Abstract
Escherichia coli MelR protein is a transcription activator that is essential for melibiose-dependent expression of the melAB genes. We have used chromatin immunoprecipitation to study the binding of MelR and RNA polymerase to the melAB promoter in vivo. Our results show that MelR is associated with promoter DNA, both in the absence and presence of the inducer melibiose. In contrast, RNA polymerase is recruited to the melAB promoter only in the presence of inducer. The MelR DK261 positive control mutant binds to the melAB promoter but cannot recruit RNA polymerase. Further analysis of immunoprecipitated DNA, by using an Affymetrix GeneChip array, showed that the melAB promoter is the major, if not the sole, target in E. coli for MelR. This was confirmed by a transcriptomics experiment to analyze RNA in cells either with or without melR.
Expression of the Escherichia coli melAB genes, which encode proteins necessary for transport and metabolism of the disaccharide melibiose, is dependent on the transcription activator, MelR, encoded by the adjacent melR gene (13). Previous studies have shown that transcription from the melAB promoter is activated by MelR and have focused on using biochemistry to understand the mechanism of activation (1, 4, 7, 12). Recent work has shown that MelR activates transcription by direct interaction with the RNA polymerase σ subunit via residue D261 (5). Although activation requires the inducer melibiose, in vitro studies have shown that MelR can bind to the melAB promoter both in the presence and absence of melibiose (1). In the experiments presented here, we have exploited novel chromatin immunoprecipitation (ChIP) and microarray technologies to study the interactions of MelR in vivo. ChIP has been used to investigate MelR and RNA polymerase binding to the melAB regulatory region in vivo, while microarrays have been used to show that the melAB promoter is the principal target for MelR in E. coli.
MATERIALS AND METHODS
E. coli strains, plasmids, and oligonucleotides.
Bacterial strains, plasmids, and oligonucleotides used in this work are listed in Table 1. In all experiments, E. coli strains WAM131, WAM132, or MG1655, carrying plasmids as appropriate, were grown to mid-exponential phase (optical density at 650 nm of 0.4 to 0.6) in minimal M63 medium, supplemented with fructose and amino acids, either with or without melibiose, according to the same method used previously in studies of the regulation of the E. coli mel operon (13).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Characteristics or description | Reference, source, or use |
|---|---|---|
| E. coli strain | ||
| WAM131 | GM-1 cured of F′ episome Δlac | 1 |
| WAM132 | WAM131 ΔmelR Δlac | 1 |
| MG1655 | F− λ−ilvG rfb-50 rph-1 | 2 |
| Plasmid | ||
| pAA121 | Cloning vector derived from pBR322 | 8 |
| pJW15 | pAA121 derivative carrying melR | 14 |
| pJW15 (FY53, KR182) | Derivative of pJW15 encoding FY and KR substitutions at codons 53 and 182 of melR respectively | C. Webster (unpublished) |
| pLGΔRS | Derivative of pLG339 carrying a deletion that results in the loss of tet− | 1 |
| pLG314 | Derivative of pLG339 carrying melR under the control of the activator-independent galP2 promoter | 12 |
| pLG314 (DK261) | Derivative of pLG314 encoding a DK substitution at codon 261 of melR | 5 |
| Oligonucleotides | ||
| D31738 | 5′-CACCACCACTCCATGGCGAATACAGATACGTTTATGGCCAGCAGCGACGAAAAACAG-3′ | |
| D31739 | 5′-GTTAGTTGGTGCGGTGTAGCGGCCCAGACCAGCGTG-3′ | Used to detect melAB promoter DNA |
| D42581 | 5′-CCTCCGTGGCCCGTGGTCTAATTTATGATTAACAG-3′ | |
| D42582 | 5′-CCAATGATAATCACGTCACTTGATTGCGAGTCGC-3′ | Used to detect glp promoter DNA |
| D42986 | 5′-GCCAGGTCGTGAGGATTTGATTG-3′ | |
| D42987 | 5′-GAATGCCATTAGCATCAACCAG-3′ | Used to detect melA DNA |
| D44685 | 5′-GGTGCGGCTGTCGAACAGTAAATAG-3′ | |
| D44686 | 5′-GCCCGCCGAATGGGAAACCCTCAG-3′ | Used to detect yabN DNA |
| D44566 | 5′-CGCCATATACCGCTGGTTCGGTG-3′ | |
| D44567 | 5′-CTGGCAATTCTTCGTCATGTTCGG-3′ | Used to detect bglF DNA |
ChIP.
In all experiments, in vivo cross-linking of bacterial nucleoprotein was initiated by the addition of formaldehyde (final concentration of 1%) to cultures. After 20 min, cross-linking was quenched by the addition of glycine (final concentration of 0.5 M). Typically, cells were then harvested from 10 ml of culture by centrifugation, washed twice with Tris-buffered-saline (pH 7.5), resuspended in 1 ml of lysis buffer (10 mM Tris [pH 8.0], 20% sucrose, 50 mM NaCl, 10 mM EDTA, 10 mg of lysozyme per ml) and incubated at 37°C for 30 min. Following lysis, 4 ml of immunoprecipitation buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) and phenylmethylsulfonyl fluoride (final concentration of 1 mM) were added. Cellular DNA was then sheared by sonication to an average size of 500 to 1,000 bp. Cell debris was removed by centrifugation and the supernatant was retained for use as the input sample in immunoprecipitation experiments.
An 800-μl aliquot of the input sample was used for each immunoprecipitation experiment. The sample was incubated with 20 μl of Ultralink protein A/G beads (catalogue no. 53132; Pierce) and 5 μl of serum containing RNA polymerase β subunit mouse monoclonal antibody (Neoclone; Madison, Wis.) or MelR rabbit polyclonal antibody (E. Tamai, Kagawa University) for 90 min at room temperature on a rotating wheel. An immunoprecipitation experiment without antibody was also set up as a negative control. The beads were collected from each sample by using Spin-X centrifuge tube filters (catalogue no. MFG003247333; VWR-International) and washed twice with immunoprecipitation buffer, once with immunoprecipitation buffer plus 500 mM NaCl, once with wash buffer (10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 1 mM EDTA, 0.5% Nonidet-P40, 0.5% sodium deoxycholate) and once with Tris-EDTA buffer (pH 7.5). Immunoprecipitated complexes were then removed from the beads by treatment with elution buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1% SDS) at 65°C for 10 min. Immunoprecipitated samples were uncross-linked by incubation for 2 h at 42°C and 6 h at 65°C in 0.5× elution buffer plus 0.8 mg of pronase per ml. Prior to analysis, DNA was purified from the immunoprecipitate by using a PCR purification kit (QIAGEN) and resuspended in 200 μl of water. All ChIP assays were repeated at least twice, and results were found to be reproducible within an error margin of 20%.
Following purification, PCR was used to analyze immunoprecipitated DNA; 2.5-μl DNA samples were used in a 50-μl reaction mix containing a 1 μM concentration of each oligonucleotide primer. DNA amplification was catalyzed by Expand DNA polymerase (Roche), and the PCR was allowed to proceed for 25 to 28 cycles before 5 μl of the reaction was analyzed by electrophoresis on a 7.5% polyacrylamide gel. Fewer than 25 PCR cycles typically yielded insufficient DNA for visualization by ethidium bromide staining, whereas too many PCR cycles allowed each reaction to reach an end point, preventing the observation of differences in amplification.
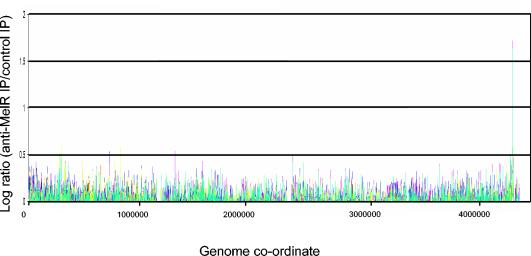
For the array analysis, independent duplicate genomic DNA samples, immunoprecipitated with anti-MelR, were amplified by PCR by using random oligonucleotide primers. Approximately 5 μg of amplified DNA was terminally labeled with biotin-ddUTP and hybridized to a GeneChip E. coli antisense genome array (Affymetrix) that was then washed, stained, and scanned according to the manufacturer's instructions. Signals generated from the arrays were subtracted from background values and normalized by applying the LOWESS algorithm and the R software package (www.bioconductor.org). The sequence of each probe present in the array was compared to the MG1655 genome sequence. Signals generated by probes with sequences that matched more than one locus in the MG1655 genome were discarded. Additionally, where several probes matched the same section of the MG1655 genome, the signal generated by one overlapping probe was randomly selected, whereas the signals from the other overlapping probes were discarded. The data set generated from each of the anti-MelR-immunoprecipitated DNA samples was compared to data sets generated from DNA immunoprecipitated with anti-LexA antibodies (Upstate) and anti-Gal4 antibodies (Santa-Cruz), which were used to calculate a background value for each probe. We compared these data sets and, for each probe, calculated the logarithm of the anti-MelR:anti-LexA or anti-MelR:anti-Gal4 signal ratio. Large positive signals represent sections of the genome found more frequently in the anti-MelR-immunoprecipitated DNA than the anti-LexA- and anti-Gal4-immunoprecipitated DNA. Data from the microarray experiment (see Fig. 3) show that a 500-bp window moved progressively through the MG1655 genome.
FIG. 3.
The genome-wide DNA binding profile of MelR. The graph shows the results of a microarray experiment designed to determine the DNA composition of nucleoprotein precipitated with anti-MelR. The signal derived from each of two independent anti-MelR immunoprecipitations was compared to that generated from anti-LexA immunoprecipitations or anti-Gal4 immunoprecipitations. The data plotted on the graph are the log of the anti-MelR/anti-LexA (yellow and pink) and anti-MelR/anti-Gal4 (dark blue and cyan) ratios.
Microarray analysis: transcriptomics.
Samples (12 ml) from triplicate independent cultures were mixed with 24 ml of RNA Protect (QIAGEN) and immediately centrifuged for 10 min at 5,000 × g. Total RNA was extracted from the bacterial pellets by using an RNeasy kit (QIAGEN) and reverse transcribed and fluorescently labeled with Cy dyes by using a CyScribe postlabeling kit (Amersham). Samples from cultures of cells carrying melR were labeled with Cy5 NHS (N-hydroxysuccinimide) ester dye, while samples from the control ΔmelR strain were labeled with Cy3 NHS ester (Amersham). Lucidea Universal Scorecard spike mRNA reference controls (Amersham) were added to the reverse transcriptase reactions for both samples.
Labeled cDNAs were analyzed by using arrays printed onto Corning GAPSII glass slides with the 6,112 70-mer oligonucleotides of the Operon Array Ready E. coli set 1.0 (Operon; QIAGEN) as described by Zhang et al. (15). For each experiment, equal quantities (80 pmol) of Cy5- and Cy3-labeled cDNA were added to a final volume of 80 μl of hybridization solution containing 25% formamide, 10 mg of bovine serum albumin (fraction V) per ml, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS, 8 μg of poly(A), and 1× Denhardt's solution. The cDNA probes were denatured at 95°C for 3 min and hybridized for 16 h at 42°C. Slides were then washed at 42°C with 2× SSC-0.1% SDS for 2 min and at room temperature with 0.2× SSC and twice with 0.05× SSC for 2 min. Slides were dried by low speed centrifugation and scanned by using an Axon 4000A scanner. The signal intensity of each spot in the microarray was quantified by using GenePix software (Axon).
Data were analyzed by using GeneSpring software (Silicon Genetics). Raw signal intensity data generated by GenePix were first transformed by using an intensity dependent LOWESS normalization to eliminate Cy dye bias. Spots with an intensity value lower than the error model cutoff value were then filtered out. Data from the three independent experiments were further filtered by using the Students' t test with a cutoff value of 0.05.
RESULTS AND DISCUSSION
ChIP can be used to monitor binding of MelR and RNA polymerase at the E. coli melAB operon.
MelR is a melibiose-triggered transcription activator that is essential for melibiose-dependent induction of the E. coli melAB operon. Previous studies identified DNA sites for MelR at the melAB promoter and showed that, in the presence of melibiose, MelR activates transcription initiation at the melAB promoter by making a direct interaction with the RNA polymerase σ subunit (1, 5). Our in vitro studies showed that MelR can bind to the melAB promoter in the absence of melibiose.
In the present work, we have exploited ChIP assays to measure MelR binding to the melAB regulatory region and melibiose-induced recruitment of RNA polymerase to the melAB operon. In ChIP experiments, a cell's nucleoprotein is cross-linked with formaldehyde, extracted, and then fragmented by sonication such that the average DNA fragment is 500 bp. Antibodies directed against the protein of interest are then used to select protein cross-linked DNA fragments that are analyzed by PCR or array technology. Hence, in our first experiments we used WAM131, an E. coli K-12 strain grown in the presence or absence of melibiose and carrying a wild-type mel operon that had been used in previous studies of MelR (1). Cells were treated with formaldehyde and lysed, and the chromosomal DNA was sheared by sonication. Anti-MelR polyclonal antibody was then used to immunoprecipitate DNA fragments attached to MelR, with a parallel sample without antibody run as a control.
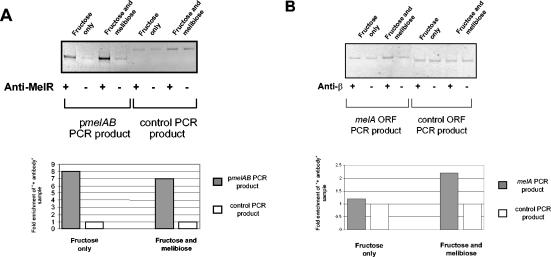
Figure 1A shows the PCR analysis of the DNA immunoprecipitated by using primers that are specific for the melAB promoter and, as a control, primers for the bglF promoter (Table 1). With the melAB promoter primers, it is clear that melAB DNA is enriched in the anti-MelR immunoprecipitate, compared to the control (without antibody) sample. To check that the anti-MelR immunoprecipitate was specifically enriched in melAB promoter DNA, the bglF primers were used to amplify a control region of the chromosome. With these primers, the same signal is seen with both the anti-MelR and control immunoprecipitates, presumably because MelR does not bind to any target near bglF.
FIG. 1.
ChIP analysis of MelR and RNA polymerase binding to the melAB promoter. (A) The figure illustrates the results of a ChIP experiment designed to monitor the binding of MelR to the melAB promoter in the presence and absence of melibiose. The top panel shows a gel on which PCR products, generated with primers designed to detect either melAB promoter DNA or control bglF open reading frame DNA in each immunoprecipitate, were analyzed. The bottom panel is a quantitative representation of the data and shows the ratio of the PCR signal in immunoprecipitate with antibody to immunoprecipitate without antibody, generated by mel or bglF primers in both sets of growth conditions. (B) The figure shows the results of a ChIP experiment designed to monitor the binding of RNA polymerase to the melAB promoter in the presence and absence of melibiose. The top panel depicts a gel on which PCR products, generated with primers designed to detect either melA DNA or control yabN open reading frame DNA in each immunoprecipitate, were analyzed. The bottom panel is a quantitative representation of the raw data and shows the ratio of the PCR signal in immunoprecipitate with antibody to immunoprecipitate without antibody, generated by melA and yabN primers in both sets of growth conditions.
The enrichment of melAB promoter DNA detected in anti-MelR immunoprecipitates must be due to the binding of MelR to the melAB regulatory region in vivo. Results shown in Fig. 1 have similar signals from cells grown in both the absence and presence of melibiose. We interpret this as evidence that MelR binding is independent of the inducer, melibiose. Since we know that the induction of transcription of the melAB genes is dependent on melibiose (13), we repeated the immunoprecipitation assays by using antibody directed against the RNA polymerase β subunit. Immunoprecipitates were analyzed by PCR by using primers (Table 1) specific for an internal segment of the melA gene or, as a control, the yabN gene, which is known not to be expressed in laboratory conditions. Data shown in Fig. 1B indicate that melA DNA is enriched in the immunoprecipitate with anti-β antibody only with cells grown with melibiose. This result argues that melibiose increases the association of the RNA polymerase β subunit (hence, RNA polymerase) with the melA gene due to melibiose-induced activation of transcription at the melAB promoter. The key control is the mock precipitate obtained in the absence of antibody. Thus, the immunoprecipitate with anti-β antibody is enriched for melA DNA compared to the no-antibody control only when derived from cells grown with melibiose. With the control yabN probes, as expected, the signal is not increased by the anti-β antibody, presumably because this gene is not transcribed.
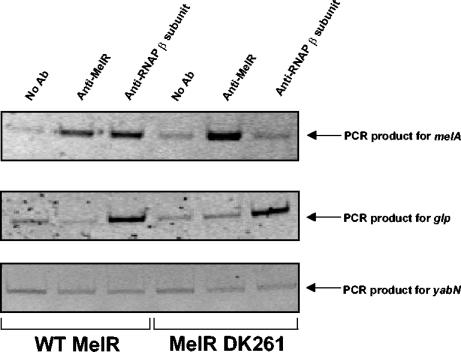
In previous work, it was shown that melibiose-dependent induction of melAB transcription is due to a direct interaction between MelR residue D261 and the RNA polymerase σ subunit, and, hence, MelR carrying the DK261 substitution can bind to target sites at the melAB promoter but is unable to activate transcription (5). Thus, here we sought to repeat the immunoprecipitation experiments in the presence of MelR DK261. To do this we used strain WAM132, a ΔmelR derivative of WAM131, carrying plasmid pLG314, encoding either wild-type MelR or mutant MelR DK261 (Table 1). Figure 2 shows the results of analyses of immunoprecipitates obtained from cells grown with melibiose in the presence of wild-type MelR or the MelR DK261 mutant with anti-MelR antibodies, anti-RNA polymerase β subunit antibodies, or no antibody as a control. Samples were analyzed with PCR primers specific for melA, glp, or yabN (Table 1).
FIG. 2.
The effect of a positive control substitution on the binding of MelR and RNA polymerase to the melAB promoter in vivo. Shown are the results of a ChIP experiment designed to monitor the effects of the MelR DK261 positive control substitution on the binding of MelR and RNA polymerase to the melAB promoter. The top panel is a gel on which PCR products, generated with melA primers and the anti-MelR, anti-RNA polymerase, or no-antibody control immunoprecipitate, were analyzed. The middle and bottom panels show the results of PCRs where the immunoprecipitates were reanalyzed by using primers to detect the glp or yabN genes.
The results of the experiment with the melA primers are shown in the top panel of Fig. 2. The signal with anti-MelR antibodies is enhanced compared to the control (no antibody) with immunoprecipitates from cells carrying both wild-type MelR and mutant MelR DK261. We interpret this as due to binding of both wild-type and mutant MelR to the melA regulatory region, as expected. In contrast, the signal with anti-β antibodies is enhanced compared to the no-antibody control only with immunoprecipitates from cells carrying wild-type MelR. Presumably, this is because RNA polymerase is recruited to transcribe the mel operon by wild-type MelR but not by the mutant MelR DK261. Results of two control experiments are shown in the other parts of Fig. 2. The middle panel shows analyses of the immunoprecipitates generated by using primers to probe the glp promoter. This is a MelR-independent promoter that is active in the minimal medium used in our experiments. Thus, the glp DNA signal is enhanced compared to the no-antibody control in immunoprecipitates generated by using the anti-β antibodies but not the anti-MelR antibodies. We interpret this as due to binding of RNA polymerase but not MelR to the glp promoter. Note that, as expected, the MelR DK261 mutation does not alter the binding of RNA polymerase to the glp promoter. The lower panel of Fig. 2 shows analyses of the immunoprecipitates generated by using primers to probe the nonexpressed yabN gene. The results show that the yabN signal is not enhanced, compared to the no-antibody control, in immunoprecipitates generated with either the anti-β antibodies or the anti-MelR antibodies. Thus, yabN provides a useful baseline control segment of the E. coli chromosome to which neither MelR nor RNA polymerase binds.
Genomic studies on MelR.
As well as its use in reporting binding of proteins to specific chromosomal targets, ChIP assays can be used to investigate the range of different binding sites of proteins throughout genomes. Thus, immunoprecipitates generated with MelR antibodies will contain DNA fragments corresponding to all DNA targets to which MelR is bound at the moment of cross-linking. To analyze the range of DNA fragments in our immunoprecipitates, we exploited array technology, using the Affymetrix E. coli antisense genome array of 170,000 25-mer oligonucleotides. Because the Affymetrix array is based on the E. coli K-12 MG1655 strain, we remade immunoprecipitates by using MG1655 grown with melibiose, after having checked that induction of the melAB operon in MG1655 is similar if not identical to induction in strain WAM131.
Figure 3 shows a graphic representation of the data set after DNA that had been immunoprecipitated by the anti-MelR antibody was labeled and hybridized to the array. The data show one clear peak centered at position 4339220 of the MG1655 chromosome. Since the melA open reading frame starts at position 4339489, we interpret the signal as due to MelR binding at the mel operon regulatory region. From this experiment we can conclude that there is only one major target for MelR in the MG1655 chromosome. We can make no conclusion about signals due to secondary weaker sites at other locations since any such signals would be lost in the background noise of the experiment.
To cross-check the conclusion that the principal, and possibly sole, target for MelR in E. coli is at the mel operon promoter region, we performed a transcriptomics experiment by using array technology to compare the RNA species made in cells with and without a functional melR gene. A complication arises from the need to include melibiose in cultures in order to induce MelR-dependent transcription activation. In order to avoid secondary effects on transcription due to the metabolism of melibiose following MelR-dependent induction of the mel operon, we used the newly isolated FY53 KR182 MelR mutant that is nearly fully active in the absence of melibiose (C. L. Webster and S. Busby, unpublished results). RNA was isolated from WAM132 ΔmelR cells grown in the absence of melibiose and carrying plasmid pJW15 encoding mutant MelR FY53 KR182 or a plasmid lacking melR as a control (Table 1). The RNA was labeled and hybridized to an array printed with the 6,112 70-mer oligonucleotides of the Operon Array Ready E. coli set that covers each open reading frame as described by Zhang et al. (submitted). Table 2 shows that in our experiments, the levels of transcripts of three genes (apart from melR) are increased by MelR. As expected, the levels of melA and melB were greatly increased, while we found a much smaller unexpected induction of citB RNA. Notwithstanding the citB result, which we cannot explain and which is unsupported by the Affymetrix data, we conclude that the mel operon is the principal, if not the sole, target for MelR in E. coli.
TABLE 2.
Transcriptome analysis of effects of MelR KR182 FY53a
| Gene | Product | melR*/ΔmelR | P |
|---|---|---|---|
| melR | Melibiose operon regulatory protein | 63.987 | 0.013 |
| melA | Alpha-galactosidase | 44.323 | 0.004 |
| melB | Melibiose carrier protein | 31.015 | 0.012 |
| citB | Transcriptional regulatory protein | 2.255 | 0.030 |
The table summarizes data from three independent experiments to investigate the effects of MelR FY53 KR182 on the transcript profile of WAM132 ΔmelR cells by using microarray analysis. The table lists the names and identities of genes that are up-regulated by more than twofold by MelR FY53 KR182. The third column lists the ratio of the level of each transcript in the strain carrying melR FY53 KR182 (melR*) to the control strain lacking melR. P values were determined by a t test.
Conclusions.
The ChIP assay has been widely exploited to study interactions between eukaryotic proteins and their DNA targets for a number of years (3), but its use in bacteria has been limited, one of the first major studies being that by L. Shapiro, H. McAdams, and colleagues of Caulobacter crescentus (9). Here we have shown that ChIP can easily be applied to studying E. coli gene regulatory elements. We have focused on the E. coli melAB promoter, a complex promoter that is activated by MelR protein in response to an inducer ligand, melibiose. We used ChIP directly to monitor MelR and RNA polymerase binding to this promoter in vivo. Our results show that MelR can bind at the melAB promoter in both the presence and absence of melibiose and that melibiose stimulates association of RNA polymerase, presumably because it is recruited to the melAB promoter. We were able to show that association of RNA polymerase is prevented by the DK261 substitution in MelR, likely because residue D261 is one of the key points of contact between MelR and RNA polymerase during transcription activation. Thus, ChIP can be used directly to measure the comings and goings of proteins at any target in vivo. Note that the only other available method is in vivo footprinting, which is contingent on finding an appropriate signature signal and, thus, is not universally applicable (3). Our results argue that MelR binds to the melAB promoter and, when triggered by melibiose, activates transcription by recruitment of RNA polymerase. This is in contrast to the prerecruitment mechanism recently suggested for the related SoxS and MarA activators that appear to bind to free RNA polymerase before binding to target promoters (6, 10).
In the second part of the study we have shown how ChIP analysis can be extended to study the global distribution of a transcription factor. Recall that many E. coli transcription factors bind multiple sites, while ∼20% of factors are thought to bind at a single locus (11). Clearly, ChIP combined with array technology provides a direct method to catalogue binding targets independently of their consequences on gene expression. Our results show that the mel operon is the major target, if not the sole target, for binding of the MelR protein, and this conclusion was supported by transcriptome analysis. To our knowledge, this is the first report of the use of ChIP, together with arrays and transcriptomics, to study transcriptional regulation in E. coli. Although our target in this study, MelR, is relatively simple, it is clear that these technologies can easily be extended to aid the study of far more complex factors.
Acknowledgments
We thank BBSRC for funding this work with grants G15075 and EGA16107.
REFERENCES
- 1.Belyaeva, T., J. Wade, C. Webster, V. Howard, M. Thomas, E. Hyde, and S. Busby. 2000. Transcription activation at the Escherichia coli melAB promoter: the role of MelR and the cyclic AMP receptor protein. Mol. Microbiol. 36:211-222. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mao, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Farnham, P. 2002. In vivo assays to examine transcription factor localisation. Methods 26:1-2. [DOI] [PubMed] [Google Scholar]
- 4.Grainger, D., T. Belyaeva, D. Lee, E. Hyde, and S. Busby. 2003. Binding of the Escherichia coli MelR protein to the melAB promoter: orientation of MelR subunits and investigation of MelR-DNA contacts. Mol. Microbiol. 48:335-348. [DOI] [PubMed] [Google Scholar]
- 5.Grainger, D., C. Webster, T. Belyaeva, E. Hyde, and S. Busby. 2004. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with its DNA target site and with domain 4 of the RNA polymerase σ subunit. Mol. Microbiol. 51:1297-1309. [DOI] [PubMed] [Google Scholar]
- 6.Griffith, K., I. Shah, T. Myers, M. O' Neill, and R. Wolf. 2002. Evidence for “pre-recruitment” as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem. Biophys. Res. Commun. 291:979-986. [DOI] [PubMed] [Google Scholar]
- 7.Howard, V., T. Belyaeva, S. Busby, and E. Hyde. 2002. DNA binding of the transcription activator protein MelR from Escherichia coli and its C-terminal domain. Nucleic Acids Res. 30:2692-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelsall, A., C. Evans, and S. Busby. 1985. A plasmid vector that allows fusion of the E. coli galactokinase gene to the translation startpoint of other genes. FEBS Lett. 180:155-159. [Google Scholar]
- 9.Laub, M., S. Chen, L. Shapiro, and H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 99:4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin, R., W. Gillette, N. Martin, and J. Rosner. 2002. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in E. coli. Mol. Microbiol. 43:355-370. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Antonio, A., and J. Collado Vides. 2000. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6:1-8. [DOI] [PubMed] [Google Scholar]
- 12.Wade, J., T. Belyaeva, E. Hyde, and S. Busby. 2001. A simple mechanism for co-dependence on two activators at an Escherichia coli promoter: the role of MelR and the cyclic AMP receptor protein. EMBO J. 20:7160-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webster, C., K. Kempsell, I. Booth, and S. Busby. 1987. Organisation of the regulatory region of the Escherichia coli melibiose operon. Gene 59:253-263. [DOI] [PubMed] [Google Scholar]
- 14.Williams, J., C. Michán, C. Webster, and S. Busby. 1994. Interactions between the E. coli MelR transcription activator protein and operator sequences at the melAB promoter. Biochem. J. 300:757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, A. J. Roe, I Vlisidou, R. K. Shaw, M. P. Stevens, et al. Regulators encoded in the ETT2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]