent, whereas those shown in uppercase are not accessible to the solvent. Ccre, C. crescentus; Xfas, Xyllela fastidiosa; Bsub, B. subtilis; Bthe, B. thermoglucosidasius; Efae, Enterococcus faecalis; Thal, Tetragenococcus halophilus; Llac, Lactococcus lactis; Spyo, Streptococcus pyogenes; Cjej, Campylobacter jejuni; Mpul, Mycoplasma pulmonis; Ecol, E. coli; Tmar, Thermotoga maritima; Ssp, Synechococcus sp. (PCC 7942); Ssol, Sulfolobus solfataricus; Cdip, Corynebacterium diphtheriae; Paer, Pseudomonas aeruginosa; Hsap, Homo sapiens; Saur, Staphylococcus aureus.
In addition, when the HrcA protein sequences are compared to other protein families in databases such as InterPro, SMART, Pfam, and PROSITE, the results invariably show similarity of HrcA with proteins of the LexA and DeoR families of DNA-binding proteins. This similarity is restricted to the N-terminal domain of HrcA, coinciding in part with the conserved sequences described above, and includes the helix-turn-helix (HTH) motifs present in LexA and DeoR (12, 35). Figure 4A shows a ClustalW alignment of 10 distinct HrcA protein sequences with DeoR and LexA from E. coli. Despite differences in amino acid sequence, the recognition helix in the HTH motifs of LexA and DeoR always aligned with the same region in the different HrcA proteins analyzed. Thus, we suggest that this sequence (Fig. 4A) constitutes the recognition helix of the HrcA proteins. The same region was recently proposed as the recognition helix of HrcA for Bacillus thermoglucosidasius and B. subtilis (19, 36). Figure 4A also shows the putative stabilization helix of the HTH motif (amino acids 42 to 48 in C. crescentus) in the various HrcA proteins.
To gain further insight into the structure of HrcA N-terminal domain, we used the FUGUE system of analysis (http://www-cryst.bioc.cam.ac.uk/∼fugue/) (33). The results obtained with the ClustalW alignment of 97 HrcA sequences indicated a winged helix template for the N-terminal domain of HrcA in 90% of the alignments achieved, with the structure of LexA N-terminal domain (Protein Data Bank entry 1LEA) as the top score, a similarity previously noticed for Mycoplasma genitalium HrcA (20). The FUGUE alignments of the HTH region of 11 winged helix proteins (representing over 60% of the winged helix templates) and C. crescentus HrcA are shown in Fig. 4B. The alignments of HrcA sequences with these templates suggest that most HrcA homologues have a HTH motif longer than usual, with a few exceptions. The FUGUE analysis also indicated that the anti-parallel β-sheet of the winged helix domain is most probably located in the region corresponding to positions 70 to 83 of C. crescentus HrcA (Fig. 4).
MEME analysis also detected a C-terminal conserved region as the third-most-conserved motif in HrcA (consensus sequence, ggrXvGtlgviGPtRMdYsrviplvdXva). This region encompasses amino acids 321 to 349 in C. crescentus HrcA (data not shown). Comparisons of C. crescentus HrcA and the B. subtilis homolog, both of which have been demonstrated to interact with GroEL (reference 29 and this work), shows 45% of amino acid identity for this region, a higher value than expected only by the conservation observed for this motif among the 97 HrcA sequences analyzed. For instance, conserved region 1 presents 46% amino acid identity between the two HrcA proteins; for conserved region 2, this value is only 35%. The region that interacts with GroEL is still unknown; the possibility that the C-terminal conserved region could be involved in this interaction is currently under investigation.
Effect of mutant HrcA proteins on GroEL levels.
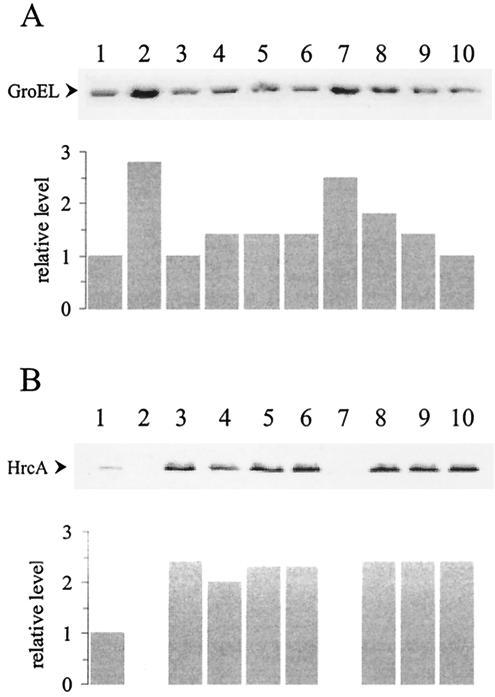
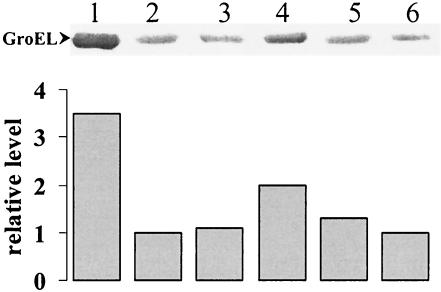
Based on the identification of the putative DNA-binding winged helix domain of C. crescentus HrcA, we constructed several mutant proteins carrying single site-directed substitutions in conserved amino acid residues located in this region of the protein. The mutated genes encoding these HrcA proteins were cloned into the plasmid vector pMR20, which replicates in C. crescentus, and the resulting constructs were used to complement the hrcA null mutant strain LS2293. The hrcA null strain presented higher levels (about 2.5-fold) of GroEL than the parental strain NA1000, as shown in Fig. 5A. When the null mutant was complemented with the wild-type hrcA gene cloned into pMR20 (pRR312-3), wild-type levels of GroEL were observed (Fig. 5A). Complementation experiments of the hrcA null mutant strain with different constructs encoding the mutant HrcA proteins were carried out, and the results are shown in Fig. 6A. The amino acids Pro39, Ser42, Ser56, Arg58, Pro81, and Arg87, which are located in or near the putative winged helix domain (Fig. 4) were each replaced by Ala. Figure 5B shows that all mutant proteins were detected in C. crescentus, except for HrcA Arg58Ala, whose corresponding full-length band was not observed, probably due to endogenous proteolysis. In agreement with its absence, the construct encoding HrcA Arg58Ala does not complement the hrcA null mutation, and GroEL levels are high in cells carrying pArg58Ala. As depicted in Fig. 5A, all other mutant proteins tested only partially complemented the null mutant phenotype, with HrcA Pro81Ala complementing the least (GroEL levels were highest). These results indicate that all the amino acids replaced should have a role in maintaining an active HrcA repressor protein. As a control, amino acid Glu301, which is poorly conserved (i.e., found only in about 20% of the 97 HrcA protein analyzed) and located outside the putative winged helix domain, was also replaced by Ala. As shown in Fig. 5, the mutated protein was produced in normal amounts and GroEL presented wild-type levels, indicating that full complementation occurred in this case. This result suggests that Glu301Ala substitution does not affect HrcA activity.
FIG. 5.
Complementation of the hrcA null mutation with plasmidial copies of the hrcA gene harboring different point mutations. Total cell extracts were obtained from bacterial cultures (OD600 nm = 0.5) of strains NA1000 pMR20 (lane 1), LS2293 pMR20 (lane 2), LS2293 pRR312-3 (lane 3), LS2293 pPro39Ala (lane 4), LS2293 pSer42Ala (lane 5), LS2293 pSer56Ala (lane 6), LS2293 pArg58Ala (lane 7), LS2293 pPro81Ala (lane 8), LS2293 pArg87Ala (lane 9), and LS2293 pGlu301Ala (lane 10). Proteins were resolved through SDS-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with anti-GroEL (A) or anti-HrcA (B) antibodies, as described in Material and Methods. The same amount of total protein was applied to each lane.
FIG. 6.
Pro81 and Arg87 are important for HrcA binding activity. Purified HrcA Arg87Ala (lanes 1 to 3), wild-type HrcA (lanes 4 to 6), and HrcA Pro81Ala (lanes 7 to 9), in concentrations ranging from 5 mg (lanes 1, 4, and 7), 10 mg (lanes 2, 5, and 8), to 20 mg (lanes 3, 6, and 9) were incubated with the 32P-labeled groESL regulatory region and analyzed by nondenaturing gel electrophoresis.
To investigate if the failure to fully complement the hrcA null mutation could be associated with the inability to bind the CIRCE element, we expressed two of the mutant proteins in E. coli and used them in gel shift assays. As shown in Fig. 6, complexes 1 and 2 were not detected when HrcA Pro81Ala and HrcA Arg87Ala were used in the binding assays. Furthermore, even though complex 3 was observed with both mutant proteins, it was present in much smaller amounts than those observed with the wild-type HrcA. These results agree well with the weak complementation capacity of each mutant protein, revealed in Fig. 5.
Mutant HrcA Ser56Ala produces a dominant negative phenotype.
The symmetrical structure of the CIRCE element suggests that HrcA binds as a dimer. Furthermore, HrcA from Streptococcus thermophilus has been shown to be a dimer (23). Thus, the presence of a mutant HrcA protein in a cell also producing the wild-type protein could induce the formation of heterodimers (or higher-order oligomers). Since some amino acid substitutions in HrcA can cause diminished binding of the protein to CIRCE (Fig. 6), the formation of hetero-oligomers between these mutant proteins and the wild-type HrcA could produce a dominant negative phenotype.
To investigate this possibility, plasmid constructs expressing either wild-type HrcA or mutant HrcA proteins (HrcA Ser56Ala, HrcA Pro81Ala, and HrcA Arg87Ala) were introduced into wild-type strain NA1000. Cell extracts of the resulting bacterial cultures were obtained, and GroEL levels were analyzed by immunoblot assays with the hrcA null strain LS2293 and the parental strain NA1000 as controls. As observed (Fig. 7), when the wild-type hrcA gene was present in trans (NA1000 pRR312-3), GroEL levels were equivalent to those detected in the parental strain NA1000. Similarly, GroEL levels in strains carrying the constructs encoding HrcA Arg87Ala and HrcA Pro81Ala were also nearly identical to the wild-type levels. Only when HrcA Ser56Ala was expressed in NA1000 did GroEL levels increase about twofold compared to the wild-type levels. Since all three mutant proteins analyzed were partially defective (Fig. 5), the results shown in Fig. 7 suggest that only HrcA Ser56Ala should form a hetero-oligomer with wild-type HrcA. Thus, the substitutions Arg87Ala and Pro81Ala could prevent proper oligomerization.
FIG. 7.
HrcA Ser56Ala produces a dominant negative phenotype. Relative levels of GroEL were determined in immunoblots of cell extracts obtained from cultures of strains LS2293 (lane 1), NA1000 (lane 2), NA1000 pRR312-3 (lane 3), NA1000 pSer56Ala (lane 4), NA1000 pArg87Ala (lane 5), and NA1000 pPro81Ala (lane 6). The same amount of total protein was applied to each lane.
Concluding remarks.
We show here that C. crescentus HrcA binds specifically to the CIRCE element in a dose-dependent fashion and that GroES/GroEL facilitates this binding. Furthermore, direct interaction between HrcA and GroEL was demonstrated in vitro by affinity chromatography. In addition, overexpression of GroES/GroEL in vivo was shown to negatively affect transcription directed by the wild-type groESL promoter region.
These results constitute the first demonstration of the positive role of GroES/GroEL in HrcA activity in gram-negative bacteria, similar to what has been proposed for B. subtilis and S. thermophilus (23, 25). Nevertheless, in these gram-positive bacteria, HrcA-CIRCE is involved in heat shock induction, and the proposed model states that upon heat stress, accumulation of unfolded proteins within the cell would sequester GroES/GroEL, which then would not be available to interact with HrcA, causing its inactivation. In C. crescentus, where the HrcA-CIRCE system is not involved in heat shock induction, a feedback control of GroES/GroEL production could occur at normal temperatures.
The putative DNA-binding domain of C. crescentus HrcA was identified based on extensive comparative protein sequence-structure analysis and was localized in the N-terminal region of the protein. The domain presents significant similarity to the winged helix DNA-binding domain of LexA of E. coli and has a noncanonical HTH motif as well, indicating that HrcA and LexA share a common fold. The HrcA DNA-binding domain comprises at least 82 amino acid residues, since the C terminus of the β-sheet in the wing is most likely located at residue 82 or 83 (Fig. 4). There is probably also a helical region C terminal to this β-sheet, extending from residue 87 or 88 possibly to as far as residue 137 in the C. crescentus protein (data not shown). It is not known if any portion of this region is part of the N-terminal domain, but both the FUGUE alignments and the amino acid substitutions Pro81Ala and Arg87Ala suggest the possible involvement of this region in the oligomerization of HrcA.
Computational analysis of the amino acid sequences of a set of 97 HrcA proteins indicated that these proteins show three conserved regions, two located in the N-terminal winged helix domain and another less-conserved region in the C-terminal domain of the protein. One possibility is that this C-terminal region may be involved in the interaction of HrcA with binding partners such as GroEL. Direct experimental evidence is needed, however, to establish the function of this region.
Several mutated HrcA proteins containing single amino acid substitutions in the proposed DNA-binding domain were obtained and shown to be deficient in binding to CIRCE, both in vitro and in vivo. Amino acid residues Pro39, Ser42, Ser56, Pro81, and Arg87 were all shown to be important for HrcA activity. Furthermore, HrcA Ser56Ala expressed together with the wild-type protein within the same cell produced a dominant negative phenotype, indicating that C. crescentus HrcA binds to CIRCE in an oligomeric form, most likely as a dimer.
Acknowledgments
This work was supported by a grant to S.L.G. from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). M.F.S. and H.R.P. are predoctoral fellows of FAPESP, R.L.B. was supported by a postdoctoral fellowship from FAPESP, and S.L.G. was partially supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
We thank A. C. R. da Silva for the kind gift of GroES and GroEL from E. coli and N. L. Hülle for his participation in the early stages of this study.
REFERENCES
- 1.Avedissian, M., and S. L. Gomes. 1996. Expression of the groESL operon is cell cycle controlled in Caulobacter crescentus. Mol. Microbiol. 19:79-89. [DOI] [PubMed] [Google Scholar]
- 2.Avedissian, M., D. Lessing, J. W. Gober, L. Shapiro, and S. L. Gomes. 1995. Regulation of the Caulobacter crescentus dnaKJ operon. J. Bacteriol. 177:3479-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 4.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. [DOI] [PubMed] [Google Scholar]
- 5.Baldini, R. L., M. Avedissian, and S. L. Gomes. 1998. The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J. Bacteriol. 180:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucca, G., G. Ferina, A. M. Puglia, and C. Smith. 1995. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol. Microbiol. 17:663-674. [DOI] [PubMed] [Google Scholar]
- 7.da Silva, A. C. A., R. C. G. Simão, M. F. Susin, R. L. Baldini, M. Avedissian, and S. L. Gomes. 2003. Down-regulation of the heat shock response is independent of DnaK and σ32 levels in Caulobacter crescentus. Mol. Microbiol. 49:541-553. [DOI] [PubMed] [Google Scholar]
- 8.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 9.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, B., G. Rummel, P. Aldridge, and U. Jenal. 2002. The FtsH protease is involved in development, stress response and heat shock control in Caulobacter crescentus. Mol. Microbiol. 44:461-478. [DOI] [PubMed] [Google Scholar]
- 12.Fogh, R. H., G. Ottleben, H. Ruterjans, M. Schnarr, R. Boelens, and R. Kaptein. 1994. Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy. EMBO J. 13:3936-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. dissertation. Cambridge University, Cambridge, United Kingdom.
- 14.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes, S. L., J. W. Gober, and L. Shapiro. 1990. Expression of the Caulobacter heat shock gene dnaK is developmentally controlled during growth at normal temperatures. J. Bacteriol. 172:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes, S. L., M. H. Juliani, J. C. Maia, and A. M. Silva. 1986. Heat shock protein synthesis during development in Caulobacter crescentus. J. Bacteriol. 168:923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general stress-response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 19.Hitomi, M., H. Nishimura, Y. Tsujimoto, H. Matsui, and K. Watanabe. 2003. Identification of a helix-turn-helix motif of Bacillus thermoglucosidasius HrcA essential for binding to the CIRCE element and thermostability of the HrcA-CIRCE complex, indicating a role as a thermosensor. J. Bacteriol. 185:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynen, M., T. Doerks, F. Eisenhaber, C. Orengo, S. Sunyaev, Y. Yuan, and P. Bork. 1998. Homology-based fold predictions for Mycoplasma genitalium proteins. J. Mol. Biol. 280:323-326. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 22.Makarova, K. S., A. A. Mironov, and M. S. Gelfand. 2001. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2:13.1-13.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martirani, L., R. Raniello, G. Naclerio, E. Ricca, and M. Felice. 2001. Identification of the DNA-binding protein, HrcA, of Streptococcus thermophilus. FEMS Microbiol Lett. 198:177-182. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. M. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 27.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinsenauer, A., C. D. Mohr, and L. Shapiro. 1996. Regulation of a heat shock σ32 gene homolog in Caulobacter crescentus. J. Bacteriol. 178:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reischl, S., T. Wiegert, and W. Schumann. 2002. Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J. Biol. Chem. 277:32659-32667. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, R., C. Toochinda, M. Avedissian, R. L. Baldini, S. L. Gomes, and L. Shapiro. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J. Bacteriol. 178:1829-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Segal, G., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 33.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243-257. [DOI] [PubMed] [Google Scholar]
- 34.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BioTechnology 1:784-790. [Google Scholar]
- 35.Valentin-Hansen, P., P. Hojrup, and S. Short. 1985. The primary structure of the DeoR repressor from Escherichia coli K-12. Nucleic Acids Res. 13:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiegert, T., and W. Schumann. 2003. Analysis of a DNA-binding motif of the Bacillus subtilis HrcA repressor protein. FEMS Microbiol. Lett. 223:101-106. [DOI] [PubMed] [Google Scholar]
- 37.Wright, R., C. Stephens, G. Zweiger, L. Shapiro, and M. R. Alley. 1996. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 10:1532-1542. [DOI] [PubMed] [Google Scholar]
- 38.Wu, J., and A. Newton. 1996. Isolation, identification, and transcriptional specificity of heat shock sigma factor σ32 from Caulobacter crescentus. J. Bacteriol. 178:2094-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.