Abstract
The Aer protein in Escherichia coli is a membrane-bound, FAD-containing aerotaxis and energy sensor that putatively monitors the redox state of the electron transport system. Binding of FAD to Aer requires the N-terminal PAS domain and residues in the F1 region and C-terminal HAMP domain. The PAS domains of other PAS proteins are soluble in water. To investigate properties of the PAS domain, we subcloned segments of the aer gene from E. coli that encode the PAS domain with and without His6 tags and expressed the PAS peptides in E. coli. The 20-kDa His6-Aer2-166 PAS-F1 fragment was purified as an 800-kDa complex by gel filtration chromatography, and the associating protein was identified by N-terminal sequencing as the chaperone protein GroEL. None of the N-terminal fragments of Aer found in the soluble fraction was released from GroEL, suggesting that these peptides do not fold correctly in an aqueous environment and require a motif external to the PAS domain for proper folding. Consistent with this model, peptide fragments that included the membrane binding region and part (Aer2-231) or all (Aer2-285) of the HAMP domain inserted into the membrane, indicating that they were released by GroEL. Aer2-285, but not Aer2-231, bound FAD, confirming the requirement for the HAMP domain in stabilizing FAD binding. The results raise an interesting possibility that residues outside the PAS domain that are required for FAD binding are essential for formation of the PAS native fold.
PAS domains are sensory input domains and protein-protein interaction sites that have been identified recently in a large family of sensory proteins from all kingdoms of life (34, 44, 50, 51; Simple Modular Architecture Research Tool [28, 38; http://smart.embl-heidelberg.de/]). The stimuli detected by PAS domains are diverse and in bacteria include light, oxygen, redox potential, and voltage (for reviews, see references 16, 21, and 44). PAS domains face the cytoplasm, unlike other sensory input domains that project into the periplasm or onto the external surface of a cell (44). These domains have a characteristic three-dimensional fold (20, 33) and bind a variety of cofactors, including 4-hydroxycinnamic acid, FMN, FAD, and heme (7, 8, 13, 20, 22, 35, 37). In eukaryotes, PAS domains have been shown to direct circadian rhythms (18), hypoxia responses (39), ion channel function (30), and development (40) (for a review, see reference 44). Considering the prominence of PAS domains in sensory proteins, surprisingly little is known about their role in signal transduction.
We are interested in PAS sensing and signaling in Aer, an FAD-containing aerotaxis receptor in Escherichia coli (6, 7, 35, 36). Aerotaxis is a rapid behavioral response to an oxygen gradient that guides cells to a region where the oxygen concentration is optimal for growth of the species (42, 43, 45). The Aer protein has an N-terminal PAS domain, an F1 segment, a 38-amino-acid hydrophobic sequence, a HAMP domain, and a C-terminal signaling domain (6, 36) (Fig. 1). The HAMP domain and highly conserved signaling region are common to all bacterial chemoreceptors, whereas the PAS domain and extended hydrophobic region are unique to Aer (7, 35). Residues essential for FAD binding have been identified in the PAS, F1, and HAMP domains, but it has not been established whether the essential F1 and HAMP residues bind to the FAD molecule or maintain the protein conformation for FAD binding (6, 36). The crystal structure of the LOV2 domain from the Adiantum phytochrome-phototropin chimeric photoreceptor, Phy3, was recently solved, and the FMN cofactor is bound within a pocket in the PAS domain (17). By analogy to phototropin and the established role of bound cofactors in the FixL and photoactive yellow protein (PYP) PAS domains, it is likely that the isoalloxazine ring of FAD binds within the PAS domain. However, the Phy3 structure cannot be used as a model to infer whether the adenine group of FAD also binds within the PAS domain. The adenine moiety might associate with the F1 region, the HAMP domain or, considering the labile nature of FAD binding, extrude into the cytosol.
FIG. 1.
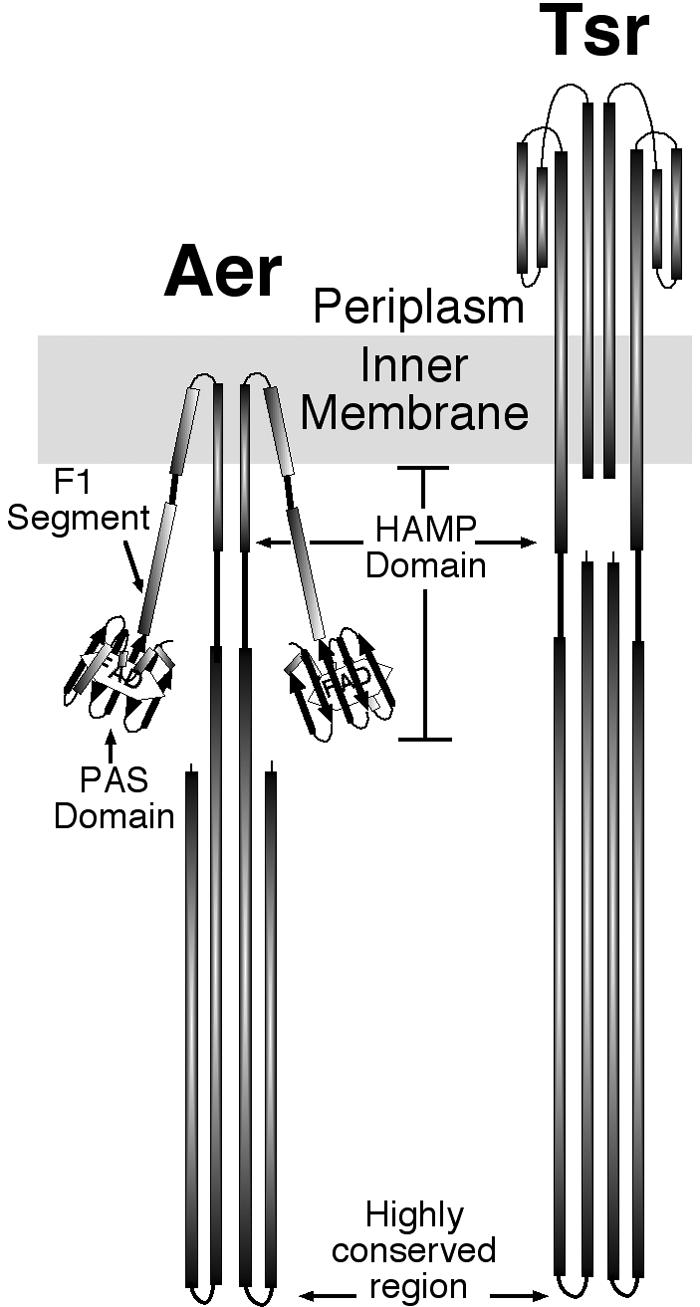
Cartoon of dimeric Aer and Tsr proteins. The periplasmic and cytosolic structures of Tar and Tsr, respectively, have been resolved by X-ray crystallography (12, 24). The cytosolic domains of two monomers form a four-helix bundle, with distal “U-turns” comprised of a highly conserved sequence that signals to the chemotactic machinery. The structure of the HAMP domain has not been resolved. Its topology has been predicted from cysteine scanning of the Tar receptor (9). The PAS domain of Aer was modeled as described earlier (36). The F1 region (6) and HAMP domain (5) of Aer were modeled from their predicted secondary structures (PSIPRED [http://insulin.brunel.ac.uk/psipred/]). The membrane binding region was predicted from hydropathy plots generated by the dense alignment surface method (DAS) (http://www.sbc.su.se/%7Emiklos/DAS/) and the C terminus was modeled from the predicted secondary structure as well as from the resolved X-ray crystal structure of the C-terminal region of Tsr (24).
The FAD cofactor in Aer does not sense oxygen directly but senses changes in the redox state of the electron transport system (45). Although residues that are critical for FAD binding and signaling have been identified in the PAS domain of Aer (6, 36), the mechanism by which the signal is transmitted from the N-terminal PAS domain to the C-terminal signaling domain remains unknown (Fig. 1). The HAMP domain in Aer is a signal transduction domain recently defined by in silico analysis as a member of a domain family present in histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis receptors, and phosphatases (5, 29; Q. Ma, B. Taylor, and I. Zhulin, unpublished observation; Protein Families Database of Alignment and HMMs [http://www.sanger.ac.uk/cgi-bin/pfam]). Although experimental data from which structure can be inferred only exist for Tar (9), members of the HAMP family are thought to contain two putative amphipathic sequences, AS1 and AS2, separated by an unstructured connector (3-5, 27, 48). The amphipathic helices likely have discrete roles and do not appear to act as simple rigid rods between sensing and signaling regions (4). It has been postulated that the HAMP domain serves as a negative regulator of the signaling region, but there is presently no mechanistic model consistent with current data (4).
The typical chemoreceptor, such as Tar or Tsr (Fig. 1), has a periplasmic sensing and cytoplasmic signaling domain. The binding of aspartate to the periplasmic domain of Tar causes a conformational change that moves the α4 helix toward the cytoplasm (11, 31). The movement of the α4 helix is transmitted to the cytoplasmic signaling domain by a helix that links α4 to the second transmembrane helix and the HAMP domain. In the Aer protein, where the N-terminal and C-terminal domains are cytosolic (Fig. 1), the signal from the FAD-PAS domain could be transmitted through the transmembrane helices or by direct interaction between the PAS domain and the C-terminal domain. In the course of investigating anomalous properties of truncated fragments of the Aer protein during purification, we determined that the membrane and/or HAMP domain sequences are essential for correct folding of the Aer PAS domain and, possibly, the F1 region.
MATERIALS AND METHODS
Materials.
High-fidelity PCR was performed using the Expand high-fidelity PCR system (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Site-specific mutagenesis was done using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, Calif.). The restriction enzymes and T4 DNA ligase were purchased from New England Biolabs, Inc. (Beverly, Mass.). Cobalt columns were from Clontech (Palo Alto, Calif.), Ni-nitrilotriacetic acid-agarose resin was from QIAGEN (Valencia, Calif.), n-dodecyl-maltoside, Triton X-100, Sephadex G-200, and Sephacryl 500-S-HR were from Sigma Chemical (St. Louis, Mo.), and cation (CM52) and anion (DE53) exchange resins were from Whatman, Inc. (Clifton, N.J.).
Bacterial strains.
E. coli XL1-Blue (Stratagene) and BL21(DE3) (Novagen, Madison, Wis.) were used for cloning and expression of constructs, respectively. BL21(DE3) is deficient in the OmpT and Lon proteases and contains a λ prophage that carries the T7 RNA polymerase gene. Isogenic strains UU1117 (aer [7]), RP5882 (tsr [10]) and BT3312 (aer tsr [36]) are derivatives of the E. coli K-12 RP437 strain (32).
Plasmids.
The plasmid pT-GroE has a chloramphenicol marker and expresses GroES and GroEL under the control of the T7 promoter (kindly provided by the Burnham Institute in San Diego [49]). The parental plasmids for Aer were pGH1 and pAVR2. Both are isopropyl-d-thiogalactopyranoside (IPTG)-inducible trc expression vectors containing the aer gene, but pAVR2 contains an N-terminal His6 tag (2, 35, 36). Plasmid pGH5 (codons 1 to 166) was constructed by high-fidelity PCR synthesis (primers: sense, 5′-CCCTGGCGCCCAATACGCAAAC-3′; antisense, 5′GCTCTAGACTAACGCGCCCGCCAGCGAAGCG-3′) from the pGH1 plasmid and subcloned into the KasI and XbaI site of vector pTrc99a. Plasmids pAVR66 (codons 20 to 119; sense, 5′-CCCCCGGCGCCCTGATGTCCACTACCGATCTG-3′; antisense, 5′-CCCCCAAGCTTCTACGTCGCCCGGGTACGAATCG-3′) and pAVR67 (codons 20 to 131; sense, 5′-CCCCCGGCGCCCTGATGTCCACTACCGATCTG-3′; antisense, 5′-CCCCCAAGCTTCTATTTGTACAGCGGCTCCACCG-3′) were constructed by high-fidelity PCR synthesis from the pGH1 plasmid and subcloned into the EheI-HindIII sites of the pTrc99a expression vector.
The parental plasmid for the His6-tagged Aer constructs was pAVR2, a His6-tagged, IPTG-inducible trc expression vector containing the aer gene (36). Plasmid pAVR63 (including aer codons 2 to 146) was constructed by using PCR (primers: sense, 5′-CCAGGCGCCTCTTCTCATCCGTATGTC-3′; antisense, 5′-CTCGTCGACCTAAGGCCTTTATGAATACGC-3′) to introduce a stop codon and XbaI restriction site after codon 146, and an EheI-XbaI fragment containing codons 2 to 146 of aer was subcloned into the pProEX HTa expression vector (Gibco BRL, Life Technologies, Gaithersburg, Md.). Plasmids pAVR64 (codons 20 to 119) and pAVR65 (codons 20 to 131) were constructed as described for pAVR66 and pAVR67, except PCR products were cloned into the pProEX HTa expression vector. Plasmids pQH7 (codons 2 to 231; sense, 5′-AGCGGATAACAATTTCACACAGGA-3′; antisense, 5′-AAAACTGCAGTTACTCAACACTATTACGTTCTCCGGTCGCCAC-3′) and pQH10 (codons 2 to 285; sense, 5′-AGCGGATAACAATTTCACACAGGA-3′; antisense, 5′-AAACTGCAGTTAGGTATGTTCGTTCAGTTCATC-3′) were similarly constructed by PCR and subcloned into the EheI-PstI sites of pProEX HTa. Plasmid pQH8 (codons 209 to 285; sense, 5′-GCAAGGCCTGTGCGCCCGATAGAAAAT-3′; antisense, 5′-AAACTGCAGTTAGGTATGTTCGTTCAGTTCATC-3′) was constructed by cloning a StuI-PstI-digested PCR fragment into the EheI-PstI sites of pProEX HTa. Plasmid pAVR1 (codons 2 to 166) was described earlier (36). All constructs (Fig. 2) were confirmed by sequencing the entire coding region of Aer. Table 1 summarizes the relevant properties of strains and plasmids used in this study.
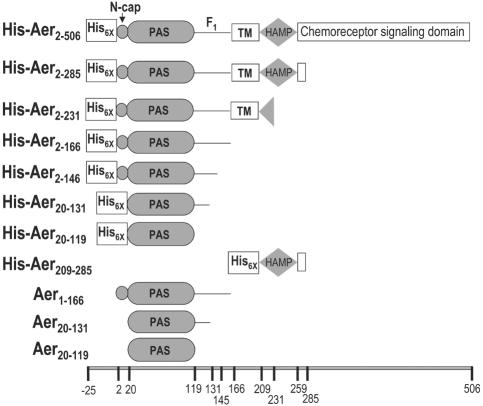
FIG. 2.
Predicted domain structure of truncated Aer constructs used for folding and FAD binding studies. The relative positions of amino acid residues referenced in the text are indicated on a scale at the bottom of the figure. Abbreviations: N-cap, N-terminal cap of the PAS domain (27); TM, hydrophobic sequence predicted to cross the membrane twice.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant properties | Reference and/or source |
|---|---|---|
| Strains | ||
| XL1-Blue | Cloning strain | Stratagene |
| BL-21 (DE3) | Cloning and expression strain; ompT gal[dcm][Ion] hsdSB(rB− mB−) and λ prophage carrying T7 RNA polymerase gene | Novagen and reference 41 |
| RP437 | thr-1 leuB6 his-4 metF159 eda-50 (chemotaxis wild type) | 32 |
| UU117 | Δaer-1 | 7 |
| RP5882 | Δtsr-7028 | 10 |
| BT3312 | Δaer-1 Δtsr-7028 | 36 |
| Plasmids | ||
| pProEXHTa | IPTG-inducible His6-tagged Ptrc expression vector | Gibco |
| pAVR1 | pProEXHTa Aer [2-166] | 36 |
| pAVR2 | pProEXHTa Aer+ [2-506] | 36 |
| pAVR63 | pProEXHTa Aer [2-146] | This study |
| pAVR64 | pProEXHTa Aer [20-119] | This study |
| pAVR65 | pProEXHTa Aer [20-131] | This study |
| pQH7 | pProEXHTa Aer [2-231] | This study |
| pQH8 | pProEXHTa Aer [209-285] | This study |
| pQH10 | pProEXHTa Aer [2-285] | This study |
| pTrc99A | IPTG-inducible Ptrc expression vector | Pharmacia and reference 2 |
| pGH1 | pTrc99A Aer+ [1-506] | 35 |
| pGH5 | pTrc99A Aer [1-166] | This study |
| pAVR66 | pTrc99A Aer [20-119] | This study |
| pAVR67 | pTrc99A Aer [20-131] | This study |
| pT-GroE | pACYC-T7 GroES+ GroEL+ | 49 |
Western blotting.
Aer was identified by Western blotting with semipurified antisera against His6-Aer2-166. Antiserum, obtained as described previously (36), was passed through a column containing 2 ml of Sepharose-4B (Sigma Chemical) to which whole-cell extract from E. coli UU1117 had been covalently bound. Western blotting using a 1:50,000 dilution of the semipurified antiserum against His6-Aer2-166 as the primary antibody and a 1:5,000 dilution of the secondary antibody (goat anti-mouse antibody conjugated to horseradish peroxidase [HRP; Bio-Rad Laboratories, Hercules, Calif.]) yielded a single band for Aer.
Fractionation of cell extract.
E. coli BL21(DE3) cells were transformed with plasmids expressing the appropriate Aer fragments and grown at 30°C to mid-log phase (optical density at 600 nm [OD600] = 0.4 to 0.6) in 250 ml of Luria-Bertani medium containing thiamine (l μM) and ampicillin (100 μg ml−1). IPTG was added to a final concentration of 1 mM, and the cells were grown for an additional 3 h to induce peptide synthesis. Cells were pelleted at 9,000 × g for 15 min, resuspended in 13 ml of lysis buffer (10 mM sodium phosphate [pH 7.5], 300 mM NaCl, 10% [wt/vol] glycerol), and lysed by seven passes through a French press at 10,000 lb/in2. Cellular debris and inclusion bodies were removed by centrifugation at 12,000 × g for 30 min. The membranes were pelleted by centrifugation at 485,000 × g for 30 min. Both the membrane fraction and soluble high-speed supernatant were stored at −20°C for up to 1 month.
HPLC.
A 100-μl aliquot of high-speed supernatant, prefiltered through a 0.2-μm microfilterfuge tube (nylon-66; Rainin Instrument Co., Emeryville, Calif.), was injected onto a size-exclusion high-performance liquid chromatography (HPLC) column (Supelco TSK-Gel; G-2000SW or G-4000SW) and eluted with buffer (10 mM sodium phosphate [pH 7.0], 300 mM NaCl, 10% [wt/vol] glycerol, and 0.02% sodium azide) at a flow rate of 1 ml min−1. Fractions were collected every minute and analyzed in a Western blot assay using rabbit sera raised against the His6-Aer2-166 peptide. When samples contained detergent, 0.02% n-dodecyl-maltoside was included in the mobile phase.
Purification of the His6-Aer2-166 complex.
Twelve milliliters of the high-speed supernatant from BL21(DE3)/pAVR1 (9 mg of protein ml−1; 108 mg of total protein) was loaded onto a Sephadex G-200 or Sephacryl 500-S-HR column (95 by 2 cm) and developed with 10 mM potassium phosphate buffer (pH 7.5) at a flow rate of 0.28 ml min−1. Fractions (100 drops) were collected and analyzed by Western blotting. The peak fraction containing His6-Aer2-166 was applied to 15 lanes of a native mini-gel and electrophoresed. One lane was cut out for Western blotting; the remaining 14 lanes were stained with Coomassie brilliant blue. One prominent band near the top of the gel was evident. His6-Aer2-166 comigrated with this prominent band, as identified by Western blotting. Subsequently, the band was excised from the 14 lanes and loaded into one well of a denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel for denaturing electrophoresis.
Detergent extraction of Aer fragments from membranes.
Membranes from E. coli BL21(DE3) cells transformed with pAVR1, pQH7, and pQH10 were prepared as described above in “Fractionation of cell extract, ” except that cell debris and inclusion bodies were removed by centrifugation at 12,000 × g for 1.5 h rather than for 30 min. Membranes were resuspended in 50 mM Tris (pH 8.1) at 4°C with HCl, 300 mM NaCl, 10% glycerol, and 2% Triton X-100 by sonication at 20% power for 15 s and incubated on ice for 15 min. Samples were recentrifuged at 485,000 × g for 30 min, and the pellet was resuspended by sonication in the same buffer. This process was repeated twice more to yield four sequential membrane extractions with detergent. Aliquots from the pellets and supernatant fractions of each step were transferred to SDS sample buffer for SDS-PAGE and Western blotting.
Detergent-dependent release of the His6-Aer2-166 fragment from GroEL.
Five hundred milliliters of BL21(DE3) cells transformed with pAVR1 was grown to an OD600 of 0.6, induced with 1 mM IPTG for 3 h, harvested at 8,000 × g for 15 min, resuspended in 12 ml of buffer (10 mM potassium phosphate [pH 7.0], 300 mM NaCl, 10% [wt/vol] glycerol), and lysed by seven passes through a French pressure cell (10,000 lb/in2). Cellular debris and inclusion bodies were removed by six sequential centrifugations at 12,000 × g for 20 min (discarding the pellet each time) until no pellet was visible. The final supernatant was then centrifuged at 485,000 × g for 30 min, and this final supernatant, if not used immediately, was aliquoted and frozen at −20°C for up to 1 month. Five hundred-microliter aliquots of the high-spin supernatant containing 3.5 mg of protein were incubated for 30 min with 0.25, 0.5, 0.75, or 1% SDS, 1 or 2% n-dodecyl-maltoside, or 1, 2, 3, 4, or 5% Triton X-100. Separation of His6-Aer2-166 from GroEL was assayed by HPLC size-exclusion chromatography as described above and by adsorption to a nickel resin, which presumably required separation from GroEL to bind the His6 tag. For nickel binding assays, 200 μl of nickel resin was prewashed three times in 1 ml of 50 mM Tris (pH 8.0) at 4°C with HCl, 300 mM NaCl, 10% glycerol, and a detergent concentration matching that used during the extraction of the specific high-spin supernatant. Samples and resin were incubated at 4°C for 30 min in a microcentrifuge tube. The unadsorbed fraction, washes, and elutions were separated from the resin by centrifugation for 1 min at 10,000 × g. After removing the unadsorbed fraction, the resin was washed three times with 1-ml aliquots of the same buffer, and the adsorbed proteins were eluted three times with 200-μl aliquots of 300 mM imidazole in the same buffer. His6-Aer2-166 was identified by Western blotting after SDS-PAGE.
ATP-dependent release of Aer fragments from GroEL.
ATP (5 mM) and MgCl2 (20 mM) were added to the soluble fraction of His6-Aer2-166 overexpressed in E. coli BL21(DE3). After 15 min, the sample was injected onto a gel filtration HPLC column and fractions were collected and analyzed using a Western blot to identify His6-Aer2-166.
Coexpression of His6-Aer2-166 and His6-Aer209-285.
Plasmids pAVR1 and pQH13 were transformed via heat shock into strain BT3312. One colony was grown overnight, subcultured, and grown to mid-log phase in Luria-Bertani medium supplemented with thiamine (1 μM), ampicillin (100 μg ml−1), and chloramphenicol (50 μg ml−1) before inducing for 3 h with 1 mM IPTG. After harvesting, cell pellets were suspended in 50 mM Tris (pH 8.0 with HCl at 4°C), 300 mM NaCl, and 10% (wt/vol) glycerol and lysed by five freeze-thaw cycles in lysozyme (0.1 mg ml−1) followed by two sonication cycles for 30 s at 50% power. The cell extract was centrifuged at 12,000 × g for 30 min, and the resulting supernatant was centrifuged at 485,000 × g for 30 min. Samples were separated by SDS-PAGE, and the Aer fragments were identified by Western blotting using the INDIA HisProbe-HRP (1:10,000 dilution), as opposed to His6-Aer2-166 antiserum, which does not recognize His6-Aer209-285. After washing off the unbound INDIA HisProbe (containing HRP-conjugated nickel), the SuperSignal chemiluminescent substrate was immediately added without the need of secondary antibodies.
FAD binding measurements.
FAD binding to the Aer protein was inferred by measuring the increase in membrane FAD when Aer was overproduced as described previously (7, 36). Direct association of FAD and His6-Aer2-285 was confirmed by the visible absorbance maxima of the nickel-purified protein, which showed peaks at approximately 450 nm (free FAD) and 470 nm (presumably His6-Aer2-285-FAD), similar to that reported for the full construct (6). A heme contaminant that absorbed at 420 nm was removed with three 1-ml washes containing 5 mM histidine, and His6-Aer2-285-FAD was eluted with four 100-μl washes containing 100 mM histidine. The FAD cofactor was identified by HPLC after extracting it from the purified protein in 8 M urea as previously described (36).
Behavioral assays.
E. coli RP5882 cells transformed with pQH8 (His6-Aer209-285) were grown at 30°C in Luria-Bertani medium supplemented with thiamine (0.5 μg ml−1) and ampicillin (100 μg ml−1) to an OD600 of 0.20 to 0.25. IPTG was added to a final concentration of 1 mM, and the cells were grown until the OD600 was 0.40 to 0.45. Aerotactic responses were quantitated with a temporal assay as described previously (26). Chemotaxis to serine and aspartate was determined in a tryptone swarm plate assay (1).
RESULTS
Anomalous properties of the Aer PAS domain.
Our laboratory showed previously that FAD readily dissociates from holo-Aer during purification in the presence of detergent (36), and we sought to determine whether FAD bound to soluble Aer peptides with higher affinity in the absence of detergent. The pAVR1 plasmid expresses an Aer2-166 peptide fragment fused to an N-terminal His6 tag (His6-Aer2-166) under the control of the trc promoter (36). His6-Aer2-166 contains the complete PAS domain and F1 sequence (Fig. 2) but terminates before the membrane binding region of Aer. Hydropathy plots (Kyte-Doolittle) showed no extended hydrophobic region, and we predicted that the fragment should be soluble. His6-Aer2-166 was overexpressed in E. coli BL21(DE3) to generate a soluble PAS domain for purification (see Materials and Methods). Approximately 20% of the overexpressed construct partitioned into the soluble fraction (supernatant from the 485,000 × g centrifugation [data not shown]), and this percentage increased to approximately 40% when the chaperone peptides, GroES and GroEL, were overexpressed in the same strain (Fig. 3). The remainder of the fragment formed inclusion bodies that were not soluble in detergent (see Materials and Methods).
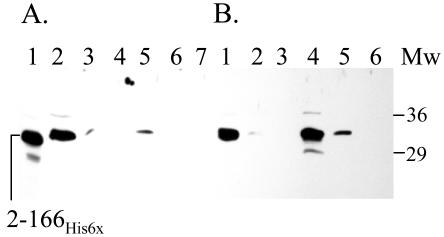
FIG. 3.
Western blot showing typical partitioning of Aer fragments into soluble and insoluble fractions when cooverexpressed with GroEL. BL21(DE3) cells transformed with GroEL (pT-GroE) and Aer peptide fragments were grown in Luria broth supplemented with thiamine (1 μM), ampicillin (100 μg/ml), and chloramphenicol (25 μg/ml) to mid-log phase (OD600 = 0.4 to 0.5), induced with 1 mM IPTG for 3 h, pelleted, suspended in buffer with 0.3 mg of lysozyme/ml, freeze-thawed five times, and sonicated with three 30-s pulses at 70% power and centrifuged at 20,800 × g for 15 min. The Aer peptides present in the pellet and supernatant fractions were separated by SDS-PAGE and identified by Western blotting as described in Materials and Methods. (A) Supernatant, “soluble ” fractions. Lanes: 1, Aer20-131; 2, Aer20-131-His6; 3, Aer20-119-His6; 4, Aer2-166-His6. Mw, molecular weight standards. (B) Pellet, “insoluble” fractions. Lanes: 1, Aer20-131; 2, Aer20-131-His6; 3, Aer20-119-His6; 4, Aer2-166-His6.
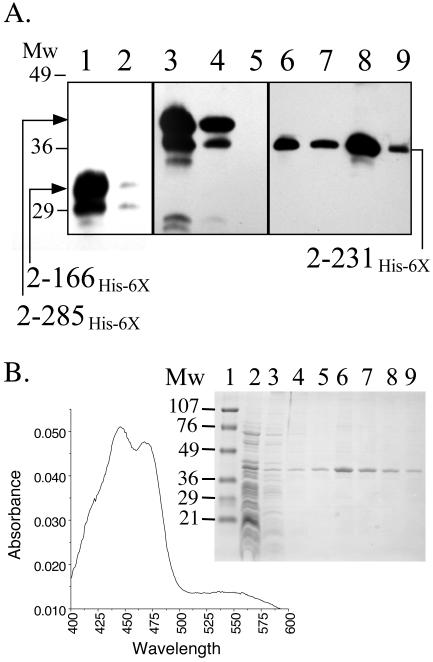
His6-Aer2-166 from the soluble fraction did not bind well to a Ni2+ or Co2+ affinity column but did bind after the protein was treated with mild detergents such as n-dodecyl-maltoside (Fig. 4) or Triton X-100 at concentrations of 1% (wt/vol) or higher, or after it was denatured in 0.75% SDS or 6 M urea (data not shown), as judged by Western blot analysis (36). This suggested that in the absence of detergent the His6 tag was sequestered from the aqueous phase. The molecular weight of His6-Aer2-166 is 22,000, and the pI was estimated to be 9.6 from the amino acid sequence (www.expasy.ch/tools/pi_tool.html). However, untreated His6-Aer2-166 did not bind to a cation exchange resin (CM52) at pH 7 as predicted, but it bound to an anion exchange resin (DE53) at pH 6 (data not shown). Thus, His6-Aer2-166 behaved as an acidic rather than a basic protein. This peptide also eluted from an HPLC gel filtration column (G-2000SW; molecular mass cutoff, ∼100,000 Da) at the void volume (data not shown). When chromatographed through resins with a larger pore size, this putative PAS complex eluted at the void volume in a Sephadex G-200 column and at a fraction representing a molecular weight of ∼800,000 in a Sepharose 500-S-HR column (Fig. 5A and B).
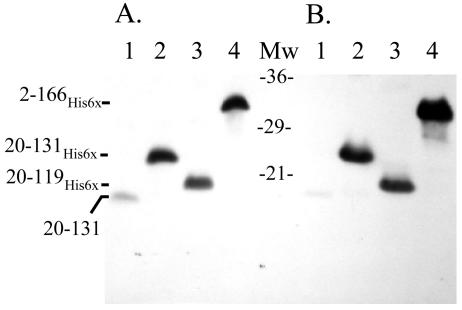
FIG. 4.
Soluble His6-Aer2-166 binds poorly to nickel resin in the absence of detergent. The supernatant of the high-speed centrifugation of the extract from BL21(DE3)/pAVR1 cells overexpressing the His6-Aer2-166 peptide was prepared, and an aliquot was adsorbed to Ni-NTA agarose and eluted as described in Materials and Methods, with or without 1% n-dodecyl-maltoside. Fractions were separated by SDS-PAGE, and Aer peptides were identified by Western blotting as described in the legend to Fig. 3. (A) Without detergent. Lanes: 1, supernatant before adsorption to resin; 2, fraction that did not adsorb to resin; 3, first wash; 4, second wash; 5, eluate from first elution; 6, second eluate; 7, third eluate. (B) With 1% n-dodecyl-maltoside. Lanes: 1, fraction that did not adsorb to resin; 2, first wash; 3, second wash; 4, first eluate; 5, second eluate; 6, third eluate. Mw, molecular weight standards.
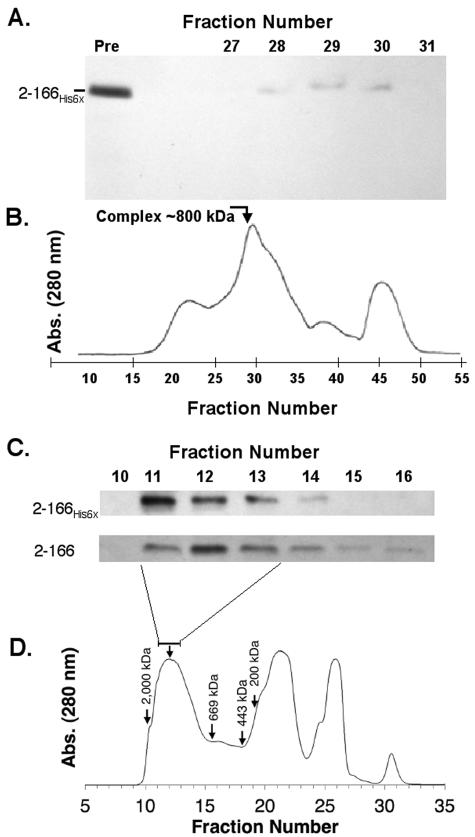
FIG. 5.
The His6-Aer2-166 and Aer2-166 peptides elute as high-molecular-weight complexes from gel filtration columns. The high-speed supernatant containing His6-Aer2-166 or Aer1-166 was prepared as described in the legend to Fig. 4. (A and B) Western blot and chromatographic profile of His6-Aer2-166 fractionated on a Sephacryl 500-S-HR column as described in Materials and Methods. Twelve milliliters of high-spin supernatant was applied to the column, and fractions (100 drops) were collected and analyzed by SDS-PAGE and by Western blotting as described in the legend to Fig. 3. These fractions (both panels) were used for further purification. (C and D) Western blots and a typical chromatographic profile of His6-Aer2-166 and Aer2-166 peptides fractionated by HPLC on a TSK-Gel G-4000SW size-exclusion column. One hundred microliters of high-spin supernatant was injected in the column, and 1-ml fractions were collected and analyzed. Both columns were calibrated using a molecular weight marker kit (GF-1000; Sigma).
Characterization of the Aer PAS complex.
After fractionating the His6-Aer2-166 complex on a Sepharose 500-S-HR column (Fig. 5), the eluted fractions containing the highest concentration of PAS domain peptide were analyzed by PAGE under nondenaturing conditions. The one prominent band that was present in the gel (Fig. 6) was excised, macerated, loaded into a well of an SDS-polyacrylamide gel, and electrophoresed. A major band with an apparent molecular weight of 68,000 and a smaller 32,000-Da band were observed after staining with Coomassie blue (Fig. 6). The 32-kDa band was identified as His6-Aer2-166 by Western blotting. The His6 tag altered the migration of the PAS peptide on SDS gels, increasing the apparent size from 22 to 32 kDa. Two bands of lower molecular weight were present, but they did not cross-react with His6-Aer2-166 antiserum. The antiserum did, however, show some cross-reactivity with a 71-kDa protein (Fig. 6). The 68-kDa band was excised from the gel, and the N terminus was sequenced by automated peptide sequencing. The sequence (AAKDVKFGNDARVKMLRGVN) was identified by a BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) as residues 2 to 21 of the chaperone protein GroEL. The native form of GroEL is a 14-subunit protein made up of 60-kDa monomers. We conclude that the 800-kDa Aer PAS complex is composed of His6-Aer2-166 peptide and GroEL. The stoichiometry of these bands suggested that approximately 80% of the semipurified GroEL was not bound to protein, while 20% was bound to His6-Aer2-166 and the unidentified peptides. The GroEL chaperone is required for correct folding of 10 to 15% of proteins in E. coli (34). A nascent or misfolded protein enters the hydrophobic interior of the cylindrical GroEL protein, where it refolds into the native conformation. Release of a folded protein from GroEL requires the GroES cochaperone and ATP (46).
FIG. 6.
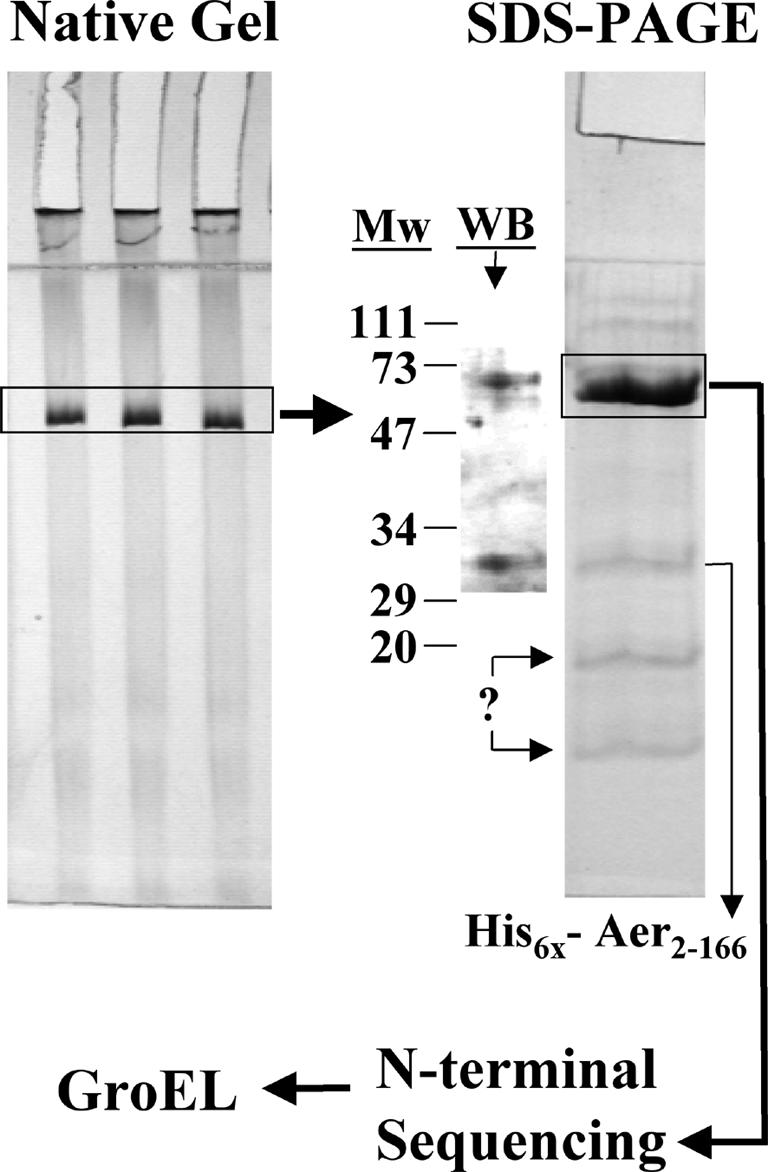
Identification of GroEL as the major component in the Aer complex. The Aer complex shown in Fig. 5A (fraction 29) was electrophoresed by PAGE under nondenaturing conditions, and the band containing the complex was excised and transferred to a denaturing SDS-PAGE gel as described in Materials and Methods. The 68-kDa band was excised and identified by N-terminal sequencing as GroEL. Both gels shown were stained with Coommassie blue. The His6-Aer2-166 band was confirmed by Western blotting with purified antisera against His6-Aer2-166. Two other unidentified low-molecular-weight proteins were also extracted from the GroEL band. Abbreviations: Mw, molecular weight standards; WB, Western blot.
Conditions favoring dissociation of the PAS fragment from the GroEL-His6-Aer2-166 complex were investigated. SDS at concentrations at or above 0.75% and 1% n-dodecyl-maltoside or Triton X-100 partially released His6-Aer2-166 from GroEL as judged by a size-exclusion HPLC column (TSK-Gel; G-2000SW) (data not shown) and a nickel column (e.g., Fig. 4). However, in a soluble cell extract, the presence of GroES and a large excess of ATP did not release the His6-Aer2-166 peptide from the complex (see Materials and Methods) (data not shown). The PAS fragment appeared to form a nondissociable complex with GroEL. The observation that the soluble His6-Aer2-166 peptide was isolated predominately in a GroEL complex suggested that the native fold of the His6-Aer2-166 fragment denatured in an aqueous environment. In contrast, the Aer holoprotein distributed into inclusion bodies, the membrane fraction and, to a small extent, into the soluble phase. The presence of a small fraction of soluble Aer, presumably in a complex with GroEL, suggests that the holoprotein may be folded by GroEL before it is released for insertion into the membrane. If the complete Aer protein dissociates from GroEL but the Aer PAS peptide does not dissociate, it is likely that the Aer protein has an internal determinant for PAS domain folding that is outside the PAS domain.
Sequence essential for PAS domain folding.
We examined Aer peptide fragments of varying lengths to identify the domains of Aer that are essential for releasing the PAS domain from GroEL (Fig. 2). Fragments His6-Aer20-119 and His6-Aer20-131 lacked the highly variable and flexible PAS N-terminal Cap (15), which is not required for folding of the PYP-PAS domain (47). His6-Aer2-146 included the entire PAS domain as well as ∼25 residues from the F1 region. The fraction of these His6-tagged fragments that was partitioned into the soluble fraction failed to bind well to a Ni2+ or Co2+ column and chromatographed as a large complex on HPLC size-exclusion chromatography, indicating that they, like His6-Aer2-166, were bound to GroEL (data not shown). When overexpressed, His6-Aer2-231, which includes the membrane binding region (residues 167 to 204) and part of the HAMP domain (residues 205 to 253), and His6-Aer2-285, which includes the entire HAMP domain (Fig. 1 and 2), were present in the membrane fraction, inclusion bodies, and to some extent in the soluble fraction. These fragments could be extracted from the membrane fraction with 1% Triton X-100 [Fig. 7A, lanes 4, 5, 7, and 9; results were similar in BL21(DE3) cells (shown) and XL1-blue and BT3312 cells (data not shown)]. The fragment His6-Aer2-166 was also present in the membrane fraction, but it was not extracted by this method (Fig. 7A, lanes 1 and 2). Therefore, it is likely that this fraction contained inclusion bodies of His6-Aer2-166 that had not pelleted at 12,000 × g. In the absence of detergent, a portion of His6-Aer2-231 and His6-Aer2-285 failed to pellet at 485,000 × g for 30 min (e.g., Fig. 7A, lane 6). This suggests that these peptides complex with GroEL, as did the shorter peptides that lack the membrane binding sequence. However, since His6-Aer2-231 and His6-Aer2-285 could be extracted from this membrane fraction with mild detergent, they were not irreversibly bound to GroEL and had inserted into the membrane. Of these two membrane-bound fragments, we observed no significant increase in bound FAD when His6-Aer2-231 was overexpressed, indicating that His6-Aer2-231 did not bind FAD. When His6-Aer2-285 was overexpressed from pQH10 in strain BT3312 by the addition of 1 mM IPTG, there was a 20-fold increase in membrane-bound FAD, indicating that His6-Aer2-285 bound FAD at levels similar to those with holo-Aer. When His6-Aer2-285 was purified on a nickel resin, it exhibited two prominent absorption maxima in the visible region at 447 and 470 nm (Fig. 7B), similar to that reported previously for Aer-FAD (6). Although not yet confirmed, the 447- and 470-nm peaks likely represent the free and bound forms of FAD, respectively (6). The identification of the FAD cofactor was confirmed by HPLC after extracting it from purified His6-Aer2-285 in 8 M urea (data not shown). These results suggest that FAD binding requires the region of the HAMP domain that includes residues between 231 and 285.
FIG. 7.
Western blot showing detergent solubilization of membrane-bound His6-Aer2-231 and His6-Aer2-285, but not membrane-bound His6-Aer2-166. (A) Membrane fractions of BL21(DE3) cells containing Aer peptide fragments were isolated, extracted, and reextracted four times with 1% Triton X-100 before Western blotting as described in Materials and Methods. Mw, molecular weight standards. Lanes: 1, membrane fraction from BL21(DE3)/pAVR1 cells expressing His6-Aer2-166 before adding detergent (lower band is a proteolytic product); 2, high-spin supernatant from sample in lane 1 after incubating for 30 min with 1% Triton X-100 and centrifuging; 3, membrane fraction from BL21(DE3)/pQH10 cells expressing His6-Aer2-285 before adding detergent; 4, high-spin supernatant from sample in lane 3 after incubating for 30 min with 1% Triton X-100 and centrifuging; 5, high-spin supernatant after reextracting pellet (produced by lane 4) with 1% detergent and recentrifuging; 6, high-spin supernatant of BL21(DE3)/pQH7 cells expressing Aer2-231 before adding detergent; 7, high-spin supernatant from sample in lane 8 after incubating for 30 min with 1% Triton X-100 and centrifuging; 8, membrane fraction from BL21(DE3)/pQH7 cells expressing His6-Aer2-231 before adding detergent; 9, high-spin supernatant after reextracting pellet (produced by lane 6) with 1% detergent and recentrifuging. (B) Purification and visible absorbance spectrum of His6-Aer2-285. Membranes containing His6-Aer2-285 were solubilized and purified on a nickel resin as described in Materials and Methods. The purity of individual fractions is shown in the SDS-PAGE gel, which was stained with Coommassie blue. Lanes: 1, molecular weight standards; 2, unadsorbed fraction; 3, first wash with 5 mM histidine; 4, second wash; 5, third wash; 6, first elution with 100 mM histidine; 7, second elution; 8, third elution; 9, fourth elution. The visible absorption spectrum of the elution from lane 6 is shown to the left of the gel.
Contribution of His6 tag.
Our group's previous study showing that His6-Aer2-506 signals normally (36) and the present study demonstrating that His6-Aer2-285 binds FAD suggest that the His6 tag does not dramatically influence the folding of these peptides. However, it was still possible that the aberrant folding properties exhibited by the other truncated His6-Aer constructs were artifacts caused by the His6 tag and/or 20-amino-acid leader sequence. To test this possibility, the high-spin soluble supernatant fractions from cells overexpressing Aer fragments lacking a His6 tag or leader sequence were chromatographed through a size-exclusion column capable of separating molecular masses of 20 to 7,000 kDa (TSK-Gel G-4000SW). Fragments Aer20-119 and Aer20-131 ran as large-molecular-mass species of ∼800,000 Da, similar to that shown for Aer1-166 and His6-Aer2-166 in Fig. 5C and D. This indicated that the association of these N-terminal peptides with GroEL was independent of the His6 tag.
Overexpression of the Aer HAMP domain fragment.
Considering that inclusion of the HAMP domain enabled native folding of the PAS domain and FAD binding, it was possible that folding was stabilized by peptide-peptide interactions between the PAS and HAMP domains. We hypothesized that if such interactions occurred, they would also be involved in signaling between the N-terminal and C-terminal domains. To investigate this, His6-Aer209-285, which encodes the HAMP domain, was expressed from pQH8 in strain RP5882 (aer+ tsr-deficient). When expression was induced with 1 mM IPTG, the cells exhibited diminished responses to an increase or decrease in oxygen concentration. The magnitude of the responses was approximately 60% of that observed for the same strain without induction or for control strains lacking the His6-Aer209-285 construct (Table 2). This result suggested that the overexpressed Aer HAMP domain fragment competes with the HAMP domain of wild-type Aer in protein-protein interactions. This could be HAMP-PAS interactions, as hypothesized, but could also be HAMP-HAMP interactions within the Aer dimer.
TABLE 2.
Effect of overexpressing His6-Aer209-285 on aerotaxis activitya
| Strain/plasmid | Relevant properties | IPTG (mM) | Response timeb
|
|
|---|---|---|---|---|
| O2 increase (0-21%) | O2 decrease (21-0%) | |||
| RP5882/pAVR2 | tsr, His6-Aer2-506 | 0 | >180 | 25.5 ± 3.5 |
| RP5882/pAVR2 | tsr, His6-Aer2-506 | 1.0 | >240 | >180 |
| RP5882/pProEXHTa | tsr, vector | 0 | 96.5 ± 6.7 | 24.0 ± 3.2 |
| RP5882/pProEXHTa | tsr, vector | 1.0 | 99.7 ± 8.6 | 23.5 ± 5.4 |
| RP5882/pQH8 | tsr, His6-Aer209-285 | 0 | 98.7 ± 11.8 | 26.0 ± 1.7 |
| RP5882/pQH8 | tsr, His6-Aer209-285 | 1.0 | 56.8 ± 8.7 | 16.5 ± 1.9 |
| BT3312/pProEX | tsr aer, vector | 0 | NRc | NR |
| BT3312/pProEX | tsr aer, vector | 1.0 | NR | NR |
Cells were prepared as described in Materials and Methods; temporal assays were performed as described previously (26).
The data represent the mean ± the standard deviation for three trials from two independent experiments.
NR, no response.
The inhibitory effect of overexpression of His6-Aer209-285 on aerotaxis is apparently not due to the nonspecific effects on the cell of overproducing the HAMP domain. No change in growth, motility, or the shape of induced cells was observed when His6-Aer209-285 was overexpressed. In contrast, when Aer or an Aer fragment that included the membrane domain (His6-Aer2-231 and His6-Aer2-285) was induced with 1 mM IPTG, cell growth and motility were defective; the cells were filamentous and swam poorly. Overexpression of His6-Aer209-285 appeared not to affect the functions of other chemoreceptors of E. coli, since RP437 cells (aer+ tsr+), when transformed with pQH8, formed normal serine and aspartate rings on tryptone swarm plates supplemented with 100, 200, or 500 μM IPTG (data not shown). This suggests that inhibition by the HAMP domain fragment was specific for the Aer-mediated aerotactic pathway.
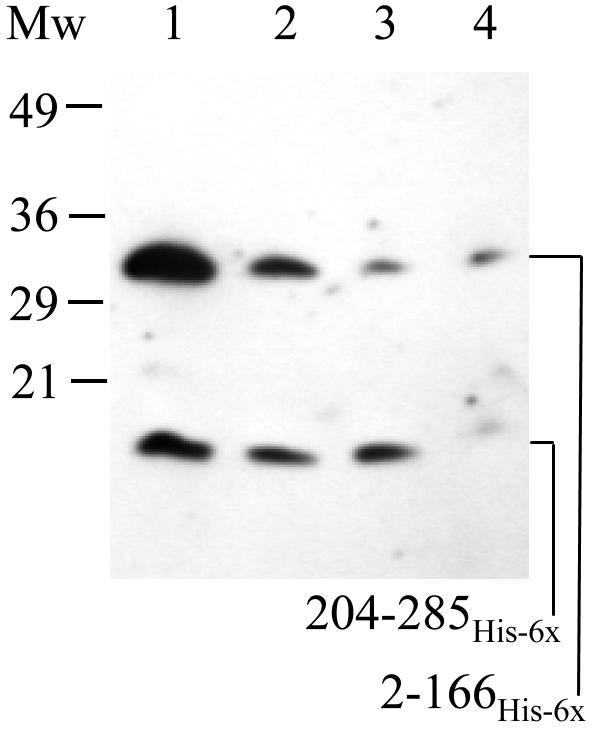
To determine whether the His6-Aer209-285 fragment could complement proper folding of the His6-Aer2-166 construct, pQH8 and pAVR1 were cotransformed into BL21(DE3) cells and coexpressed. His6-Aer209-285 distributed primarily into the soluble fraction, but little of His6-Aer2-166 partitioned into the soluble fraction (Fig. 8). When the soluble extracts were separated by size-exclusion HPLC, His6-Aer2-166 eluted as a large complex, whereas the His6-HAMP209-285 construct eluted as a low-molecular-weight species (data not shown). There was no evidence of an association between the two peptide fragments and no evidence that the HAMP fragment could complement native folding of the PAS domain in His6-Aer2-166.
FIG. 8.
Western blot of fractionated samples containing His6-Aer2-166 and His6-Aer 209-285 fragments, which were coexpressed in BT3312 cells. Mw, molecular weight standards. Lanes: 1, whole-cell extract; 2, low-spin supernatant; 3, high-spin supernatant; 4, high-spin pellet, including membrane fraction and inclusion bodies incompletely pelleted during low spin. The blot was visualized using the INDIA probe as described in Materials and Methods.
DISCUSSION
The totality of these results (summarized in Fig. 9) indicates that N-terminal peptides of Aer have low solubility in water and either precipitate into cytosolic inclusion bodies or form a stable complex with the GroEL chaperone. Insolubility may reflect exposed hydrophobic residues on the surface of the folded peptide that are shielded in the native protein. When the membrane binding region (residues ∼167 to 204) and part of the HAMP domain (residues 205 to 231) were included in the peptide, it was released from GroEL and inserted into the membrane. However, FAD binding required an additional segment of the HAMP domain located between residues 231 and 285. It is evident that amino acid residues in the HAMP domain of Aer stabilize FAD binding to the N terminus. This supports former studies showing that the point mutations R235C, E238K, and L249F in the HAMP domain of Aer abolished aerotaxis and destabilized FAD binding (6, 29, 36). Residues Y130 and L161 within the F1 region are also important for FAD binding (6). Previous mutagenesis of the Aer PAS domain, combined with the established cofactor binding properties of the PAS superfamily, suggests that FAD binds to the PAS domain of the N terminus (6, 29, 36). An important conclusion from this investigation is that residues outside the PAS domain are required to stabilize the native conformation of the PAS domain. It is possible that the F1 and HAMP residues that are essential for FAD binding stabilize the conformation of the PAS fold.
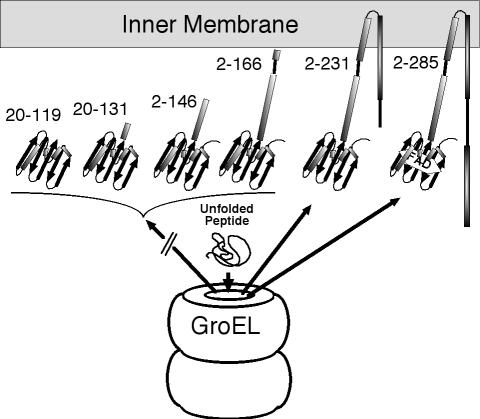
FIG. 9.
Cartoon summarizing the data, showing Aer fragments that formed a stable complex with GroEL and those that were released from GroEL. Initial complexation of GroEL and the His6-Aer2-231 and His6-Aer2-285 fragments was inferred by the presence of each in the soluble fraction (see e.g., Fig. 7A, lane 6). Fragments that included the membrane binding region and part of the HAMP domain were released from GroEL. FAD binding required a complete HAMP domain.
Aberrant physico-chemical properties of the Aer N-terminal peptide included an apparent pI several pH units lower than predicted, a His6 tag unable to bind nickel or cobalt, and a molecular weight equivalent to that of a 40-mer. These properties were explained by formation of the His6-Aer2-166-GroEL complex (Fig. 6). GroEL likely shielded the peptide residues from the solvent. The complex was stable and remained so in cell extracts that included GroES and an excess of added ATP.
Peptides that included the membrane binding domain and part (His6-Aer2-231) or all (His6-Aer2-285) of the HAMP domain inserted into the membrane in a form that was readily extractable with mild detergent, indicating that they had not folded into insoluble aggregates or precipitates. Since a small fraction of these membrane-bound Aer fragments were also found in the soluble fraction, they may also be folded by GroEL prior to membrane insertion.
Inclusion bodies would be expected when protein overexpression overwhelms the GroEL pool and allows hydrophobic moieties from one monomer to associate with hydrophobic moieties from another monomer before folding is complete. However, we expected that a fraction of these peptides would be able to fold properly when GroES and GroEL were overexpressed (Fig. 3), since other PAS domains are soluble (13, 20, 22, 30, 33, 37). Furthermore, solubility was not improved when cells were grown at 15°C for 48 h at lower induction levels (IPTG, 0.05 mM). N-terminal Aer peptides appeared unable to fold properly with or without the His6 tag, the N-terminal cap (residues 1 to 19), or the F1 region. All formed a stable complex with GroEL, and this complex represented the entire soluble fraction of these peptides.
One possible interpretation of these results is that there is a direct association between residues in the N-terminal region and those in the membrane anchor and/or in the HAMP domain. In this scenario, N-terminal hydrophobic residues would be shielded from the solvent by residues in the membrane binding or HAMP regions. A similar interdomain structural dependency occurs in the lens protein ζ-crystallin, which precipitates into opaque cataracts, despite overexpression of chaperones when the N-terminal, NADPH-binding Rossman fold is deleted (19).
Another possibility is that FAD binding is required to stabilize proper PAS folding. Since the HAMP domain is required for FAD binding, a peptide without the HAMP domain would lose FAD and compromise the native PAS structure. Other apo-PAS domains have been shown to be unstructured in solution. The apo form of PYP is more accessible to deuterium exchange and prone to precipitation (23, 25), and the unliganded form of the PAS B domain of the aryl hydrocarbon receptor binds to hsp90 but can be displaced by certain aromatic compounds that bind to the cofactor binding pocket (14). Our data could not eliminate such a scenario for Aer. His6-Aer2-231 did not bind FAD, but it was released from GroEL and partitioned into the membrane fraction in a form that was readily solubilized by mild nonionic detergents. This indicates that His6-Aer2-231 did not fold into aggregates or inclusion bodies, but the data do not prove that His6-Aer2-231 folds into the native structure.
By itself, the HAMP domain was soluble and not associated with GroEL as judged by high-speed centrifugation and by size-exclusion HPLC. However, attempts to solubilize His6-Aer2-166 by coexpression with the His6-HAMP209-285 fragment failed.
Bibikov et al. (6) reported that the HAMP domain was required for FAD binding in Aer, and they included the possibility that this domain stabilizes the structure in the N terminus. Our data support this model and suggest that the HAMP domain may be required for proper folding of the N terminus. Inspection of a putative model of the Aer PAS domain (36) revealed six hydrophobic residues (Val-55, Met-56, Phe-71, Thr-82, Ile-83, and Val-93) that are potentially solvent accessible. Although it remains to be shown, these residues might contribute to precipitation of the PAS domain. In addition, a map of the electrostatic potential of the Aer PAS domain shows a strongly basic patch that is not present in the soluble PAS domains of FixL, PYP, and HERG (A. Repik, M. Johnson, and B. Taylor, unpublished data). Whether this electrostatic potential also contributes to the unique folding requirements of the Aer PAS domain is currently under investigation.
Acknowledgments
We thank Galina Repik and Sheena Fry for technical assistance with protein purification, Gordon Harding for help with plasmid construction, and J. S. Parkinson and Kylie Watts for many helpful discussions.
This work was supported by grants from the National Institute of General Medical Sciences (GM29481) and Loma Linda University to B.L.T.
REFERENCES
- 1.Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. [DOI] [PubMed] [Google Scholar]
- 2.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 3.Appleman, J. A., L. L. Chen, and V. Stewart. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 185:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 6.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgstahl, G. E., D. R. Williams, and E. D. Getzoff. 1995. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry 34:6278-6287. [DOI] [PubMed] [Google Scholar]
- 9.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callahan, A. M., B. L. Frazier, and J. S. Parkinson. 1987. Chemotaxis in Escherichia coli: construction and properties of lambda tsr transducing phage. J. Bacteriol. 169:1246-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chervitz, S. A., and J. J. Falke. 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. USA 93:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi, Y. I., H. Yokota, and S. H. Kim. 1997. Apo structure of the ligand-binding domain of aspartate receptor from Escherichia coli and its comparison with ligand-bound or pseudoligand-bound structures. FEBS Lett. 414:327-332. [DOI] [PubMed] [Google Scholar]
- 13.Christie, J. M., M. Salomon, K. Nozue, M. Wada, and W. R. Briggs. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96:8779-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coumailleau, P., L. Poellinger, J. A. Gustafsson, and M. L. Whitelaw. 1995. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J. Biol. Chem. 270:25291-25300. [DOI] [PubMed] [Google Scholar]
- 15.Craven, C. J., N. M. Derix, J. Hendriks, R. Boelens, K. J. Hellingwerf, and R. Kaptein. 2000. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in solution by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry 39:14392-14399. [DOI] [PubMed] [Google Scholar]
- 16.Crews, S. T., and C. M. Fan. 1999. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr. Opin. Genet. Dev. 9:580-587. [DOI] [PubMed] [Google Scholar]
- 17.Crosson, S., and K. Moffat. 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98:2995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 19.Goenka, S., and C. M. Rao. 2000. Inability of chaperones to fold mutant zeta crystallin, an aggregation-prone eye lens protein. Mol. Vis. 6:232-236. [PubMed] [Google Scholar]
- 20.Gong, W., B. Hao, S. S. Mansy, G. Gonzalez, M. A. Gilles-Gonzalez, and M. K. Chan. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 95:15177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu, Y. Z., J. B. Hogenesch, and C. A. Bradfield. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40:519-561. [DOI] [PubMed] [Google Scholar]
- 22.Hill, S., S. Austin, T. Eydmann, T. Jones, and R. Dixon. 1996. Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc. Natl. Acad. Sci. USA 93:2143-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoff, W. D., A. Xie, I. H. Van Stokkum, X. J. Tang, J. Gural, A. R. Kroon, and K. J. Hellingwerf. 1999. Global conformational changes upon receptor stimulation in photoactive yellow protein. Biochemistry 38:1009-1017. [DOI] [PubMed] [Google Scholar]
- 24.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 25.Kroon, A. R., W. D. Hoff, H. P. Fennema, J. Gijzen, G. J. Koomen, J. W. Verhoeven, W. Crielaard, and K. J. Hellingwerf. 1996. Spectral tuning, fluorescence, and photoactivity in hybrids of photoactive yellow protein, reconstituted with native or modified chromophores. J. Biol. Chem. 271:31949-31956. [DOI] [PubMed] [Google Scholar]
- 26.Laszlo, D. J., and B. L. Taylor. 1981. Aerotaxis in Salmonella typhimurium: role of electron transport. J. Bacteriol. 145:990-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568-585. [DOI] [PubMed] [Google Scholar]
- 28.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, Q. 2001. HAMP domain and signaling mechanism of the Aer protein. Ph.D. dissertation. Loma Linda University, Loma Linda, Calif.
- 30.Morais Cabral, J. H., A. Lee, S. L. Cohen, B. T. Chait, M. Li, and R. Mackinnon. 1998. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell 95:649-655. [DOI] [PubMed] [Google Scholar]
- 31.Ottemann, K. M., W. Xiao, Y. K. Shin, and D. E. Koshland, Jr. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751-1754. [DOI] [PubMed] [Google Scholar]
- 32.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellequer, J. L., K. A. Wager-Smith, S. A. Kay, and E. D. Getzoff. 1998. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. USA 95:5884-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:R674-R677. [DOI] [PubMed] [Google Scholar]
- 35.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz, R. A. 1997. NifL of Klebsiella pneumoniae carries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol. Lett. 157:313-318. [DOI] [PubMed] [Google Scholar]
- 38.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semenza, G. L. 1999. Perspectives on oxygen sensing. Cell 98:281-284. [DOI] [PubMed] [Google Scholar]
- 40.Sonnenfeld, M., M. Ward, G. Nystrom, J. Mosher, S. Stahl, and S. Crews. 1997. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development 124:4571-4582. [DOI] [PubMed] [Google Scholar]
- 41.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, B. L., A. Rebbapragada, and M. S. Johnson. 2001. The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid. Redox Signal. 3:867-879. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, B. L., and I. B. Zhulin. 1998. In search of higher energy: metabolism-dependent behaviour in bacteria. Mol. Microbiol. 28:683-690. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 46.Thain, A., K. Gaston, O. Jenkins, and A. R. Clarke. 1996. A method for the separation of GST fusion proteins from co-purifying GroEL. Trends Genet. 12:209-210. [DOI] [PubMed] [Google Scholar]
- 47.Vreede, J., M. A. van der Horst, K. J. Hellingwerf, W. Crielaard, and D. M. van Aalten. 2003. PAS domains. Common structure and common flexibility. J. Biol. Chem. 278:18434-18439. [DOI] [PubMed] [Google Scholar]
- 48.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 49.Yasukawa, T., C. Kanei-Ishii, T. Maekawa, J. Fujimoto, T. Yamamoto, and S. Ishii. 1995. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J. Biol. Chem. 270:25328-25331. [DOI] [PubMed] [Google Scholar]
- 50.Yeh, K. C., and J. C. Lagarias. 1998. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95:13976-13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]