Abstract
In a process called quorum sensing, bacteria communicate using extracellular signal molecules termed autoinducers. Two parallel quorum-sensing systems have been identified in the marine bacterium Vibrio harveyi. System 1 consists of the LuxM-dependent autoinducer HAI-1 and the HAI-1 sensor, LuxN. System 2 consists of the LuxS-dependent autoinducer AI-2 and the AI-2 detector, LuxPQ. The related bacterium, Vibrio cholerae, a human pathogen, possesses System 2 (LuxS, AI-2, and LuxPQ) but does not have obvious homologues of V. harveyi System 1. Rather, System 1 of V. cholerae is made up of the CqsA-dependent autoinducer CAI-1 and a sensor called CqsS. Using a V. cholerae CAI-1 reporter strain we show that many other marine bacteria, including V. harveyi, produce CAI-1 activity. Genetic analysis of V. harveyi reveals cqsA and cqsS, and phenotypic analysis of V. harveyi cqsA and cqsS mutants shows that these functions comprise a third V. harveyi quorum-sensing system that acts in parallel to Systems 1 and 2. Together these communication systems act as a three-way coincidence detector in the regulation of a variety of genes, including those responsible for bioluminescence, type III secretion, and metalloprotease production.
Many bacteria coordinate gene expression using a process of cell-cell communication termed quorum sensing. Quorum sensing involves the production of, detection of, and response to extracellular signaling molecules known as autoinducers. Quorum-sensing systems allow bacteria to monitor their population density and to induce or repress particular genes in response to changes in cell number. Specifically, as a bacterial population grows, the extracellular concentration of autoinducer increases. When a critical autoinducer concentration is achieved, the bacteria respond as a group to coordinately alter target gene expression. Quorum-sensing systems have been identified in both gram-negative and gram-positive species of bacteria, and they are used to regulate diverse functions, such as bioluminescence, conjugation, virulence, biofilm formation, and antibiotic production (reviewed in reference 24).
Vibrio harveyi is a bioluminescent marine bacterium that lives in several milieus: it can exist free swimming in seawater, adhered to abiotic surfaces, as a constituent of biofilm consortia in marine animals, and in pathogenic associations with marine hosts (28). Two quorum-sensing systems have been identified in V. harveyi, and together they control functions such as bioluminescence (2, 3), type III secretion (TTS) (18), and siderophore (21), polysaccharide (21), and metalloprotease production (26) (Fig. 1A). The System 1 autoinducer, N-(3-hydroxybutanoyl) homoserine lactone (HSL) (denoted HAI-1 for harveyi autoinducer 1), is produced by LuxM and is detected by LuxN (2, 6). The System 2 autoinducer, called AI-2,is 3A-methyl-5,6-dihydro-furo(2,3-d)(1,3,2)dioxaborole-2,2,6,6A-tetraol, and the unborated precursor is made by LuxS (7, 31, 35). Two proteins, LuxP and LuxQ, function together as the AI-2 sensor (3). LuxP is similar to periplasmic ribose binding proteins. Both LuxN and LuxQ are hybrid two-component proteins that contain sensor kinase and response regulator domains. The remainder of the signaling cascade is shared by System 1 and System 2 (Fig. 1A).
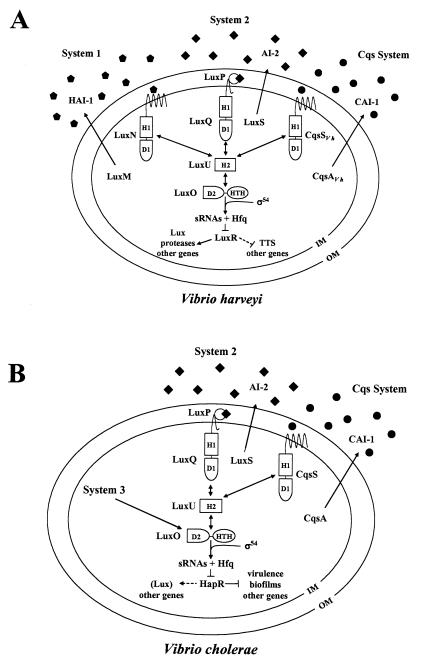
FIG. 1.
Models of the V. harveyi and V. cholerae quorum-sensing systems. (A) V. harveyi has three parallel quorum-sensing systems that regulate the genes encoding bioluminescence (Lux), TTS, a secreted metalloprotease (VhpA), and other quorum-sensing-activated and -repressed genes. (B) V. cholerae possesses the CAI-1-CqsS system and the AI-2-LuxPQ system. V. cholerae also has an unidentified third circuit (System 3) that acts through LuxO. Symbols: pentagons, HAI-1; diamonds, AI-2; circles, CAI-1. Dotted lines denote targets for which regulation has not been shown to be direct. In both panels, phosphate flows toward LuxO at low cell density, and the flow of phosphate is reversed at high cell density (double-headed arrows). Details of the phosphorelay mechanism are provided in the text. Abbreviations: H, histidine; D, aspartate; HTH, helix-turn-helix; IM, inner membrane; OM, outer membrane.
In V. harveyi, at low cell density (i.e., in the absence of the autoinducers), LuxN and LuxQ autophosphorylate and transfer phosphate to the shared phosphotransferase protein LuxU, which in turn passes it to the response regulator, LuxO (12-14). Phosphorylated LuxO is active and, in conjunction with σ54, activates the expression of genes encoding small regulatory RNAs (sRNAs) that, together with the RNA chaperone Hfq, destabilize the mRNA encoding the activator protein LuxR (20, 21). LuxR is required for expression of the target genes in the quorum-sensing regulon (Fig. 1A) (18, 20, 22, 32). Because LuxR is required for transcription of luxCDABE (encoding luciferase), no light is produced under this condition. At high cell density, (i.e., in the presence of autoinducers), LuxN and LuxQ switch from kinases to phosphatases (14). Phosphate flow through the system is reversed, draining LuxO of phosphate (12). Unphosphorylated LuxO is inactive and thus cannot promote the expression of the sRNAs, leading to translation of luxR mRNA (20). LuxR binds the luxCDABE promoter and activates its expression, and V. harveyi produces light. Many other genes are controlled by quorum sensing in V. harveyi. While all require LuxR for their regulation, some of them, like lux, are induced at high cell densities, while others are repressed at high cell densities (Fig. 1A) (18, 26).
Miller et al. identified and characterized two two-component quorum-sensing circuits controlling quorum sensing in the human pathogen Vibrio cholerae (Fig. 1B) (25). Analysis of the completed genome of V. cholerae revealed that the V. harveyi-like System 2 (luxS and luxPQ) was present as well as luxU and luxO, and a genetic screen identified a novel autoinducer synthase named CqsA (for cholerae quorum-sensing autoinducer) and a cognate sensor named CqsS (for cholerae quorum-sensing sensor). The CqsA-dependent autoinducer is called CAI-1 (for cholerae autoinducer 1), and its structure has not been determined. The V. cholerae quorum-sensing cascade functions analogously to that of V. harveyi, and the downstream regulators LuxU, LuxO, sRNA, Hfq, and HapR (which is homologous to LuxR of V. harveyi) function identically to those of V. harveyi (Fig. 1B) (20, 25). However, many of the functions controlled by quorum sensing in V. cholerae (for example, those specifying virulence determinants) are not identical to those regulated by quorum sensing in V. harveyi (for example, bioluminescence and type III secretion).
LuxS and AI-2 production are widespread in the bacterial kingdom, and AI-2 is proposed to be used for interspecies communication (1). In contrast, LuxM and HAI-1 activity appear to be restricted to V. harveyi and the closely related bacterium Vibrio parahaemolyticus, indicating that this signal molecule is relatively species specific (1). The newly identified V. cholerae CAI-1 synthase, CqsA, is not similar to LuxM nor any other known autoinducer synthase. Additionally, LuxN does not recognize CAI-1 (M. B. Miller and B. L. Bassler, unpublished data). These findings led us to speculate that CAI-1 is a novel autoinducer. We wondered if other bacteria produce CAI-1 or if this signal is unique to V. cholerae. In this report we show that several vibrios, including V. harveyi, produce CAI-1 activity. Genetic analysis in V. harveyi reveals that the Cqs system is present and acts as a third quorum-sensing system that, in parallel with Systems 1 and 2, controls the quorum-sensing target genes. All three autoinducers display synergistic activity, and thus the V. harveyi circuit is a three-way coincidence detector for the signals HAI-1, AI-2, and CAI-1. However, the V. harveyi CAI-1-CqsS system appears to differ from Systems 1 and 2 by responding to autoinducer at much lower cell densities.
MATERIALS AND METHODS
Bacterial strains and media.
The relevant genotypes of all V. harveyi and V. cholerae strains and plasmids are listed in Table 1. Other species of bacteria and their strain names are listed in Table 2. Escherichia coli strain DH5α (F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1) (Promega) was used for cloning. E. coli strains were grown at 37°C with aeration. The V. cholerae bioreporter MM920 was grown in Luria-Bertani (LB) medium with tetracycline (TET) at 30°C with shaking. Tetracycline was added to all cultures with pLAFR for cosmid maintenance. V. harveyi cultures were grown in Luria-Marine (LM) medium for all genetic experiments and conjugations. Cell-free culture (14- to 16-h) fluids were prepared as described (2). Thirty-percent LM cell-free culture fluids were added to MM920, and 10% autoinducer bioassay (AB) cell-free culture fluids were added to V. harveyi strains JMH670 and JMH674. AB medium was used for bioluminescence and β-galactosidase (β-Gal) assays. The recipes for AB medium and LM medium have been reported previously (16). Cells were plated on LM agar plates for CFU determinations. Antibiotic (Sigma) concentrations used were as follows: ampicillin (AMP), gentamicin (GM), and kanamycin (KAN), 100 mg/liter; chloramphenicol (CM), and TET, 10 mg/liter; streptomycin (SM), 1,000 mg/liter; polymyxin B (PB), 50 U/liter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| V. harveyi strains | ||
| BB120 | Wild type | 1 |
| BB170 | luxN::Tn5 | 2 |
| BB721 | luxO::Tn5 | 4 |
| BB886 | luxPQ::Tn5 | 3 |
| JAF375 | luxN::CmrluxQ::Kanr | 12 |
| JAF483 | luxOD47A linked to Kanr | 12 |
| JAF536 | luxU::Tn5 | 13 |
| JAF548 | luxOD47E linked to Kanr | 12 |
| JAF633 | ΔluxM linked to Kanr | 14 |
| KM387 | ΔluxS | This study |
| KM357 | luxR::Tn5 | 18 |
| KM413 | ΔluxS ΔluxM | This study |
| JMH597 | luxN::Tn5 cqsS::Cmr | This study |
| JMH598 | cqsS::Cmr | This study |
| JMH603 | cqsA::Cmr | This study |
| JMH605 | ΔluxM linked to Kanr; cqsA::Cmr | This study |
| JMH606 | ΔluxS cqsA::Cmr | This study |
| JMH612 | luxPQ::Tn5 cqsS::Cmr | This study |
| JMH621 | luxU::Tn5 cqsA::Cmr | This study |
| JMH625 | ΔluxN luxQ::Tn5 | This study |
| JMH626 | ΔluxN luxQ::Tn5 cqsA::Cmr | This study |
| JMH628 | ΔluxN luxQ::Tn5 cqsS::Cmr | This study |
| JMH634 | ΔluxM ΔluxS cqsA::Cmr | This study |
| JMH670 | ΔluxM ΔluxS cqsA::CmrvhpA::Tn5lacZ | This study |
| JMH674 | ΔluxM ΔluxS cqsA::CmrvcrD::Tn5lacZ | This study |
| JMH763 | cqsAS::Cmr | This study |
| JMH774 | luxOD47E cqsA::Cmr | This study |
| V. cholerae strains | ||
| KSK1052 | ΔluxS | 25 |
| MM893 | ΔcqsA | 25 |
| Plasmids | ||
| pLAFR2 | Broad-host-range cosmid; mob, Tetr | 15 |
| pRK2013 | Broad host range, tra, Kanr | 9 |
| pPHIJ1 | Broad host range, tra, mob, Gmr | 5 |
| pGEM-T | Cloning vector; Ampr | Promega |
| pALTER Amp | Ampr | Promega |
| pALTER Tet | Cloning vector; Tetr | Promega |
| pBluescript SK | Cloning vector; Ampr | Stratagene |
| pKD3 | PCR template for Wanner deletions; Cmr | 8 |
| pCP20 | Ts FLP recombinase plasmid; Ampr, Cmr | 8 |
| pKAS46 | R6K replication origin; Smr | 33 |
| pBB1 | pLAFR with V. harveyi luxCDABE locus | |
| pBB154 | pLAFR with luxM and luxN locus | 2 |
| pMM467 | Tetr pALTER with cqsA | This study |
| pKM556 | pLAFR2 with ΔluxM | 26 |
| pKM755 | pLAFR2 with vhpA::Tn5lacZ Kanr | This study |
| pJMH17F7 | pLAFR2 with cqsAVh and cqsSVh locus; Tetr | This study |
| pJMH25 | pLAFR2 with type III secretion locus; Tetr | 18 |
| pJMH159 | pJMH17F7 cqsAVh::Tn5lacZ Kanr | This study |
| pJMH163 | pJMH17F7 cqsAVh::Tn5lacZ Kanr | This study |
| pJMH164 | pJMH17F7 cqsAVh::Tn5lacZ Kanr | This study |
| pJMH240 | pJMH17F7 with cqsAVh::Cmr | This study |
| pJMH244 | pJMH17F7 with cqsSVh::Cmr | This study |
| pJMH253 | pGEM-T with cqsAVh | This study |
| pJMH273 | pLAFR with cqsAVh | This study |
| pJMH280 | pGEM-T with cqsSVh | This study |
| pJMH282 | pLAFR with cqsSVh | This study |
| pJMH300 | pBB154 with ΔluxN | This study |
| pJMH325 | pJMH25 with vcrD::Tn5lacZ Kanr | This study |
| pJMH422 | pJMH17F7 with cqsASVh::Cmr | This study |
TABLE 2.
Induction of bioluminescence in the V. cholerae CAI-1 reporter strain MM920 by cell-free culture fluids prepared from other bacteria
| Species and strain | CAI-1 activitya |
|---|---|
| Vibrio cholerae CG706 | 11,531 ± 1,558 |
| Vibrio harveyi BB120 | 3,707 ± 298 |
| Vibrio anguillarum 19264 | 2,198 ± 165 |
| Vibrio alginolyticus 118 | 1,723 ± 206 |
| Vibrio parahaemolyticus BB22 | 1,417 ± 36 |
| Vibrio furnissii 1514 | 1,417 ± 76 |
| Vibrio angustum 70 | 288 ± 18 |
| Photobacterium leiognathi 721 | 46 ± 6 |
| Photobacterium phosphoreum NZ-11-D | 43 ± 3 |
| Vibrio proteolyticus 145 | 10 ± 2 |
| Vibrio vulnificus JJ077 | 3.6 ± 0.8 |
| Vibrio natriegens 77 | 1.5 ± 0.9 |
| Vibrio fischeri MJ1 | 1.1 ± 0.1 |
| Vibrio logei SR6 | 1.0 ± 0.1 |
| Escherichia coli MG1655 | 1.1 ± 0.2 |
| Salmonella typhimurium PS1 | 0.9 ± 0.03 |
CAI-1 activity is defined as the fold increase in induction of bioluminescence that occurs in V. cholerae MM920 in the presence of 30% cell-free culture fluid over that that occurs when medium alone is added.
DNA manipulations.
All DNA manipulations were performed using standard procedures (30). Oligonucleotides used for PCR and sequencing reactions can be found in the supplemental material (see Table S1). At the National Center for Biotechnology Information, sequencing data for the V. harveyi Cqs locus can be found under the accession number AY625893, and sequencing data for the vhpA locus is under the accession number AYG30354.
CAI-1 activity assay.
V. harveyi overnight cultures were diluted and grown to an optical density at 600 nm (OD600) of 1. Cell-free culture fluids were prepared from these cultures as previously described. A dense overnight culture of V. cholerae MM920 (ΔcqsA ΔluxQ pBB1) was diluted 1:10 in LB with TET. Thirty microliters of cell-free culture fluid was added to 70 μl of diluted V. cholerae reporter MM920 in a sterile 96-well microtiter dish (Corning) and incubated at 30°C with shaking (225 rpm). Luminescence from the reporter strain was measured with a Wallac microbeta multiplate scintillation counter every half hour until the luminescence of the control (reporter with medium alone) increased. The fold induction of the reporter was measured as counts per second (cps) of reporter plus culture fluid divided by cps of the control MM920 reporter with medium alone. For the measurement of V. harveyi CAI-1 accumulation over time, an overnight culture of V. harveyi strain BB120 was diluted 1:1,000 in fresh LM medium and grown in a flask at 30°C for 22 h. Aliquots were removed over time, the OD600 was measured, and cell-free culture fluids were prepared. A standard CAI-1 (MM920) activity assay was performed on all the cell-free culture fluids after 22 h.
Screen for V. harveyi CAI-1 synthase.
A V. harveyi genomic library carried in the E. coli luxS mutant strain DH5α (2) was grown overnight in 96-well microtiter dishes in 150 μl of LB with TET. An overnight culture of MM920 was diluted 1:50 in fresh LB with TET. E. coli overnight cultures were added to 96-well microtiter dishes containing 100 μl of the diluted MM920 V. cholerae reporter by stamping. These cells were coincubated for approximately 5 h at 30°C with shaking, and the resulting bioluminescence from each well was measured using the Wallac microbeta multiplate scintillation counter. MM1467 was used as a positive control for CAI-1 activity. E. coli clones that caused 500- to 1,000-fold induction of bioluminescence in the MM920 reporter were chosen for further analysis. To identify the CAI-1 synthase gene, λTn5lacZ mutagenesis was performed on cosmid pJMH17F7 as described previously (2). Mutagenized cosmid pools were transformed into E. coli DH5α, and recombinants were selected with KAN and TET. Recombinants were arrayed in microtiter dishes and were assayed for loss of CAI-1 activity, indicated by the inability to induce luminescence in MM920, as described above.
Cloning and sequencing the V. harveyi cqsA and cqsS genes.
Tn5lacZ mutagenized pJMH17F7 cosmids were isolated from E. coli strains lacking CAI-1 activity and subsequently digested with EcoRI and shotgun cloned into the pBluescript SK and pALTER Ampr vectors, selecting for resistance to AMP and KAN, respectively. Positive clones were sequenced using an outwardly directed primer (Tn50ISR) that hybridized to the arm of Tn5lacZ. The locus was also cloned by digesting cosmid pJMH17F7 with EcoRI, HindIII, or PstI. Fragments were shotgun cloned into pBluescript SK and transformed into E. coli DH5α. Transformants were arrayed in microtiter dishes, stamped into MM920 reporter diluted 1:50, and screened for CAI-1 activity. The inserts in plasmids from transformants that were able to induce luminescence in the MM920 reporter were sequenced. DNA sequence data were analyzed and aligned using the Sequencher 4.1 program (Gene Codes) and the National Center for Biotechnology Information's blastx program.
V. harveyi cqsA and cqsS deletion constructions.
Deletions of cqsAVh (cqsA from V. harveyi), cqsSVh, (cqsS from V. harveyi), and the double deletion of the cqsAVh and cqsSVh open reading frames (ORFs) in cosmid pJMH17F7 were constructed by the method of Datsenko and Wanner (8). The Cmr cassette from vector pKD3 was amplified with primers CqsAf-1 and CqsAr-2 for the cqsAVh ORF deletion and CqsSf1 and CqsSr-2 for the cqsSVh deletion, and primers CqsAf-1 and VhCqsSAWannerr were used to construct the deletion of cqsASVh in pJMH17F7. The resulting pJMH17F7 deletion cosmids pJMH240 (ΔcqsAVh::Cmr), pJMH244 (ΔcqsSVh::Cmr), and pJMH422 (ΔcqsASVh::Cmr) were conjugated into V. harveyi strains by using a triparental mating procedure as previously reported. The Cmr-marked deletions were subsequently transferred to the V. harveyi chromosome by an allelic exchange protocol (2). The deletions were verified by PCR using the primers cqsSAflankf and cqsSAflankr and, in the case of cqsAVh deletions, by observation of loss of CAI-1 activity in the MM920 reporter assay.
Cloning of cqsAVh and cqsSVh.
cqsSVh and cqsAVh were PCR amplified from the chromosome of wild-type (WT) V. harveyi BB120 using the following primer pairs: fCqsSXbaI and rCqsSXbaI for cqsSVh and fCqsAXbaI and rCqsAXbaI for cqsAVh. Both products were subcloned into the cloning vector pGEM-T (Promega) at the XbaI site and sequenced. The resulting vectors pJMH253 and pJMH280 were digested with XbaI, and the inserts were cloned into the broad-host-range cosmid pLAFR. These constructions were transformed into E. coli DH5α to produce cosmids pJMH273 and pJMH282, respectively.
V. harveyi strain construction.
An unmarked luxN deletion in cosmid pBB1154 was constructed by the method of Datsenko and Wanner using the oligonucleotides WannerluxNf and WannerluxNr (8). Deletions were confirmed with the flanking primers LuxNflankf and LuxNflankr. The unmarked luxN deletion cosmid pJMH300 was used for an allelic replacement of the Cmr marker harbored in luxN in V. harveyi JAF375 (luxN::Cmr luxQ::Tn5 Kanr) to make strain JMH625 (ΔluxN luxQ::Tn5 Kanr). KM387 (ΔluxS) was constructed by the method of Skorupski and Taylor (33) using PCR primer pairs LuxS1-KpnI-LuxS2-NotI and LuxS3-NotI-LuxS4-EcoRI. Both PCR products were ligated simultaneously into pKAS46, and the deletion was moved onto the chromosome of a spontaneous streptomycin-resistant strain of WT V. harveyi to construct KM387 (ΔluxS). We moved the unmarked luxM deletion from cosmid pKM556 onto the chromosome of KM387 and screened for dark mutants in microtiter dishes to make KM413 (ΔluxM ΔluxS). The Cmr marker from the cosmids pJMH244 (cqsSVh::Cmr) and pJMH1240 (cqsAVh::Cmr) was introduced onto the chromosome of JMH625 by allelic replacement to construct the triple mutants JMH628 (luxN luxQ cqsSVh) and JMH626 (luxN luxQ cqsAVh), respectively. The Cmr markers from pJMH240 (cqsAVh::Cmr) and pJMH244 (cqsSVh::Cmr) were subsequently transferred to the chromosome of KM413 (ΔluxM ΔluxS) to make the triple mutant JMH634 (ΔluxM ΔluxS cqsAVh::Cmr). The vcrD::Tn5 Kanr construct pJMH325 and the vhpA::Tn5 Kanr construct pKM755 were mobilized onto the chromosome of the triple synthase mutant JMH634 (ΔluxM ΔluxS cqsAVh::Cmr) by allelic exchange with selection for Kanr to produce V. harveyi strains JMH674 (ΔluxM ΔluxS cqsAVh vcrD::Tn5 Kanr) and JMH670 (ΔluxM ΔluxS cqsAVh vhpA::Tn5 Kanr), respectively.
Bioluminescence assays.
All bioluminescence assays were performed by using a previously reported protocol (2). Data are reported as relative light units (RLU) (counts min−1 ml−1 × 1,000/CFU ml−1). To measure bioluminescence at very low cell density, the overnight cultures were diluted 1:500,000 at the start of the assay, and both luminescence and cell density were measured every hour for 14 h. For single-time-point assays, overnight cultures were diluted 1:500 in AB medium and grown for approximately 8 h. At this time, the light and cell density of the cultures were measured. All single-time-point experiments were performed in triplicate.
β-Galactosidase assays.
V. harveyi strains were grown overnight in the presence of 10% cell-free culture fluids containing various combinations of autoinducers. A 1.5-ml sample of culture was resuspended in 1 ml of Z buffer, and β-Gal assays were performed in triplicate by a previously described method (34). β-Gal units are reported as (Vmax × dilution factor)/OD600.
RESULTS
V. harveyi produces CAI-1 activity.
To test for production of CAI-1 in different species of bacteria, we used a bioluminescent reporter strain of V. cholerae. This strain, named MM920, is ΔcqsA and ΔluxQ and harbors a vector containing the V. harveyi luciferase operon. MM920, while unable to produce CAI-1 or respond to AI-2, can detect and respond to exogenously added CAI-1 because CqsS is intact (25). To demonstrate that the reporter acts as expected, we show that addition of cell-free culture fluids harvested from high-cell-density V. cholerae ΔluxS (AI-2−) stimulates a 7,000-fold increase in light production, while cell-free fluids prepared from a V. cholerae ΔcqsA (CAI-1−) strain fail to induce light production in MM920 (Fig. 2A) (25).
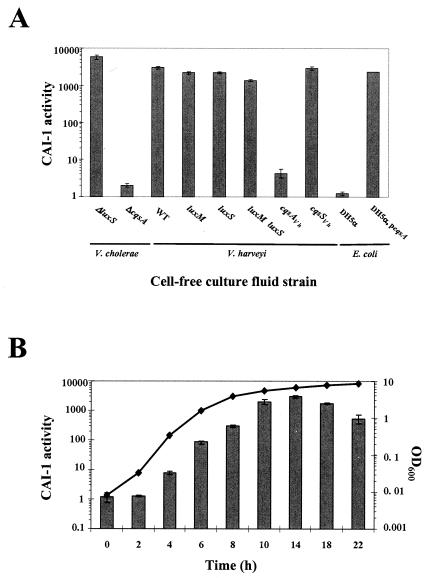
FIG. 2.
Analysis of cell-free culture fluids for CAI-1 activity. (A) Cell-free culture fluids were prepared from the strains shown and added at 30% (vol/vol) to V. cholerae MM920. Light production by MM920 was measured. Strain names are as follows (left to right): KSK1052, MM893, BB120, JAF633, KM387, KM413, JMH603, JMH598, DH5α, MM1467. (B) CAI-1 production over the growth curve is shown for WT V. harveyi strain BB120. Samples were collected every 2 h, the OD600s were measured, and cell-free culture fluids were prepared and assayed for CAI-1 activity using MM920. CAI-1 activity is reported as the fold induction of light production in MM920.
A variety of bacterial species were grown to stationary phase, cell-free culture fluids from these species were harvested and added to MM920, and light production was measured. The preparations from V. cholerae, V. harveyi, Vibrio anguillarum, Vibrio alginolyticus, V. parahaemolyticus, and Vibrio furnissii, induced an over 1,000-fold increase in bioluminescence in MM920, suggesting that these vibrios produce CAI-1 activity (Table 2 and Fig. 2A), while other bacteria tested either produced an activity that induced bioluminescence in the reporter to a much lesser extent or did not produce any activity (Table 2).
To show that, at least in the case of V. harveyi, the known autoinducers are not responsible for stimulation of the reporter strain, we assayed cell-free culture fluids from the V. harveyi WT and single (luxM or luxS) and double (luxM luxS) autoinducer synthase mutants in the V. cholerae CAI-1 reporter assay. All three strains produce a high level of activity capable of inducing light production in MM920 (Fig. 2A). Neither HAI-1 nor AI-2 prepared by in vitro procedures induces light production in MM920 (Miller and Bassler, data not shown). Together, these results suggest that an additional autoinducer synthase exists in V. harveyi and is responsible for producing the CAI-1 activity. Interestingly, we noticed that while the double luxM luxS mutant produces the CAI-1 activity, it consistently produces approximately fivefold less of this activity than does either the single luxM mutant or the single luxS mutant. Figure 2B shows that similar to most other autoinducers, in V. harveyi, CAI-1 activity accumulates in the culture fluids over time, and maximal CAI-1 activity is observed in early stationary phase. Finally, V. harveyi consistently produces less CAI-1 activity than does V. cholerae (Table 2 and Fig. 2). We do not know whether V. harveyi produces a CAI-1 molecule that differs from and is detected less efficiently by V. cholerae than the endogenous CAI-1 or if V. harveyi simply produces less of the identical CAI-1 molecule that V. cholerae produces.
Identification of the V. harveyi CAI-1 synthase gene.
To devise a genetic screen to identify the V. harveyi gene encoding the putative CAI-1 synthase, we made use of the following two observations. First, cell-free culture fluids prepared from E. coli strain DH5α do not induce luminescence in the MM920 reporter, indicating that they lack CAI-1 activity (Fig. 2A, bar labeled DH5α). Second, culture fluids prepared from E. coli DH5α carrying the cloned V. cholerae CAI-1 synthase gene cqsA induce bioluminescence in the reporter, indicating that introduction of the cqsA gene into E. coli is sufficient for CAI-1 production and release (Fig. 2A, bar labeled DH5α, pcqsAVh). We reasoned that we could identify the V. harveyi CAI-1 synthase gene by introducing a V. harveyi genomic cosmid library into E. coli DH5α and assaying cell-free culture fluids from the transformants for the ability to induce luminescence in the CAI-1 reporter MM920.
One thousand E. coli DH5α transformants, each containing a cosmid carrying ∼25 kb of V. harveyi genomic DNA, were tested for activation of light production in V. cholerae MM920. Six positive transformants were identified, and restriction mapping and Southern blot analyses revealed that each of the six contained a set of overlapping fragments of V. harveyi genomic DNA (data not shown). One of these cosmids, pJMH17F7, was chosen for further analysis. Cell-free culture fluids from E. coli DH5α carrying pJMH17F7 induce luminescence 1,120-fold in MM920 (Fig. 3A), whereas preparations from E. coli DH5α carrying the vector alone do not (Fig. 3A, bar labeled DH5α, pLAFR). The amount of CAI-1 activity produced by DH5α carrying pJMH17F7 is comparable to that made by E. coli DH5α carrying the V. cholerae cqsA gene in trans (over 1,000-fold stimulation of the reporter) (Fig. 2A).
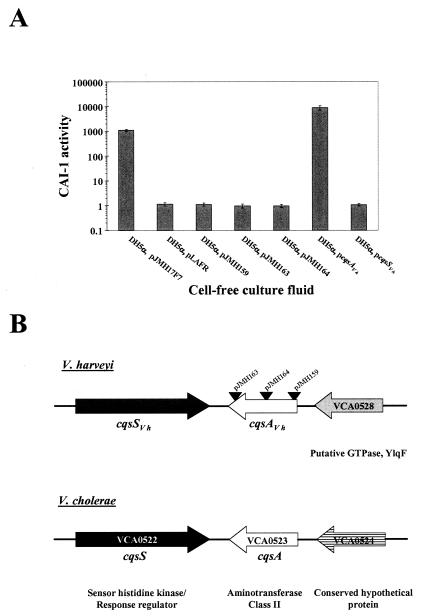
FIG. 3.
The cqs locus in V. harveyi. (A) CAI-1 activity is shown for recombinant E. coli DH5α strains carrying either the cosmid pLAFR or pLAFR containing vibrio DNA. pJMH17F7 carries 25 kb of V. harveyi genomic DNA containing the gene responsible for CAI-1 activity. pJMH159, pJMH163, and pJMH164 contain Tn5lacZ insertions in pJMH17F7. pJMH273 carries only the cqsAVh gene, and pJMH282 carries only the cqsSVh gene. (B) Map of the cqs locus of V. harveyi and V. cholerae. Black triangles indicate the sites of Tn5lacZ insertions in pJMH163, pJMH164 and pJMH159. The cqsS and cqsA genes are shown in black and white, respectively. The downstream ORF in V. harveyi has amino acid homology to VCA0528 and is shown in gray. This gene was not fully sequenced. The downstream ORF in V. cholerae, denoted VCA0524, is shown in stripes.
The cosmid pJMH17F7 was subjected to Tn5lacZ mutagenesis to identify the locus responsible for producing the CAI-1 activity. Pools of transposon-mutagenized pJMH17F7 cosmid DNA were introduced into E. coli DH5α. Transformants were arrayed in 96-well plates and coincubated with the V. cholerae reporter strain MM920. Subsequently, light production by MM920 was quantitated. Three E. coli strains that did not stimulate light production in the reporter were identified, indicating that the cosmids they carried (pJMH159, pJMH163, and pJMH164) had acquired Tn5lacZ insertions that inactivated some function required for CAI-1 production (Fig. 3A).
Sequencing of the DNA flanking the Tn5lacZ insertion sites in pJMH159, pJMH163, and pJMH164 revealed that all three transposon insertions were located in the same ORF (Fig. 3B). This ORF encodes a protein with 57% identity to V. cholerae VCA0523, recently designated cqsA (25), and so we designate this ORF cqsAVh. Our sequence analysis showed that a gene encoding a two-component hybrid sensor kinase with 48% identity to V. cholerae VCA0522 (designated cqsS) was situated 3′ to cqsAVh with the same orientation as in V. cholerae (Fig. 3B). We designate this second gene cqsSVh. The loci adjacent to the 5′ termini of the V. harveyi and V. cholerae cqsA genes are not similar and encode a conserved hypothetical protein (V. cholerae) and a putative GTPase annotated as YlqF (V. harveyi). Database analysis shows that cqsAVh and cqsSVh share 82 and 78% identity with VPA0711 and VPA0710 in V. parahaemolyticus. This latter result is not surprising given that V. parahaemolyticus also produces CAI-1 activity (Table 2).
The cqsAVh promoter and ORF were amplified from the chromosome of WT V. harveyi and cloned onto the low-copy-number pLAFR cosmid. This construct was introduced into E. coli DH5α, and the strain was tested for CAI-1 production (Fig. 3A, bar labeled DH5α pcqsAVh). The results show that the V. harveyi cqsA gene confers the ability to produce CAI-1 to E. coli. Not surprisingly, introduction of cqsSVh (encoding the putative CAI-1 sensor) into E. coli does not promote CAI-1 production (Fig. 3A, bar labeled DH5α, pcqsSVh). Interestingly, the cosmid carrying the cqsAVh gene without any flanking V. harveyi genomic DNA produces substantially greater activity than does the parent cosmid carrying the cqsAVh gene on a 25-kb genomic insert. This result suggests that a repressor of cqsAVh expression or CAI-1 production could be linked to the cqsAVh gene.
Although the V. harveyi genome sequence is not available, in the closely related bacterium V. parahaemolyticus, two putative transcriptional regulators, VPA0716 and VPA0717, are located five genes downstream of the cqsA homologue (annotated as VPA0711). VPA0716 is annotated as a putative transcriptional regulator protein sharing homology with AraC regulators. VPA0717 is annotated as a LysR-like transcriptional regulator. These loci, if present on the V. harveyi genomic cosmid carrying cqsAVh, could be involved directly or indirectly in negatively regulating CAI-1 production. We are investigating this possibility now.
To examine the in vivo roles of cqsAVh and cqsSVh in CAI-1 production, each gene was deleted from the chromosome of V. harveyi, and subsequently CAI-1 production was measured by using the V. cholerae reporter MM920. V. harveyi with a deletion of cqsAVh does not make CAI-1, whereas V. harveyi with a deletion of cqsSVh produces WT levels of CAI-1 (Fig. 2A, bars labeled cqsAVh and cqsSVh). The function of CqsSVh is examined further in a subsequent section.
CAI-1 acts synergistically with HAI-1 and AI-2 to control quorum-sensing genes in V. harveyi.
The above results show that cqsAVh encodes the V. harveyi CAI-1 synthase, and based on our earlier findings in V. cholerae, we predict that CAI-1 controls quorum-sensing target gene expression in V. harveyi. To test this prediction, we constructed the three V. harveyi single autoinducer synthase deletions, all the combinations of double synthase mutants, and the triple autoinducer synthase mutant and assayed light production in each. In this experiment, we are using bioluminescence at high cell density as the representative readout for the quorum-sensing regulon.
Deletion of any single autoinducer synthase severely decreases light production in V. harveyi (Fig. 4A). Specifically, Fig. 4 shows that the cqsAVh mutant (CAI-1−) makes 17%, the luxS mutant (AI-2−) produces 4%, and the luxM mutant (HAI-1−) produces only 0.1% of the light produced by the WT (note the log scale). The double and triple synthase mutants produce less light than the single mutants. The levels of light measured here are at the limit of detection in our assay, and so we do not know if the small differences in light production by the double and triple synthase mutants are meaningful. Nonetheless, we make three conclusions from this experiment. First, the CAI-1 autoinducer is a signal that is critical for regulation of bioluminescence expression in V. harveyi. Second, under these experimental conditions, the influence of HAI-1 on light production is stronger than that of AI-2, which is stronger than that of CAI-1. Third, the three autoinducers act synergistically because elimination of even the weakest signal (CAI-1) results in an 83% reduction in light production. Thus, by the logic presented previously (26), the V. harveyi Lux circuit appears to act as a three-way coincidence detector.
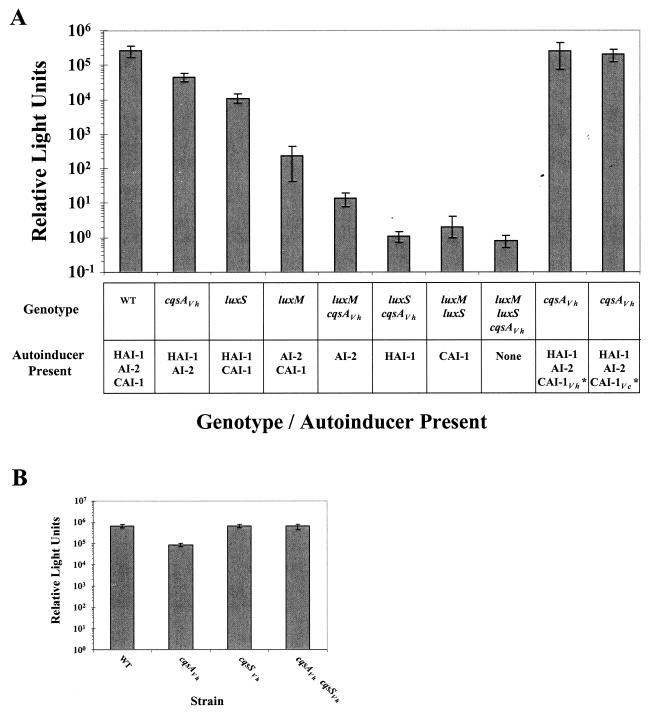
FIG. 4.
V. harveyi single, double, and triple autoinducer synthase mutant phenotypes. (A) Light production per cell (RLU) was measured from high-cell-density V. harveyi cultures. The V. harveyi strains are (from left to right) BB120 (WT), JMH603 (cqsAVh), KM387 (luxS), JAF633 (luxM), JMH605 (luxM cqsAVh), JMH606 (luxS cqsAVh), KM413 (luxM luxS), and JMH634 (luxM luxS cqsAVh), and the rightmost two bars are JMH603 (cqsAVh). In these latter two cases, JMH603 (cqsAVh) was grown in the presence of 30% V. harveyi or V. cholerae WT culture fluids. The asterisks denote exogenously added autoinducer. (B) CqsAVh and CqsSVh epistasis analysis. RLU were measured at high cell density in the following strains (left to right): BB120 (WT), JMH603 (cqsAVh), JMH598 (cqsSVh), JMH763 (cqsAVh cqsSVh). Relative light units are defined as counts min−1 ml−1 × 1,000/CFU ml−1.
The absence of the CAI-1 signal is responsible for the reduction in light production observed in the cqsAVh mutant because exogenous addition of CAI-1 complements the defect. Specifically, addition of cell-free culture fluids prepared from a luxM luxS V. harveyi strain (i.e., HAI-1− AI-2− CAI-1+) or a luxS V. cholerae strain (i.e., AI-2− CAI-1+) restores WT bioluminescence to the cqsAVh (i.e., CAI-1−) mutant (Fig. 4A). We have shown previously that exogenous addition of HAI-1 and AI-2 restores light production in the luxM and luxS mutants, respectively (2, 3).
CqsS is the sensor for CAI-1.
CqsS is the sensor for CAI-1 in V. cholerae and thus acts downstream of the CAI-1 signal (25). To test if this is also the case in V. harveyi, we performed an epistasis analysis with single and double cqsAVh and cqsSVh mutants. Figure 4B shows that the cqsAVh mutant produces approximately 13% as much light as the WT. In contrast, the cqsSVh mutant produces nearly WT levels of light. We assume that this latter phenotype stems from Systems 1 and 2 remaining operational in the cqsSVh mutant. Importantly, the cqsAVh cqsSVh double mutant has a phenotype similar to that of the single cqsSVh mutant (i.e., it produces WT levels of light), demonstrating that CqsS acts downstream of CAI-1. Based on these results, we assert that the information supplied by CAI-1 cannot be transduced to the lux operon in the absence of CqsS, and therefore, CqsS is the sensor for the CAI-1 autoinducer in V. harveyi.
LuxU and LuxO are required to transduce the CAI-1 signal in V. harveyi.
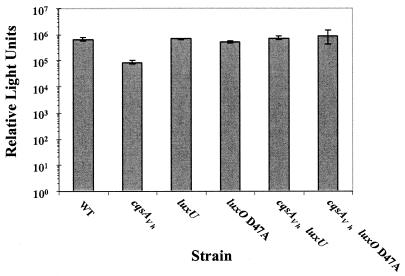
We wondered whether CAI-1 signaling is mediated through LuxU and LuxO or through other regulatory functions. We again performed epistasis tests to examine the roles of LuxU and LuxO. In this case we constructed single and double cqsAVh, luxU, and luxO null mutants and compared their bioluminescence phenotypes. We used a transposon insertion in luxU to analyze the luxU null phenotype and used the luxOD47A missense mutation to test the luxO null phenotype. We chose this particular luxO allele because, while it confers the luxO null phenotype, it is not polar on luxU (luxO and luxU are in an operon) (12). Thus, this strategy allowed us to individually test the effects of luxO and luxU null mutations on CAI-1 signal transduction. Like Fig. 4, Fig. 5 shows that the cqsAVh single mutant produces approximately 13% of the light produced by the WT. The luxU null and luxOD47A single mutants are constitutively bright and produce WT levels of light. The epistasis tests show that the cqsAVh luxU and cqsAVh luxOD47A double mutants have phenotypes like those of the luxU and luxO single mutants (constitutive bright). Therefore, both LuxU and LuxO are required for CAI-1 signal relay, and therefore, both act downstream of CAI-1 and CqsS in the V. harveyi quorum-sensing circuit (Fig. 1).
FIG. 5.
LuxU and LuxO are epistatic to CqsA. RLU were measured from high-cell-density V. harveyi cultures of the following strains (left to right): BB120 (WT), JMH603 (cqsAVh), JAF536 (luxU), JAF483 (luxOD47A), JMH621 (luxU cqsAVh), JMH774 (luxOD47A cqsAVh).
Three autoinducers control the quorum-sensing regulon.
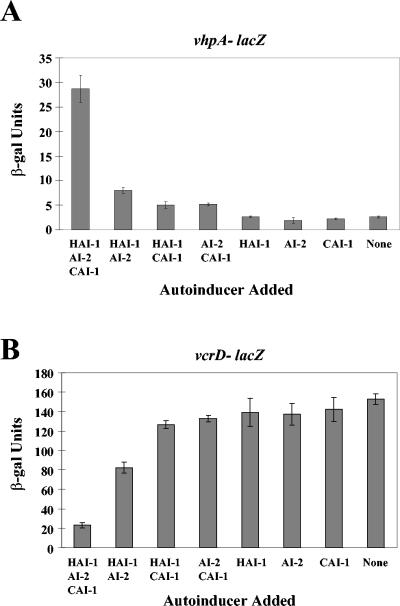
We have shown that HAI-1 and AI-2 act synergistically to regulate Lux and many other quorum-sensing targets, such as a TTS system (18) and a secreted metalloprotease (26). The results illustrated in Fig. 4A show that with respect to lux expression, three autoinducers contribute to this synergy. These findings, coupled with the finding that CAI-1 acts through LuxU and LuxO (Fig. 5), suggest that CAI-1 regulation is not exclusive to bioluminescence but can be extended to other members of the quorum-sensing regulon. To test this assumption, we analyzed CAI-1 regulation of the expression of two genes that we have previously shown to be jointly controlled by HAI-1 and AI-2. Here we describe our examination of one representative quorum-sensing-activated gene (encoding a metalloprotease we have named VhpA [V. harveyi protease A]) (26) and one representative quorum-sensing-repressed gene (encoding the TTS system protein VcrD) (18). lacZ transcriptional fusions to these two genes were made and introduced onto the chromosome of a V. harveyi triple autoinducer mutant (i.e., luxM luxS cqsAVh). Cell-free culture fluids containing either HAI-1, AI-2, CAI-1, or combinations of these signals were added to the fusion strains, and LacZ activity was subsequently measured.
Induction of the vhpA-lacZ fusion occurs when cell-free culture fluids containing all three autoinducers are added (Fig. 6A, leftmost bar); however, much reduced expression occurs following the addition of the preparations containing any two, any one, or no autoinducers (Fig. 6A, remaining bars). The quorum-sensing-repressed target vcrD shows an opposite pattern of expression (Fig. 6B). In this case, β-Gal activity is low (20 U) when all three autoinducers are present (Fig. 6B, leftmost bar) but is high (80 to 150 U) in the presence of none or any one or two autoinducers (Fig. 6B, remaining bars). Thus, for bioluminescence, metalloprotease production, and type III secretion, all three autoinducers act in concert.
FIG. 6.
All three autoinducers are required for quorum-sensing-controlled gene regulation. β-Galactosidase assays were performed on V. harveyi recipient strains (high cell density) grown overnight in the presence of 10% cell-free culture fluids containing various autoinducers. Cell-free culture fluids were prepared from the following strains (left to right): BB120 (WT), JMH603 (cqsAVh), KM387 (luxS), JAF633 (luxM), JMH606 (luxS cqsAVh), JMH605 (luxM cqsAVh), KM413 (luxM luxS), and JMH634 (luxM luxS cqsAVh). (A) The recipient strain was V. harveyi JMH670 (vhpA::Tn5lacZ luxM luxS cqsAVh). (B) The recipient strain was JMH674 (vcrD::Tn5lacZ luxM luxS cqsAVh). β-Galactosidase units are reported as (Vmax × dilution factor)/OD600.
The CAI-1-CqsS system functions at very low cell densities.
The results of this study demonstrate that HAI-1, AI-2, and CAI-1 act synergistically through the Lux network to control gene expression. To begin to analyze the relative contribution of each sensory system, we constructed combinations of quorum-sensing mutants and assayed their resulting bioluminescence phenotypes. This strategy allowed us to examine both the timing and the intensity of each of the three inputs.
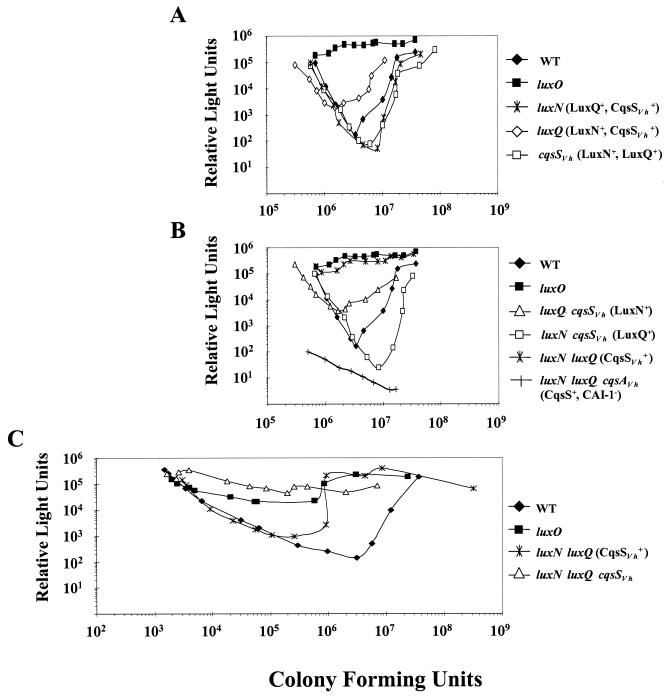
Figure 7A shows the characteristic WT V. harveyi density-dependent bioluminescence curve. Following dilution of the culture into fresh medium, light production sharply decreases because no autoinducers are present. Under these conditions, the sensors act as kinases and funnel phosphate to LuxO, and this results in Lux repression (12). Light production begins increasing at approximately 2.5 × 106 cells/ml because by the time the cells have grown to this cell density, the autoinducers have accumulated to the threshold level required to induce the sensors to switch from net kinase to net phosphatase. Removal of phosphate from LuxO terminates repression and allows light production to occur (Fig. 1A and see the introduction). A luxO mutant is constitutively bright (Fig. 7A) because LuxO is required (indirectly) for Lux repression (21). Mutation of any one of the three sensors alters the shape of the Lux curves slightly; however, each sensor mutant (luxN, luxQ, and cqsSVh) retains density-dependent expression of bioluminescence, presumably due to the input from the remaining two systems.
FIG. 7.
V. harveyi single, double, and triple autoinducer sensor mutant phenotypes. Density-dependent bioluminescence assays were performed on (A) BB120 (WT), BB721 (luxO), BB170 (luxN), BB886 (luxQ), and JMH598 (cqsSVh); (B) BB120 (WT), BB721 (luxO), JMH612 (luxQ cqsSVh), JMH597 (luxN cqsSVh), JMH625 (luxN luxQ), and JMH626 (luxN luxQ cqsAVh); and (C) BB120 (WT), BB721 (luxO), JMH625 (luxN luxQ), and JMH628 (luxN luxQ cqsSVh). Relative light units are reported as counts min−1 ml−1 × 1,000/CFU ml−1. In panels A and B, cells were diluted 1:5,000, and in panel C, cells were diluted 1:500,000 prior to the start of the experiment.
The phenotypes of the double sensor mutants are compared to those of the WT and the constitutively bright luxO null strain, shown in Fig. 7B. The luxQ cqsSVh double mutant that has only LuxN remaining transitions from decreasing to increasing light production at a lower cell density than in the WT. By contrast, the luxN cqsSVh double mutant that has only has LuxQ remaining transitions to increasing light output that is delayed relative to that of the WT. Together these first two results suggest that normally, LuxQ remains a kinase beyond the point when LuxN has switched to phosphatase. Finally, a luxN luxQ double mutant that has only CqsSVh remaining is nearly constitutively bright.
Because the luxN luxQ double mutant phenotype is nearly identical to the luxO mutant phenotype (constitutive light production), we hypothesized that either CqsSVh switches from kinase to phosphatase at a very low cell density or CqsSVh contributes neither kinase nor phosphatase activity to the Lux circuit. To distinguish between these possibilities we examined a luxN luxQ cqsAVh triple mutant. In this mutant, the only sensor present is CqsSVh, but because no CAI-1 is produced due to the cqsAVh mutation, according to our model, CqsSVh should be locked in kinase mode. The luxN luxQ cqsAVh triple mutant is completely dark (Fig. 7B), demonstrating that CqsSVh indeed acts as a kinase that shuttles phosphate to LuxO to produce full Lux repression. We therefore conclude that CAI-1 and CqsSVh contribute to quorum-sensing gene regulation at very low cell densities.
We suspect that the input from CqsSVh is not apparent under our usual experimental conditions. Specifically, we hypothesize that at the cell densities at which we started the experiments, the transition of CqsS from kinase to phosphatase had already occurred. If so, this could explain why we never identified CAI-1 and CqsSVh as components of the Lux quorum-sensing cascade. To test this possibility, we performed a density-dependent bioluminescence assay using V. harveyi strains diluted 1:500,000 instead of using the usual 1:5,000 dilution (Fig. 7C). At these very low cell densities, the luxN luxQ double mutant that has only CqsS remaining exhibits density-dependent bioluminescence and has a phenotype that is quite distinct from the luxO mutant. Importantly, unlike the double luxN luxQ sensor mutant, under this condition the triple luxN luxQ cqsSVh sensor mutant is constitutively bright, showing that in WT cells, the CqsSVh sensor switches from a kinase to a phosphatase at very low cell densities (∼1 × 105 cells/ml), and this switch occurs earlier than that of LuxN and LuxQ. The results of this experiment also show that the buildup of the critical concentrations of the autoinducers required to switch their cognate sensors from kinase mode to phosphatase mode occurs in the following order: CAI-1, then HAI-1, then AI-2.
DISCUSSION
This report describes the identification and analysis of a quorum-sensing system in V. harveyi that acts in parallel with the two previously identified quorum-sensing systems (Fig. 1A). V. harveyi and other closely related species produce an autoinducer activity that mimics the CAI-1 activity of V. cholerae (Table 2). As in V. cholerae, in V. harveyi the genes cqsA and cqsS are required for production and detection of CAI-1, respectively (Fig. 3, 4, and 7). CAI-1 acts in conjunction with the V. harveyi HAI-1 and AI-2 autoinducers to control a common set of target genes (Fig. 5 to 7). However, the CAI-1-CqsS system switches from kinase mode to phosphatase mode at a much lower cell density than do the HAI-1-LuxN and AI-2-LuxQ circuits (Fig. 7). While it remains possible that genes responsive to only one or only two of the three autoinducers exist in V. harveyi, proper regulation of the quorum-sensing targets identified thus far requires the simultaneous presence of all three autoinducers (Fig. 4 and 7), suggesting that the V. harveyi quorum-sensing circuit likely functions as a three-way coincidence detector.
The two major classes of autoinducers, HSLs in gram-negative bacteria and oligopeptides in gram-positive bacteria, function in intraspecies communication (reviewed in reference 24). By contrast, the autoinducer AI-2 and its synthase LuxS are broadly conserved in the bacterial kingdom, and are proposed to function in interspecies communication (reviewed in references 11 and 37). V. harveyi CAI-1 can control gene expression in V. cholerae and vice versa, demonstrating that CAI-1-mediated cell-cell communication need not be restricted to within a species. Thus, CAI-1 represents a new activity that fits under the classification of interspecies communication signal. Importantly, however, in contrast to LuxS and AI-2, CqsS and its product CAI-1 are not exceptionally widespread. Rather, our preliminary analysis suggests that they are only present in closely related marine bacteria. Thus, CAI-1, while capable of promoting communication across species boundaries, might be restricted to use exclusively within the genus Vibrio. Database analysis shows that while V. cholerae and V. parahaemolyticus possess cqsA and make CAI-1, Vibrio vulnificus and Vibrio fischeri do not. We predict the existence of a cqsA gene in the genomes of V. anguillarum, V. alginolyticus, and V. furnissii based on our ability to detect CAI-1 activity in their cell-free culture fluids (Table 2). Conceivably, the vibrio species that produce and detect CAI-1 are ones that inhabit the same niches and communicate with one another therein.
We do not know the identity of CAI-1, whether the CAI-1 molecule made by different bacteria is identical, or whether more than one molecule has CAI-1 activity, nor do we know the role of CqsA in CAI-1 production. The CqsA protein is similar to class II aminotransferases which catalyze the transamination of the terminal amino groups of basic α-amino acids, diamines, and ω-amino acids (23). Interestingly, aminotransfer reactions are also involved in HSL autoinducer production by LuxI-type autoinducer synthase enzymes. Specifically, LuxI-type enzymes couple the acyl group from acyl-acyl carrier proteins to the amino moiety of methionine on S-adenosylmethionine to produce specific HSLs (17, 27, 29, 36). It is possible that enzymes in addition to CqsA are required for CAI-1 production. However, because E. coli containing either the V. cholerae or the V. harveyi cqsA gene produces high levels of CAI-1 activity (Fig. 3), either CqsA is the only protein required to make CAI-1 from substrates existing in E. coli, or, if several steps are required, then E. coli must possess the additional enzymes in the pathway.
We are presently attempting to purify CAI-1 from cell-free culture fluids of V. cholerae and V. harveyi and from recombinant E. coli. These studies should reveal whether V. harveyi and V. cholerae make identical CAI-1 or not. Our preliminary evidence suggests that CAI-1 is not an HSL-type autoinducer because it has no activity in the commonly used HSL reporters that detect a variety of HSL molecules, and coincubation of cell-free culture fluids containing CAI-1 with a Bacillus species that secretes a nonspecific homoserine lactonase (AiiA) (10) causes no loss of CAI-1 activity (data not shown). Verification of this supposition awaits structural identification of CAI-1.
Our work demonstrates that the simultaneous presence or simultaneous absence of HAI-1, AI-2, and CAI-1 is required for maximal impact on gene expression in V. harveyi (Fig. 4 and 7); however, the strengths of the different autoinducer signals are not equivalent. The different signal strengths are represented in the relative kinase-to-phosphatase ratio of the corresponding receptors (LuxN > LuxQ > CqsS). In natural settings, the strength of input from each system could vary depending on the prevailing environmental conditions. Supporting this idea, we have observed that on agar surfaces, the luxS mutant produces WT levels of light, the cqsA mutant is dim, and the luxM mutant is dark. Thus, under this condition, the relative signal strengths must be as follows: HAI-1 > CAI-1 > AI-2. This observation implies that, unlike in liquid where the relative kinase-phosphatase inputs are greatest for LuxN, followed by LuxQ and then CqsS, on a surface the activities are greatest for LuxN, followed by CqsS and then LuxQ (data not shown). We are presently studying the regulation of the three sensors and synthases on agar surfaces to understand the molecular basis for the alteration in signal strength that occurs between liquid and surface environments.
Importantly, whereas in liquid cultures of V. harveyi the CqsS input is the weakest, in V. cholerae it is the dominant input (25). Furthermore, the CqsS switch from kinase mode to phosphatase mode occurs at a much higher cell density in V. cholerae than it does in V. harveyi. We suspect that the differences in the relative strengths and timing of these inputs in V. harveyi and V. cholerae reflect each species's fine-tuning of the signals for optimized survival in the distinct niches in which each resides. Thus, while V. harveyi and V. cholerae likely have some niches in common, they obviously have distinct niches as well. For example, V. cholerae colonizes the human intestinal tract (for a review, see reference 19), while V. harveyi does not. While we do not know the underlying molecular mechanism giving rise to the differences in the individual signal input strengths in V. harveyi and V. cholerae, an obvious possibility is that the sensors simply have different relative kinase and/or phosphatase activities or different affinities for CAI-1.
Why does V. harveyi use three quorum-sensing circuits that converge to control the same set of genes? Using multiple signals in the coincidence detection scheme could protect the quorum-sensing system from noise due to the presence of molecules that are similar or identical to one of the autoinducers or from the disappearance of one of the signals due to degradation or consumption by other species of bacteria. Alternatively, if the system does not act as a coincidence detector under natural conditions, one obvious rationale for having multiple autoinducers is that three signals provide eight possible input states, whereas only four states are achievable with two signals and only two states are possible with one signal. The ability to monitor fluctuations in many signals could be critical for the survival of V. harveyi in different niches because the cell numbers and/or species compositions of the quorums required for commensal living, biofilm formation, and pathogenesis could vary greatly. Irrespective of the detection logic used, this careful probing of the environment could ensure that the decision to transition into or out of quorum-sensing mode is made with high fidelity.
Supplementary Material
Acknowledgments
This work was supported by NSF grant MCB-0343821, NIH grant 5RO1 GM065859, and ONR grant N00014-03-0183.
We thank the Princeton Syn/Seq facility for DNA sequencing. We thank Kenny Mok and Melissa Miller for construction of strains used in this study. We especially thank Melissa Miller and the entire Bassler lab for insightful discussions. We are grateful to L. Zhang for supplying the lactonase assay strain.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. [DOI] [PubMed] [Google Scholar]
- 5.Beringer, J. E., J. L. Beynon, A. V. Buchanan-Wollaston, and A. W. B. Johnston. 1978. Transfer of the drug resistance transposon Tn5 to Rhizobium. Nature 276:633-634. [Google Scholar]
- 6.Cao, J. G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 7.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Federle, M. J., and B. L. Bassler. 2003. Interspecies communication in bacteria. J. Clin. Investig. 112:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 13.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman, J. A., B. N. Lilley, and B. L. Bassler. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol. Microbiol. 35:139-149. [DOI] [PubMed] [Google Scholar]
- 15.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg, E. P., J. W. Hastings, and S. Ulitzer. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 179:87-91. [Google Scholar]
- 17.Hanzelka, B. L., and E. P. Greenberg. 1996. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J. Bacteriol. 178:5291-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose, K. E. 2001. Regulation of virulence in Vibrio cholerae. Int. J. Med. Microbiol. 291:81-88. [DOI] [PubMed] [Google Scholar]
- 20.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 21.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 22.Martin, M., R. Showalter, and M. Silverman. 1989. Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J. Bacteriol. 171:2406-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehta, P., T. Hale, and P. Christen. 1993. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214:549-561. [DOI] [PubMed] [Google Scholar]
- 24.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 25.Miller, M. B., K. Skorupski, D. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 26.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.More, M. I., L. D. Finger, J. L. Stryker, C. Fuqua, A. Eberhard, and S. C. Winans. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272:1655-1658. [DOI] [PubMed] [Google Scholar]
- 28.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 32.Showalter, R. E., M. O. Martin, and M. R. Silverman. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 172:2946-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 34.Slauch, J. M., and T. J. Silhavy. 1991. cis-Acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Val, D. L., and J. E. Cronan, Jr. 1998. In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer synthases. J. Bacteriol. 180:2644-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.