Abstract
In the pathway that controls σB activity, the RsbR-RsbS complex plays an important role by trapping RsbT, a positive regulator of σB of Bacillus subtilis. We have proposed that at the onset of stress, RsbR becomes phosphorylated, resulting in an enhanced activity of RsbT towards RsbS. RsbT is then free to interact with and activate RsbU, which in turn ultimately activates σB. In this study with purified proteins, we used mutant RsbR proteins to analyze the role of its phosphorylatable threonine residues. The results show that the phosphorylation of either of the two RsbT-phosphorylatable threonine residues (T171 and T205) in RsbR enhanced the kinase activity of RsbT towards RsbS. However, it appeared that RsbT preferentially phosphorylates T171. We also present in vitro evidence that identifies RsbX as a potential phosphatase for RsbR T205.
In Bacillus subtilis, the general response to stress is controlled by the activity of the alternative sigma factor, σB (7, 13, 15, 16, 24, 35), which, when active, associates with RNA polymerase and allows the transcription of a set of genes conferring multiple resistances on the cell (23-25). In the absence of stress, the activity of σB is inhibited by the formation of a complex with RsbW (5), an anti-sigma factor which is also a kinase for RsbV (5, 6, 11, 12). A partner-switching mechanism involving these three proteins, RsbV, RsbW, and σB, has been shown to control the activity of σB (3). A variety of stresses triggers the release of σB from RsbW, which then forms an alternative complex with the newly dephosphorylated RsbV. The phosphorylation state of RsbV is therefore the key factor for the regulation of σB. It is controlled by the balance between the kinase activity of RsbW and the activities of two specific phosphatases for phosphorylated RsbV (RsbV-P), namely, RsbP (32) and RsbU (34, 36). In the absence of stress, RsbV is found in its phosphorylated form. Depending on the nature of the stress, one of the two phosphatases, RsbP or RsbU, is activated. An energy stress caused by starvation for oxygen, carbon, or phosphate (for example, by entry into the stationary phase of growth) leads ultimately to a decrease in ATP concentration. The energy stress signal is transduced to RsbP, which contains a PAS (Per-Amt-Sim) domain and requires RsbQ (a hydrolase or acyltransferase of the alpha/beta fold superfamily [4]) for its activation by a mechanism that remains to be determined (8, 37). In the case of heat shock or changes in pH or salinity (7), known as environmental stresses, the signal is transduced to RsbU via its interaction with RsbT (36). How the signal from environmental stress is transduced to RsbT is currently unknown; for example, no specific receptors for these types of stresses have been reported.
Lying in the sigB operon, upstream of rsbT and rsbU, the rsbR and rsbS genes have been shown to be involved in the environmental stress response (2, 14, 19). It has recently been shown that RsbR and RsbS form a high-molecular-mass complex (∼1 MDa) in vitro, and there is evidence for its existence in vivo (9). Unlike its constituent individual proteins, the RsbR-RsbS complex is capable of binding to RsbT, forming a stable complex (RsbR-RsbS-RsbT) in which RsbT is trapped and unable to associate with and activate RsbU. The kinase activity of RsbT towards RsbS is counterbalanced by the phosphatase activity of RsbX, the phosphatase for RsbS-P (9). It has also been observed that if RsbR is phosphorylated by RsbT before the formation of the complex with RsbS, then the kinase activity of RsbT towards RsbS in the RsbR-P-RsbS complex is increased and the balance between the kinase activity of RsbT and the phosphatase activity of RsbX tilts in favor of RsbT (9). Under these conditions, RsbS-P accumulates, and RsbT, which has no affinity for the RsbR-P-RsbS-P complex, is released and can associate with and activate RsbU. It has therefore been suggested that a key step in the transduction of the signal is the phosphorylation of RsbR. RsbR can be phosphorylated by RsbT on two threonine residues, T171 and T205, and genetic studies have revealed that these two residues have an important role in the activation of σB (2, 14). These residues are highly conserved in RsbR homologues in close relatives and also in three paralogs encoded by the B. subtilis genome, namely, YkoB, YojH, and YqhA (1).
In this paper we report the effect of the phosphorylation of each of the threonine residues of RsbR in the RsbR-RsbS complex on the kinase activity of RsbT towards RsbS. We have also discovered that RsbX, which has been described as a specific phosphatase for RsbS (36), is also a phosphatase for phosphorylated T205 of RsbR in the RsbR-RsbS complex.
MATERIALS AND METHODS
Site-directed mutagenesis of RsbR.
Site-directed mutagenesis of RsbR was carried out by the method described in reference 17, in which Pfu DNA polymerase (Stratagene) was used in PCR. Two overlapping DNA fragments were amplified by PCR from genomic DNA (B. subtilis SG38) with two sets of primers. The first fragment was amplified with a forward primer corresponding to the 5′ end of the rsbR gene and an added NdeI site and a reverse, internal primer carrying the mutated gene codon; the second fragment was amplified with a forward primer overlapping the 3′ end of the first amplified fragment and carrying the same mutated codon and a reverse primer corresponding to the 3′ end of the rsbR gene, which also adds a BamHI site. A final PCR was carried out with the two end primers and the two fragments generated from the first PCR as the DNA template. The fragment was further digested by NdeI and BamHI and cloned into pET-11a (Novagen). The resulting DNA fragments were cloned into pET-11a, and the presence of the different mutations was confirmed by sequencing.
Purification of the proteins and gel filtration experiment.
Escherichia coli BL21 (DE3) was used for the overexpression of all proteins, which was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a 1 mM final concentration. RsbR, RsbS, RsbT, RsbU, and RsbX were purified as described in reference 9, and RsbR mutants were purified as described for RsbR. The phosphorylated forms of RsbR and mutant variants were obtained as described in reference 9.
Gel filtration.
Purified mutant RsbRs were tested for their ability to form high-molecular-weight complexes by the use of a gel filtration analytical column (Superose-12; Amersham Biosciences) essentially as described in reference 9. Briefly, 300-μg portions of RsbS, RsbT, and each mutant RsbR were mixed and incubated for a few minutes at ambient temperature before being loaded onto the column. Fractions collected were analyzed by sodium dodecyl sulfate (SDS)-13.5% polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie blue.
This technique was also applied to isolate complexes of RsbS and mutated versions of RsbR used in this study. Since the complexes are composed of an uncertain number of units (between 8 and 10) believed to be three RsbR molecules complexed with one RsbS molecule, the mass concentration of the complex was converted to a molar concentration by using the molecular weight of one unit.
Enzymatic measurements.
The kinase activity of RsbT towards RsbS and RsbR was measured as described previously (9) for the wild-type RsbR-RsbS complex and for complexes made of mutant RsbRs and RsbS. Each complex consisting of 2 μM RsbS was incubated with 0.2 μM RsbT at 30°C in phosphorylation buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol) supplemented with 0.1 mM ATP and 20 μCi of [γ-32P]ATP. At time intervals, reactions were stopped by adding 3× SDS loading buffer and heating the mixtures for 3 min at 95°C. Samples were loaded onto SDS-13.5% PAGE gels, and the gels were exposed to a phosphor screen. The bands were quantified by the Image Gauge program, version 3.3 (FLA-3000 phosphorimager; FujiFilm).
Dephosphorylation reactions were carried out essentially as described in reference 22. Complexes of RsbS and RsbR-P and mutant RsbR-P proteins were incubated with phosphatases in dephosphorylation buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2, and 1 mM MnCl2). The reactions were performed at 30°C and were stopped at intervals by mixing samples with loading buffer containing 8 M urea, 5% β-mercaptoethanol, 10 mM Tris-HCl (pH 8.5), and 1% bromophenol blue. Samples were subjected to alkaline urea-PAGE. Gels were prepared with 200 mM Tris-HCl buffer (pH 8.9)-4% acrylamide-8 M urea for the stacking gel and with 8% acrylamide-4 M urea for the separating gel. This technique was used to denature the RsbR-RsbS complex before electrophoresis, because the complex is too large to allow it to enter a usual native gel. Gel electrophoresis was conducted in Tris-glycine buffer (pH 8.5) at 150 V for 3 h at room temperature. Protein bands were visualized by Coomassie blue staining, and purified RsbR was used as a marker. Gels were scanned (Adobe Photoshop 5.0), and the intensities of the bands were measured using Scion Image Beta 4.02 software.
RESULTS
Electrophoretic mobilities of mutant RsbRs.
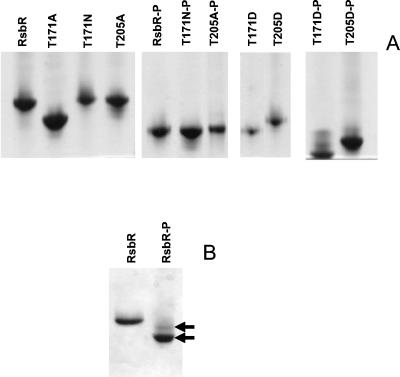
We introduced mutations in the rsbR sequence (i) to convert the threonine residues at positions 171 and 205 to alanine to prevent phosphorylation and (ii) to convert the same residues to aspartic acid to mimic the phosphorylated forms. The resulting altered RsbR proteins, RsbRT171A, RsbRT205A, RsbRT171D, and RsbRT205D, were purified in the same way as RsbR (9), and their electrophoretic mobilities were analyzed by native gel electrophoresis (Fig. 1A). When compared to RsbR (for alanine substitutions) or RsbR-P (for aspartic acid substitutions), only the electrophoretic mobilities of RsbRT171A (Fig. 1A, lane 2) and RsbRT205D (Fig. 1A, lane 9) differed significantly from that of wild-type RsbR, possibly because of a conformational change in RsbR as a result of these mutations. RsbRT171A was also affected in its ability to act as a coantagonist with RsbS in binding to RsbT (see Fig. 2 and Table 1). We therefore made an additional mutant RsbR in which we replaced T171 with an asparagine residue (RsbRT171N). Electrophoretic mobilities of RsbRT205A and RsbRT171N after phosphorylation by RsbT were identical to that of RsbR-P.
FIG. 1.
Analysis of the electrophoretic mobilities of RsbR mutants by nondenaturing 10% PAGE. (A) Each lane contains between 3 and 10 μg of purified RsbR or mutant RsbRs. Phosphorylated proteins were prepared by overnight incubation with RsbT and ATP. (B) Most preparations of RsbR-P showed two bands, as indicated by the arrows.
FIG. 2.
Mutant RsbRs (labeled at the tops of the gels) were mixed with RsbS and RsbT, and the mixture was applied to an analytical gel filtration column (Superose-12; Pharmacia). The first fractions (numbered 1 to 5), which contain the high-molecular-weight complex formed by RsbR and RsbS, were analyzed by SDS-PAGE to reveal whether RsbT was present. Table 1 shows the presence (+) or the absence (−) of RsbT in the different combinations of complexes of RsbS and RsbR or mutant RsbRs.
TABLE 1.
Presence or absence of binding to RsbT in different combinations of complexes of RsbS and RsbR or RsbR mutantsa
| Complex | Binding to RsbTb |
|---|---|
| RsbR-RsbS | + |
| RsbR-P-RsbS | + |
| RsbRT171A-RsbS | − |
| RsbRT171N-RsbS | + |
| RsbRT171N-P-RsbS | + |
| RsbRT205A-RsbS | + |
| RsbRT205A-P-RsbS | + |
| RsbRT171D-RsbS | + |
| RsbRT171D-P-RsbS | − |
| RsbRT205D-RsbS | + |
| RsbRT205D-P-RsbS | − |
Experimental procedures were as described for Fig. 2.
+, present; −, absent.
We noted that most of our preparations of RsbR-P also showed a minor band that migrated to the same position as RsbRT205D (Fig. 1B). Given that RsbR is phosphorylatable at two threonines, we had previously assumed that the two bands corresponded to monophosphorylated and diphosphorylated RsbR. The present result (Fig. 1) leads to a different interpretation: the faster-running band might correspond to the phosphorylation only of T171, and the slower-running band might correspond to the phosphorylation only of T205. Indeed, RsbRT171D and RsbRT171N-P (Fig. 1A) have the same mobility as most of RsbR-P (Fig. 1B), whereas RsbRT205D has the same mobility as the minor band in RsbR-P. It is therefore possible that RsbT phosphorylates RsbR mainly (∼90%) at T171 and to only a slight extent (∼10%) at T205. This conclusion is reinforced by the results obtained with the mimics of a doubly phosphorylated RsbR protein, RsbRT171D-P and RsbRT205D-P, which migrate faster than wild-type RsbR-P (Fig. 1A), suggesting again that only one threonine residue of RsbR is phosphorylated. The same conclusion is also in accordance with the results of mass spectroscopic analysis of wild-type RsbR-P, which clearly showed the presence of only one phosphate group per mole (results not shown). However, we cannot discount the possibility that the phosphorylation of RsbR might be different when RsbR is in the high-molecular-weight complex with RsbS (9).
RsbR-RsbS-RsbT complex formation with mutated versions of RsbR.
The role of the RsbR-RsbS complex is to trap RsbT prior to stress (9). We first checked the ability of the RsbR mutants to form a high-molecular-weight complex with RsbS only. All the RsbR mutants, including their phosphorylated counterparts, could form high-molecular-weight complexes with RsbS (data not shown). We then investigated the ability of all the complexes containing RsbS and either the mutant RsbRs or their phosphorylated forms to bind to RsbT. The absence or presence of RsbT in the complexes was analyzed by SDS-PAGE of the first fractions of the Superose-12 gel filtration column, which contain the high-molecular-weight species. Several examples are shown in Fig. 2, and all the results are summarized in Table 1.
The only complex of nonphosphorylated proteins that could not bind RsbT was RsbRT171A-RsbS. Furthermore, the electrophoretic mobility of RsbRT171A did not resemble that of wild-type RsbR (Fig. 1A), and thus we have to conclude that this mutation is deleterious to the structure of RsbR. Consequently, RsbRT171A was not considered further in this study. Mutant RsbRs in which one threonine residue had been converted into aspartic acid (RsbRT171D and RsbRT205D) were still able to bind to RsbT (Fig. 2). However, RsbRT171D-P and RsbRT205D-P, although able to form a complex with RsbS, could not associate with RsbT (Fig. 2). Contrary to what has previously been reported (9), this experiment suggested that the phosphorylation state of RsbR in the RsbR-RsbS complex can affect binding to RsbT, but only when RsbR is doubly phosphorylated. Given this, and given that RsbT was found to bind to the RsbR-P-RsbS complex, we conclude that when RsbR is phosphorylated in vitro by RsbT, the phosphorylation occurs mainly at one threonine residue, most likely T171.
Effect of the phosphorylation of RsbR on the kinase activity of RsbT.
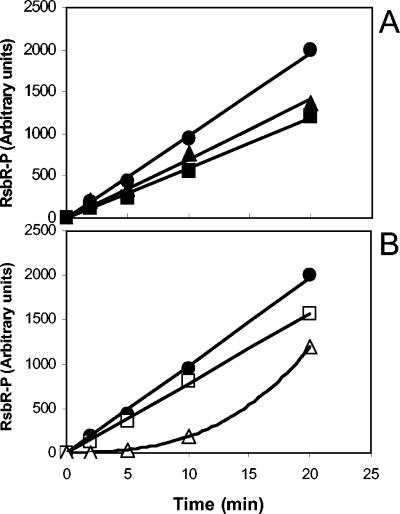
It was previously demonstrated that the kinase activity of RsbT towards RsbS was enhanced by a factor of about 5 when RsbR in the RsbR-RsbS complex was phosphorylated (9). To establish whether the phosphorylation of one threonine residue was sufficient for this effect (and if so, which one), we used the mutant versions of RsbR described above in RsbT kinase assays. Complexes for all of the altered RsbRs with RsbS were obtained and purified by gel filtration and then mixed with a standardized amount of RsbT and radiolabeled with ATP. The results revealed that the kinase activity of RsbT towards RsbS was enhanced when RsbS was complexed with the phosphorylated proteins RsbR-P, RsbRT205A-P, and RsbRT171N-P (Fig. 3). All these versions of RsbR are monophosphorylated, and therefore we conclude that, whichever threonine residue is phosphorylated in RsbR, be it T171 or T205, the same enhanced activity of RsbT towards RsbS is observed. This finding also implies that the phosphorylation of one threonine residue of RsbR is sufficient to affect the rate of phosphorylation of RsbS by RsbT.
FIG. 3.
Effect of the phosphorylation of RsbR T205 or T171 on the phosphorylation rate of RsbS by RsbT. RsbR-RsbS complexes formed with wild-type RsbR (⧫) or wild-type RsbR-P (□) (A), RsbRT205A (•) or RsbRT205A-P (○) (B), and RsbRT171N (▴) or RsbRT171N-P (▵) (C) were mixed with RsbT and [γ-32P]ATP. Samples taken at the times indicated on the graphs were subjected to SDS-PAGE, and gels were exposed to a phosphor screen.
We performed similar analyses with complexes that mimic diphosphate RsbRs, i.e., RsbRT205D-P, RsbRT171D-P, and RsbRT171D,T205D. These data revealed that the addition of two negative charges to RsbR abolished the enhancement of the activity of RsbT (data not shown), which was even slower than in the unphosphorylated RsbR-RsbS complex. This double modification of RsbR also prevents the RsbR-RsbS complex from trapping RsbT (Fig. 2), which suggests either that RsbT is repelled by these additional negative charges or that the phosphorylation of the two threonines affects the structure of RsbR-RsbS in such a way that it no longer binds to RsbT.
Phosphorylation rates of RsbR T171 and T205 in the RsbR-RsbS complex.
We have demonstrated that when RsbR is not in a complex with RsbS, RsbT preferentially phosphorylates T171 of RsbR (Fig. 1). As the formation of the RsbR-RsbS complex might alter the structure of RsbR, we wondered if the formation of the RsbR-RsbS complex might affect the preference of RsbT for T171 over T205. We therefore used complexes formed with RsbS and RsbRT171N or RsbRT205A, which left only one threonine available for phosphorylation, T205 or T171, respectively. Figure 4A shows that the rates of phosphorylation of the two threonines of RsbR, T171 and T205, were almost identical to and only slightly lower than that of wild-type RsbR.
FIG. 4.
Phosphorylation rates of RsbR T171 and T205 in the RsbR-RsbS complex. The incorporation of radiolabeled phosphate in RsbR was measured as described for Fig. 3. Complexes were made of RsbS and wild-type RsbR (•), RsbRT171N (▴), or RsbRT205A (▪) (A) and RsbS and wild-type RsbR (○), RsbRT171D (▵), or RsbRT205D (□) (B).
We then asked if the rate of phosphorylation of one threonine residue of RsbR could be affected by the phosphorylation state of the other. The results (Fig. 4B) showed that in complexes formed with RsbRT205D, the rate of phosphorylation of T171 was almost identical to that observed in complexes formed with wild-type RsbR. However, with complexes formed with RsbRT171D, the phosphorylation of RsbR T205 by RsbT appeared to be delayed. The proximity of these two residues may explain this delay.
The C-terminal domain of RsbR is similar in sequence to the whole of SpoIIAA. RsbR T205 corresponds to S58 of SpoIIAA (the phosphorylatable residue in SpoIIAA), and RsbR T171 corresponds to H24. The solution structure of SpoIIAA has been solved by nuclear magnetic resonance (21), and it has been found that H24 and S58 face each other. It has recently been shown by 1H nuclear magnetic resonance pH titration of H24 in SpoIIAA that H24 interacts with the phosphate group of S58-P (10). As previously suggested (14), if the overall structure of the C-terminal domain of RsbR is comparable to that of SpoIIAA, an interaction between RsbR T205 and T171 is highly probable. This could explain the negative effect on the phosphorylation rate of T205 in RsbRT171D, which could be reinforced by the two negative charges on a phosphothreonine residue. The single negative charge of an aspartic acid might only partially mimic phosphorylation. The inhibition of the phosphorylation of T205 by T171-P is consistent with our previous conclusion that RsbR is phosphorylated mainly on one residue, T171.
Dephosphorylation of RsbR-P by RsbX in the RsbR-P-RsbS complex.
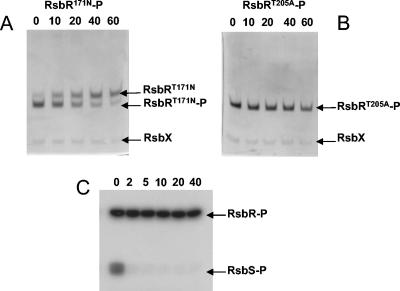
RsbR can be phosphorylated by RsbT (14), but to date no phosphatase for RsbR has been reported. Here we present evidence that RsbR in the RsbR-RsbS complex can be dephosphorylated by RsbX, the phosphatase for RsbS. The RsbRT171N-P-RsbS complex was mixed with RsbX, and samples of the reaction were analyzed by urea-alkaline gel electrophoresis. The intensity of the band corresponding to RsbRT171N-P gradually diminished, while a band corresponding to RsbRT171N appeared (Fig. 5A). The experiment was repeated with purified RsbRT171N-P alone (i.e., not complexed with RsbS), but no dephosphorylation was observed (data not shown). Hence, the dephosphorylation of T205 of RsbR by RsbX can occur only when RsbR is in a complex with RsbS. The fact that RsbX has previously been reported to be unable to dephosphorylate RsbR (14) can perhaps be explained by the need for RsbR to be in a complex with RsbS for RsbX to be active. Control experiments performed with either RsbU or the catalytic domain of SpoIIE as the phosphatase and using the RsbRT171N-P-RsbS complex as the substrate failed to produce the unphosphorylated form of RsbRT171N (data not shown). This result demonstrates, first, that the appearance of a new band was not due to the instability of the phosphorylated form of RsbR, and second, that the dephosphorylation of T205 of RsbR was due to RsbX specifically.
FIG. 5.
Dephosphorylation of RsbR-P by RsbX. (A) Dephosphorylation of 10 μM RsbRT171N-P-RsbS by 2.5 μM RsbX. At the time intervals (in minutes) indicated at the top of the gel, samples of the reaction mixture were subjected to alkaline urea-PAGE, and the gel was stained by Coomassie blue. (B) Dephosphorylation of 10 μM RsbRT205A-P-RsbS by 2.5 μM RsbX. Samples were treated as described for panel A. (C) After phosphorylation of the RsbR-RsbS complex by RsbT in the presence of 20 μCi of [γ-32P]ATP, a 10 μM concentration of the radiolabeled RsbR-RsbS complex was mixed with 2.5 μM RsbX. At the indicated time intervals (in minutes), reaction samples were mixed with 3× SDS loading buffer and heated for 3 min at 95°C. Samples were analyzed by SDS-13.5% PAGE and revealed by the use of a phosphor screen.
From these experiments, a turnover number of 0.08 × 10−2 s−1 can be derived for the RsbX dephosphorylation activity towards T205 of RsbR (Fig. 5A), which is much lower than that of other PP2C phosphatases of B. subtilis, such as RsbU or SpoIIE. Under the same conditions (buffer, Mg2+ and Mn2+ concentrations, and temperature), the turnover number for RsbU, the phosphatase for RsbV-P, in the presence of an equimolar amount of its activator RsbT reaches 6 × 10−2 s−1 (O. Delumeau, M. D. Yudkin, and R. J. Lewis, unpublished data) and that for SpoIIE, the phosphatase for SpoIIAA-P, is 7 − 10−2 s−1 (22). The rate of dephosphorylation of T205 of RsbR by RsbX is therefore 75 times lower than those of SpoIIE and activated RsbU. However, the in vitro phosphatase activity of purified RsbX towards RsbR T205-P (turnover number, 0.08 × 10−2 s−1) is more comparable to the activity of inactivated RsbU towards RsbV-P (0.3 × 10−2 s−1) than to that of RsbU activated by a molar equivalent of RsbT (O. Delumeau, M. D. Yudkin, and R. J. Lewis, unpublished data). To determine whether phosphorylation of the other threonine, T171, affected the rate of dephosphorylation of T205, we used the RsbRT171D-P-RsbS complex, but we found that the rate of dephosphorylation was increased only by a factor of 2 (data not shown). Given that RsbT activates RsbU by more than 20-fold, we are inclined to think that a 2-fold increase in RsbX activity is probably not physiologically significant.
We then assayed the dephosphorylation of T171-P using the RsbRT205A-P-RsbS complex. No dephosphorylation of T171-P was observed, either in the presence of RsbX (Fig. 5B) or with a large amount of RsbU and its activator RsbT (data not shown). We conclude that only T205-P in RsbR can be dephosphorylated by RsbX and that no phosphatase tested so far can dephosphorylate T171-P.
We exploited the fact that RsbX can dephosphorylate T205-P of RsbR to test whether RsbX could dephosphorylate RsbR-P in a phosphorylated RsbR-RsbS complex. This complex was prepared by incubating RsbR-RsbS with RsbT and [γ-32P]ATP, leading to phosphorylation of both RsbR and RsbS, before purifying the complex by gel filtration and mixing it with RsbX. While RsbX was active towards RsbS-P, whose band disappeared in less than 2 min, the intensity of the band corresponding to RsbR-P remained constant throughout the experiment (Fig. 5C). The fact that RsbX could not dephosphorylate RsbR-P, even in a complex with RsbS, suggests that the phosphorylation site of RsbR in the RsbR-RsbS complex is predominantly T171 and not T205.
DISCUSSION
This study shows that the phosphorylation of either of the two threonines (T171 or T205) of RsbR has a positive effect on the phosphorylation rate of RsbS by RsbT and therefore is expected to have a positive effect on σB activity in vivo. It has been shown previously that RsbT, a positive regulator of σB, is trapped by the RsbR-RsbS complex unless RsbS is phosphorylated (9). The role of RsbR in the control of σB activity can therefore be seen as twofold: first, it takes part in the trapping of RsbT by forming a complex with the antagonist RsbS, and second, it exerts an inhibition on the kinase activity of RsbT towards RsbS. This inhibition, although not complete, is probably sufficient to keep RsbT trapped by the RsbR-RsbS complex (9). The phosphorylation of either of the two threonine residues of RsbR abolishes the inhibition of RsbT kinase activity. However, this study suggests that RsbT appears to phosphorylate preferentially T171 of RsbR, although predictions based on alignments of the C-terminal domain of RsbR with the three other members of the STAS family (4) of B. subtilis (SpoIIAA, RsbV, and RsbS) had suggested that T205 of RsbR is the preferred phosphorylation site, as this residue corresponds to the phosphorylatable serines of SpoIIAA (S58), RsbS (S59), and RsbV (S56). The fact that the preferred phosphorylation site is RsbR T171 contrasts with our discovery that RsbX, the known phosphatase for RsbS, dephosphorylates T205-P, and not T171-P, of RsbR. Whether this phosphatase activity is effective in vivo needs further investigation.
RsbX is overexpressed following the imposition of stress, and it plays a role in restricting the activity of σB and curtailing its activation after stress (18, 30, 33). In our study, RsbX phosphatase activity appeared to be very weak, and it would certainly need to be enhanced in vivo to play a physiological role. It has been proposed that RsbX is activated by stress signals, as the cellular levels of RsbX are insufficient to limit σB activation (28, 29). The phosphatases involved in the stress response, RsbU, RsbP, and RsbX, in addition to the related phosphatase SpoIIE, are extremely specific for their substrates and do not cross talk, despite the obvious sequence similarities between their substrates (31, 35). RsbU, RsbP, and SpoIIE are regulated by a variety of mechanisms, and perhaps RsbX is also under some means of control, the biochemical basis for which remains unknown.
Taken together, the results presented in this study suggest that phosphorylation of RsbR by a stress-dependent mechanism could be the most upstream event in the environmental stress-signaling pathway defined by the Rsb proteins. The facts that the preferred phosphorylation site for RsbT is RsbR T171 and that the dephosphorylation by RsbX occurs only on T205 of RsbR also suggest either that there is an additional, unknown protein kinase which phosphorylates T205 of RsbR upon stress or that additional components of the stress-signaling pathway modify the preference of RsbT so that it phosphorylates T205 of RsbR in the case of stress. For example, the presence of paralogs of RsbR, YkoB and YojH, in RsbR-RsbS complexes purified in vivo (C.-C. Chen, et al., unpublished data) might affect this preference and/or allow interactions with other signaling partners. For instance, Obg, an essential GTP-binding protein, has been shown to be important in σB activation by stress, and a possible interaction between Obg and RsbT has been revealed by the yeast two-hybrid system (26, 27, 31).
The results of this in vitro study suggest a biochemical mechanism by which the stress signal might enter the σB activation pathway. The early event would be the stress-dependent phosphorylation of RsbR in the RsbR-RsbS complex, which would relieve the inhibition exerted on RsbT by RsbR and allow RsbT to phosphorylate RsbS efficiently. Once RsbS is phosphorylated, RsbT could bind to and activate RsbU, the positive regulator of σB activity. Our results predict that, if RsbT is the only kinase for RsbR, then the phosphorylation of the latter should occur on the T171 residue. How RsbX, for which we have demonstrated a phosphatase activity towards T205-P of RsbR in the RsbR-RsbS complex, acts to fulfil its role as the negative feedback phosphatase is still unclear. Nevertheless, a recent independent study of the system in vivo (20) confirms our in vitro results in three respects: that the phosphorylation of RsbR is crucial for the transduction of the signal, that RsbR is phosphorylated mainly at T171, and that RsbR dephosphorylation is dependent on RsbX.
Acknowledgments
We are grateful to Joanna Clarkson and Helen Prescott for providing us with the purified SpoIIE catalytic domain and to Neil Oldham for mass spectroscopic analysis. We also thank Richard J. Lewis for useful discussions.
This work was supported by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, S., C. M. Kang, T. A. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567-578. [DOI] [PubMed] [Google Scholar]
- 3.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 4.Aravind, L., and E. V. Koonin. 2000. The STAS domain—a link between anion transporters and antisigma-factor antagonists. Curr. Biol. 10:R53-R55. [DOI] [PubMed] [Google Scholar]
- 5.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis sigma B is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, A. K., and W. G. Haldenwang. 1992. Characterization of a regulatory network that controls σB expression in Bacillus subtilis. J. Bacteriol. 174:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an α/β hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657-1669. [DOI] [PubMed] [Google Scholar]
- 10.Clarkson, J., I. D. Campbell, and M. D. Yudkin. 2003. Phosphorylation induces subtle structural changes in SpoIIAA, a key regulator of sporulation. Biochem. J. 372:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delumeau, O., R. J. Lewis, and M. D. Yudkin. 2002. Protein-protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. J. Bacteriol. 184:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaidenko, T. A., X. Yang, Y. M. Lee, and C. W. Price. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J. Mol. Biol. 288:29-39. [DOI] [PubMed] [Google Scholar]
- 15.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 16.Hecker, M., and U. Volker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, T.-J., T. A. Gaidenko, and C. W. Price. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135-150. [DOI] [PubMed] [Google Scholar]
- 21.Kovacs, H., D. Comfort, M. Lord, I. D. Campbell, and M. D. Yudkin. 1998. Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 95:5067-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucet, I., R. Borriss, and M. D. Yudkin. 1999. Purification, kinetic properties, and intracellular concentration of SpoIIE, an integral membrane protein that regulates sporulation in Bacillus subtilis. J. Bacteriol. 181:3242-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein et al. (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 25.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 26.Scott, J. M., and W. G. Haldenwang. 1999. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor σB. J. Bacteriol. 181:4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott, J. M., J. Ju, T. Mitchell, and W. G. Haldenwang. 2000. The Bacillus subtilis GTP binding protein Obg and regulators of the σB stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 182:2771-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott, J. M., T. Mitchell, and W. G. Haldenwang. 2000. Stress triggers a process that limits activation of the Bacillus subtilis stress transcription factor σB. J. Bacteriol. 182:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott, J. M., N. Smirnova, and W. G. Haldenwang. 1999. A Bacillus-specific factor is needed to trigger the stress-activated phosphatase/kinase cascade of sigmaB induction. Biochem. Biophys. Res. Commun. 257:106-110. [DOI] [PubMed] [Google Scholar]
- 30.Smirnova, N., J. Scott, U. Voelker, and W. G. Haldenwang. 1998. Isolation and characterization of Bacillus subtilis sigB operon mutations that suppress the loss of the negative regulator RsbX. J. Bacteriol. 180:3671-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trach, K., and J. A. Hoch. 1989. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171:1362-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 33.Voelker, U., T. Luo, N. Smirnova, and W. Haldenwang. 1997. Stress activation of Bacillus subtilis σB can occur in the absence of the sigma B negative regulator RsbX. J. Bacteriol. 179:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-σB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, S., and W. G. Haldenwang. 2003. RelA is a component of the nutritional stress activation pathway of the Bacillus subtilis transcription factor σB. J. Bacteriol. 185:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]