Abstract
In this paper, we report the synthesis of Pseudomonas aeruginosa cDNA in the presence of oligo(dT) primers. Hybridization of oligonucleotide DNA microarrays indicates that under the experimental conditions used, at least 43.7% of the expressed genes from P. aeruginosa PAO1, representing many different functional classes, can be detected by using oligo(dT)-primed cDNAs.
There is growing evidence for the occurrence of polyadenylated mRNAs in a variety of prokaryotes (1, 2, 3, 4, 5, 14, 15, 17), though the phenomenon has been historically regarded as a feature of eukaryotic mRNA processing. Poly(A) tracts in eukaryotic mRNA typically range between 80 and 200 nucleotides, whereas poly(A) tracts in prokaryotes range between 14 and 60 nucleotides (18). The functions of bacterial RNA polyadenylation are variable and range from control of plasmid replication to modulation of mRNA stability to mRNA degradation, and it may play a role in mRNA translation (18). We report here the occurrence of polyadenylation of mRNA in Pseudomonas aeruginosa as evidenced by the oligo(dT)-dependent cDNA synthesis and reverse transcription-PCR (RT-PCR) analysis of different strains of P. aeruginosa. Furthermore, we used oligonucleotide DNA microarrays to analyze the extent of mRNA polyadenylation in P. aeruginosa PAO1.
RT-PCR of selected genes with oligo(dT)-primed cDNA from different P. aeruginosa strains.
PAO1, a wound isolate and genetic reference strain (9, 20), was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Strains 20265 (wild type) (8, 22) and TB (21) were isolated from cystic fibrosis patients and maintained in our laboratory collection. P. aeruginosa strains were grown in modified Vogel-Bonner medium (3.3 mM MgSO4, 10 mM citric acid, 28 mM NaNH4HPO4, 37 mM K2HPO4, 214 mM potassium d-gluconate [pH 7.2]), and RNA was prepared exactly as recently described (22). For the RT-PCR, 2 μg of the total RNA and 150 ng of a random primer (Invitrogen) were incubated at 65°C for 10 min. Then, 4 μl of fivefold-concentrated first-strand buffer of Expand reverse transcriptase (2 μl of 10 mM dithiothreitol, 2 μl of a 10 mM concentration of deoxynucleoside triphosphates, 1 μl [1 U] of Expand reverse transcriptase [all of these components were purchased from Roche]) was added and incubated at 43°C for 60 min. PCR was done with 2 μl of the above-described reaction mixture, and the following components were added: 2 μl of 10× PCR buffer (Roche), 2 μl of a 2 mM concentration of each deoxynucleoside triphosphate (dATP, dGTP, dTTP, and dCTP), 2 μl of the 5 pM forward primer (for rpoD, 5′-GGG GAT CAA CGT ATT CGA GA-3′; for feoB, 5′-TGT ACC ATG TGC CAC ACC TT-3′), 2 μl of the 5 pM reverse primer (for rpoD, 5′-TCA TCG GGA TCG ATA TAG CC-3′; for feoB, 5′-GGT CGT AGC CTT CGT AGT CG-3′), 0.3 U of Expand Hifi enzyme (Roche), and 8 μl of double-distilled water. The expected amplification products of both genes were 356 bp.
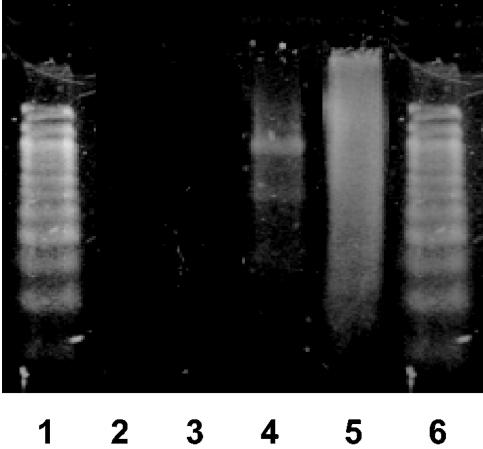
As shown in Fig. 1, a continuous smear of P. aeruginosa PAO1 cDNA could be observed with oligo(dT) primers as well as with random hexamer primers, whereas no bands were observed in the negative controls lacking RNA or oligo(dT) primers. The yield of cDNA resulting from the oligo(dT)-primed reaction was approximately five times less than that resulting from random-primed reactions with the same amounts of RNA used, as determined by UV spectrophotometry.
FIG. 1.
cDNA synthesis in the presence of oligo(dT) primers or random hexamer primers with RNA isolated from PAO1. Lanes: 1, 100-bp DNA ladder; 2, cDNA mixture without RNA (negative control); 3, cDNA mixture without oligo(dT) primers (negative control); 4, cDNA preparation made by using oligo(dT) primers; 5, cDNA preparation made with random hexamer primers (positive control); 6, 100-bp DNA ladder. Preparations were run on a 2% agarose gel and stained with SYBR green I.
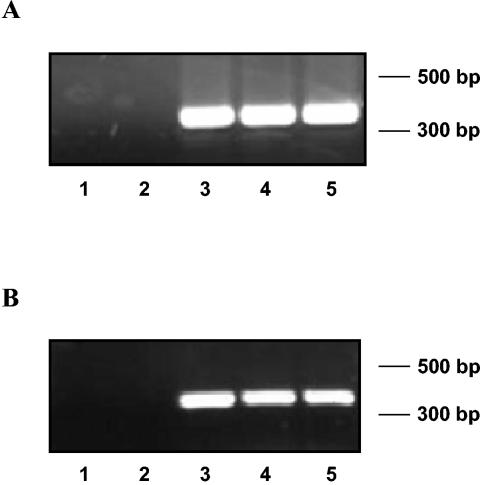
In order to determine if the phenomenon of polyadenylation was prevalent among different strains of P. aeruginosa, rpoD was amplified from cDNA synthesized with oligo(dT) primers from strain PAO1 and two clinical isolates, namely, strain TB and strain 20265. Figure 2A shows, as expected, a 356-bp amplification product corresponding to rpoD of strain TB. It was confirmed by sequencing the eluted PCR products from these three reactions that the amplification product of rpoD with cDNA prepared in the presence of oligo(dT) primers was identical to the product obtained by use of genomic DNA or the cDNA generated by use of random hexamer primers as templates. The same results were obtained for strains 20265 and PAO1 (data not shown). Additionally, an mRNA transcript of a feoB homologue in P. aeruginosa, PA4358, was also amplified by RT-PCR in strain 20265 by using gene-specific primers, and amplification products could be detected in cDNA preparations made in the presence of oligo(dT) primers (Fig. 2B). Again, the size and sequence identity of the RT-PCR product were found to match those of the product amplified from the genomic DNA. Thus, the ability to synthesize cDNA by using oligo(dT) primers and the occurrence of polyadenylation in mRNAs were clearly demonstrated in different strains of P. aeruginosa.
FIG. 2.
(A) RT-PCR of rpoD with oligo(dT)- and random-primed cDNA from P. aeruginosa TB. Lanes: 1, cDNA preparation without reverse transcriptase as a template; 2, cDNA preparation made without oligo(dT) or random primers; 3, rpoD amplified by using genomic DNA from strain TB; 4, random-primed cDNA from strain TB; 5, oligo(dT)-primed cDNA from strain TB. (B) RT-PCR of feoB homologue (PA4358) with oligo(dT)- and random-primed cDNA from P. aeruginosa 20265. Lanes: 1, cDNA preparation without reverse transcriptase as a template; 2, cDNA preparation made without oligo(dT) and random primers; 3, feoB amplified by using genomic DNA; 4, random-primed cDNA; 5, oligo(dT)-primed cDNA.
DNA microarray analysis with oligo(dT)-primed cDNA from P. aeruginosa PAO1.
To confirm and evaluate the phenomenon of polyadenylation of P. aeruginosa mRNA, P. aeruginosa PAO1 RNA isolated from cells grown in Vogel-Bonner medium was reverse transcribed with either the oligo(dT) primer or the random hexamer primers. Both resultant cDNAs were hybridized to a P. aeruginosa PAO1 GeneChip (Affymetrix). Procedures for cDNA synthesis and DNA microarray analysis using the P. aeruginosa GeneChip were performed as recently described (22). cDNA synthesized with oligo(dT) primers and random, arbitrary primers were hybridized to two chips each in order to have replicates for the analysis (each chip was hybridized with an independent cDNA prepared with independent RNA pools). Primary data analysis was performed with Microarray Suite (MAS) 5.0 software supplied by Affymetrix. Subsequent analysis was performed using Microsoft Access and Microsoft Excel. When primary data were analyzed, a signal for every gene was considered to be present only if there was strong presence or marginal presence of signals (as computed by Affymetrix MAS 5.0 with default parameters) on both replicate chips hybridized with the same cDNA. When a signal was absent in only one of the replicate chips, the gene itself was considered to be undetected. The annotations of the genes were based on the most recent annotations available from the PseudoCAP website (http://www.pseudomonas.com).
A comparative DNA microarray analysis was used to identify the specific genes whose RNA transcripts were polyadenylated, giving some estimates of the extent of polyadenylation and the type of transcripts that were polyadenylated. A total of 2,739 genes with significant signal intensities (according to the statistical algorithms of MAS 5.0) were detected by using random-primed cDNA synthesis. This figure represents 49.3% of the total 5,549 P. aeruginosa PAO1 coding sequences on the chip (Table 1). Out of these 2,739 genes, 1,199, representing 21.6% of the 5,549 coding sequences, were found to have polyadenylation of their mRNA transcripts because they were also detected in cDNA preparations made from oligo(dT)-primed reactions (Table 1). These genes included rpoD and feoB, whose transcripts were found to be polyadenylated by RT-PCR. Thus, 43.7% of the genes detected by random-primed cDNA synthesis showed polyadenylation of their corresponding mRNAs. Signals for only 16 genes were detected exclusively by oligo(dT)-primed cDNA (Table 1). None of the 16 genes were detected in random-primed cDNA preparations in the present study, and, interestingly, almost all of them belonged to the functional class of hypothetical proteins. However, mRNA transcripts of these 16 genes had been detected in earlier hybridizations on microarrays with random-primed cDNA from strains 20265 (22) and PAO1 (16), indicating that cDNAs for these genes were not primed exclusively by oligo(dT), as revealed by the present study.
TABLE 1.
Detection of genes on a P. aeruginosa GeneChip by using cDNAs prepared with either oligo(dT) primers or random hexamer primers
| Parameter | No. (%) of genes |
|---|---|
| Total detected by random primers | 2,739 (49.3) |
| Detected by random primers but not by oligo(dT) primers | 1,540 (27.7) |
| Detected by both | 1,199 (21.6) |
| Total detected by oligo(dT) primers | 1,215 (21.8) |
| Detected by oligo(dT) primers but not by random primers | 16 (0.1) |
| Detected by both | 1,199 (21.6) |
| Total detected by oligo(dT) and random primers | 2,755 (49.6) |
| Total not detected | 2,794 (50.3) |
| Total (coding sequences) on GeneChip | 5,549 (100) |
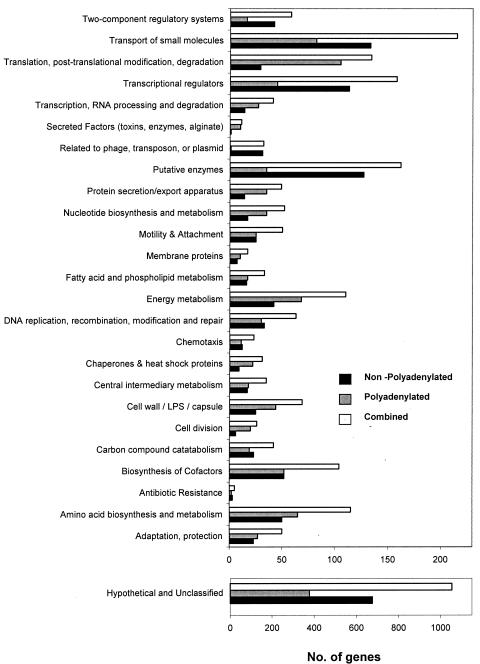
Genes detected by oligo(dT)-primed cDNA belong to almost all 26 of the major functional classes (Fig. 3). Significantly fewer or no genes were detected in the functional classes of two-component regulatory systems, transcriptional regulators, phage-related proteins, and putative enzymes. The number of genes detected by oligo(dT)-primed cDNA exceeded the number of genes detected by random-primed cDNA in 14 of the 26 major functional classes.
FIG. 3.
Comparison of the numbers of genes, whose mRNA transcripts were polyadenylated or nonpolyadenylated, as detected by GeneChip analysis. The genes analyzed are categorized into functional classes based on the annotation available from the P. aeruginosa PAO1 genome sequence. Combined, the combined total of polyadenylated and nonpolyadenylated transcripts identified in each functional class.
To the best of our knowledge, there has been no report demonstrating the occurrence of polyadenylation in any pseudomonad. DNA microarray analysis has been used to demonstrate the extent of polyadenylation of mycobacterial mRNAs (12). It has been shown that 30% of genes which could be detected by mycobacterial cDNA primed with genome-directed oligonucleotides could also be detected by oligo(dT)-primed cDNA, but no information on the functional classes of the genes detected was provided (12). It is possible that the number of mRNAs primed by oligo(dT) was underestimated because the large amounts of polyadenylated rRNA may have depressed cDNA synthesis from mRNA templates. Indeed, there were intense signals for the rRNA genes as well when the GeneChips were hybridized with oligo(dT)-primed cDNAs (data not shown). The priming efficiency of the random hexamer primers to generate representative cDNAs was unrivaled by the oligo(dT) primers. Therefore, use of oligo(dT) primers to generate cDNA representative of the total RNA preparation is largely relevant to the detection of the polyadenylated species of RNAs among a heterogenous mixture of total RNA preparation and not to complement detection using random hexamer-primed cDNA.
The chief enzymes that are responsible for polyadenylation are poly(A) polymerases. Even though the poly(A) polymerases of Escherichia coli have been well studied, their counterparts in P. aeruginosa have been annotated purely on the basis of their sequence homologies. pcnB encodes the major poly(A) polymerase PapI in E. coli. Poly(A) polymerase (ATP:polyribonucleotide adenylyltransferase) catalyzes the template-independent sequential addition of adenylate residues to the 3′-hydroxyl termini of RNA molecules. pnp encodes a 3′ exonuclease, polynucleotide phosphorylase, and rne encodes an endonuclease, RNase E, and both of these are implicated in the turnover of poly(A) tracts and the in vivo modulation of polyadenylation in E. coli (13). It is known that the S1 ribosomal protein is a poly(A) binding protein in E. coli (11). hfq is known to stimulate elongation of poly(A) tails by poly(A) polymerase I in E. coli (7). As further evidence for the occurrence of polyadenylation in P. aeruginosa, expression signals of pnp (PA4740), rne (PA2976), and pcnB (PA4727) could be detected in the array analysis. In addition, high levels of the putative poly(A) binding protein PA3162 were detected. hfq (PA4944) was yet another gene with a possible involvement in polyadenylation, whose transcripts were detected with significant signal intensities (data not shown). The future construction of knockouts of these P. aeruginosa genes will help to elucidate their role in polyadenylation.
Conclusions.
Our study indicates that polyadenylation seems to occur in the mRNAs of all the major functional classes of genes in P. aeruginosa and is not restricted to any one or group of them. Much of our knowledge about bacterial polyadenylation has been derived from works about E. coli (18, 19). In E. coli, polyadenylation of some genes depends on growth rate (10) and growth phase (6), with increased polyadenylation observed in slowly growing cells. It remains to be investigated whether there exist any differences in polyadenylation with respect to growth rate and growth phases in P. aeruginosa as well. Another question to be investigated is whether the polyadenylation in P. aeruginosa has the same functions as those described for polyadenylation in E. coli, i.e., marking mRNA transcripts for degradation by RNases (19), which is important for maintaining equilibrium of the normal RNA turnover, or if it has some novel functions. Understanding the role of P. aeruginosa mRNA polyadenylation in the overall gene expression and the enzymes involved in these processes might be crucial to enhance our understanding of the versatile lifestyle of this opportunistic pathogen.
Acknowledgments
This work was supported by the European Graduate College on Pseudomonas aeruginosa: Pathogenicity and Biotechnology sponsored by the Deutsche Forschungsgemeinschaft (DFG).
We are most grateful for the interest and strong support of Dieter Bitter-Suermann and for the excellent technical assistance of Tanja Toepfer.
REFERENCES
- 1.Adilakshmi, T., P. D. Ayling, and C. Ratledge. 2000. Polyadenylylation in mycobacteria: evidence for oligo(dT)-primed cDNA synthesis. Microbiology 146:633-638. [DOI] [PubMed] [Google Scholar]
- 2.Bralley, P., and G. H. Jones. 2001. Poly(A) polymerase activity and RNA polyadenylation in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:1155-1164. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. W., and J. N. Reeve. 1986. Polyadenylated RNA isolated from the archaebacterium Halobacterium halobium. J. Bacteriol. 166:686-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, G. J., and N. Sarkar. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. USA 89:10380-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, G. J., and N. Sarkar. 1993. Poly(A) RNA in Bacillus subtilis: identification of the polyadenylylation site of flagellin mRNA. FEMS Microbiol. Lett. 108:281-285. [DOI] [PubMed] [Google Scholar]
- 6.Cao, G. J., and N. Sarkar. 1997. Stationary phase-specific mRNAs in Escherichia coli are polyadenylated. Biochem. Biophys. Res. Commun. 239:46-50. [DOI] [PubMed] [Google Scholar]
- 7.Hajnsdorf, E., and P. Regnier. 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. USA 97:1501-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Häussler, S., I. Ziegler, A. Lottel, F. von Götz, M. Rohde, D. Wehmhöner, S. Saravanamuthu, B. Tümmler, and I. Steinmetz. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295-301. [DOI] [PubMed] [Google Scholar]
- 9.Holloway, B. W., U. Römling, and B. Tümmler. 1994. Genomic mapping of Pseudomonas aeruginosa PAO1. Microbiology 140:2907-2929. [DOI] [PubMed] [Google Scholar]
- 10.Jasiecki, J., and G. Wegrzyn. 2003. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Rep. 4:172-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalapos, M. P., H. Paulus, and N. Sarkar. 1997. Identification of ribosomal protein S1 as a poly(A) binding protein in Escherichia coli. Biochimie 79:493-502. [DOI] [PubMed] [Google Scholar]
- 12.Lakey, D. L., Y. Zhang, A. M. Talaat, B. Samten, L. E. DesJardin, K. D. Eisenach, S. A. Johnston, and P. F. Barnes. 2002. Priming reverse transcription with oligo(dT) does not yield representative samples of Mycobacterium tuberculosis cDNA. Microbiology 148:2567-2572. [DOI] [PubMed] [Google Scholar]
- 13.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol. Microbiol. 36:982-994. [DOI] [PubMed] [Google Scholar]
- 14.Ohta, N., M. Sanders, and A. Newton. 1978. Characterization of unstable poly (A)-RNA in Caulobacter crescentus. Biochim. Biophys. Acta 517:65-75. [DOI] [PubMed] [Google Scholar]
- 15.Rindi, L., N. Lari, M. G. Gil, and C. Garzelli. 1998. Oligo(dT)-primed synthesis of cDNA by reverse transcriptase in mycobacteria. Biochem. Biophys. Res. Commun. 248:216-218. [DOI] [PubMed] [Google Scholar]
- 16.Salunkhe, P., F. von Götz, L. Wiehlmann, J. Lauber, J. Buer, and B. Tümmler. 2002. GeneChip expression analysis of the response of Pseudomonas aeruginosa to paraquat-induced superoxide stress. Genome Lett. 4:165-174. [Google Scholar]
- 17.Sarkar, N. 1996. Polyadenylation of mRNA in bacteria. Microbiology 142:3125-3133. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar, N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66:173-197. [DOI] [PubMed] [Google Scholar]
- 19.Steege, D. A. 2000. Emerging features of mRNA decay in bacteria. RNA 6:1079-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 21.Tümmler, B., J. Bosshammer, S. Breitenstein, I. Brockhausen, P. Gudowius, C. Herrmann, S. Herrmann, T. Heuer, P. Kubesch, F. Mekus, U. Römling, K. D. Schmidt, C. Spangenberg, and S. Walter. 1997. Infections with Pseudomonas aeruginosa in patients with cystic fibrosis. Behring Inst. Mitt. 249-255. [PubMed]
- 22.von Götz, F., S. Häussler, D. Jordan, S. Saravanamuthu, D. Wehmhöner, A. Strüβmann, J. Lauber, I. Attree, J. Buer, B. Tümmler, and I. Steinmetz. 2004. Expression analysis of a highly adherent and cytotoxic small colony variant of Pseudomonas aeruginosa isolated from a lung of a patient with cystic fibrosis. J. Bacteriol. 186:3837-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]