Abstract
Mycoplasma pneumoniae lacks a cell wall but has internal cytoskeleton-like structures that are assumed to support the attachment organelle and asymmetric cell shape of this bacterium. To explore the fine details of the attachment organelle and the cytoskeleton-like structures, a fluorescent-protein tagging technique was applied to visualize the protein components of these structures. The focus was on the four proteins—P65, HMW2, P41, and P24—that are encoded in the crl operon (for “cytadherence regulatory locus”), which is known to be essential for the adherence of M. pneumoniae to host cells. When the P65 and HMW2 proteins were fused to enhanced yellow fluorescent protein (EYFP), a variant of green fluorescent protein, the fused proteins became localized at the attachment organelle, enabling visualization of the organelles of living cells by fluorescence microscopy. The leading end of gliding M. pneumoniae cells, expressing the EYFP-P65 fusion, was observed as a focus of fluorescence. On the other hand, when the P41 and P24 proteins were labeled with EYFP, the fluorescence signals of these proteins were observed at the proximal end of the attachment organelle. Coexpression of the P65 protein labeled with enhanced cyan fluorescent protein clearly showed that the sites of localization of P41 and P24 did not overlap that of P65. The localization of P41 and P24 suggested that they are also cytoskeletal proteins that function in the formation of unknown structures at the proximal end of the attachment organelle. The fluorescent-protein fusion technique may serve as a powerful tool for identifying components of cytoskeleton-like structures and the attachment organelle. It can also be used to analyze their assembly.
Mycoplasma pneumoniae, one of the smallest self-replicating bacteria known, is a causative agent of bronchitis and primary atypical pneumonia in humans (43, 44). M. pneumoniae lacks a cell wall and hence has a pleomorphic cell shape. However, a majority of M. pneumoniae cells in cultures are filamentous and have a differentiated terminal structure at one pole. This terminal structure, the attachment organelle, is a tapered membrane protrusion responsible for the adherence of M. pneumoniae to host respiratory epithelium (cytadherence) (23, 24). The attachment organelle renders M. pneumoniae cells asymmetric and functions as a leading end for gliding motility. This organelle also may have a role in initiating cell division in M. pneumoniae, because the bifurcation of the attachment organelle seems to occur prior to the binary fission of M. pneumoniae (2, 6, 7, 25, 26, 34, 36, 48).
The attachment organelle and polar filamentous cell shape of M. pneumoniae are thought to be stabilized by intracellular cytoskeleton-like structures, which have been observed in electron micrographs of M. pneumoniae (5, 25, 33). The most remarkable architectural feature of the cytoskeleton-like structures is the electron-dense core, a rod-like structure that exists longitudinally at the center of the attachment organelle (33). This rod-like structure, measuring about 300 nm long and 80 nm thick, has a knob at the distal end (terminal button) (33, 45). A network of fibrous structures is also observed in the cytoplasm of M. pneumoniae (33). These cytoskeleton-like structures are major components of the Triton X-100-insoluble fraction of M. pneumoniae cells (Triton shell) and are thought to have a scaffold-like function upon which other cell components construct M. pneumoniae cells (45, 51).
A recent report indicated that the Triton X-100-insoluble fraction contains about 100 proteins, including most of the known proteins required for cytadherence (P1, B, C, HMW1, HMW2, and HMW3) (45). These cytadherence-related proteins are believed to be the main components of the attachment organelle and are encoded in three operons, designated p1, hmw, and crl, in the genome (24, 25). Protein P1 (encoded in the p1 operon) is a major adhesin molecule responsible for cytadherence and is densely clustered at the surface of the attachment organelle (9, 18, 26, 48). Proteins B, C, HMW1, HMW2, and HMW3, called cytadherence accessory proteins, are not adhesin molecules but are required for the formation of functional attachment organelles (2, 3, 25). Proteins B and C, also named P90 and P40 (2, 26), are products of open reading frame 6, which exists just downstream of the p1 gene in the p1 operon (19). Proteins B and C associate with protein P1 at the attachment organelle and may support the proper structural configuration of P1 (29, 30). HMW1, HMW2, and HMW3 are large proteins necessary for the localization of P1 at the attachment organelle. These HMW proteins are present in high concentrations at the attachment organelle and are thought to be the most likely components of the electron-dense core (3, 25, 48, 49, 52). HMW1 and HMW3 are encoded in the hmw operon, and the gene encoding HMW2 is in the crl operon (24). In addition to these cytadherence-related proteins, the Triton X-100-insoluble fraction contains proteins P65 and P200. P65 and P200 share a structural domain, the acidic proline-rich domain, with HMW1 and HMW3 (40, 41). The structural similarity suggests that proteins P65 and P200 have roles similar to those of HMW1 and HMW3 as components of cytoskeleton-like structures. However, it is not clear whether proteins P65 and P200 participate in cytadherence. Recent studies revealed that P65 localizes to the attachment organelle with P30, an additional adhesin protein that is an essential factor for cytadherence (2, 20, 25, 48, 49). The genes encoding P65 and P30 are located in the crl and hmw operons, respectively. The gene encoding P200 is not located in one of the three operons of cytadherence-related proteins (2, 24).
Although these candidate components of the attachment organelle and cytoskeleton-like structures have been identified, the spatial configuration and interaction between these proteins are poorly understood. Antibodies have been used to localize specific proteins to the attachment organelle (46, 48, 49, 52), but their use is limited because of the need for specificity of an antibody for a target protein and the inability to observe living systems in real time. Green fluorescent protein (GFP), an intrinsically fluorescent molecule obtained from the jellyfish Aequorea victoria, is widely used to study protein-protein interactions, cell division, and gene expression in a variety of organisms in real time (39, 50). In this study, we developed a dual GFP expression system for M. pneumoniae to study the spatial relationship of P65 to HMW2, P41, and P24, which are encoded in the crl operon (28).
MATERIALS AND METHODS
Organism and culture conditions.
The M. pneumoniae strains listed in Table 1 were cultured in PPLO medium (2.1% PPLO broth [Becton Dickinson, Sparks, Md.], 0.25% glucose, 0.002% phenol red, 0.5% yeast extract [Becton Dickinson], 10% horse serum [Gibco BRL, Rockville, Md.], 50 μg of ampicillin/ml) or in Aluotto medium (1, 38) at 37°C. For drug-resistant M. pneumoniae strains, 18 μg of gentamicin/ml or 15 μg of chloramphenicol/ml was added to the media. Escherichia coli JM83 (53), DH5α (13), and DB3.1 (Invitrogen, Carlsbad, Calif.) were used as host strains to construct plasmids and were grown in Luria-Bertani medium (47) with or without 50 μg of ampicillin/ml, 50 μg of kanamycin/ml, and 15 μg of chloramphenicol/ml at 37°C.
TABLE 1.
M. pneumoniae strains used in this studya
| Strain | Descriptionb |
|---|---|
| M129 | Wild type |
| TK2062 | M129(pISM2062.2) |
| TK155 | M129(pTK155) |
| TK161 | M129(pTK161) |
| TK162 | M129(pTK162) |
| TK164 | M129(pTK164) |
| TK165 | M129(pTK165) |
| TK210 | M129(pTK210) |
| TK2100 | TK210(pISM2062.2) |
| TK2310 | TK210(pMPN310) |
| TK2311 | TK210(pMPN311) |
| TK2312 | TK210(pMPN312) |
| TK2310T | TK210(pMPN310-tuf) |
| TK2311T | TK210(pMPN311-tuf) |
| TK2312T | TK210(pMPN312-tuf) |
| TK3310 | M129(pMPN310) |
| TK3311 | M129(pMPN311) |
| TK3312 | M129(pMPN312) |
| TK3310T | M129(pMPN310-tuf) |
| TK3311T | M129(pMPN311-tuf) |
| TK3312T | M129(pMPN312-tuf) |
All strains were designed in this study, except for the wild-type strain (31).
Tn4001 plasmids were used to transform M. pneumoniae.
Construction of fusion genes and plasmids.
The synthetic oligonucleotides used for plasmid construction are listed in Table 2. M. pneumoniae M129 genomic DNA was prepared by a conventional phenol extraction method. The p65 gene was amplified from the genomic DNA by PCR with primers P65F-Bam and P65R-Nco. To minimize mutations caused by PCR amplification, high-fidelity DNA polymerase PyroBest (Takara, Tokyo, Japan) was used. The amplified fragment was digested with BamHI and NcoI and was inserted into the BamHI-NcoI site (the 5′ end of the eyfp gene) of plasmid pEYFP (Clontech, Palo Alto, Calif.), producing a plasmid that we designated pTK150. The p65 gene was also amplified from the genomic DNA by PCR with primers P65F-Bsr and P65R-Eco. The amplified fragment was inserted into the BsrGI-EcoRI site (the 3′ end of the eyfp gene) of plasmid pEYFP after digestion with BsrGI and EcoRI, producing a plasmid that we designated pTK153. The p65-eyfp and eyfp-p65 fusion genes were excised from plasmids pTK150 and pTK153 by using PvuII and StuI and were inserted into the SmaI site of plasmid pISM2062.2 (22), producing plasmids pTK155 and pTK158, respectively (Table 3). Plasmids pTK161 and pTK162 (Table 3) were constructed by replacing the E. coli lac promoter sequence in pTK158 (derived from plasmid pEYFP) with p65 or tuf promoter fragments from M. pneumoniae at the BamHI-NcoI site. The p65 and tuf promoter fragments were obtained from M. pneumoniae genomic DNA by PCR with primers P65F-Bam and P65-PR and primers tuf-PF and tuf-PR, respectively. These promoter fragments were also used to replace the p65 gene and its promoter sequence in pTK155 at the BamHI-NcoI site. The resulting plasmids, which expressed the eyfp gene alone from the p65 and tuf promoters, were designated pTK164 and pTK165, respectively (Table 3).
TABLE 2.
Synthetic oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| P65F-Bam | GCGGGATCCTGCAGCAGCTGACAACAACATTTAGCACACT |
| P65R-Nco | CTAGCCATGGCTTCGTAAAATTCATCACCAC |
| P65F-Bsr | GAGCTGTACAAGATGGATATAAATAAACCAGG |
| P65R-Eco | TCGCGGAATTCAGCTGTTTATTCGTAAAATTCATCACCAC |
| P65-PR | CAACCCATGGCATTTATATCCATTTACTGTCT |
| tuf-PF | GTGGGATCCATTTTGCAAACTGATGACAA |
| tuf-PR | TAACCATGGGTTTAGATCGGTCAAATTT |
| CFP1F-Sma | GATCCCGGGAGCGCCCAATACGCAAACCGCC |
| CFP-R | CCTATTATTTTTGACACCAGAC |
| 165RVS | GTACGATATC |
| tuf-F | ATTTTGCAAACTGATGACAA |
| tufR-Age | TCGGACCGGTTTCTCTCTTGCCATGTGTTTG |
| MPN309-F-Gw | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGATATAAATAAACCAGGTTGAA |
| MPN309-R-Gw | GGGGACCACTTTGTACAAGAAAGCTGGGTTATTATTCGTAAAATTCATCACCAC |
| MPN310-F-Not | CCGCGGCCGCCATGAATGATACTGACAAGAAGT |
| MPN310-R-Asc | GTCGGCGCGCCCTTATTTAGCTGCTTTTTGGGC |
| MPN311-F-Gw | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGACTAATGATTACCAACAATTAAA |
| MPN311-R-Gw | GGGGACCACTTTGTACAAGAAAGCTGGGTATCCTTCATTACTTTGTTCT |
| MPN312-F-Gw | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGAAGGATAGTGCACTAACACT |
| MPN312-R-Gw | GGGGACCACTTTGTACAAGAAAGCTGGGTTACTTCTTGTAAGAAATTAAC |
Recognition sites for restriction enzymes and the att sites for recombinase are underlined.
TABLE 3.
Plasmids constructed in this studya
| Plasmid | Vector | Marker | Promoter | Gene | Expressionb | Fluorescencec | Localizationd |
|---|---|---|---|---|---|---|---|
| pTK150 | pEYFP | Apr | p65 | p65-eyfp | NT | NT | NT |
| pTK153 | pEYFP | Apr | lac (E. coli) | eyfp-p65 | NT | NT | NT |
| pTK155 | pISM2062.2 | Apr Gmr | p65 | p65-eyfp | + | + | + |
| pTK158 | pISM2062.2 | Apr Gmr | lac (E. coli) | eyfp-p65 | − | − | − |
| pTK161 | pISM2062.2 | Apr Gmr | p65 | eyfp-p65 | + | + | + |
| pTK162 | pISM2062.2 | Apr Gmr | tuf | eyfp-p65 | ++ | ++ | + |
| pTK164 | pISM2062.2 | Apr Gmr | p65 | eyfp | + | + | − |
| pTK164-D | pISM2062.2 | Apr Gmr Cmr | p65 | eyfp | NT | NT | NT |
| pTK165 | pISM2062.2 | Apr Gmr | tuf | eyfp | ++ | ++ | − |
| pTK165-D | pISM2062.2 | Apr Gmr Cmr | tuf | eyfp | NT | NT | NT |
| pTK205 | pKV104 | Apr Cmr | lac (E. coli) | ecfp | NT | NT | NT |
| pTK207 | pKV104 | Apr Cmr | tuf | ecfp | ++ | ++ | − |
| pTK207-D | pKV104 | Apr Cmr | tuf | ecfp | NT | NT | NT |
| pTK210 | pTK207-D | Apr Cmr | tuf | ecfp-p65 | ++ | ++ | + |
| pMPN310-E | pENTR/D-TOPO | Kmr | None | hmw2 | NT | NT | NT |
| pMPN311-E | pDONR201 | Kmr | None | p41 | NT | NT | NT |
| pMPN312-E | pDONR201 | Kmr | None | p24 | NT | NT | NT |
| pMPN310 | pTK164-D | Apr Gmr | p65 | eyfp-hmw2 | + | + | + |
| pMPN311 | pTK164-D | Apr Gmr | p65 | eyfp-p41 | + | + | + |
| pMPN312 | pTK164-D | Apr Gmr | p65 | eyfp-p24 | + | + | + |
| pMPN310-tuf | pTK165-D | Apr Gmr | tuf | eyfp-hmw2 | ++ | ++ | + |
| pMPN311-tuf | pTK165-D | Apr Gmr | tuf | eyfp-p41 | ++ | ++ | + |
| pMPN312-tuf | pTK165-D | Apr Gmr | tuf | eyfp-p24 | ++ | ++ | + |
M. pneumoniae strains transformed with Tn4001 plasmids were analyzed by Western blotting and fluorescence microscopy. NT, not applicable or not tested.
Expression of the fusion protein in M. pneumoniae cells was monitored by Western blotting: +, ++, and −, moderate, strong, and no expression, respectively.
Fluorescence intensity was observed by microscopy: +, ++, and −, faint, strong, and no fluorescence, respectively.
Localization of the fusion protein at cell poles was observed by microscopy; +, present; −, absent.
The ecfp gene was amplified from plasmid pECFP (Clontech) by PCR with primers CFP1F-Sma and CFP-R. The amplified ecfp fragment was inserted into the SmaI site of pKV104, producing a plasmid that we designated pTK205. pKV104 contains a chloramphenicol-resistant (Cmr) variant of Tn4001 (12) and was kindly provided by D. C. Krause of the University of Georgia. The lac promoter region of pTK205 (upstream of the ecfp gene) was replaced at the SmaI-AgeI site with the M. pneumoniae tuf promoter sequence amplified by PCR with primers tuf-F and tufR-Age, resulting in plasmid pTK207. The BsrGI site of pTK207 (the 3′ end of the ecfp gene) was converted to an EcoRV site with an oligonucleotide linker, 160RVS. Next, the Gateway vector conversion system (reading frame cassette A) (Invitrogen) was inserted into the EcoRV site, producing a plasmid that we designated pTK207-D. The p65 gene was amplified from M. pneumoniae genomic DNA with primers MPN309-F-Gw and MPN309-R-Gw, subcloned into plasmid pDONR201 (Invitrogen) by using BP clonase (Invitrogen), and then transferred to plasmid pTK207-D by using LR clonase (Invitrogen); this procedure produced plasmid pTK210 (Table 3).
Plasmids containing hmw2, p41, and p24 fusion genes were constructed as follows. The BsrGI site of plasmid pTK164 and that of plasmid pTK165 were converted to an EcoRV site by inserting an oligonucleotide linker, 160RVS. Next, the Gateway vector conversion system (reading frame cassette A) was inserted in the created EcoRV site, producing plasmids pTK164-D and pTK165-D. The hmw2 gene sequence was amplified from M. pneumoniae genomic DNA by PCR with primers MPN310-F-Not and MPN310-R-Asc. After digestion with NotI and AscI, the hmw2 gene fragment was inserted into the NotI-AscI site of plasmid pENTR/D-TOPO (Invitrogen), resulting in a plasmid that we designated pMPN310-E. The p41 and p24 gene sequences were amplified from M. pneumoniae genomic DNA by PCR with primers MPN311-F-Gw and MPN311-R-Gw and primers MPN312-F-Gw and MPN312-R-Gw, respectively. The amplified fragments were subcloned into plasmid pDONR201 by using BP clonase, producing plasmids that we designated pMPN311-E and pMPN312-E. The hmw2, p41, and p24 gene fragments of plasmids pMPN310-E, pMPN311-E, and pMPN312-E were transferred to plasmids pTK164-D and pTK165-D by using LR clonase. The resulting plasmids (pMPN and pMPN-tuf series), which are listed in Table 3, were used to transform M. pneumoniae.
Transformation of M. pneumoniae.
M. pneumoniae with the modified Tn4001 (Tn4001mod) plasmids was transformed by the electroporation method described by Hedreyda et al. (15). The transformed cells were grown in liquid PPLO medium containing 18 μg of gentamicin/ml or 15 μg of chloramphenicol/ml. The transformation efficiencies were checked by counting the transformant colonies on PPLO agar plates. To minimize the positional effect of Tn4001mod insertion in the comparisons of the transformants, we analyzed and compared the transformant strains as a whole transformed population without picking up a single colony.
Protein analysis.
M. pneumoniae cells were grown in tissue culture flasks to the mid-log phase and were scraped from the bottom of the flasks. The cells were collected by centrifugation at 20,000 × g for 15 min at 4°C and washed three times with phosphate-buffered saline. The final cell suspension, adjusted to a total protein concentration of 1 μg/μl, was lysed by adding sample loading buffer and was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) at a load of 5 or 10 μg of total protein per lane (47). For Western blot analysis, the separated proteins were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.). Monoclonal antibody JL-8 (specific for A. victoria GFP variants) (Clontech) was used at a 1:2,000 dilution to detect enhanced yellow fluorescent protein (EYFP) and enhanced cyan fluorescent protein (ECFP). Anti-P65 antiserum (49) was also used at a 1:2,000 dilution. The reacting antibodies were detected with an alkaline phosphatase-conjugated second antibody (goat anti-mouse immunoglobulin G) (Promega, Madison, Wis.) and 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) color development substrate (Promega) according to manufacturer instructions.
Microscopy.
M. pneumoniae strains were cultured in Aluotto medium at 37°C to the mid-log phase. Cytadherence-positive cells were scraped from the bottom of the culture flasks after the medium was replaced with a volume of fresh medium that was two to five times smaller. The cell suspension was passed through a 25-gauge needle several times, filtered through a membrane filter unit with a 0.45-μm-pore size (Millipore, Billerica, Mass.) to disperse aggregates (46, 48), and placed on coverslips cleaned with saturated ethanolic KOH (4). The coverslips with the cell suspensions were incubated at 37°C for 0.5 to 1 h and were mounted on glass slides after excess cell suspensions were removed. To observe ECFP fluorescence, coverslips were washed twice with phosphate-buffered saline before being mounted on glass slides to reduce background fluorescence. The cells were observed with a BX51 fluorescence microscope equipped with YFP and CFP filter units (U-MYFPHQ and U-MCFPHQ, respectively) and a phase-contrast setup (Olympus, Tokyo, Japan). The images were digitized by using a Photometrics CoolSNAPcf charge-coupled device camera (Roper Scientific, Atlanta, Ga.) and LuminaVision software (Mitani Corp., Tokyo, Japan); signals were adjusted to obtain proper intensities. The fluorescence images were pseudocolored by using the LuminaVision software. The images were also processed by using Adobe Photoshop software, versions 6.0 and 7.0 (Adobe Systems, San Jose, Calif.).
To observe gliding, cells of strain TK162 were suspended in saline containing 20% horse serum. The cell suspension was inserted into a tunnel that was 12 mm wide, 18 mm long, and 0.06 mm high and that was assembled from a glass slide, a coverslip, and two pieces of double-sided tape; the cells were incubated in this tunnel for 10 min at 37°C. The cells then were observed with the fluorescence microscope at 37°C; this temperature was achieved by attaching a heating system to the sample stage and the objective lens. Cell images were recorded by using a charge-coupled device camera (WV-BP510; Panasonic, Osaka, Japan) and a digital videocassette recorder (WV-D9000; Sony, Tokyo, Japan) and were digitized as described previously (37).
RESULTS
Construction of the p65 fusion genes and their expression in M. pneumoniae cells.
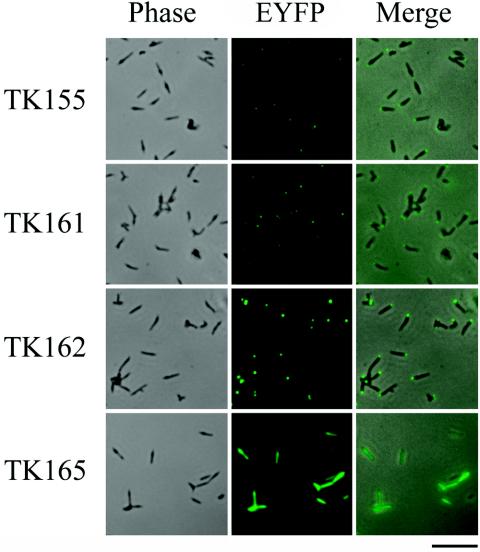
We chose the P65 protein as the initial target for the fluorescent-protein tagging strategy by virtue of the location of its gene just downstream of the promoter of the crl operon (28) (Fig. 1A); this promoter can be used to express recombinant p65 genes. The eyfp gene, encoding EYFP, which is a yellow-green-shifted variant of GFP that gives a stronger fluorescence emission than the wild type (10), was fused to the 3′ or 5′ end of the p65 gene. The fusion genes were under the control of the native p65 promoter and were inserted into the SmaI site of Tn4001mod vector plasmid pISM2062.2 (22) (Fig. 1B). The plasmids that we designated pTK155 and pTK161 carry the p65-eyfp and eyfp-p65 fusion genes, respectively (Table 3). These plasmids were introduced into M. pneumoniae M129 by electroporation to deliver fusion genes to the chromosome by the transposition of Tn4001mod. Transformants TK155 and TK161 (named for plasmids pTK155 and pTK161, respectively) were obtained and were examined by fluorescence microscopy, which detected faint fluorescent signals from both strains. In most cells, the signals were located at one pole (Fig. 2), suggesting that the P65-EYFP and EYFP-P65 fusions were produced in these strains and localized at the attachment organelle. The intensities of the fluorescence signals were similar between the strains but slightly stronger in TK161 (Fig. 2).
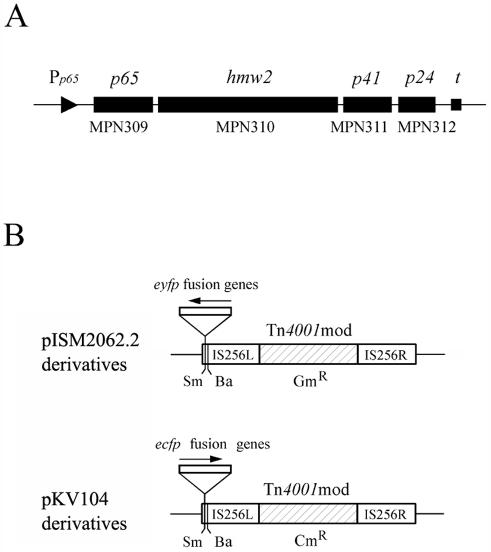
FIG. 1.
(A) Schematic illustration of the crl operon of M. pneumoniae. The four rectangular bars indicate the p65, hmw2, p41, and p24 genes in this operon. These genes are also designated MPN309, MPN310, MPN311, and MPN312 according to the serial numbering system of the M. pneumoniae genome project (8). The triangle and the square represent the p65 promoter (Pp65) and the terminator (t) of this operon, respectively. The figure is not drawn precisely to scale. (B) Structures of modified staphylococcal transposon Tn4001mod vectors (11, 22). Plasmid pISM2062.2 carries a Gmr version of Tn4001mod. Plasmid pKV104 carries a Cmr version of Tn4001mod. The IS256L, IS256R, and drug resistance genes (Gmr and Cmr) of Tn4001mod are illustrated. Cloning sites (Sm, SmaI; Ba, BamHI) in Tn4001mod are indicated. The eyfp fusion genes were inserted into the SmaI site of pISM2062.2. The ecfp fusion genes were inserted into the SmaI site of pKV104. The arrows indicate the directions in which the fusion genes were inserted.
FIG. 2.
Subcellular localization of EYFP fusions. The left and middle panels in each row show the same cells observed by phase-contrast microscopy and fluorescence microscopy, respectively. The right panel in each row shows the merged image of the left and middle panels. The transformants are named at left. Bar, 5 μm.
To enhance the fluorescence intensity, we chose the eyfp-p65 fusion gene, which exhibited slightly brighter fluorescence, and we tested a promoter sequence of the tuf gene of M. pneumoniae instead of the p65 promoter. Plasmid pTK162, which carries the eyfp-p65 fusion gene under the control of the tuf promoter, was constructed and used to transform M. pneumoniae M129. The strain obtained, TK162, showed strong fluorescent signals at the cell poles (Fig. 2), suggesting higher production of the EYFP-P65 fusion in TK162 than in TK161. To confirm whether the polar localization of fusion proteins depends on the P65 moiety, we constructed plasmids pTK164 and pTK165, which carried the eyfp gene alone under the control of the p65 or the tuf promoter (Table 3). The transformants with these plasmids, TK164 and TK165, showed fluorescence throughout the whole cell body (Fig. 2; only TK165 is shown), indicating that the polar localization of the P65-EYFP and EYFP-P65 fusions is caused by the properties of the P65 moiety. The intensity of EYFP fluorescence was strong in TK165 but faint in TK164, corresponding to their promoter activities.
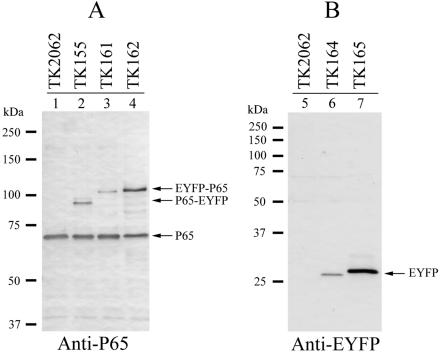
We next analyzed transformant strains by Western blot analysis to examine the levels of expression of fusion proteins. By using anti-P65 antiserum, we detected P65-EYFP in TK155 and EYFP-P65 in both TK161 and TK162 (Fig. 3). TK155 and TK161 both had lower levels of fusion proteins than of native P65 (Fig. 3, lanes 2 and 3). On the other hand, the level of expression of EYFP-P65 in TK162 was comparable to that of native P65 (Fig. 3, lane 4). The size difference between P65-EYFP and EYFP-P65 (Fig. 3, lanes 2, 3, and 4) was caused by the addition of a short amino acid sequence at the N terminus of EYFP-P65, which resulted from the construction of the promoter fusion. The anti-EYFP antibody detected EYFP in both TK164 and TK165 (Fig. 3, lanes 6 and 7). The level of EYFP was low in TK164 but high in TK165, reflecting the activities of the p65 and tuf promoters. The size difference for EYFP between TK164 and TK165 was attributed to the construction of the promoter fusion. These results indicated that the fluorescence intensities of the transformants correlated with their levels of EYFP fusion expression.
FIG. 3.
Expression of fusions of P65 and EYFP in M. pneumoniae cells. (A) Western blot analysis of M. pneumoniae transformants with an anti-P65 antibody. Lysates of M. pneumoniae transformant cells (5 μg of total protein) were separated by SDS-8% PAGE, transferred to a nitrocellulose membrane, and probed with an anti-P65 antibody. The positions of the detected P65-EYFP, EYFP-P65, and native P65 are indicated by arrows. Molecular sizes are shown at left. The analyzed transformants are shown above the lanes. (B) Detection of EYFP expression by Western blot analysis. Lysates of M. pneumoniae transformant cells (5 μg of total protein) were separated by SDS-12% PAGE. The position of EYFP detected by an anti-EYFP (anti-GFP variant) antibody is indicated by an arrow. Molecular sizes are shown at left. The analyzed transformants are shown above the lanes.
The fluorescence diminished in 10 s even in TK165 cells, with the highest intensity. This rather rapid bleaching may be attributable to the small number of fluorescent molecules caused by the small dimensions of M. pneumoniae cells. An M. pneumoniae cell is about 2 μm in length and 0.2 μm in diameter. The total volume is estimated to be 25 times lower than that of an E. coli cell. The signal from the small number of fluorescent molecules may easily fall below the detection limit as a result of photobleaching of the molecules.
The cytadherence ability of the transformants was also analyzed by a standard hemadsorption (HA) assay (14, 27). Of the colonies tested, 97% showed HA activity, suggesting that the expression of the EYFP fusions did not disturb the cytadherence processes of M. pneumoniae. However, 3% of the colonies did not show HA activity. This frequency of HA-negative colonies, obviously higher than that caused by spontaneous mutation (27), may have been caused by the insertion of Tn4001 into the cytadherence-related genes.
Observation of gliding cells.
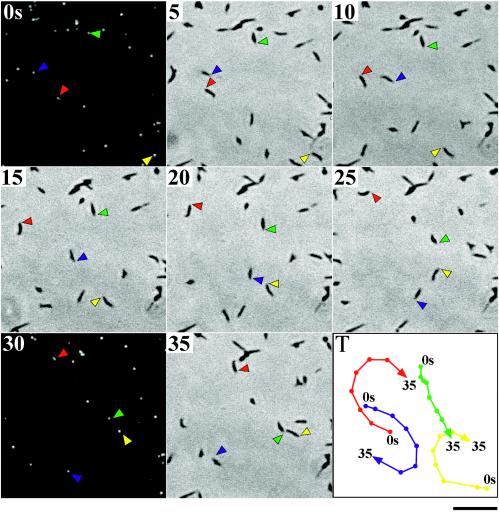
Bredt (6, 7) and Radestock and Bredt (42) studied the gliding motility of M. pneumoniae cells by phase-contrast microscopy and concluded that the attachment organelle functions as the leading end of gliding cells. If EYFP-P65 fusions are properly incorporated into the attachment organelle, then the fluorescent foci must be observed at the leading end of gliding M. pneumoniae cells. To confirm this notion directly, we studied the gliding motility of TK162 cells, which exhibited the brightest fluorescent foci among the TK strains. To do so, we made slight modifications to a method used to observe the gliding of Mycoplasma mobile cells (35). TK162 cells were suspended in saline including 20% horse serum and were inserted into a tunnel assembled from a glass slide and a coverslip. We excluded heart infusion broth and yeast extract from the cell suspension, because these components of mycoplasma growth medium cause strong background fluorescence. The coverslip was maintained at 37°C on the microscope stage. Gliding cells on the glass surface and fluorescent foci were observed by phase-contrast microscopy and fluorescence microscopy, respectively (Fig. 4). As expected, the fluorescent focus was always positioned ahead of the gliding cell, indicating the localization of EYFP-P65 at the attachment organelle. In gliding cells, the attachment organelle (fluorescent focus) always moved smoothly, while the other part of the cell body often showed lateral wobble motion, consistent with previous observations (6, 7, 42). These results suggested that the organelle kept contact with the glass surface and that the other part of the cell body detached from the surface during gliding.
FIG. 4.
Gliding motility of M. pneumoniae cells whose attachment organelles are fluorescently labeled with the EYFP-P65 fusion. Strain TK162 was observed by phase-contrast microscopy and fluorescence microscopy at 37°C. The phase-contrast image was recorded continuously with a video recorder. The microscope was shifted to the fluorescence setup for 2 s at 28-s intervals. The time intervals between images in this figure are 5 s. The positions of attachment organelles of four typical cells are indicated by colored arrowheads. The tracks of cell movement (positions of attachment organelles) are shown by colored lines in the bottom right panel (T). Bar, 5 μm.
The proportion of gliding cells was apparently higher in saline containing 20% serum than in growth medium (unpublished data). Nutrient-starved conditions may accelerate the movement, as observed in other gliding bacteria (32). The addition of 1 to 5% gelatin was not needed with our conditions, although it has been reported to be essential for keeping cells on the glass surface (42). This difference may be related to differences in glass surface conditions between previous studies and our investigation.
The average speed of the gliding shown in Fig. 4 was calculated to be 0.40 μm/s, consistent with previous observations (6, 21, 42). We tried multiple times to observe the cell division process of M. pneumoniae during 30 min of continuous video recording but failed to find cells that exhibited nascent attachment organelle formation or cytokinesis.
Subcellular localization of the HMW2, P41, and P24 proteins.
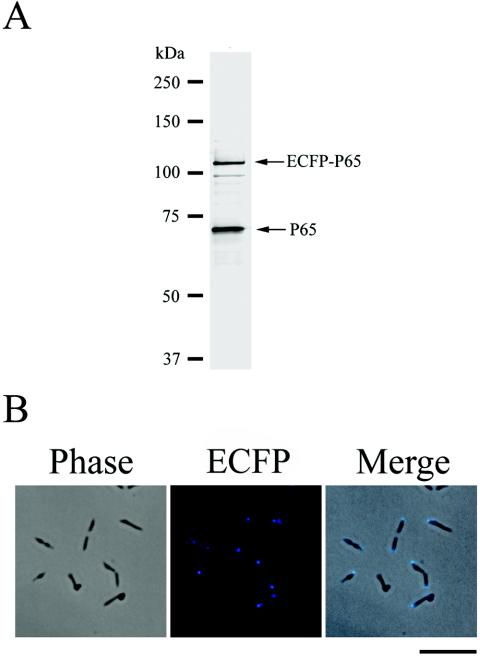
We extended the fluorescent-protein tagging strategy to the other gene products of the crl operon (the HMW2, P41, and P24 proteins) (Fig. 1A). Although the polar localization of HMW2 at the attachment organelle was reported recently by Balish et al. (3), the localization of P41 and P24 was unknown. For these experiments, we introduced a second fluorescent protein, ECFP (a blue-colored derivative of GFP) (10), to mark the positions of the attachment organelles of living cells. The ecfp gene was fused to the 5′ end of the p65 gene and was under the control of the tuf promoter. This ecfp-p65 fusion gene was introduced into M. pneumoniae M129 by use of a Cmr derivative of Tn4001mod (11) (Fig. 1B). The resulting Cmr transformant, which we designated TK210, expressed ECFP-P65 at a level slightly lower than that of native P65 (Fig. 5A) and exhibited blue fluorescent signals at the cell poles (attachment organelle) (Fig. 5B). We used TK210 as a host strain to examine the subcellular localization of HMW2, P41, and P24.
FIG. 5.
Western blot analysis and fluorescence microscopy of M. pneumoniae TK210. (A) Western blot analysis of M. pneumoniae strain TK210. Lysates of M. pneumoniae TK210 cells (5 μg of total protein) were separated by SDS-8% PAGE, transferred to a nitrocellulose membrane, and probed with an anti-P65 antibody. The positions of the detected ECFP-P65 and native P65 are indicated by arrows. Molecular sizes are shown at left. (B) Subcellular localization of the ECFP-P65 fusion. The left and middle panels show the same cells observed by phase-contrast microscopy and fluorescence microscopy, respectively. The right panel shows the merged image of the left and middle panels. Bar, 5 μm.
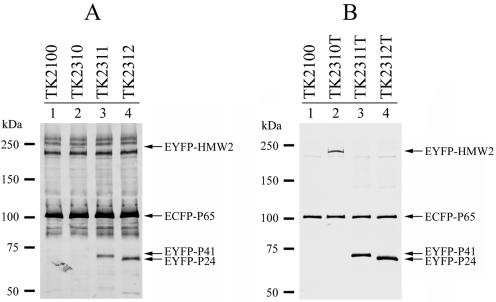
We constructed two groups of plasmids for the expression of EYFP fusions of HMW2, P41, and P24 by modifying plasmids pTK164 and pTK165 (Table 3). The first group of plasmids was designated pMPN (derivatives of pTK164); each of these plasmids possessed the eyfp-hmw2, eyfp-p41, or eyfp-p24 fusion genes under the control of the p65 promoter for low-level expression (Table 3). The second group of plasmids, designated pMPN-tuf (derivatives of pTK165) (Table 3), contained the eyfp-hmw2, eyfp-p41, or eyfp-p24 fusion genes under the control of the tuf promoter for high-level expression. These plasmids were introduced into strain TK210, and Cmr-Gmr transformants were obtained. Western blot analysis for the fusion proteins confirmed that transformants TK2310, TK2311, and TK2312 (created with pMPN plasmids) expressed EYFP-HMW2, EYFP-P41, and EYFP-P24 at low levels (Fig. 6A), while transformants TK2310T, TK2311T, and TK2312T (created with pMPN-tuf plasmids) expressed them at higher levels (Fig. 6B). The EYFP fluorescent signals of strains with low-level expression were weak and were captured with an exposure time longer than that of strains with high-level expression (Fig. 7).
FIG. 6.
Expression of EYFP-HMW2, EYFP-P41, and EYFP-P24 fusions in M. pneumoniae cells. (A) Western blot analysis of low-level expression transformants of EYFP fusions. Lysates of M. pneumoniae transformant cells (TK2100, TK2310, TK2311, and TK2312) (10 μg of total protein) were separated by SDS-5 to 10% gradient PAGE, transferred to a nitrocellulose membrane, and probed with an anti-GFP variant antibody. The positions of detected EYFP-HMW2, EYFP-P41, EYFP-P24, and ECFP-P65 are indicated by arrows. Molecular sizes are shown at left. (B) Western blot analysis of high-level expression transformants. Lysates of M. pneumoniae transformant cells (TK2100, TK2310T, TK2311T, and TK2310T) (5 μg of total protein) were analyzed under the same conditions in those used in panel A. The positions of detected fusion proteins are indicated by arrows.
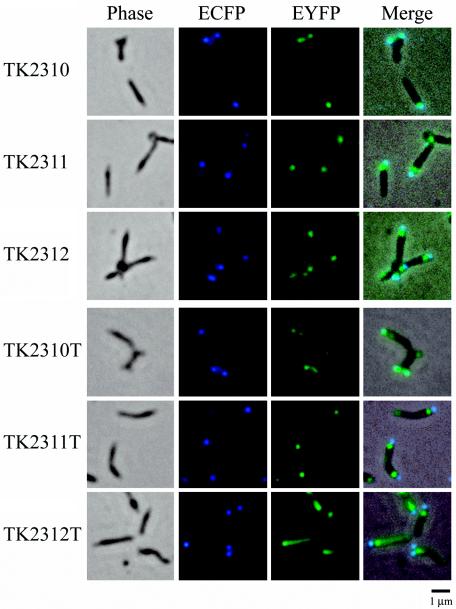
FIG. 7.
Subcellular localization of EYFP-HMW2, EYFP-P41, and EYFP-P24 fusions in the M. pneumoniae TK210 cell background. Images of six M. pneumoniae transformants (names at left) are shown. The first panel in each row shows the phase-contrast image of the cells. The second and third panels in each row show ECFP and EYFP fluorescence images of the same cells, respectively. The fourth panel in each row shows the merged image of the phase-contrast and fluorescence images. Transformants TK2310, TK2311, and TK2312 show low levels of expression of EYFP-HMW2, EYFP-P41, and EYFP-P24, respectively; transformants TK2310T, TK2311T, and TK2312T show high levels of expression. Bar, 1 μm.
Strains TK2310 and TK2310T expressing EYFP-HMW2 showed fluorescent signals for EYFP at their cell poles. The distribution patterns for fluorescent foci were basically identical between the two strains, regardless of the level of expression of EYFP-HMW2, but the fluorescence signal intensities reflected the expression levels, as indicated by the difference in brightness between the focus signals and the background of the EYFP images (Fig. 7). In both strains, the fluorescent foci of EYFP-HMW2 almost overlapped those of ECFP-P65 (Fig. 7), indicating that the HMW2 protein was localized at the attachment organelle. These results confirmed the findings of Balish et al. (3).
Transformants TK2311, TK2312, TK2311T, and TK2312T revealed that EYFP-P41 and EYFP-P24 formed fluorescent foci in M. pneumoniae cells (Fig. 7). Unlike EYFP-HMW2, the foci of EYFP-P41 and EYFP-P24 were located mainly at the proximal region of the attachment organelle and did not overlap those of ECFP-P65. In strains showing low-level expression of EYFP-P41 or EYFP-P24 (TK2311 and TK2312), the fluorescent foci of these proteins were confined to the proximal end of the attachment organelle (Fig. 7), and the profiles of these proteins were very similar. However, in strains showing high-level expression (TK2311T and TK2312T), the distribution patterns for fluorescent signals were not identical between EYFP-P41 and EYFP-P24 (Fig. 7). In strain TK2311T, additional fluorescent foci for EYFP-P41 were frequently observed at the opposite end of the attachment organelle, i.e., at the cell tail (Fig. 7). On the other hand, the fluorescence signals for EYFP-P24 were diffused from the proximal end of the organelle to the cell tail (Fig. 7, TK2312T). These nonidentical distribution patterns for EYFP-P41 and EYFP-P24 in strains showing high-level expression suggested that these proteins had different properties in the cells. However, even in these strains showing high-level expression, the strongest signals were located at the proximal end of the organelle, suggesting that this site is the preferential localization site for both P41 and P24.
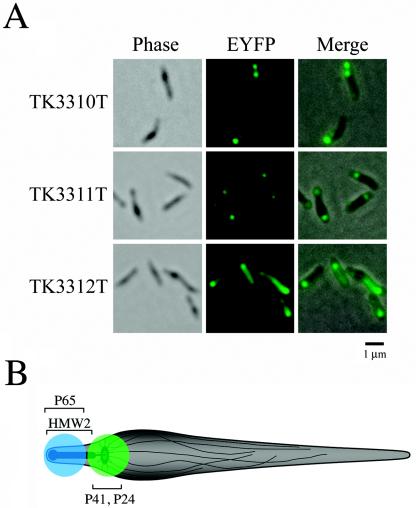
We also transformed M. pneumoniae M129 with the pMPN and pMPN-tuf plasmids, and the eyfp-hmw2, eyfp-p41, and eyfp-p24 fusion genes were expressed (strains TK3310, TK3311, TK3312, TK3310T, TK3311T, and TK3312T). We confirmed that the localization patterns for EYFP-HMW2, EYFP-P41, and EYFP-P24 in the background of strain M129 were identical to those in the background of strain TK210 (Fig. 8A; only the images for TK3310T, TK3311T, and TK3312T are shown); these results indicate that the presence of ECFP-P65 does not affect the localization of these EYFP fusions.
FIG. 8.
(A) Subcellular localization of EYFP-HMW2, EYFP-P41, and EYFP-P24 fusions in the wild-type M. pneumoniae M129 cell background (without ECFP-P65). The left and middle panels in each row show images of the same cells observed by phase-contrast microscopy and fluorescence microscopy, respectively. The right panel in each row shows the merged image of the left and middle panels. The transformants are named at left. Bar, 1 μm. (B) Schematic illustration of an M. pneumoniae cell. Cytoskeleton-like structures within the M. pneumoniae cell (electron-dense core, wheel-like structure, and fibrous network) are illustrated (see the text). Approximate positions of the fluorescent signals observed by microscopy are shown with colors (blue, ECFP-P65; green, EYFP-P41 and EYFP-P24). Positions of sites of localization of P65, HMW2, P41, and P24 are indicated.
DISCUSSION
Fluorescent-protein tagging is a widely used strategy for visualizing proteins in living cells. In this study, we have constructed vectors for fluorescent-protein tagging in M. pneumoniae and used them to visualize the protein components of cytoskeleton-like structures. The vectors constructed in this study are based on the Tn4001mod vector system (11, 22) and possess M. pneumoniae p65 or tuf promoters for the expression of fluorescent target proteins. These two promoters allow for high and low levels of expression of target proteins and are helpful for enhancing the resolution of fluorescent images and assessing the patterns of localization of target proteins. We used EYFP and ECFP as fluorescent-protein tags and designed a coexpression procedure for these proteins. This procedure enabled dual labeling of two target proteins in living M. pneumoniae cells. Since M. pneumoniae cells are pleomorphic, it is sometimes difficult to judge the position of the attachment organelle in the cell. The dual-labeling method made it easier to ascertain the position of the organelle relative to the target protein. The first fluorescent protein can be used to label the organelle, while the second is used for the other target proteins.
Using our fluorescent-protein tagging method, we visualized the four proteins—P65, HMW2, P41, and P24—that are encoded in the crl operon (Fig. 1A). The P65 protein labeled with EYFP was localized at the attachment organelle (Fig. 2), confirming previous observations obtained by immunofluorescence microscopy (20, 48, 49). The localization of P65 at the organelle indicates that P65 is a component of the attachment organelle. However, neither the function of P65 nor its involvement in cytadherence is fully understood, mainly because of the lack of P65 mutant strains. It is known that P65 is present at reduced steady-state levels in mutant strains that lack any of the cytadherence accessory proteins—HMW1, HMW2, HMW3, and P30. In these mutant strains, the polar localization of P65 is partially disrupted, depending on the extent to which the P65 levels are reduced (2, 20, 52). The stability and polar localization of P65 are thought to be correlated. To stabilize P65, it may be necessary to incorporate it into the stable localization site at the organelle. This stable localization site may be provided by the other cytoskeletal proteins (25). Unincorporated P65 tends to be degraded by proteolysis. Consistent with this model, in strain TK162, expressing a high level of EYFP-P65, the level of native P65 was lower than the levels in the other strains (Fig. 3, lane 4). It is likely that the localization site for P65 at the organelle was occupied by an excess of EYFP-P65 in this strain and that unincorporated native P65 and EYFP-P65 were degraded. This scenario may also explain the clear focal fluorescence signals of EYFP-P65 at the attachment organelle and the lesser amounts of additional fluorescence in other parts of M. pneumoniae cells, even with high-level expression of EYFP-P65 (Fig. 2). Because EYFP-P65 gave clear focal fluorescence signals at the organelle, we also labeled P65 with ECFP and used ECFP-P65 as a positional marker of the organelle for examining the localizations of the other proteins (Fig. 5).
The HMW2 protein is a critical factor for cytadherence. It is thought to function in the early stage of assembly of the attachment organelle, together with the HMW1 protein (25). The loss of HMW2 affects the stability and polar localization of most of the other cytadherence accessory proteins, but HMW2 itself is also less stable in the absence of HMW1 (2). The EYFP-HMW2 fusion was localized at the attachment organelle when expressed at both high and low levels (Fig. 7), supporting the observations of Balish et al. (3). The localization sites for EYFP-HMW2 were almost identical to those for ECFP-P65 coexpressed in the same cells. However, in a considerable number of these cells, the fluorescence signals from EYFP-HMW2 extended slightly farther toward the proximal end than did the signals from ECFP-P65, which were relatively limited to the distal end of the organelle (data not shown). These observations agree with previous ones (49) and with the current structural model of the organelle, which proposes that P65 is localized at the surface of the distal end of the attachment organelle and that HMW2 is the most probable component of the electron-dense core (25).
Little is known about the P41 and P24 proteins (25, 28). A homologous gene for P41 is present in the closely related species Mycoplasma genitalium (17), but no homologous gene has been found for P24. Although the functions of P41 and P24 are unknown, both attract considerable interest as cytoskeletal proteins, since they are encoded in the crl operon, together with P65 and HMW2, and are associated with the Triton shell (H. Ogaki et al., unpublished data). In addition, the P41 protein is predicted to contain a coiled-coil structure that has been observed in the other cytoskeletal proteins. Therefore, we analyzed the subcellular localization of these proteins by fluorescent-protein tagging and demonstrated that EYFP-P41 and EYFP-P24 were preferentially localized at the proximal end of the attachment organelle in M. pneumoniae cells, suggesting that they are cytoskeletal proteins that form unknown structures at this site. However, it should be noted that the high-level expression of both of these proteins exhibited additional fluorescent signals in the cells (Fig. 7). We thought that these additional localization patterns were caused by accumulations of excess proteins in the cells, but it remains possible that these localization patterns in cells with high-level expression reflect the native localization patterns for P41 and P24. This point must be assessed by using another method, such as immunofluorescence. If P41 and P24 really do localize to the proximal end of the attachment organelle, then what structures are present at this site? Recently, the presence of a wheel-like complex that might be part of a cytoskeleton-like structure was suggested at the proximal end of the electron-dense core by transmission electron microscopy of an ultrathin section of M. pneumoniae cells (16). This wheel-like complex is structurally similar to the flagellar motor and might be connected to fibrous structures extending into the cytoplasm of M. pneumoniae cells. The detailed structure of this wheel-like complex has yet to be elucidated, but its position (at the proximal end of the electron-dense core) corresponds to the P41 and P24 localization site (Fig. 8B).
We also used the fluorescent-protein tagging technique to observe gliding M. pneumoniae cells (Fig. 4). The expression of EYFP-P65 in M. pneumoniae allowed real-time visualization of the attachment organelle of gliding cells by phase-contrast and fluorescence microscopy. The successful labeling of the attachment organelle of living M. pneumoniae cells indicated that this technique should be applicable to the direct observation of the cell division processes in M. pneumoniae (i.e., nascent organelle formation, the migration of one of the organelles to the opposite end, and cytokinesis) (34, 36, 48). However, an attempt at such an application was not successful in this study. The major reason for this result might have been the nutrient conditions of M. pneumoniae cells used for microscopy. We used saline containing 20% horse serum to suspend cells for fluorescence microscopy in order to reduce background fluorescence. Such low-nutrient conditions might not be sufficient to support cell division in M. pneumoniae. If cell division did somehow occur under these conditions, then a longer observation time might have been required, since the doubling time of M. pneumoniae M129 is estimated to be about 10 h, even under optimal conditions (31). In future studies, these situations could be improved by reducing the background fluorescence in the medium or by using fusion proteins with more intense signals.
Acknowledgments
We are grateful to D. C. Krause of the University of Georgia for providing plasmid pKV104 and to C. Citti of the University of Veterinary Medicine, Vienna, Austria, for providing plasmid pISM2062.2. We thank Y. Arakawa of the National Institute of Infectious Diseases, Tokyo, Japan, for helpful input.
This work was supported in part by a Grant-in-Aid for JSPS Fellows from the Japan Society for the Promotion of Science (to S.S.) and by Grants-in-Aid for Young Scientists (to A.H.), for Scientific Research (to M.M.), and for Science Research on Priority Areas (motor proteins, genome science, and infection and host response) (to M.M.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 2.Balish, M. F., and D. C. Krause. 2002. Cytadherence and cytoskeleton, p. 491-518. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 3.Balish, M. F., R. T. Santurri, A. M. Ricci, K. K. Lee, and D. C. Krause. 2003. Localization of Mycoplasma pneumoniae cytadherence-associated protein HMW2 by fusion with green fluorescent protein: implications for attachment organelle structure. Mol. Microbiol. 47:49-60. [DOI] [PubMed] [Google Scholar]
- 4.Berg, H. C., and L. Turner. 1993. Torque generated by the flagellar motor of Escherichia coli. Biophys. J. 65:2201-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biberfeld, G., and P. Biberfeld. 1970. Ultrastructural features of Mycoplasma pneumoniae. J. Bacteriol. 102:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. (Basel) 32:321-326. [DOI] [PubMed] [Google Scholar]
- 7.Bredt, W. 1973. Motility of mycoplasmas. Ann. N. Y. Acad. Sci. 225:246-250. [Google Scholar]
- 8.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldner, J., U. Gobel, and W. Bredt. 1982. Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature 298:765-767. [DOI] [PubMed] [Google Scholar]
- 10.Green, G., S. R. Kain, and B. Angres. 2000. Dual color detection of cyan and yellow derivatives of green fluorescent protein using conventional fluorescence microscopy and 35-mm photography. Methods Enzymol. 327:89-94. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, T. W., E. A. Mothershed, R. H. Waldo, and D. C. Krause. 1999. Construction and analysis of a modified Tn4001 conferring chloramphenicol resistance in Mycoplasma pneumoniae. Plasmid 41:120-124. [DOI] [PubMed] [Google Scholar]
- 12.Hahn, T. W., M. J. Willby, and D. C. Krause. 1998. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J. Bacteriol. 180:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, E. J., R. M. Wilson, and J. B. Baseman. 1979. Isolation of mutants of Mycoplasma pneumoniae defective in hemadsorption. Infect. Immun. 23:903-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170-175. [DOI] [PubMed] [Google Scholar]
- 16.Hegermann, J., R. Herrmann, and F. Mayer. 2002. Cytoskeletal elements in the bacterium Mycoplasma pneumoniae. Naturwissenschaften 89:453-458. [DOI] [PubMed] [Google Scholar]
- 17.Himmelreich, R., H. Plagens, H. Hilbert, B. Reiner, and R. Herrmann. 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, P. C., R. M. Cole, Y. S. Huang, J. A. Graham, D. E. Gardner, A. M. Collier, and W. A. Clyde, Jr. 1982. Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216:313-315. [DOI] [PubMed] [Google Scholar]
- 19.Inamine, J. M., S. Loechel, and P. C. Hu. 1988. Analysis of the nucleotide sequence of the P1 operon of Mycoplasma pneumoniae. Gene 73:175-183. [DOI] [PubMed] [Google Scholar]
- 20.Jordan, J. L., K. M. Berry, M. F. Balish, and D. C. Krause. 2001. Stability and subcellular localization of cytadherence-associated protein P65 in Mycoplasma pneumoniae. J. Bacteriol. 183:7387-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchhoff, H. 1992. Motility, p. 473-489. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 22.Knudtson, K. L., and F. C. Minion. 1993. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene 137:217-222. [DOI] [PubMed] [Google Scholar]
- 23.Krause, D. C. 1998. Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol. 6:15-18. [DOI] [PubMed] [Google Scholar]
- 24.Krause, D. C. 1996. Mycoplasma pneumoniae cytadherence: unravelling the tie that binds. Mol. Microbiol. 20:247-253. [DOI] [PubMed] [Google Scholar]
- 25.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 26.Krause, D. C., and M. F. Balish. 2001. Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 198:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause, D. C., T. Proft, C. T. Hedreyda, H. Hilbert, H. Plagens, and R. Herrmann. 1997. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J. Bacteriol. 179:2668-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Layh-Schmitt, G., and R. Herrmann. 1992. Localization and biochemical characterization of the ORF6 gene product of the Mycoplasma pneumoniae P1 operon. Infect. Immun. 60:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Layh-Schmitt, G., A. Podtelejnikov, and M. Mann. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology 146:741-747. [DOI] [PubMed] [Google Scholar]
- 31.Lipman, R. P., W. A. Clyde, Jr., and F. W. Denny. 1969. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J. Bacteriol. 100:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 33.Meng, K. E., and R. M. Pfister. 1980. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J. Bacteriol. 144:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyata, M. 2002. Cell division, p. 117-130. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 35.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyata, M., and S. Seto. 1999. Cell reproduction cycle of mycoplasma. Biochimie 81:873-878. [DOI] [PubMed] [Google Scholar]
- 37.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of the gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 38.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 39.Phillips, G. J. 2001. Green fluorescent protein—a bright idea for the study of bacterial protein localization. FEMS Microbiol. Lett. 204:9-18. [DOI] [PubMed] [Google Scholar]
- 40.Proft, T., H. Hilbert, G. Layh-Schmitt, and R. Herrmann. 1995. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J. Bacteriol. 177:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proft, T., H. Hilbert, H. Plagens, and R. Herrmann. 1996. The P200 protein of Mycoplasma pneumoniae shows common features with the cytadherence-associated proteins HMW1 and HMW3. Gene 171:79-82. [DOI] [PubMed] [Google Scholar]
- 42.Radestock, U., and W. Bredt. 1977. Motility of Mycoplasma pneumoniae. J. Bacteriol. 129:1495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 44.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regula, J. T., G. Boguth, A. Gorg, J. Hegermann, F. Mayer, R. Frank, and R. Herrmann. 2001. Defining the mycoplasma ′cytoskeleton': the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147:1045-1057. [DOI] [PubMed] [Google Scholar]
- 46.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Southward, C. M., and M. G. Surette. 2002. The dynamic microbe: green fluorescent protein brings bacteria to light. Mol. Microbiol. 45:1191-1196. [DOI] [PubMed] [Google Scholar]
- 51.Stevens, M. K., and D. C. Krause. 1991. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike Triton shell. J. Bacteriol. 173:1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willby, M. J., and D. C. Krause. 2002. Characterization of a Mycoplasma pneumoniae hmw3 mutant: implications for attachment organelle assembly. J. Bacteriol. 184:3061-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]