Abstract
Several species of mycobacteria express abundant glycopeptidolipids (GPLs) on the surfaces of their cells. The GPLs are glycolipids that contain modified sugars including acetylated 6-deoxy-talose and methylated rhamnose. Four methyltransferases have been implicated in the synthesis of the GPLs of Mycobacterium smegmatis and Mycobacterium avium. A rhamnosyl 3-O-methytransferase and a fatty acid methyltransferase of M. smegmatis have been previously characterized. In this paper, we characterize the methyltransferases that are responsible for modifying the hydroxyl groups at positions 2 and 4 of rhamnose and propose the biosynthetic sequence of GPL trimethylrhamnose formation. The analysis of M. avium genes through the creation of specific mutants is technically difficult; therefore, an alternative approach to determine the function of putative methyltransferases of M. avium was undertaken. Complementation of M. smegmatis methyltransferase mutants with M. avium genes revealed that MtfC and MtfB of the latter species have 4-O-methyltransferase activity and that MtfD is a 3-O-methyltransferase which can modify rhamnose of GPLs in M. smegmatis.
Mycobacteria are acid-fast bacteria that have characteristic lipid-rich cell envelopes that afford the cells protection from desiccation, chemical disinfectants, and some antibiotics. The cell envelopes of pathogenic mycobacteria are one of the major virulence factors that assist the bacteria to live within host macrophages and withstand the bactericidal defenses of such cells. The mycobacterial cell envelope has a multilaminar structure, comprising an inner layer of peptidoglycan and arabinogalactan polysaccharides and mycolic acids and an outer layer made of species-specific glycolipids and phospholipids. Among the lipids of the outer layer are the glycopeptidolipids (GPLs) that are characteristic of some nontuberculosis mycobacteria such as Mycobacterium avium and Mycobacterium smegmatis. The core structure of most GPLs comprises a long-chain fatty acid (3-hydroxy and 3-methoxy C26-34) amidated with a tripeptide-amino alcohol (d-Phe-d-allo-Thr-d-Ala-l-alaninol). In most cases, the hydroxyl groups of the allo-Thr and/or alaninol are modified with 6-deoxytalose (dTal) or rhamnose (Rha), respectively. In M. smegmatis, the dTal is variably acetylated, while the Rha residue can be modified with up to three methyl groups. Heterogeneity in the methylation of both amide-linked fatty acids and the Rha residue results in the appearance of four main GPL species in M. smegmatis. GPL-1 and GPL-2 have 3-methoxy C26-34 fatty acids but differ from each other in the degree of methylation of the terminal Rha, in that GPL-1 has 2,3,4-tri-O-Me-Rha and GPL-2 has 3,4-di-O-Me-Rha. GPL-1a and GPL-2a have tri- and di-Me-Rha as above; however, these GPLs have 3-hydroxy C26-34 fatty acids. We have simplified our nomenclature of the GPLs from that used in Patterson et al. (9).
M. avium GPLs have the same lipopeptide core as M. smegmatis GPLs, but the alaninol is glycosylated with 3-O-Me-Rha or 3,4-di-O-Me-Rha, and the dTal can be extended with additional sugars. A locus designated ser2 contains some of the genes encoding enzymes required for synthesis of the haptenic disaccharide of GPLs of serotype 2 M. avium. Eckstein et al. showed that the simpler GPLs naturally found in M. smegmatis could serve as intermediates in the biosynthesis of a ser2-like locus encoding GPL in recombinant M. smegmatis (3). We have identified an equivalent region in M. smegmatis, shown that the mps gene encodes a peptide synthetase that makes the GPL peptide, and assigned the roles to two of the four potential methyltransferases encoded in the GPL biosynthetic locus (1, 7, 9). Mtf1 is a rhamnosyl 3-O-methyltransferase, and mtf2 encodes a fatty acid O-methyltransferase that modifies the hydroxyl at C3 of the GPL fatty acid (7, 9). For the purposes of clarity, we propose that the M. smegmatis genes formerly designated mtf1, mtf2, mtf3, and mtf4 (GenBank accession no. AY138899) be renamed rmt3 for rhamnosyl 3-O-methyltransferase, fmt for fatty acid O-methyltransferase, rmt4 for rhamnosyl 4-O-methyltransferase, and rmt2 for rhamnosyl 2-O-methyltransferase, respectively. As we show, this nomenclature reflects the functions of the respective methyltransferases. In this paper, we define the roles of the remaining two putative methyltransferase genes, rmt4 and rmt2, and identify the functional homologues from M. avium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA manipulation.
Bacterial strains and their derivatives are listed in Table 1. Escherichia coli was grown on Luria medium, and M. smegmatis and M. avium were grown on Middlebrook 7H9 or 7H10 medium (Difco). Antibiotics were added at the following concentrations as required: ampicillin, 100 μg/ml; kanamycin, 20 μg/ml; hygromycin, 200 μg/ml; and streptomycin, 20 μg/ml. Sucrose (10% [wt/vol]) was included in media for sacB counter selection. DNA manipulation was performed according to standard protocols (12). PCRs were carried out with an MJ Research Peltier Thermal cycler. Plasmid DNA was introduced into bacterial cells with a Bio-Rad gene pulser. The manufacturer's recommendations were followed for preparation and transformation of E. coli, whereas the protocols of Jacobs et al. were used for M. smegmatis (6).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant feature | Source |
|---|---|---|
| Strains | ||
| M. smegmatis | ||
| mc2155 | Parent strain used for mutagenesis | 14 |
| Myco29 | rmt3::Tn611, Kanr Strr | 9 |
| Myco493 | fmt::str | 7 |
| Myco657 | Δrmt4, Kanr | This study |
| Myco694 | Δrmt2, Kanr | This study |
| E. coli DH5α | Host for cloning | Stratagene |
| M. avium TMC 724 | Source of mtfA to -D | |
| Plasmids | ||
| pGEM-T Easy | Vector for efficient cloning of PCR products | Promega |
| pBluescript SK(+) | Cloning vector | Stratagene |
| pK18mobsacB | aph sacB | 13 |
| pK18mob | aph | 13 |
| pHBJ395 | pBluescript SK(+) containing aph | This study |
| pHBJ428 | pBluescript SK(+) containing str and sacB | This study |
| pHBJ430 | pHBJ428, Δrmt4 aph | This study |
| pHBJ488 | pHBJ428, Δrmt2 aph | This study |
| pVV16 | Mycobacterial-E. coli shuttle vector, hyg | T. Eckstein |
| pHBJ415 | pVV16, rmt4 | This study |
| pHBJ409 | pVV16, rmt2 | This study |
| pHBJ475 | pVV16, mtfA (M. avium) | This study |
| pHBJ512 | pVV16, mtfB (M. avium) | This study |
| pHBJ505 | pVV16, mtfC (M. avium) | This study |
| pHBJ502 | pVV16, mtfD (M. avium) | This study |
Preparation of mycobacterial genomic DNA.
Genomic DNA was extracted from mycobacteria as follows. Mycobacterial cells were harvested from liquid cultures by centrifugation and then incubated for 1 h at 37°C in 1 ml of Tris-EDTA buffer containing 200 μg of proteinase K/ml, 10 mg of lysozyme/ml, and 100 μg of RNase A/ml. The suspensions were centrifuged, and the cell pellet was resuspended in 750 μl of 4 M guanidine thiocyanate, 25 M sodium citrate, and 0.5% Sarkosyl; 200 mg of 100-μm diameter glass beads was added to each tube. Cells were lysed with a tissue homogenizer (Biospec), and then the beads and unlysed cells were removed by centrifugation. The lysate was extracted three times with equal volumes of phenol:chloroform:isoamyl alcohol (25:24:1 [vol/vol]) and then once with chloroform:isoamyl alcohol (25:1 [vol/vol]). The genomic DNA was precipitated with sodium acetate and ethanol and then resuspended in Tris-EDTA buffer.
Southern blotting.
Purified genomic DNA was digested with restriction endonucleases and resolved by electrophoresis in 1% (wt/vol) agarose in 1× TAE buffer and then transferred to nylon membranes (Hybond) for Southern blotting (12). DNA probes for hybridization were prepared by PCR amplification of the rmt4 open reading frame (ORF) with primers 383 and 384 on an M. smegmatis genomic DNA template (Table 2). The PCR product was purified with a QIAquick PCR purification kit (QIAGEN) and digoxigenin labeled with a DIG DNA labeling kit (Roche). Membranes were incubated with labeled probes and processed according to the manufacturer's protocols.
TABLE 2.
Oligonucleotide primers
| Primer | Target or purpose | Sequence (5′-3′) |
|---|---|---|
| 299 | SOE to delete rmt4 | CCCTCTAGACGTTTCACACTGACCGTAC |
| 300 | SOE to delete rmt4 | CGTGCTTGGCACGGTAATGGCCATAACGTGCGCGTGCTCG |
| 301 | SOE to delete rmt4 | ACGAGCACGCGCACGTTATGGCCATTACCGTGCCAAGCACG |
| 302 | SOE to delete rmT4 | AGCTCTAGACAAACTGAGCTGATCATCG |
| 310 | sacB | GGGGTACCGCGTCGCTTGGTCGGTCA |
| 382 | sacB | TTGGTACCTTCTGAGCGGACTCT |
| 383 | rmt4 | TTTGGATCCGACTGATCGCGATAGCCGA |
| 384 | rmt4 | CCCGGATCCATCTCGGCGATCTTCACTC |
| 388 | rmt2 | TTTGGATCCGGGGTGGGCTCGGTCGTC |
| 389 | rmt2 | GCTGGATCCAATATATGAGCGGGCAGACC |
| 428 | PCR to delete rmt2 | ATCTAGAGGGTTCCGGCCTCTCA |
| 442 | PCR to delete rmt2 | TTTCTAGAGTGGTGGGTACAACGCAACTCCTC |
| 443 | PCR to delete rmt2 | CTACATTCGTCGACTGGCCAAACCTCGGCTGTATCACCAAG |
| 444 | PCR to delete rmt2 | ATACAGCCGAGGTTTGGCCAGTCGACGAATGTAGCGATGAA |
| 458 | mtfA | TGAATCGGTGGGCATTGTCG |
| 459 | mtfA | GTGCGGCACGGCGAGTTTG |
| 496 | mtfB | GGATCCGACTGATCGTGACATGCGATC |
| 497 | mtfB | AAGCTTGCGCCACAGCACGCCG |
| 498 | mtfC | GGATCCGACCGATCACGACACGCGG |
| 499 | mtfC | AAGCTTGGGCTTGCGCCACAGCACAC |
| 500 | mtfD | GGATCCGCACGCCGATGTTATGGTTG |
| 501 | mtfD | AAGCTTGCGGGCCGGCGTTGATGAG |
Construction of sacB-str suicide vector.
The vector for delivery of allelic exchange cassettes was based on pBluescript SK(+) (Stratagene) which is unable to replicate in mycobacteria. This suicide vector contained a streptomycin resistance marker (str) for positive selection of transformants in which the plasmid had integrated into the host chromosome. It also contained the levansucrase gene sacB so that sucrose could be used for counter selection of single-crossover (SCO) mutants (10). The vector was constructed as follows. The sacB gene was PCR amplified from pK18mobsacB (13) with primers 382 and 310 (Table 2), ligated into pGEM-T Easy (Promega) and then subcloned as an EcoRI fragment into pBluescript SK(+). A str gene was excised from pHP45 (11) with BamHI and was cloned into the BamHI site of the above plasmid, resulting in the sacB-str vector, pHBJ428.
Deletion of rmt4 and rmt2.
The rmt4 and rmt2 genes were separately deleted from M. smegmatis by homologous recombination between the chromosome and allelic exchange cassettes carried by the sacB-str vector pHBJ428. The cassettes comprised two 1-kb sequences corresponding to regions from the genome on either side of the target gene, and a kanamycin resistance marker (aph) replaced the target gene.
For deletion of rmt4, the two flanking sequences were amplified and spliced together by splice overlap extension (SOE) PCR with primers 299, 300, 301, and 302 (Table 2) (16). An MscI site was created at the junction of the fragments, and XbaI sites occurred at the termini. The spliced product was cloned into pGEM-T Easy (Promega). The aph gene was excised from pK18mob by digestion with NheI and BstBI, cloned, and ligated into the MscI site of the cassette. The cassette was excised from pGEM-T Easy by digestion with XbaI and cloned into an XbaI site on the sacB-str vector, resulting in the final construct, pHBJ430. For the deletion of rmt2, the upstream and downstream sequences were amplified with primers 442 and 443 and with 444 and 428, respectively (Table 2), joined with an MscI site at the junction, and cloned into pGEM-T Easy. The aph gene was cloned into the MscI site, and then the cassette was excised from the pGEM-T Easy by digestion of NotI and cloned into the NotI-digested sacB-str vector, resulting in pHBJ488. Both pHBJ430 and pHBJ488 had the target gene replaced with aph, which was cloned so that the ORF was in the same direction as the original target gene had been, allowing transcription to proceed beyond the cassette to minimize the chances of polar effects on downstream genes.
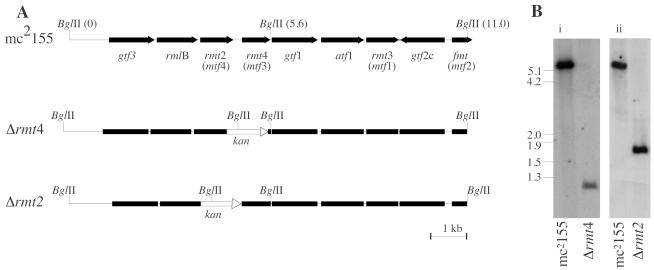
The plasmids pHBJ430 and pHBJ488 were introduced into M. smegmatis, and then SCO mutants were isolated by selection with kanamycin and streptomycin in 7H10 agar. An SCO mutant was grown in 7H9 broth without antibiotics for 3 days and then plated on 7H10 agar containing kanamycin and sucrose. Survivors were screened for sensitivity to streptomycin, and double-crossover (DXO) mutants (kanamycin-sucrose resistant, streptomycin sensitive) were isolated. The DXO mutants were confirmed by Southern blotting as follows. BglII-digested genomic DNA from a putative DXO mutant was blotted onto a nylon membrane. A probe was prepared by PCR amplification of rmt4 with primers 383 and 384 (Table 2) and digoxigenin labeled as described above for Southern hybridization. The resulting hybridization pattern (Fig. 1) showed that rmt4 occurred on a 5.6-kb BglII fragment in the parent strain, mc2155, whereas the band from the DXO mutant was smaller (∼0.95 kb), corresponding to the size expected if rmt4 was replaced by aph and a BglII site was introduced (Fig. 1A). Hybridization occurred between the DXO mutant and the rmt4 probe because the mutant retained 75 bp of rmt4. The mutant was named Myco657 (Δrmt4). A putative DXO rmt2 deletion mutant was obtained and Southern blotted as described for Myco657. Replacement of the rmt2 gene with aph introduced a BglII site; consequently, the hybridizing BglII band was smaller (∼1.7 kb) than in the parent strain (5.6 kb). Southern blotting confirmed that rmt2 had been deleted, and the mutant was designated Myco694 (Δrmt2).
FIG. 1.
Restriction maps and Southern blot of GPL locus of Myco657 (Δrmt4) and Myco694 (Δrmt2). (A) Genetic arrangement of part of the GPL locus containing rmt2, rmt4, rmt3, and fmt. Positions of the BglII site are shown. The gene names used previously are shown in parentheses. The position where aph was inserted is designated kan. (B) Southern blot of BglII-digested genomic DNA extracted from M. smegmatis mc2155, Myco657 (Δrmt4), and Myco694 (Δrmt2). The blot was probed with digoxigenin-labeled DNA corresponding to rmt4. Gene replacement with aph introduced an additional BglII site, resulting in smaller hybridizing bands for the deletion mutants.
Complementation.
A plasmid carrying rmt4 was created to test complementation of Myco657. The rmt4 ORF was PCR amplified from M. smegmatis genomic DNA with primers 383 and 384, which incorporated BamHI sites at the ends (Table 2). The PCR product was cloned into the BamHI site of pVV16 to produce plasmid pHBJ415. Myco657 (Δrmt4) and mc2155 were then transformed with pHBJ415. The vector pVV16 is a derivative of pMV261 (15) and carries a hygromycin resistance marker. It allows expression of recombinant proteins from cloned genes from the GroEL promoter as fusions with a carboxyl-terminal hexahistidine epitope tag, although the tag was not utilized in these experiments. A plasmid for complementation of Myco694 (Δrmt2) was constructed as follows. The rmt2 ORF was PCR amplified with primers 388 and 389, which incorporate terminal BamHI sites (Table 2). The PCR fragment was cloned into the BamHI site of pVV16 to create plasmid pHBJ409, which was then introduced into mc2155 and Myco694 (Δrmt2). Controls were transformed with pVV16. Transformants were used in cell compositional analyses.
Cloning M. avium mtf genes.
The genes mtfA to mtfD of M. avium were PCR amplified from genomic DNA of M. avium TMC 724 with the primer pairs 458 and 459 (mtfA), 496 (BamHI) and 497 (HindIII) (mtfB), 498 (BamHI) and 499 (HindIII) (mtfC), and 500 (BamHI) and 501 (HindIII) (mtfD) (Table 2). Restriction sites incorporated into the primers are indicated above. In the cases of mtfB, mtfC, and mtfD, the PCR product was cloned into BamHI-HindIII-digested pVV16, whereas the mtfA fragment was blunt cloned into the PvuII site. Each plasmid and the vector control were transferred into mc2155, and each M. smegmatis methyltransferase mutant, specifically Myco29 (rmt3::Tn611) (9), Myco493 (fmt::str) (7), Myco657 (Δrmt4) (this study), and Myco694 (Δrmt2) (this study). The GPL profile of each of the transformed strains was analyzed by high-performance thin-layer chromatography (HPTLC) to determine which of the M. avium methyltranfserase genes could complement the M. smegmatis mutations and therefore indicate which methyltransferases were functional homologues.
Sequence analysis.
A neighbor-joining tree showing the relationship of authentic and putative methyltransferases was constructed using the protdist and neighbor-joining tree programs from PHYLIP (Phylogeny Inference Package) with the default parameters (J. Felsenstein, PHYLIP [Phylogeny Inference Package] version 3.5c, Department of Genetics, University of Washington, Seattle). Bootstrap values were determined using the seqboot program with1,000 bootstraps.
Extraction and compositional analysis of cell wall components.
Lipids were extracted from mycobacteria by Folch partitioning as previously described (4, 9). Cell wall lipids were recovered in the chloroform phase and subjected to alkaline methanolysis to cleave ester-linked fatty acids followed by partitioning between water-saturated 1-butanol (4 volumes) and water (2 volumes). The alkali-stable GPLs were analyzed by HPTLC with aluminum-backed Silica Gel 60 HPTLC plates (Merck) and a liquid phase containing chloroform:methanol (9:1 [vol/vol]). Plates were developed with orcinol-H2SO4 and charring at 100°C. For monosaccharide compositional analysis, fractions containing GPLs were hydrolyzed in 2 M trifluoroacetic acid (2 h at 100°C), and the acid was removed under a stream of nitrogen. Released monosaccharides were subsequently converted to their alditol acetate derivatives by reduction in sodium borodeuteride (NaBD4) and acetylation in acetic anhydride with 1-methyl-imidazole as a catalyst (5). For gas chromatography-mass spectrometry (GC-MS) analysis, conditions were as previously described (9).
RESULTS
Deletion of rmt2 and rmt4.
The rmt2 and rmt4 genes were individually deleted from M. smegmatis, resulting in two mutants, Myco657 (Δrmt4) and Myco694 (Δrmt2). Each mutant had deep rough-colony morphology compared to the parent strain, which is fairly flat and waxy when grown on Middlebrook 7H10 agar. This phenotype is typical of mutants that have defects in GPL synthesis (1). Complementation of these mutants with rmt4 and rmt2, respectively, restored the wild-type colony morphology and mature GPL synthesis (see below) suggesting that the phenotype is directly related to the gene deletion rather than to any polar effects on downstream genes.
Analysis of GPL content of M. smegmatis Myco657 (Δrmt4) and Myco694 (Δrmt2).
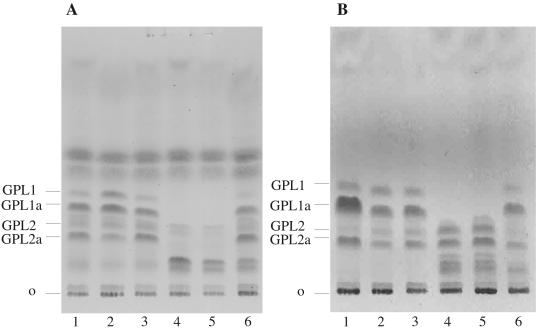
To determine whether the colony morphologies of Myco657 and Myco694 were due to changes in GPL levels or structures, the base-treated GPLs of the wild-type and mutant strains were analyzed by HPTLC (Fig. 2). As shown previously (9) wild-type M. smegmatis expresses two major GPL species that migrate as doublets on HPTLC, due to chain length heterogeneity in the amide-linked fatty acid (Fig. 2A, lane 1). Myco657 no longer expressed the GPL-1 doublet and contained very low levels of the GPL-2 doublet. However, a new GPL doublet with slower HPTLC mobility accumulated in this mutant (Fig. 2A, lane 4). Complementation of Myco657 with rmt4 largely restored synthesis of GPI-1 and GPL-2 to wild-type levels, although low levels of the polar GPL were still detected in the complemented cell line (Fig. 2A, lane 6). To further characterize the nature of the GPL defect in Myco657, the base-treated GPLs were acid hydrolyzed, and the released monosaccharides were analyzed as their alditol acetates by GC-MS. As previously shown, the major GPL species of wild-type M. smegmatis contained dTal; 3,4-di-O-Me-Rha; and 2,3,4-tri-O-Me-Rha (Fig. 3A) (9). In contrast, the Myco657 GPLs contained dTal and 3-O-Me-Rha as major sugars and 2,3-di-O-Me-Rha as a minor sugar (Fig. 3B). No 2,3,4-tri-O-Me-Rha was detected in the Myco657 GPLs (Fig. 3B). Complementation of Myco657 with rmt4 resulted in the appearance of GPLs with 3-O-Me-Rha, 2,3-di-O-Me-Rha, and 2,3,4-tri-O-Me-Rha (Fig. 3C). These data strongly suggest that the polar GPLs in Myco657 are modified with 3-O-Me-Rha and that the enzyme activity associated with Rha 4-O-methylation is missing in this Myco657. The presence of low levels of 2,3-di-O-Me-Rha in the Myco657 GPLs is consistent with the presence of low levels of GPL-1-GPL-1a in the mutant strain (Fig. 2A). The decrease in dimethylated GPLs in this mutant was consistently observed, suggesting that abrogation of Rha 4-O-methylation may reduce the efficiency of Rha 2-O-methylation.
FIG. 2.
HPTLC analysis of base-treated GPLs. Lipid extracts of wild-type M. smegmatis (lanes 1 to 3) and rmt mutants (lanes 4 to 6) were base treated and analyzed by HPTLC. (A) GPL extracts of parent strain M. smegmatis mc2155 (lane 1), mc2155 pVV16 vector control (lane 2), mc2155 complementation plasmid bearing rmt4, pHBJ415 (lane 3), Myco657 (Δrmt4) (lane 4), Δrmt4 pVV16 (lane 5), and Δrmt4 pHBJ415 (lane 6). (B) GPL extracts of parent strain M. smegmatis mc2155 (lane 1), mc2155 pVV16 vector control (lane 2), mc2155 complementation plasmid bearing rmt2, pHBJ409 (lane 3), Myco694 (Δrmt2) (lane 4), Δrmt2 pVV16 (lane 5), and Δrmt2 pHBJ409 (lane 6). Each of the mutants (lanes 4) has a novel GPL profile compared to the parent strain (lanes 1), and complementation restored mature GPL synthesis to the mutants (lanes 6).
FIG. 3.
Monosaccharide analysis of mutant GPLs. Based-treated GPLs from wild-type and mutant cell lines were subjected to acid hydrolysis, and released monosaccharides were analyzed as their corresponding alditol acetate derivatives by GC-MS. (A) M. smegmatis mc2155, (B) Δrmt4, (C) Δrmt4 pHBJ415, (D) Δrmt2, and (E) Δrmt2 pHBJ409. dTal and methylated Rha alditol acetates were identified based on their retention time compared to authentic standards and their mass spectrum. Diagnostic ions for methylated Rha derivatives were as follows: 3-O-Me-Rha, m/z 101, 130, 143, 190, 203; 2,3-di-O-Me-Rha, m/z 102, 118, 143, 203; 3,4-di-O-Me-Rha, m/z 89, 131, 190; and 2,3,4-tri-O-Me-Rha, m/z 102, 118, 131, 162, 175. Asterisks indicate reagent peaks.
The Myco694 (Δrmt2) mutant also lacked the GPL-1 doublet but expressed GPLs that comigrated with GPL-2 and a complex of bands that had a HPTLC mobility similar to that of the polar GPLs of Myco657. GC-MS analysis confirmed that Myco694 lacked trimethylated Rha and that it also contained 3-O-Me-Rha. However, levels of 3-O-Me-Rha (relative to that of dTal) were lower than in Myco657, and a 3,4-di-O-Me-Rha species was observed, rather than 2,3-di-O-Me-Rha (Fig. 3D). Traces of GPLs with 3-O-Me-Rha were still apparent in the complemented strains, suggesting that partial complementation may have occurred, possibly due to low levels of expression from the complementation plasmids. Complementation restored the synthesis of mature GPLs with trimethylated Rha (Fig. 2B, lane 6, and 3E). These data suggest that Myco694 is deficient in the enzyme activity required for Rha 2-O-methylation. Interestingly, loss of Rha 2-methylation had only a minor affect on the formation of dimethylated GPLs, suggesting that Rha 2-O-methylation is not critical for Rha 3- or 4-O-methylation.
Methyltransferase gene sequence analysis.
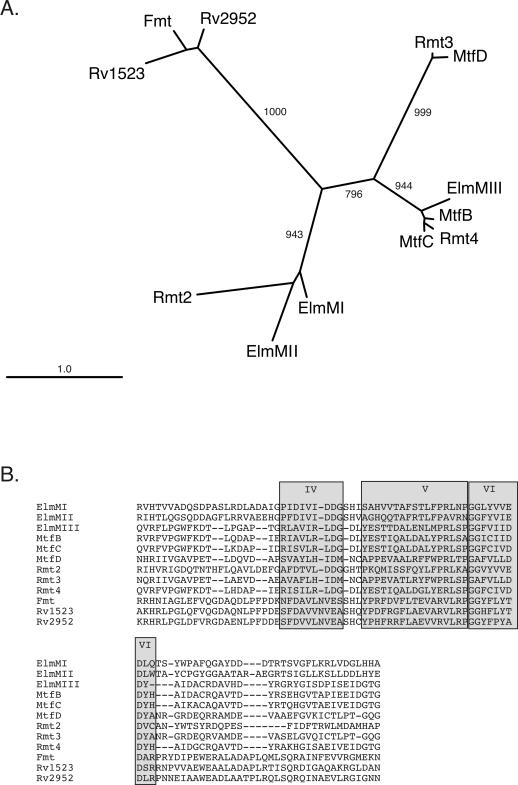
The ser2 locus of M. avium contains four ORFs predicted to encode methyltransferases that were annotated mtfA through mtfD (GenBank accession no. AF143772). Four methylation events are required to make mature ser2-encoded GPLs containing 3,4,di-O-Me-Rha and 2,3-di-O-Me-fucose. Methylation of Rha C2 does not seem to occur in M. avium. We hypothesized that the rhamnosyl 3-O-methyltransferase and rhamnosyl 4-O-methyltransferases might be conserved between M. avium and M. smegmatis. The S-adenosyl methionine binding site and three other motifs associated with methyltransferases are evident in the M. smegmatis methyltransferases and MtfB, MtfC, and MtfD of M. avium (7, 9). The methyltransferases can be grouped on the basis of similarity of motifs IV, V, and VI to create a neighbor-joining tree (Fig. 4A). The polypeptides encoded by M. avium mtfB and mtfC resemble M. smegmatis Rmt4, whereas MtfD and Rmt3 are similar. Fmt is dissimilar to the rest of the rhamnosyl methyltransferases but is similar to two putative methyltransferases of Mycobacterium tuberculosis encoded by RV2952 and RV1523 (2). M. avium MtfA is dissimilar to any of the above sequences and is not included in the tree. Other authentic rhamnosyl transferases (ElmMI, ElmMII, and ElmMIII) which methylate Rha during elloramycin synthesis in Streptomyces olivaceus were included for comparison (8). ElmMIII, a 4-O-methyltransferase, clusters with Rmt4, MtfB, and MtfC. We were interested in determining if the sequence similarity between the methyltransferases correlated with similar enzymatic activities. M. avium is difficult to genetically modify, so rather than make M. avium mutants, we tested each of the M. avium ser2-encoded methyltransferases for their ability to functionally complement our M. smegmatis methyltransferase mutants, namely Myco29 (rmt3::Tn611) (9), Myco493 (fmt::str) (7), Myco657 (Δrmt3), and Myco694 (Δrmt2).
FIG. 4.
Neighbor-joining tree showing the similarity between methyltransferase motifs IV and V in the M. smegmatis GPL rhamnosyl methyltransferases (Rmt2, Rmt3, and Rmt4), M. avium ser2 putative methyltransferases (MtfB, -C, and -D), S. olivaceus elloramycin rhamnosyl methyltransferases (ElmMI, ElmMII, and ElmMIII), and an authentic fatty acid methyltransferase (Fmt) and two putative methyltransferases from M. tuberculosis (Rv2952 and Rv1523). Amino acid sequences used for the tree are shown below. The bootstrap values are shown on the branches.
Cross-species complementation.
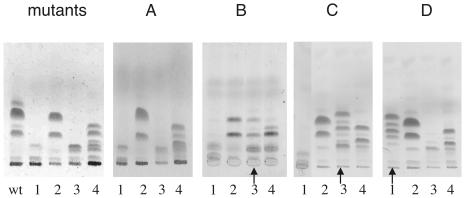
The ORFs of mtfA, -B, -C, and -D were PCR amplified from M. avium genomic DNA, and the amplicons were cloned into pVV16. The recombinant plasmids were introduced into M. smegmatis mc2155 and into each of the M. smegmatis methyltransferase mutants by transformation. The phenotypes of transformants were assessed by HPTLC analysis of base-treated GPL extracts. M. avium mtfA did not complement any of the M. smegmatis mutants (Fig. 5A), and the GPL profiles for each of the transformed mutant strains appeared the same as those for the untransformed mutant strains (Fig. 5, panel “mutants”). M. avium mtfB complemented M. smegmatis with an rmt4 defect (Myco657) (Fig. 5B, lane 3) and restored a wild-type GPL profile. The untransformed mutant makes only GPLs with monomethylated Rha (Fig. 5, panel “mutants,” lane 4) whereas the Myco657 mutant containing mtfB had mature GPLs. Interestingly, GPL-1a and GPL-2a were more abundant than GPL-1 and GPL-2 in the complemented strain. The result suggests that mtfB encodes a methyltransferase that can methylate the hydroxyl group at C4 of Rha. The result from mtfC transformations gave similar results in that it complemented the rhamnosyl 4-O-methyltransferase mutant (Δrmt4) but had no effect on the other mutants (Fig. 5C). M. avium mtfD complemented the M. smegmatis rmt3::Tn611 mutant previously characterized (9) and was shown to be a rhamnosyl 3-O-methyltransferase mutant. In each case where complementation was observed, the morphology of the complemented strains returned to the smooth-colony type. These data show that M. avium mtfB and mtfC have rhamnosyl 4-O-methyltransferase activity and that mtfD has rhamnosyl 3-O-methyltransferase activity.
FIG. 5.
HPTLC analysis of base-treated GPL extracts from M. smegmatis mutants transformed with M. avium methyltransferase genes. Lanes 1, host strain Myco29 (rmt3::Tn611); lanes 2, Myco493 fmt::str; lanes 3, Myco657 (Δrmt4); and lane 4, Myco694 (Δrmt2). Plasmids containing M. avium putative methytransferase genes (listed in parentheses after the plasmid) were introduced into each strain. (A) Strain transformed with pHBJ475 (mtfA); (B) strain transformed with pHBJ512 (mtfB); (C) strain transformed with pHBJ505 (mtfC); (D) strain transformed with pHBJ502 (mtfD). The “mutants” panel shows extracts from untransformed mutants and the parent strain (lane wt). Lanes showing a significant difference from the untransformed control are indicated with arrows.
DISCUSSION
It has previously been shown that Rmt3 is a rhamnosyl 3-O-methyltransferase, while Fmt methylates the hydroxylated fatty acid of the GPL (7, 9). In this study, we provide genetic and biochemical evidence that the genes rmt4 and rmt2 encode rhamnosyl 4-O-methyl- and rhamnosyl 2-O-methyltransferases, respectively. Interestingly, deletion of rmt2 completely inhibited 2-O-methylation of GPL Rha, but only had a minor affect on the formation of GPLs with either 3- and 3,4-di-O-methylated Rha. In contrast, deletion of rmt4 not only inhibited Rha 4-O-methylation, but also appeared to prevent efficient 2-O-methylation of Rha. Deletion of rmt2 and rmt4 had no apparent affect on 3-O-methylation of Rha. Finally, deletion of rmt3 results in complete inhibition of Rha 3-O-methylation as well as the accumulation of GPLs with unmethylated Rha (9). Collectively, these results suggest that methylation of the core GPL Rha residue is initiated by the 3-O-methyltransferase and that the product of this reaction is sequentially methylated by 4-O-methyltransferase and 2-O-methyltransferase. As the rmt3 mutant contained small amounts of a novel GPL with 4-O-methylated Rha, it is likely that the 4-O-methyltransferase can initiate GPL methylation in the absence of the 3-O-methyltransferase, but this reaction is very inefficient (9).
The only other characterized natural product containing trimethylated Rha is elloramycin from S. olivaceus (8). The methyltransferases ElmMI, ElmMII, and ElmMIII are responsible for consecutive methylation of the hydroxy groups at the 2′, 3′, and 4′ positions of Rha, respectively. All three of the S. olivaceus enzymes have some relaxed substrate specificity in that both ElmMII and ElmMIII can act on unmethylated or partially methylated Rha (8). The M. smegmatis enzymes may also have relaxed substrate specificity. Previously, it was observed that GPLs of the rmt3 mutant had unmethylated Rha and a novel 4-O-Me-Rha not observed in the parent strain (9). This mutant is the only mutant studied here that accumulated GPLs with unmodified Rha in detectable quantities. It appears that a 4-O-methyltransferase, possibly Rmt4, can use unmodified Rha as a substrate if it is available. Our data show that Rmt4 carries out methylation of the C4 hydroxy on 3-O-Me-Rha and that in the absence of Rmt4, GPLs containing a small amount of novel residue 2,3-di-O-Me-Rha can be generated, as well as the expected 3-O-Me-Rha. The presence of 2,3-di-O-Me-Rha suggests that a 2-O-methyltransferase, possibly Rmt2, can use 3-O-Me-Rha as a substrate, whereas Rmt2 usually acts on 3,4-di-O-Me-Rha.
The terminal alaninol of GPLs encoded by ser2 M. avium is glycosylated with 3,4-di-O-Me-Rha and the haptenic side chain comprised comprising 2,3-di-O-Me-fucosyl-(1→ 3)-rhamnosyl-(1→ 2)-6-deoxy talose. Four methylation events would be required for full modification of the Rha and fucose. Indeed, there are four predicted methyltransferase genes in the ser2 locus of M. avium. The polypeptide sequence of M. avium MtfD is most similar to that of M. smegmatis Rmt3, whereas MtfB, MtfBC, and Rmt4 are very similar to each other. None of the M. avium genes are similar to fmt (8). We cloned each of the M. avium methyltransferase genes (mtfA, mtfB, mtfC, and mtfD) into a shuttle vector and introduced them into wild-type M. smegmatis and each of the mutants defective in GPL methylation. MtfD expression functionally complemented our rmt3 mutant, suggesting that MtfD is a rhamnosyl 3-O-methyltransferase. Both MtfB and MtfC complemented the rmt4 mutant, suggesting that they have rhamnosyl 4-O-methyltransferase activity. These enzymes would be sufficient to fully methylate terminal Rha on M. avium GPLs. MtfA did not complement any of the M. smegmatis mutants, raising the possibility that it is involved in methylating the terminal fucose in M. avium GPLs. It is possible that MtfB, MtfC, or MtfD also has fucosyl methyltransferase activity that would not be detected in our experiments. None of the M. avium genes complemented the fmt mutant, which was not unexpected because the fatty acid is not methylated in M. avium.
Only a few O-methyltransferases that modify sugars have been identified, and those are involved in macrolide synthesis (89). The O-methyltransferases that modify Rha during GPL synthesis in M. avium and M. smegmatis expand this group considerably. Further characterization of the activities of these enzymes may show how they could be used as tools to generate bioactive compounds.
Acknowledgments
This research was supported by NH&MRC Research Program grant 215201. M.J.M. is an NH&MRC Principal Research Fellow and a Howard Hughes International Research Fellow.
REFERENCES
- 1.Billman-Jacobe, H., M. J. McConville, R. E. Haites, S. Kovacevic, and R. L. Coppel. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244-1253. [DOI] [PubMed] [Google Scholar]
- 2.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 3.Eckstein, T. M., F. S. Silbaq, D. Chatterjee, N. J. Kelly, P. J. Brennan, and J. T. Belisle. 1998. Identification and recombinant expression of a Mycobacterium avium rhamnosyltransferase gene (rtfA) involved in glycopeptidolipid biosynthesis. J. Bacteriol. 180:5567-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folch, J., M. Lee, and G. H. S. Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 5.Henry, R. J., A. B. Blakeney, P. J. Harris, and B. A. Stone. 1983. Detection of neutral and aminosugars from glycoproteins and polysaccharides as their alditol acetates. J. Chromatogr. 256:419-427. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 7.Jeevarajah, D., J. H. Patterson, M. J. McConville, and H. Billman-Jacobe. 2002. Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology 148:3079-3087. [DOI] [PubMed] [Google Scholar]
- 8.Patallo, E. P., G. Blanco, C. Fischer, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2001. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. Characterization of three methyltransferase genes. J. Biol. Chem. 276:18765-18774. [DOI] [PubMed] [Google Scholar]
- 9.Patterson, J., M. McConville, R. Haites, R. Coppel, and H. Billman-Jacobe. 2000. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J. Biol. Chem. 275:24900-24906. [DOI] [PubMed] [Google Scholar]
- 10.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J. Bacteriol. 178:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 14.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 15.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 16.Warrens, A. N., M. D. Jones, and R. I. Lechler. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29-35. [DOI] [PubMed] [Google Scholar]