Abstract
The oligomeric organization of enzymes plays an important role in many biological processes, such as allosteric regulation, conformational stability and thermal stability. α-Glucuronidases are family 67 glycosidases that cleave the α-1,2-glycosidic bond between 4-O-methyl-d-glucuronic acid and xylose units as part of an array of hemicellulose-hydrolyzing enzymes. Currently, two crystal structures of α-glucuronidases are available, those from Geobacillus stearothermophilus (AguA) and from Cellvibrio japonicus (GlcA67A). Both enzymes are homodimeric, but surprisingly their dimeric organization is different, raising questions regarding the significance of dimerization for the enzymes' activity and stability. Structural comparison of the two enzymes suggests several elements that are responsible for the different dimerization organization. Phylogenetic analysis shows that the α-glucuronidases AguA and GlcA67A can be classified into two distinct subfamilies of bacterial α-glucuronidases, where the dimer-forming residues of each enzyme are conserved only within its own subfamily. It seems that the different dimeric forms of AguA and GlcA67A represent the two alternative dimeric organizations of these subfamilies. To study the biological significance of the dimerization in α-glucuronidases, we have constructed a monomeric form of AguA by mutating three of its interface residues (W328E, R329T, and R665N). The activity of the monomer was significantly lower than the activity of the wild-type dimeric AguA, and the optimal temperature for activity of the monomer was around 35°C, compared to 65°C of the wild-type enzyme. Nevertheless, the melting temperature of the monomeric protein, 72.9°C, was almost identical to that of the wild-type, 73.4°C. It appears that the dimerization of AguA is essential for efficient catalysis and that the dissociation into monomers results in subtle conformational changes in the structure which indirectly influence the active site region and reduce the activity. Structural and mechanistic explanations for these effects are discussed.
The oligomeric organization of proteins, and especially of enzymes, provides an additional level of complexity and plays an important role in numerous biological processes. Indeed, many enzymes are found as oligomers, and changes in enzymatic activity very commonly accompany oligomer dissociation. Such changes are part of allosteric regulation mechanisms in a number of enzymes (32), while in other enzymes and proteins, the assembly of oligomers contributes to both conformational and thermal stabilities (15, 31). One of the major driving forces for protein oligomerization originates from shape complementarity between the associating molecules, brought about by a combination of hydrophobic and polar interactions (e.g., hydrogen bonds and salts bridges) (16, 17). Another common mechanism for protein oligomerization is three-dimensional domain swapping, in which a segment of the monomeric protein is replaced by an identical segment from a second monomer (2).
The importance of protein oligomerization can be studied by comparing the catalytic and biochemical properties of the monomeric and oligomeric forms. However, the conditions required to dissociate oligomeric proteins into their monomeric subunits usually lead to inactive proteins, thus preventing the seclusion of the state of oligomerization as the only parameter measured. Another approach to examine the role of quaternary structures of proteins is to artificially create the monomers by mutating specific residues that are directly involved in intersubunit interactions within the protein oligomer. By using such an approach, it has been shown that mutations of different enzymes resulted in either active or inactive monomers (1, 24, 26). One of the most studied enzymes in this regard is triosephosphate isomerase, which usually forms a stable and tightly bound dimer (4). Several studies on triosephosphate isomerases from various sources showed that different mutations in the dimer interface lead to monomers with different levels of activity. Some of these mutants showed concentration-dependent specific activity, suggesting that the inactive monomers can associate, at least to some degree, to give an active dimer (4, 19, 27, 34).
α-Glucuronidases (EC 3.2.1.139) cleave the α-1,2-glycosidic bond between 4-O-methyl-d-glucuronic acid and xylose units in short β-1,4-xylooligomers, as part of an array of hemicellulose-hydrolyzing enzymes. In the sequence-based classification of carbohydrate-active enzymes (available on the CAZy server at http://afmb.cnrs-mrs.fr/CAZY), the different bacterial and fungal α-glucuronidases are classified as glycoside hydrolase family 67 (GH-67). The enzymes of this family were found to use a single displacement-inverting mechanism for the hydrolysis of the glycosidic bond (3). Currently, complete three-dimensional structures of only two representatives of GH-67 enzymes are available, the α-glucuronidase from Cellvibrio japonicus (formerly Pseudomonas cellulosa), GlcA67A (22, 23), and the α-glucuronidase from Geobacillus stearothermophilus, AguA (12). The overall structures of these two enzymes appear to be quite similar. Each of them contains three domains, the central of which has a (β/α)8 barrel fold that accommodates the active site. The combination of biochemical analysis of mutant enzymes and structural analysis of enzymes complexed with their substrates or products enabled the identification of three carboxylic acids as the catalytic residues of these enzymes. The two enzymes were previously reported to be homodimeric proteins in solution (21, 35), but, surprisingly, according to their crystal structures, the dimeric organization of the two enzymes seems to be different. The fact that structurally and functionally very close enzymes form different oligomeric organizations raises questions regarding the significance of the dimerization for the activity and stability of these enzymes.
In the present work, we performed a sequence and structural comparison of AguA and GlcA67A and found several elements that are responsible for the different dimeric forms of these enzymes. Based on phylogenetic analysis, it seems that bacterial α-glucuronidases can be divided into two distinct subfamilies and that the different dimeric forms of AguA and GlcA67A represent the two different dimeric forms of these subfamilies. Mutagenesis of three of the dimer-forming residues of AguA resulted in the formation of a protein that was stable as a monomer in solution. This allowed us to compare the catalytic activity and thermostability of the enzyme in its dimeric and (mutated) monomeric forms. The crystal structure of AguA in complex with its reaction products (12) provides structural explanations for the different characteristics of the dimeric and monomeric forms of this enzyme.
MATERIALS AND METHODS
Mutagenesis, protein expression, and purification.
The aguA gene (GenBank accession number AF098273) from G. stearothermophilus T-6 was cloned in the pET9d vector and overexpressed in Escherichia coli BL21(DE3). Site-directed mutagenesis was performed by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The mutagenic primers for the mutant genes were as follows (the mutated nucleotides are italicized): W328E-R329T, 5′-CTGCCAGCAGGATGAGACCGATCGGACGACC-3′ and 5′-GGTCGTCCGATCGGTCTCATCCTGCTGGCAG-3′; R665N, 5′-CGAGATGTCATCAATACCTACTTTTACAACAAGTCAGGAATAGACGACC-3′ and 5′-GGTCGTCTATTCCTGACTTGTTGTAAAAGTAGGTATTGATGACATCTCG-3′. The mutated genes were sequenced to confirm that only the desired mutations were inserted. The wild-type (WT) and mutated proteins were overexpressed and purified in a procedure that included heat treatment at 60°C for 30 min and size exclusion chromatography, as previously described (35). Protein concentrations were determined by the Bradford method, with bovine serum albumin as a standard (5).
Enzyme assay, effect of temperature on activity, and thermostability.
α-Glucuronidase activity was determined by measuring the release of 4-O-methyl-α-d-glucuronic acid from aldotetraouronic acid [2-O-α-(4-O-methyl-α-d-glucuronosyl)-xylotriose] by using the Milner and Avigad assay for uronic acids (20) as described previously (35). The effect of temperature on the reaction rate was determined by performing the standard reaction at different temperatures, ranging from 20 to 75°C, in 100 mM sodium cacodylate-HCl buffer, pH 7.0. The thermal stability was determined after incubating purified enzyme solutions in 10 mM phosphate buffer solution (pH 7.0) in the presence of 1 mg of bovine serum albumin per ml for 20 min at different temperatures. The residual activity was measured under the standard assay conditions described above at 55°C for the WT enzyme or 35°C for the mutant enzyme. All activity measurements were performed with newly made dilutions of the enzymes.
Size exclusion chromatography.
Following heat treatment, the WT and mutant AguA proteins were subjected to size exclusion chromatography on a Superdex 200 26/60 gel filtration column (AKTA explorer; Pharmacia). Protein samples (10-ml) were applied on the column and eluted at room temperature with 50 mM Tris-Cl buffer (pH 7.0), 100 mM NaCl, and 0.02% sodium azide, running at 2.5 ml/min. Molecular weights were determined from regression analysis of the log relative molecular weight (Mr) of protein standards as a function of Kav. Kav, the available partition coefficient, is defined as (Ve − Vo)/Vt−Vo), where Ve is the elution volume of the protein from the column, Vo is the void number of the column, and Vt is the total column volume. The protein standards used were α-arabinofuranosidase (Mr, 343,308), β-xylosidase1 (Mr, 232,004), β-xylosidase2 (Mr, 159,788), xylanase (Mr, 43,808) and intracellular xylanase (Mr, 39,357), all from G. stearothermophilus T-6, and the cellobiohydrolase CelS (Mr, 80,796) from Clostridium thermocellum.
DSC.
The melting temperatures of the proteins were determined by using differential scanning calorimetry (DSC) (VP-DSC MicroCalorimeter; MicroCal Inc., Northampton, Mass.). Protein samples (0.5 mg/ml) were extensively dialyzed against 100 mM NaCl-50 mM Tris-Cl buffer at pH 7.0, and the actual dialysis buffer was used as the reference solution for the DSC scan. Samples were analyzed at a heating rate of 1°C/min over a temperature range of 50 to 90°C. A scan with a buffer solution in both cells was subtracted from each data set, and the molar heat capacity, Cp, was obtained. The melting temperature was defined as the temperature at the maximum molar heat capacity. Analysis of calorimetric data was carried out with Origin 5.0 (MicroCal).
RESULTS
Sequence and structural analyses of GH-67 members.
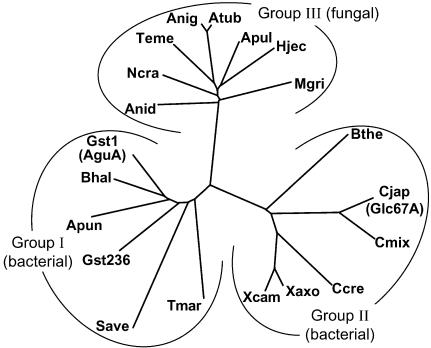
Currently, there are 20 different complete sequences of bacterial and fungal α-glucuronidases classified within the GH-67 family. The phylogenetic tree of these enzymes is presented in Fig. 1, and their partial sequence alignments are shown in Fig. 2. The phylogenetic analysis shows that GH-67 α-glucuronidases can be divided into three subfamilies: groups I and II are bacterial proteins and group III proteins are fungal. Currently, no archaeal, plant, or higher-animal α-glucuronidase genes are known. The fact that no plant α-glucuronidases have been found to date is especially surprising, since the substrate 4-O-methyl-d-glucurono-xylooligomer is part of xylan, the most abundant hemicellulose in the plant cell wall. In addition, plant genomes were found to contain significantly more glycosidase and glycosyltransferase-related genes than any organism sequenced to date, explained mainly by the complex structure of the plant cell wall (8). On the other hand, ancient plants do not contain 4-O-methyl-d-glucuronoxylan, which may imply that α-glucuronidases have evolved relatively recently, as was suggested by Nagy et al. (21). This explanation may account for the lack of α-glucuronidase genes in the currently available plant sequences. Among the bacterial α-glucuronidases, there is no obvious correlation between the two subfamilies and the bacterial 16S ribosomal RNA phylogenetic tree (7). Group I α-glucuronidases includes gram-positive bacteria (Bacillus, Geobacillus, and Streptomyces), gram-negative Proteobacteria (Aeromonas), and hyperthermophiles (Thermotoga). Group II includes mainly Proteobacteria (Cellvibrio, Xanthomonas, and Caulobacter) but also Bacteroides.
FIG. 1.
The phylogenetic tree of the GH-67 α-glucuronidases. The abbreviations for the α-glucuronidases are as follows (GenBank accession numbers of the proteins are in parentheses): GstT1 (or AguA), G. stearothermophilus T-1 (AAL32057); Gst236, G. stearothermophilus 236 (AAG09715); Bhal, Bacillus halodurans C-125 (BAB04780); Apun, Aeromonas punctata (BAA74508); Tmar, Thermotoga maritima (AAD35149); Save, Streptomyces avermitilis MA-4680 (BAC69160); Cjap (GlcA67A), Cellvibrio japonicus (AAL57752); Cmix, Cellvibrio mixtus (AAL57753); Xaxo, Xanthomonas axonopodis (AAM39062); Xcam, Xanthomonas campestris (AAM43323); Ccre, Caulobacter crescentus CB15 (AAK24775); Bthe, Bacteroides thetaiotaomicron VPI-5482 (AAO77729); Anig, Aspergillus niger CBS 120.49 (CAC38119); Atub, Aspergillus tubingensis NW756 (CAA75605); Apul, Aureobasidium pullulans (AAR87862); Teme, Talaromyces emersonii (AAL33576); Anid, Aspergillus nidulans FGSC A4 (EAA66353); Ncra, Neurospora crassa OR74A (EAA29095); Hjec, Hypocrea jecorina RutC-30 (CAA92949); Mgri, Magnaporthe grisea 70-15 (EAA53369). The tree was constructed by the GrowTree program of the GCG Wisconsin package (Accelrys Inc.) and visualized with the program TreeView (25).
FIG.2.
Partial amino acid sequence alignments of the GH-67 α-glucuronidases. The α-glucuronidases are the same as shown in Fig. 1. The alignments were generated with the PRETTY program of the GCG Wisconsin package (Accelrys Inc.), with plurality of 18 out of 20 for the consensus sequence (shown in uppercase letters). The sequences of the three subfamilies are indicated. Only the regions that are involved in the dimerization of either AguA or GlcA67A are shown, and the number of omitted residues in representative sequences of each of the subfamilies is shown. Residues involved in the dimerization of AguA or GlcA67A are highlighted in boxes, with the residue numbers of the corresponding sequence shown above them. The three catalytic residues (E285, D364, and E392 in AguA) are in bold.
The differences in the amino acid sequences of the three subfamilies are clustered mainly in the N- and C-terminal ends of the proteins. The middle part of the alignments show a higher degree of similarity between the different proteins, in agreement with the location of the active site within this domain. The three subfamilies differ also in the lengths of their amino acid sequences. On average, group II proteins are longer at the C terminus by about 22 residues compared to group I proteins. The fungal α-glucuronidases (group III) are much longer than the bacterial proteins, containing about 110 additional residues at the C terminus.
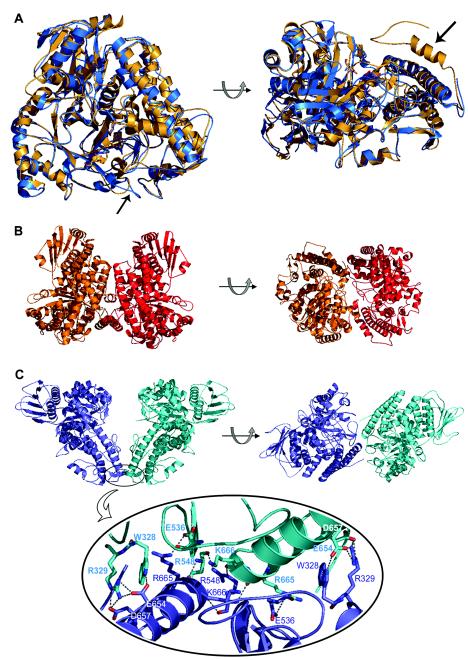
The two crystal structures of α-glucuronidases which are currently available represent the two bacterial subfamilies: the α-glucuronidase from G. stearothermophilus (AguA) belongs to group I, whereas the α-glucuronidase from C. japonicus (GlcA67A) belongs to group II. The two proteins share sequence identity of 42% and sequence similarity of 59% out of 665 aligned residues. A superposition of the AguA and GlcA67A structures is shown in Fig. 3A . In addition to the relatively high similarity at the primary structural level, considerable similarity is observed also for the tertiary structures of the two proteins. As expected, the highest structural homology between the two structures is at the central (β/α)8 domain, which contains the active site. The root mean square deviation between the two structures is 1.02 Å for all Cα atoms and 0.7 Å between Cα atoms of only the (β/α)8 domains (structural alignments were performed by using the K2 server) (29). The longer C-terminal polypeptide chain of GlcA67A is located on the outer side of the enzyme monomer, interacting with other residues of the C-terminal domain, as well as with residues of the middle (β/α)8 domain.
FIG.3.
(A) Superposition of the crystal structures of the α-glucuronidases AguA from G. stearothermophilus (blue) and GlcA67A from C. japonicus (gold), shown in two orientations. The two arrows point to the structural elements that are proposed to be responsible for the differences in the oligomerization of the two enzymes: the loop containing residues 323 to 329 in AguA (left) and the additional 29-residue polypeptide at the C terminus of GlcA67A (right). Superimposition was performed by the K2 server (29). (B) Two views of the GlcA67A dimer related by a 90° rotation (protein data bank code 1GQI). (C) Two views of the suggested AguA dimer related by a 90° rotation (protein data bank code 1L8N) (top). The lower part of the panel is an enlargement of the dimer interface region of AguA, showing the hydrogen bonds between the two subunits. The figures were generated by PyMOL (10).
As previously described, both AguA and GlcA67A exist as homodimers in solution (21, 35). Indeed, in the crystal structure of GlcA67A, a dimer was found in the crystallographic asymmetric unit (Fig. 3B) (23). In the crystal structure of AguA, however, only one molecule of the enzyme was found in the crystallographic asymmetric unit under the crystallization conditions used (12, 30). Generation of all the symmetry-related molecules in the crystallographic unit cell of AguA revealed one dimeric form which seems to be the most biologically relevant (12). The interface region of this form includes W328 and R329, located on a loop in the (β/α)8 domain, and E536, R548, E654, D657, R665, and K666, located in the C-terminal domain (Fig. 3C). The assignment of this dimerization contact is supported by mutagenesis and biochemical studies described below. Two more possible dimers were generated from symmetry-related molecules in the crystal structure of AguA (data not shown), but in both of them significantly fewer conserved residues are involved compared to the dimeric organization described above. It should be noted that none of the three crytallographically possible dimers of AguA is similar to the reported dimer organization of GlcA67A (23).
The family GH-67 sequence alignment reveals a very good correlation between the different dimerization points suggested for AguA and GlcA67A and the conservation pattern of these residues in the different GH-67 subfamilies (Fig. 2). For example, most of the dimer-forming residues of AguA, such as W328, R329, E536, E654, and R665, are completely conserved within group I enzymes but are not conserved in group II enzymes. Residues W328 and R329 in AguA are located on a loop that in the GlcA67A structure is shorter and has a different conformation (Fig. 3A). Similarly, several of the dimer-forming residues of GlcA67A, such as L163, Q220, R451, and P701, are completely (or almost completely) conserved within group II α-glucuronidases but are significantly less conserved in group I sequences. The last residue, P701, is located on the long polypeptide chain at the C terminus of GlcA67A that is absent in the group I enzymes.
It is interesting that many fungal α-glucuronidases were found to be monomeric proteins (11, 18, 33). Indeed, the residues found in the dimerization positions of both groups I and II enzymes are no longer conserved in the group III fungal α-glucuronidases, in correlation with their observed monomeric characteristics.
Construction of a monomeric form of AguA.
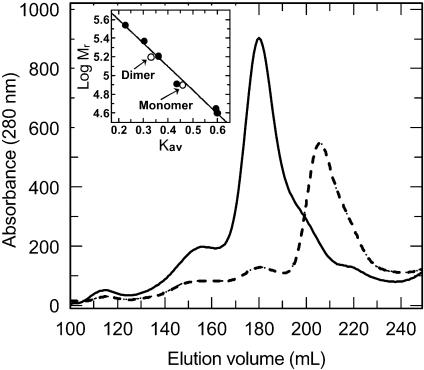
Site-directed mutagenesis was performed to test whether the suggested dimeric structure of AguA is the biologically relevant dimer of the WT enzyme in solution. Three of the interface-forming residues, W328, R329, and R665, which are conserved only within group I sequences, were mutated into the corresponding residues in the sequences of the fungal enzymes. Two mutant AguA proteins were constructed: a double mutant W328E-R329T and a triple mutant W328E-R329T-R665N. The mutant enzymes were overexpressed and purified similarly to the WT enzyme, in a procedure that included heat treatment (60°C for 30 min) and gel filtration chromatography (35). According to sodium dodecyl sulfate-polyacrylamide gel electrophoresis analyses that followed the purification steps, both mutants had Mrs of about 78.5 × 103, in agreement with their calculated Mr of 78,381 and 78,339 for the double and triple mutants, respectively. It seemed that only small amounts of the proteins precipitated after the heat treatments (results not shown); thus, the mutants retained the thermostability of the WT protein. The Mrs of the native double and triple mutant AguA proteins in solution were estimated by gel filtration chromatography calibrated by using protein markers of known molecular mass. Both mutants eluted from the column at different elution volumes than the WT AguA (Fig. 4). The elution volume of the major peaks of the mutants corresponded to proteins with a Mr of about 88 × 103, whereas the elution volume of the WT AguA corresponded to a protein with a Mr of about 180 × 103. About 10% of the double mutant protein and 5% of the triple mutant protein eluted in elution volumes similar to that of the WT protein. Further characterization therefore was performed with the triple mutant W328E-R329T-R665N. The calculated Mr of the WT dimeric enzyme is 156,986, and it hence appears that the three replacements, W328E, R329T, and R665N, were sufficient to transform almost all of the AguA molecules from their natural dimeric form to a monomeric form. These results confirm that the dimeric crystal structure which was suggested previously (12) and is presented in Fig. 3C is the true biological dimeric structure of the enzyme also in solution. Crystallization experiments for the monomeric AguA are now under way.
FIG. 4.
Gel filtration chromatograms (overlaid) of the WT dimeric AguA (solid line) and the W328E-R329T-R665N mutant AguA (broken line). The inset shows the linear regression of the protein standards used for Mr determination (filled circles) and the position of the dimeric and monomeric forms of AguA on this curve (open circles).
Catalytic and biophysical properties of the monomeric AguA.
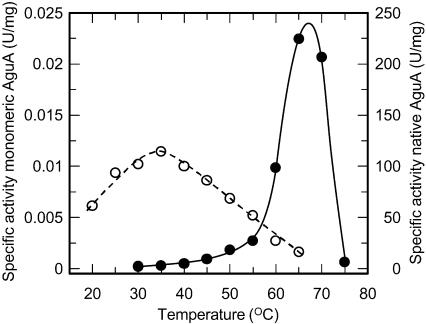
The catalytic activities of the two mutant monomeric AguA proteins W328E-R329T and W328E-R329T-R665N were similar and were significantly lower than the catalytic activity of the WT dimeric enzyme. From the temperature-dependent activity profile of the triple mutant, it seems that the optimal temperature for activity of the monomeric form had shifted to around 35°C, compared to 65°C of the WT enzyme (Fig. 5). The highest activity of the monomeric AguA (at 35°C) was 240-fold lower than the WT activity at this temperature and 2 × 104-fold lower than the maximal activity of the WT activity at 65°C.
FIG. 5.
The effect of temperature on the catalytic activity of WT AguA (filled circles and solid line) and the W328E-R329T-R665N mutant (monomeric) AguA (open circles and broken line). The results are averages of at least three independent measurements.
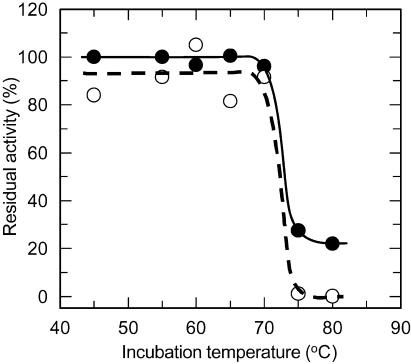
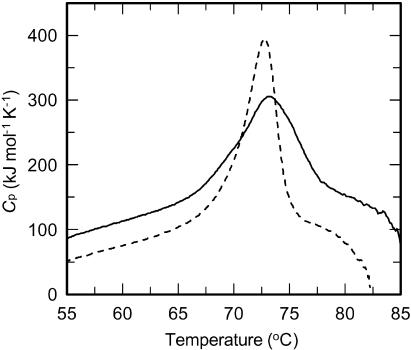
To examine whether the lower optimal temperature of the monomeric AguA results from reduced stability of the protein at high temperatures, the thermostability of the monomeric AguA was compared to that of the WT enzyme. As described above, during the purification procedure only a small amount of monomeric AguA precipitated after the heat treatment of 60°C for 30 min, indicating that most of the protein did not undergo denaturation at this temperature. The residual activity profile of the monomeric enzyme after a 20-min incubation at different temperatures was very similar to that of the WT enzyme, and both forms retained about 90% of their activity even after incubation at 70°C (Fig. 6). After incubation at 75°C, a significant decrease in the residual activity was observed for both forms, but while the WT AguA retained about 30% of its activity, the monomeric form was completely inactivated. The melting temperatures of the two proteins were determined by DSC and were also found to be similar. The melting temperature of the monomeric AguA was 72.9°C, and that of the WT dimeric form was 73.4°C (Fig. 7). Altogether, these results demonstrate that the thermostability of the protein was not reduced as a result of the three mutations and the resulting dissociation of the AguA dimer into monomers.
FIG. 6.
Thermostability of the WT (dimeric) AguA (filled circles and solid line) and its W328E-R329T-R665N mutant (monomeric) form (open circles and broken line). The enzymes were incubated for 20 min at the given temperatures and cooled to room temperature, and the residual activities were measured at 55°C for the WT enzyme and 35°C for the mutant enzyme. The residual activity is shown as a percentage of the maximal activity of each form of the enzyme.
FIG. 7.
Melting temperature of WT AguA (solid line) and the W328E-R329T-R665N mutant (monomeric) AguA (broken line). Heat capacity curves were obtained by DSC. The scans were performed at heating rates of 1°C/min. The melting temperature was defined as the temperature at the maximum molar heat capacity, Cp.
As mentioned above, a small fraction of the triple mutant AguA eluted in an elution volume identical to that of the dimeric WT enzyme (Fig. 4). This raises the possibility that monomer-dimer equilibrium takes place, where the monomer is catalytically inactive and the observed activity reflects a small fraction of active dimer. Such dimerization-dependent activity profiles have been demonstrated for other enzymes (19, 24). In these cases, since the concentration of the enzyme influences the equilibrium between monomers and oligomers, time-dependent inactivation upon enzyme dilution, as well as concentration-dependent specific activities, are expected (32). In the case of AguA, however, the specific activity of the monomeric mutant at 30°C was independent of the initial sample concentration, and the same specific activity was measured when the enzyme was diluted 5- to 1,000-fold, 5 h prior to the assay. In addition, the activity was not time dependent, and following a 100-fold dilution, the specific activity was practically constant after periods of 1 min to 20 h (data not shown). These results suggest that even if there is equilibrium between monomeric and dimeric forms of the mutant AguA, its rate is very slow. Since all activity measurements were performed with newly made dilutions of the enzymes and were carried out in time scales of up to a few hours, it seems that the activity measured for the monomeric mutant is its intrinsic activity and not the residual activity of a small fraction of the dimer.
DISCUSSION
The sequence-based classification of glycosidases into families demonstrates that enzymes grouped within one family share the same stereochemical course of the catalytic mechanism (retaining or inverting), the same identity and location of the catalytic residues in the active site, and the same overall structure (13, 14). Indeed, the two structures of GH-67 α-glucuronidases discussed here, AguA from G. stearothermophilus and GlcA67A from C. japonicus, show high structural similarity, especially in the active site and its surroundings (12, 23). Surprisingly, the organization of the two monomeric subunits into a functional homodimer appears to be completely different in each of these enzymes. In this regard, GlcA67A was reported to be an extracellular cell-associated (21) enzyme, whereas AguA was found to be an intracellular enzyme (28).
Two structural elements seem to be mainly responsible for the difference in the oligomerizations of the two enzymes (Fig. 3A). In AguA, residues W328 and R329, which play an important role in the dimerization, are located on a loop that is much longer than the corresponding loop of GlcA67A. This difference in length seems to be critical for the ability of these two residues to form efficient intermolecular contacts. Probably more important is the fact that in GlcA67A the additional polypeptide at the C terminus (29 residues) is located exactly on the surface of the molecule whereas the corresponding AguA monomer forms the dimeric intermolecular contacts. This extra polypeptide, which is absent in AguA, is therefore a major factor in the dimer organization of the two enzymes, since in the case of GlcA67A it prevents monomer-monomer interactions similar to those observed in AguA. The phylogenetic analysis of all GH-67 sequences (Fig. 2) suggests that these two dimeric forms may represent two possibilities of dimerization for bacterial α-glucuronidases. AguA and its homologous enzymes (group I) adopt one dimeric form, while GlcA67A and its homologous enzymes (group II) adopt another form. Obviously, additional bacterial α-glucuronidases should be structurally examined to validate the generality of such an explanation. It is possible also that in fungal α-glucuronidases, the much longer extra polypeptide chains at their C termini (∼110 residues) completely prevent the formation of dimers of the types observed in either AguA or GlcA67A and stabilize the monomers in solution. Such an arrangement is consistent with the reported monomeric forms of the fungal α-glucuronidases obtained by biochemical studies (11, 18, 33). It is noted that whereas all known bacterial α-glucuronidases are intracellular or cell associated, the fungal α-glucuronidases are extracellular. Thus, the monomeric forms of the fungal enzymes might be a consequence of their physiological role as extracellular enzymes.
The construction of the triple mutant AguA, which appears to be mostly monomeric, allowed us to evaluate the significance of the dimerization for the catalytic activity and stability of AguA. In the suggested dimeric structure of AguA, there seems to be only one major hydrophobic interaction between the two subunits, involving W328 and V658. In addition, the hydrophobicity of the suggested interface is not larger than the hydrophobicity of the rest of the protein (data not shown). On the other hand, there are eight amino acid residues from each AguA monomer that could participate in the dimerization by forming altogether 18 possible intersubunit hydrogen bonds. The replacements of W328, R329, and R665 would remove 12 of these potential interactions (Fig. 3C). As evident from the results of size-exclusion chromatography, the combination of the three mutations (W328E, R329T, and R665N) leads to an almost total dissociation of the original AguA WT dimer into monomers, confirming the central role of these residues for dimer formation.
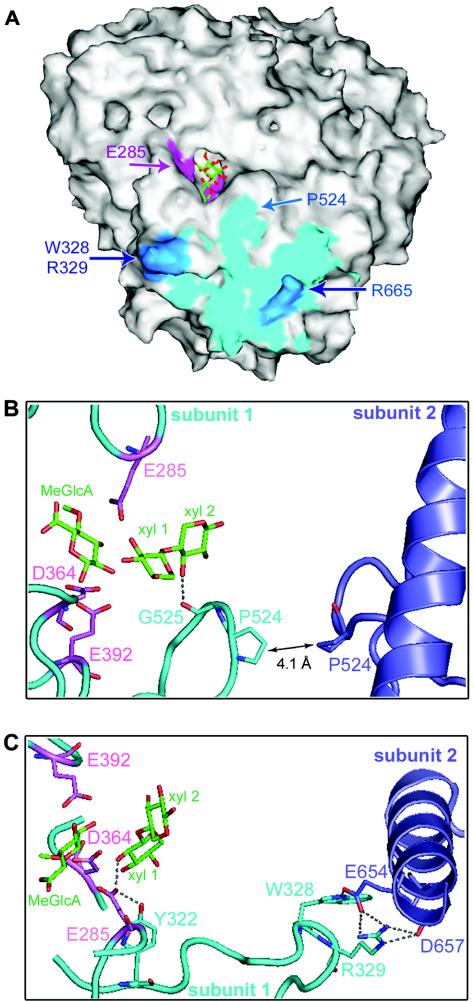
The biochemical characterization of the monomeric AguA revealed a puzzling picture: whereas the optimal temperature for activity was reduced from 65°C for the WT enzyme to 35°C for the monomeric enzyme, the thermostability of the enzyme was not changed as a result of the dimer dissociation (as judged by an almost identical melting temperature) (Fig. 7). Since the catalytic activity of the monomeric protein was measurable and showed temperature dependency, it seems that the overall three-dimensional structure of the enzyme remained generally unchanged. In addition, since the activity of the monomeric AguA was not concentration dependent, it appears that the measured activity is indeed the activity of the mutant form and not an activity of a small fraction of the mutant that was still capable of forming dimers. Thus, the reduced activity of the monomeric AguA resulted not from lower thermostability but probably from minute structural changes that affected the active site itself. The most pronounced influence of the three mutations is the dissociation of the dimeric structure into monomers. As a result, residues that were hidden in the interface between monomers became exposed to the solvent. Such exposure to the solvent could change the capability of these surface residues to form hydrogen bonds and electrostatic and hydrophobic interactions. Evidently, these changes were not significant enough to disrupt the protein structure in a way that would change its melting temperature. Figure 8A shows the dimer contact region on the AguA monomer surface. Of all the residues that are in proximity to residues from the other subunit (in a distance of less than 5 Å), P524 is the closest residue to the active site (Fig. 8B). The backbone carbon atom of P524 is 6 Å away from the xylose unit of the substrate in the +2 subsite (subsite nomenclature according to Davies et al. [9]). The exposure of P524 to the solvent could influence the activity also through the adjacent G525, whose backbone oxygen atom is an acceptor of a hydrogen bond (2.6 Å) from one of the hydroxyls of the xylose at the +2 subsite. Thus, the dimer dissociation could affect the binding of the substrate by changing this interaction. Interestingly, P524 shows the same sequence conservation pattern as the dimer-forming residues of AguA. All group I α-glucuronidases have proline in the homologue position, while other residues are found in the homologous position of the sequences of α-glucuronidases from groups II and III.
FIG. 8.
The structural elements suggested to be responsible for the reduced activity and lower optimal temperature of the monomeric AguA. (A) The solvent-accessible surface of one monomer of the dimeric AguA in complex with its reaction products. The surface of important residues is colored as follows: interface residues that are in distance of <5 Å from the second monomer are cyan; the interface residues that were mutated to form the monomeric AguA are blue; the catalytic residues are magenta. The products are shown in stick representation in green (carbons) and red (oxygens). The location of P524, which is the interface residue closest to the active site (∼6 Å), is indicated by an arrow. The figure was generated by using SPOCK (6). (B) Interactions around P524 that could potentially affect the active site of AguA upon dimer dissociation. One subunit is shown in cyan and the other subunit is in blue; potential hydrogen bonds (less than 3.5 Å long) are shown as dotted lines; the three catalytic residues of the first subunit are shown in pink; and the bound reaction products, 4-O-methyl-d-glucuronic acid (MeGlcA) and xylobiose (xyl 1 and xyl2), are shown in green. In the dimer structure, P524 of one subunit is 4.1 Å from P524 of the second subunit. The adjacent G525 can form a hydrogen bond of 2.6 Å with the 3-OH of xyl 2. (C) The interactions around W328-R329 that could affect the active site of AguA upon dimer dissociation. The color coding is the same as in panel B. W328 and R329 are located on the same loop as Y322, which can form a hydrogen bond of 2.6 Å with the catalytic acid E285. The loop on which E285 is located was previously found to be flexible, and can adopt a closed active conformation, or an open inactive one. Thus, the removal of W328 and R329 and the dimer dissociation could alter the important hydrogen bond between Y322 and E285 and consequently affect the stability of the flexible loop.
Residues W328 and R329 that were mutated to form the monomeric AguA are about 15 Å from the general acid catalytic residue E285. Such distance is definitely too long to produce a direct influence on the catalytic activity. Nevertheless, these residues are located on the same loop as Y322, a completely conserved residue that forms a hydrogen bond 2.6 Å long with the catalytic residue E285 (Fig. 8C). The loop carrying this catalytic residue (residues 283 to 287) was previously found to be flexible in the structures of AguA: it was not resolved in some of the enzyme structures, but it was visualized in its “open” (inactive) and “closed” (catalytic) conformations in other structures of AguA (12). It is possible, therefore, that the replacement of W328 and R329 altered the hydrogen-bonding network involving residues around E285, making the loop at residues 283 to 287 even more flexible. Assuming such increased flexibility in the AguA mutant, its higher activity at lower temperatures could be attributed to the slightly higher rigidity of this critical loop at lower temperatures.
In conclusion, our results indicate that the phylogenetic and biochemical analyses of family 67 α-glucuronidases are in good agreement with the structural data. Taken together, it appears that bacterial α-glucuronidases can have two different dimeric organizations, one for each of the subfamilies. The evolutionary conservation of the residues involved in the dimerization of each of the subfamilies suggests that the dimeric structure is important for the catalytic activities of these enzymes. From the crystal structure of the dimeric form of AguA, it seems that the dimerization itself does not have a direct role in catalysis since the active site is located far from the dimer interface. However, as can be seen from the biochemical comparison of the dimeric and monomeric forms of AguA, the assembly of the dimer is essential for efficient catalytic activity, in agreement with the phylogenetic analysis. It appears that the complex network of hydrogen bonds surrounding the active site is affected by the exposure of the dimer interface surface to the solvent. Currently, there are no available three-dimensional structures for fungal α-glucuronidases, which were found to function as monomers. Once a structure of one of them is available, it would be interesting to see how the extra C-terminal segment is arranged as to allow structural stability and efficient catalysis in monomeric α-glucuronidases.
Acknowledgments
The authors thank N. Adir for critical reading of the manuscript.
This study was supported by grants from the German-Israeli-Foundation for Scientific Research and Development (to Y.S. and G.S.), from the French-Israeli Association for Scientific and Technological Research (to Y.S.), and from the Israel Science Foundation (to G.S. and Y.S.). G.G. was supported by the Chorafas Fund. Additional support was provided by the Otto Meyerhof Center for Biotechnology, Technion, established by the Minerva Foundation (Munich, Germany).
REFERENCES
- 1.Beernink, P. T., and D. R. Tolan. 1996. Disruption of the aldolase A tetramer into catalytically active monomers. Proc. Natl. Acad. Sci. USA 93:5374-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, M. J., M. P. Schlunegger, and D. Eisenberg. 1995. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 4:2455-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biely, P., R. P. de Vries, M. Vrsanska, and J. Visser. 2000. Inverting character of alpha-glucuronidase A from Aspergillus tubingensis. Biochim. Biophys. Acta 1474:360-364. [DOI] [PubMed] [Google Scholar]
- 4.Borchert, T. V., J. P. Zeelen, W. Schliebs, M. Callens, W. Minke, R. Jaenicke, and R. K. Wierenga. 1995. An interface point-mutation variant of triosephosphate isomerase is compactly folded and monomeric at low protein concentrations. FEBS Lett. 367:315-318. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-252. [DOI] [PubMed] [Google Scholar]
- 6.Christopher, J. A. 1998. SPOCK, the structural properties observation and calculation kit. Center for Macromolecular Design, Texas A&M University, College Station, Tex.
- 7.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutinho, P. M., M. Stam, E. Blanc, and B. Henrissat. 2003. Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci. 8:563-565. [DOI] [PubMed] [Google Scholar]
- 9.Davies, G. J., K. S. Wilson, and B. Henrissat. 1997. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321:557-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, Calif.
- 11.de Vries, R. P., C. H. Poulsen, S. Madrid, and J. Visser. 1998. aguA, the gene encoding an extracellular α-glucuronidase from Aspergillus tubingensis, is specifically induced on xylose and not on glucuronic acid. J. Bacteriol. 180:243-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golan, G., D. Shallom, A. Teplitsky, G. Zaide, S. Shulami, T. Baasov, V. Stojanoff, A. Thompson, Y. Shoham, and G. Shoham. 2004. Crystal structures of Geobacillus stearothermophilus α-glucuronidase complexed with its substrate and products: mechanistic implications. J. Biol. Chem. 279:3014-3024. [DOI] [PubMed] [Google Scholar]
- 13.Henrissat, B., and G. Davies. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637-644. [DOI] [PubMed] [Google Scholar]
- 14.Henrissat, B., and G. J. Davies. 2000. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124:1515-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaenicke, R. 1999. Stability and folding of domain proteins. Prog. Biophys. Mol. Biol. 71:155-241. [DOI] [PubMed] [Google Scholar]
- 16.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, S., and J. M. Thornton. 1995. Protein-protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 63:31-65. [DOI] [PubMed] [Google Scholar]
- 18.Khandke, K. M., P. J. Vithayathil, and S. K. Murthy. 1989. Purification and characterization of an alpha-d-glucuronidase from a thermophilic fungus, Thermoascus aurantiacus. Arch. Biochem. Biophys. 274:511-517. [DOI] [PubMed] [Google Scholar]
- 19.Mainfroid, V., P. Terpstra, M. Beauregard, J. M. Frere, S. C. Mande, W. G. Hol, J. A. Martial, and K. Goraj. 1996. Three hTIM mutants that provide new insights on why TIM is a dimer. J. Mol. Biol. 257:441-456. [DOI] [PubMed] [Google Scholar]
- 20.Milner, Y., and G. Avigad. 1967. A copper reagent for the determination of hexuronic acids and certain ketohexoses. Carbohyd. Res. 4:359-361. [Google Scholar]
- 21.Nagy, T., K. Emami, C. M. Fontes, L. M. Ferreira, D. R. Humphry, and H. J. Gilbert. 2002. The membrane-bound α-glucuronidase from Pseudomonas cellulosa hydrolyzes 4-O-methyl-d-glucuronoxylooligosaccharides but not 4-O-methyl-d-glucuronoxylan. J. Bacteriol. 184:4925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy, T., D. Nurizzo, G. J. Davies, P. Biely, J. H. Lakey, D. N. Bolam, and H. J. Gilbert. 2003. The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of aldobiouronic acid as important substrate recognition determinants. J. Biol. Chem. 278:20286-20292. [DOI] [PubMed] [Google Scholar]
- 23.Nurizzo, D., T. Nagy, H. J. Gilbert, and G. J. Davies. 2002. The structural basis for catalysis and specificity of the Pseudomonas cellulosa alpha-glucuronidase, GlcA67A. Structure (Cambridge) 10:547-556. [DOI] [PubMed] [Google Scholar]
- 24.Ottosen, M. B., O. Bjornberg, S. Norager, S. Larsen, B. A. Palfey, and K. F. Jensen. 2002. The dimeric dihydroorotate dehydrogenase A from Lactococcus lactis dissociates reversibly into inactive monomers. Protein Sci. 11:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Zavala, J. S., and H. Weiner. 2002. Structural aspects of aldehyde dehydrogenase that influence dimer-tetramer formation. Biochemistry 41:8229-8237. [DOI] [PubMed] [Google Scholar]
- 27.Schliebs, W., N. Thanki, R. Jaenicke, and R. K. Wierenga. 1997. A double mutation at the tip of the dimer interface loop of triosephosphate isomerase generates active monomers with reduced stability. Biochemistry 36:9655-9662. [DOI] [PubMed] [Google Scholar]
- 28.Shulami, S., O. Gat, A. L. Sonenshein, and Y. Shoham. 1999. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 181:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szustakowski, J. D., and Z. Weng. 2000. Protein structure alignment using a genetic algorithm. Proteins 38:428-440. [DOI] [PubMed] [Google Scholar]
- 30.Teplitsky, A., S. Shulami, S. Moryles, G. Zaide, Y. Shoham, and G. Shoham. 1999. Crystallization and preliminary X-ray analysis of alpha-d-glucuronidase from Bacillus stearothermophilus T-6. Acta Crystallogr. Sect. D 55:869-872. [DOI] [PubMed] [Google Scholar]
- 31.Torshin, I. Y. 2002. Functional maps of the junctions between interglobular contacts and active sites in glycolytic enzymes: a comparative analysis of the biochemical and structural data. Med. Sci. Monit. 8:123-135. [PubMed] [Google Scholar]
- 32.Traut, T. W. 1994. Dissociation of enzyme oligomers: a mechanism for allosteric regulation. Crit. Rev. Biochem. Mol. Biol. 29:125-163. [DOI] [PubMed] [Google Scholar]
- 33.Uchida, H., T. Nanri, Y. Kawabata, I. Kusakabe, and K. Murakami. 1992. Purification and characterization of intracellular α-glucuronidase from Aspergillus niger 5-16. Biosci. Biotech. Biochem. 56:1608-1615. [Google Scholar]
- 34.Walden, H., G. S. Bell, R. J. Russell, B. Siebers, R. Hensel, and G. L. Taylor. 2001. Tiny TIM: a small, tetrameric, hyperthermostable triosephosphate isomerase. J. Mol. Biol. 306:745-757. [DOI] [PubMed] [Google Scholar]
- 35.Zaide, G., D. Shallom, S. Shulami, G. Zolotnitsky, G. Golan, T. Baasov, G. Shoham, and Y. Shoham. 2001. Biochemical characterization and identification of catalytic residues in alpha-glucuronidase from Bacillus stearothermophilus T-6. Eur. J. Biochem. 268:3006-3016. [DOI] [PubMed] [Google Scholar]