Abstract
A primer extension inhibition (toeprint) assay was developed using ribosomes and ribosomal subunits from Streptomyces lividans. This assay allowed the study of ribosome binding to streptomycete leaderless and leadered mRNA. Purified 30S subunits were unable to form a ternary complex on aph leaderless mRNA, whereas 70S ribosomes could form ternary complexes on this mRNA. 30S subunits formed ternary complexes on leadered aph and malE mRNA. The translation initiation factors (IF1, IF2, and IF3) from S. lividans were isolated and included in toeprint and filter binding assays with leadered and leaderless mRNA. Generally, the IFs reduced the toeprint signal on leadered mRNA; however, incubation of IF1 and IF2 with 30S subunits that had been washed under high-salt conditions promoted the formation of a ternary complex on aph leaderless mRNA. Our data suggest that, as reported for Escherichia coli, initiation complexes with leaderless mRNAs might use a novel pathway involving 70S ribosomes or 30S subunits bound by IF1 and IF2 but not IF3. Some mRNA-ribosome-initiator tRNA reactions that yielded weak or no toeprint signals still formed complexes in filter binding assays, suggesting the occurrence of interactions that are not stable in the toeprint assay.
Protein synthesis in bacteria is initiated when the small ribosomal subunit (30S), aided by the three initiation factors (IF1, IF2, and IF3), interacts with mRNA and initiator tRNA (fMet-tRNAfMet) to form a 30S initiation complex. The molecular biology of the interactions of the ribosome (or ribosomal subunits) with mRNA that lead to the formation of this translation initiation complex has not been fully characterized. Initiation is the rate-limiting step of translation and relies upon the ribosome binding to mRNA and is influenced by sequence and structural motifs in and around the ribosome binding site (RBS). The nucleotide sequences identified within bacterial mRNA that are most important for translation initiation include the start codon (i.e., AUG most commonly in Escherichia coli) and the purine-rich Shine-Dalgarno (SD) sequence; together, the start codon and the SD sequence help to identify the RBS within a bacterial mRNA. However, there is evidence that other nucleotides within the more loosely defined translation initiation region are important effectors of translational efficiency; the nucleotide sequences directly upstream and downstream of the start codon (the −1 triplet and the +2 codon), as well the spacing of a canonical SD sequence from the start codon, have been shown to affect the rate of initiation in bacteria (7). Further, an increasing number of genes that entirely lack a 5′ untranslated leader region are being identified. Sequence or structural motifs that might be important for translation initiation on leaderless messages must be contained in the region downstream of the start codon, with the start codon itself being very important for recognition by the initiating ribosomes (13, 22). Leaderless mRNAs are translated, often at high levels, and are found in the Archaea, the Bacteria, and the Eucarya (10, 24). Most of the research on leaderless messages has been performed with Escherichia coli, but this bacterium expresses only three known leaderless mRNAs: cI (17), tetR (11), and gene V (2). Genes encoding leaderless mRNA have not been identified in the E. coli chromosome. In comparison, the streptomycetes encode a relatively large number of leaderless mRNAs, and there is evidence that they may be better able than E. coli to translate certain leaderless mRNAs (25).
In E. coli, the ability to compare increased gene expression in vivo (using protein production or enzyme activity, for example) to increased ribosome binding in vitro via a primer extension inhibition assay (toeprinting) (9) makes for a compelling duo of experiments when translational efficiency of certain gene constructs is being examined (12). A similar in vitro assay would be of great utility in the study of a system in which little is known about translation initiation, namely, the high-G+C gram-positive streptomycetes. A primer extension inhibition assay developed with streptomycete components would allow characterization of the streptomycete ribosome-binding site, including the SD sequence; the sequence motifs downstream of the start codon important for translation initiation with leaderless messages could also be characterized. Additionally, the function of the streptomycete IFs and their contribution to translation initiation could be directly assessed.
To these ends, a toeprinting assay was developed by using ribosomes isolated from Streptomyces lividans. This procedure has allowed direct observation of streptomycete ribosomes interacting with both leadered and leaderless streptomycete mRNA. To further investigate ribosome binding and translation initiation in vitro, the three streptomycete IFs were overexpressed and purified. The activities of the IFs were tested by using ribosomes and ribosomal subunits isolated from S. lividans and E. coli, and the IFs were included in toeprint and nitrocellulose filter binding assays using leadered and leaderless mRNAs. 30S subunits that had been washed under high-salt conditions produced a strong toeprint on leadered aph and malE mRNA but required IF1 and IF2 to produce a toeprint signal on leaderless aph mRNA; addition of IF3 to this complex eliminated the toeprint signal on leaderless mRNA. 70S ribosomes produced toeprints on aph leaderless mRNA, but high-salt washing removed a component(s) necessary for 70S ribosomes to toeprint leaderless mRNA. Further, filter binding assays provided evidence for interactions between ribosomes, mRNA, and IFs that were not detected by toeprints and may represent different types of complexes in the pathway to translation initiation.
MATERIALS AND METHODS
Isolation of S. lividans ribosomes and ribosomal subunits.
Streptomycete ribosomes were isolated from S. lividans TK24 by a method developed for use with in vitro coupled transcription-translation systems (E. Cundliffe, personal communication). Mycelia were grown to a density of 1.5 to 2.0 g/liter in yeast extract-malt extract medium supplemented with 5 g of PEG 6000 and 1 g of MgCl2 per liter. Mycelia were chilled on ice, collected via centrifugation at 4°C, washed twice with washing buffer (10 mM HEPES-KOH [pH 7.5], 10 mM MgCl2, 1 M NH4Cl, 5 mM β-mercaptoethanol), collected by filtration with Whatman no. 1 paper on a Buchner funnel, and resuspended to a uniform cell paste in HRS buffer (10 mM HEPES-KOH [pH 7.5], 10 mM MgCl2, 50 mM NH4Cl, 5 mM β-mercaptoethanol) by using 2 ml of HRS buffer per g (wet weight) of cells. This cell paste was broken via two passes through a French pressure cell at 10,000 to 12,000 lb/in2. Alternatively, cells were lysed in a mortar and pestle after the addition of alumina (type A-5; Sigma; 2.5 g for each 1 g of cell paste). The cell paste was ground until a snapping noise was heard and was then ground for another 15 min; the ground paste was then suspended in HRS buffer (∼2 ml for each gram of cell paste). The resulting lysate from either method was treated with 20 μl of RNase-free DNase I (10 U/μl; Roche) for 30 min on ice; the slurry was agitated and triturated several times during this incubation. The cell debris was removed with two 30-min spins at 16,000 rpm and 4°C (Beckman JA-20 rotor) to obtain the S30 supernatant. The S30 was then layered over high-salt buffer (10 mM HEPES-KOH [pH 7.5], 10 mM MgCl2, 1 M NH4Cl, 5 mM β-mercaptoethanol) containing 20% sucrose and centrifuged for 5 h at 40,000 rpm and 4°C (Beckman 90 Ti rotor) in order to pellet high-salt-washed 70S ribosomes that lack IFs and elongation factors. Alternatively, the S30 was layered over a low-salt buffer (10 mM HEPES-KOH [pH 7.5], 10 mM MgCl2, 50 mM NH4Cl, 5 mM β-mercaptoethanol) containing 40% sucrose and centrifuged as before in order to pellet low-salt-washed 70S ribosomes that still contained initiation and elongation factors. 70S ribosomes were resuspended in HRS buffer plus 10% glycerol and stored at −80°C. After activation for 15 min at 37°C, these 70S ribosomes were used directly in toeprinting and IF3/IF1 dissociation assays. 30S and 50S subunits were obtained by lowering the Mg2+ concentration of 70S ribosomes in HRS storage buffer to 1 mM and layering the dissociated ribosomes over 37-ml sucrose gradients (10 to 30% sucrose in HRS plus 1 mM Mg2+). The gradients were centrifuged at 18,000 rpm (Beckman SW28 rotor) for 16 h to separate the subunits. Gradients were fractionated, and fractions containing 30S or 50S subunits were pooled; subunits were pelleted by centrifugation at 45,000 rpm and 4°C (Beckman 90 Ti rotor) for 16 h. The pellets were resuspended in 300 to 500 μl of HRS buffer; subunits were purified further by centrifugation through a second sucrose gradient and pelleting, as described above. The final 30S or 50S ribosomal subunit pellet was resuspended in HRS buffer plus 10% glycerol, flash frozen in liquid nitrogen, and stored at −80°C.
Preparation of in vitro mRNA and radiolabeled primers.
Genes of interest from Streptomyces spp. and E. coli were PCR amplified by using an upstream primer that contained the T7 promoter consensus sequence placed upstream of the transcriptional start site (leadered mRNAs) or the transcriptional/translational start site (leaderless mRNAs). mRNAs were synthesized by using T7 RNA polymerase as described previously (12). Radiolabeled oligonucleotide primers were prepared as described previously (24). The unincorporated [γ-32P]ATP was removed with a NucAway spin column (Ambion, Inc.). [α-32P]CTP-labeled mRNA was prepared in a 10-μl reaction mixture containing 15 mM dithiothreitol, a 2 mM concentration of each nucleoside triphosphate, 20 mM MgCl2, 40 mM Tris-HCl (pH 7.9), 2 mM spermidine, 500 U of T7 RNA polymerase (New England Biolabs), 1 μg of template DNA, and 50 μCi of [α-32P]CTP (3,000 Ci/mmol; NEN). The reaction mixture was incubated at 37°C for 1 h, followed by treatment with 10 U of RNase-free DNase I (Roche) for 30 min. at 37°C. The reaction product was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and the RNA was precipitated with sodium acetate (pH 6) and isopropanol. The final RNA concentration was determined spectrophotometrically.
Primer extension inhibition assays.
The conditions for primer extension inhibition (toeprint) assays using S. lividans ribosomes and ribosomal subunits were essentially as described previously (12), with some important modifications. The toeprint reactions were carried out in 1× SB-HEPES buffer (10 mM HEPES-KOH [pH 7.5], 10 mM magnesium acetate, 60 mM NH4Cl, 6 mM β-mercaptoethanol). Ten-microliter reaction mixtures typically contained 0.2 pmol of mRNA (1×) annealed to a downstream radiolabeled primer and either 2 pmol of ribosomes or ribosomal subunits (10×) or 4 pmol of ribosomes or ribosomal subunits (20×) and 15 pmol of E. coli tRNAfMet (Sigma). Reaction mixtures that included S. lividans IFs (IF1, IF2, and/or IF3) contained either 2 or 4 pmol of each IF to reflect the amount of ribosomes or ribosomal subunits plus 15 pmol of fMet-tRNAfMet or tRNAfMet where indicated. Either IFs were preincubated with ribosomes and initiator tRNA for 10 min at 37°C in 1× SB-HEPES prior to the addition of the mRNA, or IFs were added to the reaction mixture after a 10-min incubation of ribosomes and initiator with mRNA. For order-of-addition toeprint assays, after an initial 10-min incubation of the mRNA with a 30S ribosomal subunit (either alone or preincubated with IFs as described above) and initiator tRNA, the 50S subunit was then added, and the incubation was continued for an additional 10 min. Avian myeloblastosis virus reverse transcriptase (1 U; Life Sciences, Inc.) was then added, and the reaction mixture was incubated at 37°C for 15 min. The radiolabeled cDNA was precipitated and resolved on a 6% denaturing polyacrylamide gel containing 8 M urea.
Purification of methionyl-tRNA synthetase and methionyl-tRNA formyltransferase.
The plasmids pQE16-MTF, expressing the E. coli methionyl-tRNA formyltransferase (MTF) and pQE60-MetRS, expressing the E. coli methionyl tRNA-synthetase (MetRS) were kindly provided by Uttam L. RajBhandary (Department of Biology, Massachusetts Institute of Technology). Both of these plasmids encode enzymes that are expressed with C-terminal His6 tags. The enzymes were overexpressed in E. coli and purified with Talon cobalt affinity resin (BD Biosciences Clontech)
tRNAfMet charging and formylation.
Charged, formylated initiator tRNA was included in ribosome binding assays containing IF2, since the blocked αNH2 group of N-formylmethionine directs the interaction of fMet-tRNAfMet with IF2 (8). The charging and formylation reactions were performed essentially as described previously (3, 18). The charging of tRNAfMet with methionine was performed as follows: a 200-μl reaction mixture contained 20 μl of 10× aminoacylation buffer (AB) (200 mM imidazole [pH 7.5], 1 mM EDTA, 1.5 M NH4Cl, 100 μg of bovine serum albumin per ml, 100 mM MgCl2), 4 μl of 100 mM ATP, 20 μl of 1 mM methionine, 10 μl of purified MetRS (20 μg), and 50 μl of tRNAfMet (50 pmol/μl). This mixture was incubated at 37°C for 30 min. Subsequent formylation of the Met-tRNAfMet was performed by the addition of 8 μl of 10× AB, 10 μl of purified MTF (20 μg), and 3 μl of 5:10 methenyltetrahydrofolate (prepared from folinic acid [Sigma] as described previously (3). The mixture was then incubated at 37°C for an additional 5 min. Following the charging and formylation reactions, the fMet-tRNAfMet was extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated at −80°C by using 0.2 M ammonium acetate (final concentration) and an equal total volume of isopropyl alcohol. Finally, the fMet-tRNAfMet was passed over a NucAway column (Ambion), and the concentration was determined spectrophotometrically.
Cloning of inf genes from S. lividans.
The infA, infB, and infC genes were identified from the S. coelicolor genome sequence (http://www.sanger.ac.uk/Projects/S_coelicolor/). Primers designed using each gene sequence were used to PCR amplify the corresponding genes from S. lividans TK24 total DNA. For infA, the forward primer was 5′GGAATTCCATATGGCCAAGAAGCAAGGTGCCATCG3′ and the reverse primer was 5′CCCAAGCTTCTACTTGTACCGGTAGACGATC3′. The forward primer changed the predicted GTG start codon for infA to ATG. For infC, the forward primer was 5′GGAATTCCATATGAGCGCCGAGCCCCGCATCAAC3′ and the reverse primer was 5′CCCAAGCTTTCACGCTTGGGCAGGTGC3′. The forward primer changed the predicted ATC start codon for infC to ATG. The PCR products for the genes infA and infC were digested with NdeI and HindIII and were cloned into the expression vector pET17b (Novagen), which had been digested with the same enzymes. infB was not able to be amplified as a full-length product and was subsequently amplified in two fragments. The primers used for the 5′ portion of the infB gene were 5′GGAATTCCATATGGCTAAGGTCCGGGTCTACGAAC3′ (forward) and 5′ATGAAGGTGATCTTGCGCTCTTCG3′ (reverse). The forward primer changed the predicted GTG start codon for infB to ATG. The primers for the 3′ portion of the infB gene were 5′TCACCGACTTCGCGGAGAAGATCAACG3′ (forward) and 5′CCCAAGCTTTTACACCCGCGGCTTCTCGCG3′ (reverse). The 5′ infB PCR product was digested with NdeI and XhoI, the 3′ amplicon was digested with HindIII and XhoI, and the two fragments were ligated in a single step into pET17b cut with NdeI and HindIII. The resulting plasmids for each gene were transformed into E. coli BL21D(pLysS), and overexpression was accomplished via induction of the T7 RNA polymerase with isopropyl-β-d-thiogalactopyranoside (IPTG).
Overexpression and purification of S. lividans IFs.
IF3 from S. lividans was purified essentially as described previously (20), with some important updates and modifications. The appropriate culture of E. coli BL21D(pLysS), containing cloned inf genes and supplemented with 200 μg of ampicillin and 20 μg of chloramphenicol per ml, was grown to an optical density at 600 nm of ∼0.5 in L broth and was induced by the addition of 0.5 mM IPTG (final concentration). After 2 h of induction, the cells were collected by centrifugation and washed once with 1/20 volume of washing buffer (10 mM Tris-acetate [pH 7.6], 10 mM Mg acetate, 50 mM NH4Cl, 14 mM β-mercaptoethanol, 0.5 mM EDTA). The cells were weighed and lysed with a mortar and pestle plus alumina equaling 2.5 times the weight of the cells. During the grinding, 15 μl of DNase I (Roche), 20 μl of β-mercaptoethanol, 50 μl of 0.1 M phenylmethylsulfonyl fluoride (PMSF), and 50 μl of 0.1 M benzamidine were added to the cells.
The lysate containing IF3 was diluted with ∼30 ml of IF3 buffer (20 mM Tris-HCl [pH 7.7], 10 mM MgCl2, 5 mM β-mercaptoethanol, 0.1 mM PMSF, 0.1 mM benzamidine, 0.5 mM EDTA) and centrifuged at 30,000 × g (Beckman JA-20 rotor) for 30 min to obtain an S30 fraction. This S30 fraction was further clarified by a second centrifugation as described above. The S30 fraction was diluted 1:2 with IF3 buffer and loaded onto a 5-ml HiTrap SP column (Amersham Pharmacia Biotech) that had been equilibrated with 5 volumes of IF3 buffer plus 0.25 M NaCl at a flow rate of 5 ml/min. Bound proteins were eluted with a linear gradient from 0.1 to 0.7 M NaCl in IF3 buffer; the flowthrough was collected, and 1.3-ml fractions were collected during elution. A large flowthrough was detected along with three smaller, distinct protein peaks; IF3 eluted in two distinct protein peaks. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed that the two forms of IF3 were identical in size. The fractions containing the two forms of IF3 were pooled separately, aliquoted, and stored at −80°C in 10% glycerol.
Cells overexpressing IF1 were grown, collected, and lysed as above. The lysate containing IF1 was diluted with ∼30 ml of NB buffer (10 mM Tris-HCl [pH 7.7], 60 mM NH4Cl, 10 mM Mg acetate, 2 mM β-mercaptoethanol, 0.1 mM PMSF, 0.1 mM benzamidine), and an S30 fraction was obtained as described above. The S30 fraction containing IF1 was diluted 1:4 with IF1 buffer (20 mM HEPES-KOH [pH 7.1], 0.5 mM EDTA, 10% glycerol, 0.1 mM PMSF, 0.1 mM benzamidine) and loaded onto a 5-ml HiTrap SP column equilibrated with 5 volumes of IF1 buffer plus 0.1 M NH4Cl. The bound proteins were eluted with a linear gradient of 0.1 to 0.7 M NH4Cl in IF1 buffer. IF1 eluted in a single peak that contained other larger proteins as well. The fractions containing IF1 were pooled and loaded onto an S100 sizing column (Pharmacia). Fractions containing IF1 were pooled, and IF1 was concentrated with a 5 ml HiTrap SP column. IF1 was eluted by using a step gradient from 0.1 to 0.7 M NH4Cl in IF1 buffer; the fractions were pooled, aliquoted, and stored at −80°C.
IF2 was overexpressed in E. coli BL21D as an N-terminal His6 tag protein. The cleared cell lysate containing IF2 was prepared as described above, except that the alumina-cell paste slurry was resuspended in 1× equilibration-wash buffer (50 mM sodium phosphate [pH 7.0], 300 mM NaCl, 5 mM β-mercaptoethanol, 0.1 mM benzamidine, 0.1 mM PMSF). The cleared lysate was mixed with BD Biosciences Clontech Talon cobalt affinity resin per the manufacturer's instructions; the resin was then washed with equilibration buffer containing 5 mM imidazole. Bound His6-tagged protein was then eluted with a linear gradient of 5 to 150 mM imidazole in equilibration buffer. The fractions containing IF2 were determined via sodium dodecyl sulfate-polyacrylamide gel electrophoresis; these fractions were pooled and dialyzed against IF2 buffer (20 mM HEPES-KOH [pH 7.1], 0.5 mM EDTA, 5 mM β-mercaptoethanol, 0.1 mM benzamidine, 0.1 mM PMSF, 10% glycerol), aliquoted, and frozen at −80°C.
Filter binding assays.
Filter binding reactions were set up essentially as described for primer extension inhibition reactions. Ribosomes, initiator tRNA, and IFs (when present) were incubated together for 10 min prior to incubation with [α-32P]CTP-radiolabeled mRNA for 15 min at 37°C in a total volume of 10 μl. Reaction mixtures were then diluted to 500 μl with 1× SB-HEPES and filtered through a nitrocellulose membrane (0.45-μm pore size; Schleicher and Schuell) in a Mini-Fold slot blot manifold. Each well was then washed with at least 3 ml of 1× SB-HEPES. Membranes were dried at room temperature, UV cross-linked, and analyzed on a Molecular Dynamics PhosphorImager. To determine the amount (in picomoles) of radiolabeled mRNA bound to ribosomes in each reaction, a standard curve was generated with known amounts of input mRNA as follows: 50-μl reaction mixtures contained 0, 0.5, 1.0, 2.0, or 4.0 pmol of radiolabeled mRNA and 50 μg of tRNA (E. coli MRE600) as a carrier; to this, 50 μl of 10% trichloroacetic acid (TCA) was added, followed by 2 ml of 5% cold TCA. This mixture was mixed thoroughly and held on ice for 10 min. The entire mixture was filtered through 24-mm GF/A filters (Whatman) fitted onto a vacuum manifold; each filter was rinsed once with 5% TCA, once with 0.1 N HCl, and once with absolute ethanol. The filters were dried under a heat lamp and analyzed on a PhosphorImager as described above.
RESULTS
Purification of S. lividans 70S ribosomes and 30S and 50S ribosomal subunits and functional assays for S. lividans IFs.
S. lividans 30S and 50S ribosomal subunits were obtained by placing 70S ribosomes under low-Mg2+ (1 mM) conditions and separating the subunits on a 10-to-30% sucrose gradient. The majority of S. lividans 70S ribosomes dissociated, as expected, under the low-Mg2+ conditions, but a portion of the 70S ribosomes remained intact after ultracentrifugation and fractionation. This incomplete dissociation of the 70S ribosomes is in contrast to the behavior of E. coli 70S ribosomes, which completely dissociate in 1 mM Mg2+ (data not shown). The high-salt wash (1 M NH4Cl) employed during the isolation of the 70S ribosomes strips the ribosomes of associated initiation and elongation factors, while the low-salt wash (50 mM NH4Cl) allows these ancillary factors to remain associated with the isolated ribosomes (1; E. Cundliffe, personal communication).
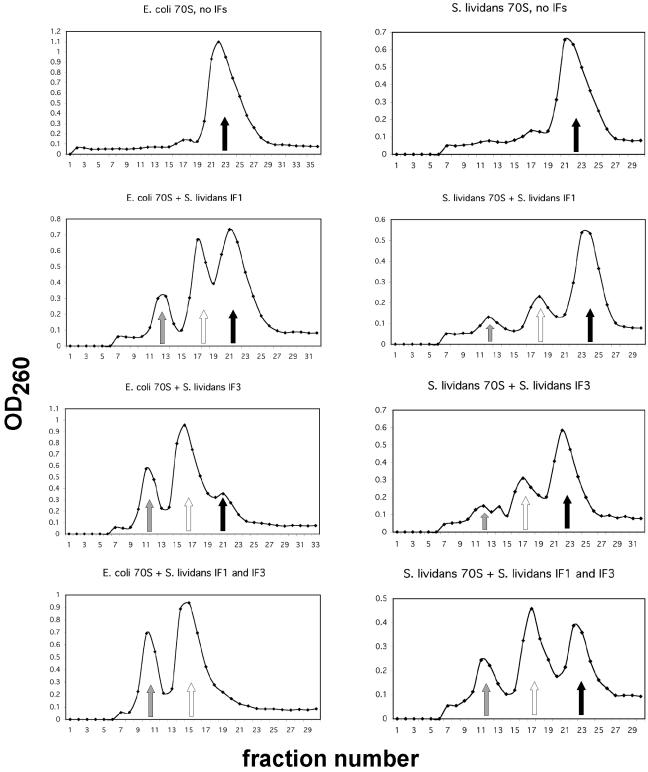
In E. coli, IF3 contributes to 70S ribosome dissociation in order to provide a pool of free 30S subunits for translation initiation (15). IF1 promotes subunit exchange in the 70S ribosome (4) and generally has been observed to enhance the functions of IF2 and IF3 (7). The ability of the streptomycete IF3 to dissociate 70S ribosomes into 30S and 50S subunits was tested directly in vitro with S. lividans and E. coli 70S ribosomes; IF1 was tested for its ability to enhance the dissociating activity of IF3. S. lividans IF3 actively dissociated S. lividans 70S ribosomes into 30S and 50S subunits, and IF1 enhanced IF3's ability to dissociate S. lividans 70S ribosomes (Fig. 1). Interestingly, a reaction in which IF1 alone was incubated with S. lividans 70S ribosomes revealed that IF1 also dissociated a portion of the S. lividans ribosomes into subunits, similar to dissociation caused by IF3 alone (Fig. 1). The incomplete dissociation of S. lividans 70S ribosomes in the presence of IF3 and of IF1 plus IF3 resembles the incomplete dissociation noted for S. lividans 70S ribosomes under low Mg2+ concentrations.
FIG. 1.
S. lividans and E. coli 70S ribosome dissociation assays. E. coli or S. lividans 70S ribosomes (100 and 88 pmol, respectively) were incubated with or without a 20-fold molar excess of S. lividans IF1, IF3, or IF1 plus IF3 and applied to 10-to-30% sucrose gradients as described in Materials and Methods. After centrifugation, the optical density at 260 of gradient fractions was measured. Control reaction mixtures contained no IFs. The positions of 30S subunits (shaded arrows), 50S subunits (open arrows), and 70S ribosomes (filled arrows) are indicated.
Identical dissociation assays were performed with S. lividans IFs and E. coli 70S ribosomes (Fig. 1). IF3 alone dissociated E. coli 70S ribosomes to a greater extent than S. lividans 70S ribosomes, with only a small percentage of ribosomes remaining intact; IF1 plus IF3 completely dissociated E. coli 70S ribosomes into 30S and 50S subunits. IF1 alone also dissociated a portion of E. coli ribosomes into 30S and 50S subunits, but to a lesser degree than IF3 alone.
Effects of IFs on ternary complex formation with leadered or leaderless mRNA.
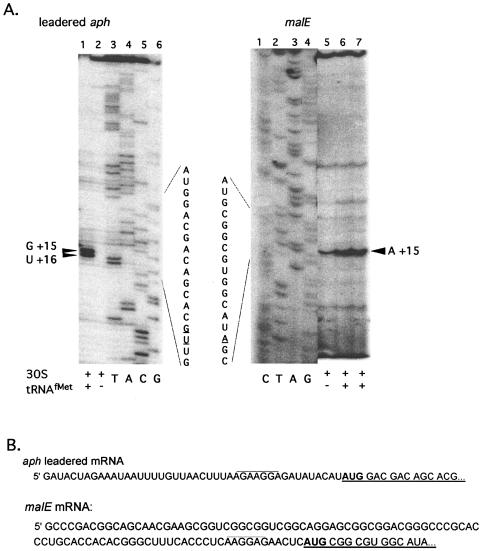
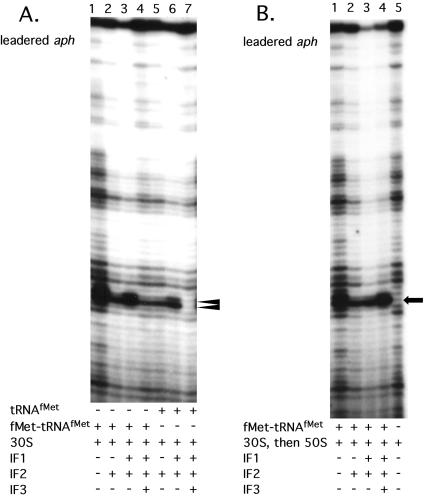
S. lividans 70S ribosomes and 30S ribosomal subunits isolated under low-salt and high-salt conditions were tested to determine their ability to produce ternary complexes in the presence of E. coli tRNAfMet on streptomycete leadered and leaderless mRNA. High-salt-washed 30S subunits produced a tRNA-dependent toeprint signal on leadered aph and malE mRNAs (Fig. 2A). The toeprint signals observed at +15 (with the A of the AUG start codon at +1) were consistent with reports for E. coli (9) indicating that the downstream boundary of the streptomycete 30S subunits on mRNA was similar to that in E. coli and that the E. coli tRNAfMet was functional in ternary complex formation with S. lividans 30S subunits. Some reactions gave a two-position toeprint signal (+15 and +16) (Fig. 2A, lane 1; Fig. 3A, lanes 5 and 6, aph mRNA).
FIG. 2.
(A) Primer extension inhibition (toeprint) assays using high-salt-washed 30S subunits with leadered aph or malE mRNAs. The two-position toeprint signal in lane 1, indicating a ternary complex with aph mRNA, S. lividans 30S subunits and tRNAfMet, is marked by a double arrowhead. Reaction mixtures contained 0.2 pmol of aph mRNA, 4 pmol of 30S subunits, and 10 pmol of initiator tRNA, if present. The toeprint signals in lanes 6 and 7 indicating a ternary complex with malE mRNA, S. lividans 30S subunits, and tRNAfMet are marked with a single arrowhead. Reaction mixtures contained 0.2 pmol of malE mRNA, 2 (lane 6) or 4 (lane 7) pmol of 30S subunits, and 10 pmol of initiator tRNA, if present. (B) mRNA sequence of the malE (23) and aph untranslated leader regions. The 5′ untranslated leader regions are shown along with the first five codons of coding sequence (underlined), including the start codon (in bold). The presumed SD sequence is overlined. The 5′ untranslated leader region added to the aph coding sequence was derived from an E. coli pT7 expression vector. The malE and leadered aph mRNAs used are 206 and 133 nucleotides, respectively.
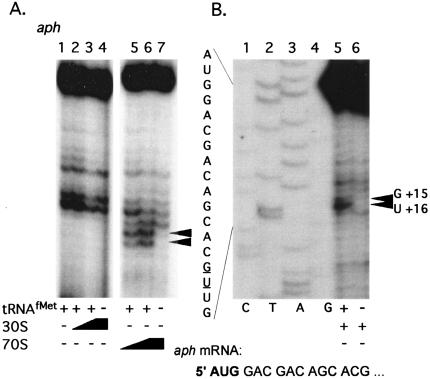
FIG. 3.
(A) Primer extension inhibition (toeprint) assays using leaderless aph mRNA. The two-position toeprint signal in lanes 5 and 6 indicating a ternary complex with aph mRNA (0.2 pmol), low-salt-washed S. lividans 70S ribosomes (2 or 4 pmol), and tRNAfMet (10 pmol) is marked with a double arrowhead. (B) Coelectrophoresis of toeprint reaction mixtures containing leaderless aph mRNA with a DNA sequencing ladder. Mixtures contained 0.2 pmol of aph mRNA, 4 pmol of 30S subunits, 4 pmol of IF1 and IF2, and 10 pmol of initiator tRNA, when present. The first five codons of aph coding sequence are indicated, with the start codon in bold. The aph mRNA used is 89 nucleotides long.
Low-salt-washed 70S ribosomes produced a tRNA-dependent toeprint signal on naturally leaderless aph mRNA; however, low-salt-washed 30S subunits and tRNAfMet did not produce a toeprint signal on leaderless aph mRNAs, even at subunit concentrations that were 20 times the mRNA concentration (Fig. 3A). High-salt-washed 70S ribosomes did not produce a toeprint signal on leaderless aph mRNA (see Fig. 7). These observed differences between high-salt- and low-salt-washed ribosomes and subunits in producing a ternary complex on leaderless mRNA suggested that the high-salt wash removed a component needed for ternary complex formation and led us to purify S. lividans IFs and to include them in ribosome binding assays in an effort to restore ternary complex formation with leaderless mRNA.
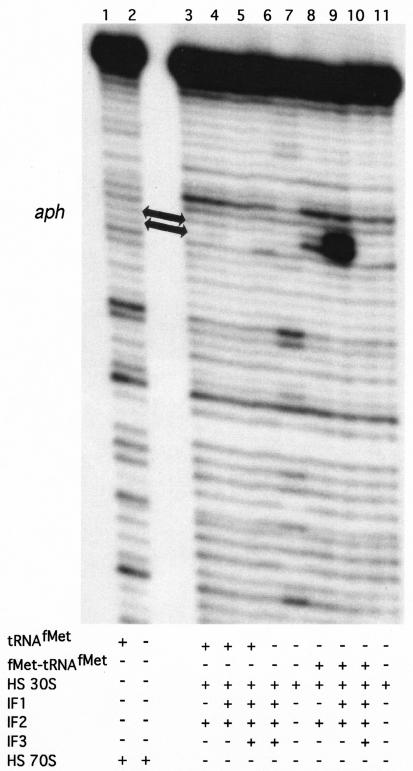
FIG. 7.
Primer extension inhibition (toeprinting) assays using 0.2 pmol of leaderless aph mRNA. High-salt-washed S. lividans 30S subunits (4 pmol) or 70S ribosomes (4 pmol) were used as indicated; 4 pmol of each IF, when present, was preincubated with 30S subunits as explained in Materials and Methods. Ten picomoles of charged (fMet-tRNAfMet) or uncharged (tRNAfMet) initiator tRNA was present as indicated. The position of the toeprint signal, indicating a ternary complex, is marked with arrows.
Ternary complex formation with IFs and leadered mRNA.
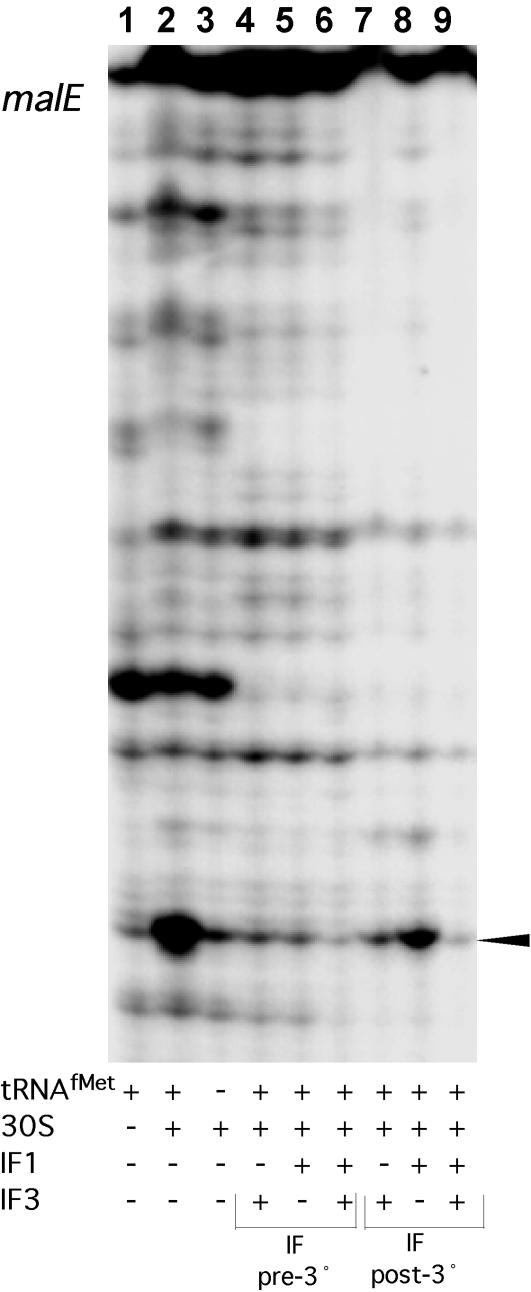
Toeprint assays were performed with high-salt-washed ribosomes or ribosomal subunits, streptomycete IFs, and charged (fMet-tRNAfMet) or uncharged (tRNAfMet) initiator tRNA on leadered aph and malE mRNAs. In toeprint assays with leadered aph mRNA, IF2 reduced the toeprint signal to below that seen when no IFs were present; this was observed when either charged or uncharged initiator tRNA was present (Fig. 4A, compare lanes 2 and 5, respectively, with lane 1). Addition of IF1 and IF2 to reaction mixtures containing fMet-tRNAfMet increased the toeprint signal over that observed with IF2 alone (Fig. 4A, compare lanes 3 and 6 with lanes 2 and 5, respectively), but not to the level observed when no IFs were present (Fig. 4A, lane 1). Interestingly, the inclusion of IF3 (i.e., IF1, IF2, and IF3) reduced the toeprint signal to an intensity comparable to that with IF2 alone in the presence of fMet-tRNAfMet (Fig. 4A, compare lanes 2 and 4) and abolished the toeprint signal in the presence of tRNAfMet (Fig. 4A, lane 7). 30S subunits that were allowed to form a ternary complex in the presence of all three IFs and fMet-tRNAfMet, followed by the addition of 50S subunits, produced a strong, single-position, tRNA-dependent toeprint signal (compare Fig. 4B, lane 4, with Fig. 4A, lane 4). IFs were also included in toeprint reactions with leadered malE mRNA. 30S subunits produced a tRNA-dependent toeprint signal with malE mRNA, as observed for aph mRNA (Fig. 5, lanes 1 to 3). 30S subunits prebound with initiator tRNA and IF1, IF3, or IF1 plus IF3 did not yield a toeprint signal with malE mRNA (Fig. 5, lanes 4 to 6). Further, addition of IF3 or IF1 plus IF3 to a preformed 30S ternary complex containing malE mRNA resulted in a complete loss of the toeprint signal; addition of IF1 alone reduced, but did not eliminate, the toeprint signal (Fig. 5, lanes 7 to 9). In general, toeprint signal intensity was reduced for 30S subunits with IFs compared to that of 30S subunits without IFs for leadered aph and malE mRNAs, suggesting a less stable ternary complex.
FIG. 4.
A. Primer extension inhibition (toeprint) assays using leadered aph mRNA (0.2 pmol). High-salt-washed S. lividans 30S subunits (4 pmol) were used in all lanes; 4 pmol of each IF, when present, was preincubated with the 30S subunits as described in Materials and Methods. Either charged (fMet-tRNAfMet) or uncharged (tRNAfMet) initiator tRNA (10 pmol) was present as indicated. The position of the toeprint signal, indicating a ternary complex, is marked with a double arrowhead. (B) Order of addition toeprint assays (30S ternary complex followed by the 50S subunit) performed as described in Materials and Methods. The toeprint signal is marked with an arrow.
FIG. 5.
Primer extension inhibition (toeprint) assays using malE mRNA (0.2 pmol). High-salt-washed S. lividans 30S subunits (4 pmol) were used in all lanes; 4 pmol of each IF, when present, was preincubated with the 30S subunits (IF pre-3°) or were added to each reaction mixture after the ternary complex had been allowed to form (IF post-3°). The toeprint signal indicating a ternary complex with malE, 30S subunits, and tRNAfMet is marked with an arrowhead.
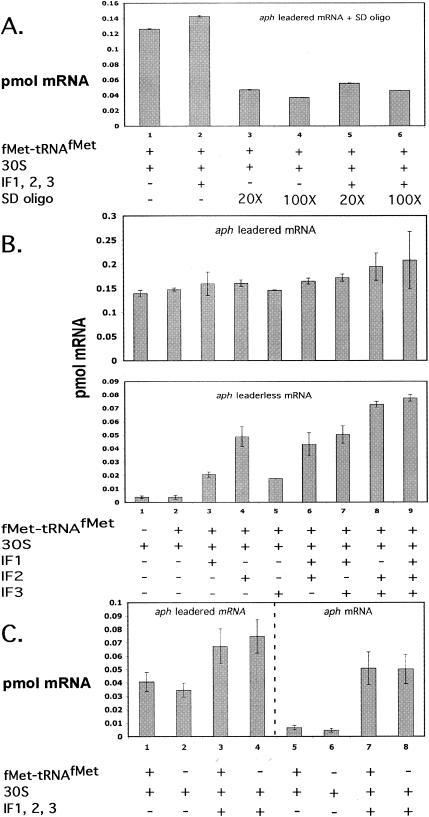
Filter binding assays were also used to study the interaction of 30S subunits, fMet-tRNAfMet, and various combinations of IFs with leadered aph mRNA. When no IFs were included, approximately 73% of input mRNA was bound in a complex with 30S subunits, while only slightly less mRNA was bound when tRNA was absent (Fig. 6B). The preincubation of the 30S subunits with a molar excess (100×) of a DNA oligonucleotide complementary to the anti-SD (ASD) sequence of S. lividans 16S rRNA reduced the amount of complex formed by 68 and 71%, respectively, in the presence and absence of IFs, suggesting that the binding was largely dependent on the SD sequence-ASD sequence interaction (Fig. 6A). The addition of IF1, IF2, or IF3 alone did not significantly affect the binding of leadered aph mRNA over binding with 30S subunits plus fMet-tRNAfMet (Fig. 6B). Essentially 100% of the input leadered mRNA was retained by the filter when all three IFs were included. Comparison of Fig. 6A and C, lanes 1 to 4, suggests that complex formation with leadered mRNA was more dependent on the SD sequence-ASD sequence interaction than on the presence of initiator tRNA. The observed reductions in toeprint ternary complexes with IFs and leadered mRNA (Fig. 4A) were not reflected in the complexes measured with filter binding assays (see the Discussion).
FIG. 6.
(A) Filter binding assay with leadered aph mRNA (0.2 pmol), 4 pmol of 30S subunits, 10 pmol of initiator tRNA, and 4 pmol of each IF (when present). In selected reactions, 30S subunits were preincubated with either 80 pmol (lanes 3 and 5) or 400 pmol (lanes 4 and 6) of an SD oligonucleotide (5′AGA AAG GAG GTG AT3′). (B) Filter binding assays with 0.2 pmol of leadered (top) and leaderless (bottom) aph mRNAs. Each bar represents the average of two reactions. Four picomoles of 30S subunits, 10 pmol of fMet-tRNAfMet, and 4 pmol of IFs (when present) were incubated as described in Materials and Methods prior to nitrocellulose filter binding. (C) Filter binding assays with 0.2 pmol of leadered (lanes 1 to 4) and leaderless (lanes 5 to 8) aph mRNA, 4 pmol of 30S subunits, 4 pmol of each IF (when present), and 10 pmol of fMet-tRNAfMet (when present).
Ternary complex formation with IFs and leaderless mRNA.
High-salt-washed 70S ribosomes did not produce ternary complexes on leaderless aph mRNA (Fig. 7). High-salt-washed 30S subunits did not produce a ternary complex on leaderless aph mRNA when IFs were provided in the presence of tRNAfMet (Fig. 7, lanes 3 to 5). When fMet-tRNAfMet was provided, however, a weak toeprint signal was observed with 30S subunits in the presence of IF2; the toeprint signal was increased significantly in the presence of IF1 and IF2 (Fig. 7, compare lanes 8 and 9). The presence of IF3 (i.e., IF1, IF2, and IF3) abolished the toeprint signal on leaderless aph mRNA (Fig. 7, lane 10). The remaining combinations of IFs (i.e., IF1 plus IF3, IF2 plus IF3, IF1, and IF3), provided in the presence of tRNAfMet or fMet-tRNAfMet, did not promote the formation of a toeprint signal on leaderless aph mRNA (data not shown).
The interaction of 30S subunits with leaderless aph mRNA in the presence of IFs was explored further with filter binding assays. In the absence of IFs, considerably less mRNA was bound than with complexes containing IFs (Fig. 6B). In fact, 30S subunits with all three IFs bound significantly more aph mRNA than 30S subunits with only IF1 and IF2. Note that 30S subunits with IF1 plus IF2 yielded the strongest toeprint signal on aph mRNA, while 30S subunits with IF1, IF2, and IF3 yielded no toeprint signal (Fig. 7). Filter binding assays indicated that 30S subunits incubated with IF2 bound the same amount of mRNA as 30S subunits incubated with IF1 plus IF2, even though toeprint assays suggested that significantly less ternary complex was formed with IF2 than with IF1 plus IF2. While 30S subunits with fMet-tRNAfMet bound less than 2% of the input leaderless aph mRNA, 30S subunits with fMet-tRNAfMet and all three IFs bound approximately 38% of input aph mRNA. As observed for leadered aph mRNA, the complex formed between aph, all three IFs, and the 30S subunit was not dependent on the presence of fMet-tRNAfMet (Fig. 6C, lanes 7 and 8). This suggests weak binding that does not involve anticodon-codon pairing or that involves some direct contribution of the IFs to mRNA binding under the conditions of the assay.
DISCUSSION
A primer extension inhibition assay has been developed by using ribosomes and ribosomal subunits from S. lividans. The utility of this assay has been demonstrated in the study of streptomycete ribosome binding and ternary complex formation; this has led us to a more thorough investigation of the effects of IFs on ternary complex formation with leadered and leaderless mRNA. In addition, interaction of ribosomes with mRNA was studied by using nitrocellulose filter binding assays. The decision to include filter binding assays in this study stemmed from the observation that some IF-ribosomal-subunit combinations produced no toeprint signals, particularly when leaderless mRNA was used, suggesting that weak or transient mRNA-ribosome complexes might not be stable to reverse transcription but might be detected in filter binding assays. The comparison of results from the filter binding and toeprint assays suggests that some complexes are not stable to reverse transcription and might represent initial or intermediate interactions that precede and lead to the formation of a translation initiation complex in the streptomycetes. Presumably, any interaction of the 30S subunit with mRNA can increase the concentration of the subunit near the mRNA's RBS. These interactions, though weak and apparently unstable in toeprinting assays, may significantly affect the subsequent formation of a bona fide ternary complex.
Leadered mRNAs (aph and malE): toeprint and filter binding assays.
The ability of the S. lividans toeprint assay to detect a ternary complex containing ribosomes and mRNA was first tested using leadered aph and malE mRNAs. The use of the leadered form of aph (Fig. 2) gave us the opportunity to test the effect of adding a leader sequence to a naturally leaderless mRNA. Comparing Fig. 2A, with Fig. 3A reveals that the leader sequence can direct the binding of the 30S subunit to aph. High-salt-washed (1 M NH4Cl) 30S subunits produced a tRNAfMet-dependent ternary complex on leadered aph and malE mRNAs, indicating that ancillary factors such as IFs were not needed for binding of ribosomes to leadered mRNA in vitro.
The inclusion of purified S. lividans IFs in the toeprint reactions produced some unexpected results with both leadered aph and malE mRNA. In general, the presence of the IFs reduced the toeprint signal on both mRNAs. The antagonistic effect of the three IFs in combination was especially strong when uncharged initiator tRNA (tRNAfMet) was included in the reactions. Ternary complexes with tRNAfMet were abolished in the presence of IF3, suggesting that IF3 may aid in destabilizing a ternary complex formed with improperly charged or uncharged tRNAfMet, even in the presence of a strong SD sequence-ASD sequence interaction. IF3 appears to be the critical factor in discriminating against a ternary complex with tRNAfMet (Fig. 4A, lane 7), and its presence generally led to a less stable complex, even when fMet-tRNAfMet was included (Fig. 4A, lane 4). The antagonistic effect of IF3 on the ternary complex is not observed when the 30S ternary complex is subsequently bound by the 50S subunit (Fig. 4B, lane 4). In toeprint assays with malE mRNA, the antagonistic effect of IF3 is noted even when IF3 is added to a preformed 30S ternary complex. These reaction mixtures contained tRNAfMet, so this effect may be analogous to the effect of IF3 on leadered aph mRNA when uncharged tRNA is present. The reduced toeprint signal noted when IF2 was included in toeprint reactions with leadered aph mRNA cannot be explained by any known effect of IF2 directly on the conformation of the 30S subunit. IF2 functions to stimulate formation of the 30S-fMet-tRNAfMet binary complex, and our results suggest that the presence of IF2 on the 30S subunit destabilizes the ternary complex, although the presence of IF1 can partially reverse this effect (Fig. 4A, lane 3). This may be a result of the nature of the toeprint assay, in which the ribosome must be bound in such a way that the elongating reverse transcriptase is impeded. A reaction that reflects the presumed state of the initiation complex in vivo (Fig. 4B, lane 4), in which all three IFs were prebound to the 30S subunit, followed by binding to mRNA and subsequent addition of a 50S subunit, indeed moved the toeprint signal to a single position (+16) and increased its intensity. Additionally, this effect of IF2 could be a function of the untranslated leader sequence added to the normally leaderless aph sequence, since IF2 plus IF1 is necessary for ternary complex formation on leaderless aph mRNA, discussed below.
Filter binding data show that a large amount of leadered aph mRNA forms binary complexes with 30S subunits (Fig. 6B). Complexes of leadered mRNA with 30S subunits, with or without IFs, were largely dependent upon the SD sequence-ASD sequence interaction (Fig. 6A). It is important to note that IF combinations that tended to reduce the toeprint signal on leadered aph mRNA (Fig. 4A) did not reduce the amount of complex measured by filter binding assays (Fig. 6B). This suggests that certain IF combinations might adjust the mRNA in such a way that interactions occurring with the 30S subunit alone contribute less to stability than interactions influenced by the IFs.
Leaderless mRNA (aph): toeprint and filter binding assays.
Low-salt-washed 30S subunits did not produce a ternary complex on leaderless aph mRNA (Fig. 3); high-salt-washed 30S subunits produced a strong toeprint signal with aph mRNA only in the presence of IF1 and IF2. This observation that low-salt-washed 30S subunits did not produce a toeprint signal on leaderless aph mRNA suggests either that 30S subunits carry IF3, thereby supporting an earlier report that IF3 negatively affects binding of E. coli 30S subunits to leaderless mRNA (21), or that the subunits carry a factor in vivo that precludes the formation of a stable ternary complex on leaderless messages. Further, it has been speculated that E. coli 30S subunits are bound by all three IFs in vivo (8), suggesting that 30S subunits with the full complement of IFs may not contribute significantly to initiation on leaderless mRNAs in a bacterial cell, perhaps as a direct consequence of IF3. Possibly analogous to 30S subunit-associated factors that affect initiation complex formation, low-salt-washed 70S ribosomes produced initiator tRNA-dependent toeprints on leaderless aph mRNA (Fig. 3A), but high-salt-washed 70S ribosomes did not (Fig. 7), thereby suggesting the removal of a factor(s) during the high-salt wash needed for 70S ribosome binding with leaderless messages.
Interestingly, filter binding data reveal that 30S subunits incubated with all three IFs plus fMet-tRNAfMet bind significantly more leaderless mRNA than 30S subunits incubated with IF1 and IF2. Furthermore, 30S subunits bound a diminishingly small amount of leaderless aph mRNA when no IFs were included in the reactions. A similar result has been seen with E. coli 30S subunits on leaderless cI34 mRNA (5). Filter binding assays suggested the occurrence of complexes containing 30S subunits, initiator tRNA, IFs, and leaderless mRNA that proved to be unstable in the toeprint assay. Indeed, the toeprint assay proved its value by revealing stable ternary complexes formed with low-salt-washed 70S ribosomes and with 30S subunits plus IF1 and IF2. The fact that the binding of 30S subunits to aph mRNA was tRNA independent in the filter binding assays suggests that these complexes are not true ternary initiation complexes, with the initiator tRNA and the start codon in the ribosomal P site, but rather may be interactions that serve in vivo to place the major components of translation initiation in close proximity to one another. It appears at least that IF3 plays an active part in discriminating against the stable association of 30S subunits with leaderless mRNA, since its presence precludes the formation of a strong ternary complex between aph and 30S subunits (Fig. 7). In possible support of this, the binding of IF3 to the E. coli 30S subunit changes the subunit's conformation in such a way that tRNA binding in the P site is affected (15, 16, 19).
S. lividans IFs.
The ribosome dissociation studies described here suggest that a subpopulation of 70S ribosomes that is stable to the predicted effects of IF3 (or IF1 plus IF3) may exist in S. lividans. The observation that S. lividans IF3 dissociates E. coli ribosomes into subunits suggests a conservation of function between the IF3 proteins in these distantly related bacteria. A conserved-domain search using the National Center for Biotechnology Information BLAST reveals that the streptomycete IF3 contains the conserved domains that define bacterial IF3. However, the streptomycete IF3 does contain a C-terminal extension that E. coli IF3 lacks.
S. lividans IF1, in addition to enhancing the ability of IF3 to dissociate E. coli and S. lividans 70S ribosomes, was able partially to dissociate both E. coli and S. lividans 70S ribosomes. E. coli IF1 has been reported to have a slight but measurable effect on the splitting of 70S ribosomes into 30S and 50S subunits (4), but S. lividans IF1 split a substantial amount of E. coli and S. lividans ribosomes into subunits. In E. coli, IF1 occupies the A site of a 30S subunit initiating translation, effectively blocking the A-site binding by the initiator tRNA, and increases the rate of subunit exchange; perhaps the binding of S. lividans IF1 to E. coli 30S subunits in this manner prevents some of the 30S and 50S subunits from associating to form 70S ribosomes.
IF1 was also observed to enhance greatly the ability of IF2 to promote binding of 30S subunits to leaderless mRNA. IF2 did enhance the binding of 30S subunits to leaderless mRNA when included alone, and it further showed a preference for properly charged and formylated initiator tRNA. The ability of IF2 to enhance ternary complex formation and translation from leaderless mRNA in E. coli has been reported (5, 6, 14).
Concluding remarks.
The toeprint assay developed for S. lividans ribosomes has proven to be valuable in dissecting the interactions of the streptomycete translational machinery with leadered and leaderless mRNAs. The study of ribosome binding in S. lividans will shed light on translation initiation in a genus that naturally carries genes for a substantial number of leaderless mRNAs. This first look at the effect of streptomycete IFs on ribosome binding has revealed possible differences in the way these IFs behave when affecting ribosome binding on leadered and leaderless mRNA. Our observations of leaderless mRNAs in two different ribosome binding assays supports earlier suggestions that mRNAs initiating with a 5′-terminal AUG may be translated via a pathway involving 70S ribosomes or 30S subunits lacking IF3 (14); it is possible that 30S subunits with all three IFs would bind leaderless mRNA weakly in vivo and that these complexes would be unable to compete with leaderless mRNA translated via a dedicated pathway of 70S ribosomes or 30S subunits with only IF1 and IF2. Additionally, where the inclusion of in vivo gene expression data with in vitro ribosome binding data (toeprint assays) helps to dissect the contribution of ternary complex formation to translation initiation and protein production, coupling toeprint assays with filter binding assays may further shed light on the interactions that precede formation of the ternary initiation complex. A more complete, albeit more complicated, picture of the molecular composition of the ribosomal subunits that bind leadered and leaderless mRNAs could result from this type of investigation. Furthermore, this investigation suggests that bacteria may deal with leaderless mRNAs differently than leadered mRNAs, with the IFs playing an important role in the stability of the initiation and perhaps preinitiation complexes. The simple fact that the streptomycetes contain several leaderless mRNAs that encode proteins involved in antibiotic resistance and differentiation tells us that these mRNAs are not insignificant in the physiology of the genus, particularly if leaderless mRNAs are translated via a dedicated pathway that would allow successful competition with leadered mRNAs, thus ensuring the expression of these important genes.
Acknowledgments
We thank Uttam RajBhandary for providing the plasmids pQE16-MTF and pQE60-MetRS, Eric Cundliffe for helpful advice on ribosome isolation protocols in the streptomycetes, and Isabella Moll for tips on nitrocellulose filter binding assays.
J.M.D. thanks the Miami University Graduate School for a Graduate Student Achievement Award and the Miami University Center for Bioinformatics and Functional Genomics for a Summer Research Fellowship. This work was supported by grant GM65120 from the National Institutes of Health.
REFERENCES
- 1.Calcutt, M. J., and E. Cundliffe. 1989. Use of a fractionated, coupled transcription-translation system in the study of ribosomal resistance mechanisms in antibiotic-producing Streptomyces. J. Gen. Microbiol. 135:1071-1081. [DOI] [PubMed] [Google Scholar]
- 2.Christie, G. E., and R. Calendar. 1985. Bacteriophage P2 late promoters II. Comparison of the four late promoter sequences. J. Mol. Biol. 181:373-382. [DOI] [PubMed] [Google Scholar]
- 3.Dubnoff, J. S., and U. Maitra. 1971. Isolation and properties of protein factors involved in polypeptide chain initiation in Escherichia coli. Methods Enzymol. 20:248-260. [DOI] [PubMed] [Google Scholar]
- 4.Godefroy-Colburn, T., A. D. Wolfe, J. Dondon, M. Grunberg-Manago, P. Dessen, and D. Pantaloni. 1975. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol. 94:461-478. [DOI] [PubMed] [Google Scholar]
- 5.Grill, S., C. O. Gualerzi, P. Londei, and U. Blasi. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 19:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grill, S., I. Moll, D. Hasenohrl, C. O. Gualerzi, U. Blasi. 2001. Modulation of ribosomal recruitment to 5′-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett. 495:167-171. [DOI] [PubMed] [Google Scholar]
- 7.Gualerzi, C. O., L. Brandi, E. Caserta, A. La Teana, R. Spurio, J. Tomšic, and C. L. Pon. 2000. Translation initiation in bacteria, p. 477-494. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 8.Gualerzi, C. O., and C. L. Pon. 1990. Initiation of mRNA translation in prokaryotes. Biochemistry 29:5881-5889. [DOI] [PubMed] [Google Scholar]
- 9.Hartz, D., D. S. McPheeters, R. Traut, and L. Gold. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164:419-425. [DOI] [PubMed] [Google Scholar]
- 10.Janssen, G. R. 1993. Eubacterial, archaeabacterial, and eucaryotic genes that encode leaderless mRNA, p. 59-67. In B. H. Balt, G. D. Hegema, and P.L. Skatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. American Society for Microbiology, Washington, D.C.
- 11.Klock, G., and W. Hillen. 1986. Expression, purification and operator binding of the transposon Tn1721-encoded Tet repressor. J. Mol. Biol. 189:633-641. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Farmer, J., and G. R. Janssen. 1999. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 31:1025-1038. [DOI] [PubMed] [Google Scholar]
- 13.O'Donnell, S. M., and G. R. Janssen. 2001. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 183:1277-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell, S. M., and G. R. Janssen. 2002. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J. Bacteriol. 184:6730-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrelli, D., A. La Teana, C. Garolo, R. Spurio, C. L. Po, and C. O. Gualerzi. 2001. Translation initiation factor IF3: two domains, five functions, one mechanism. EMBO J. 20:4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrelli, D., C. Garofalo, M. Lammil, R. Spurio, C. L. Po, C. O. Gualerz, and A. La Teana. 2003. Mapping the active sites of bacterial translation initiation factor IF3. J. Mol. Biol. 331:541-556. [DOI] [PubMed] [Google Scholar]
- 17.Ptashne, M., K. Backman, M. Z. Humayum, A. Jeffery, R. Maurer, B. Meyer, and R. T. Sauer. 1976. Autoregulation and function of a repressor in bacteriophage lambda. Science 194:156-161. [DOI] [PubMed] [Google Scholar]
- 18.Ramesh, V., S. Gite, and U. L. RajBhandary. 1998. Functional interaction of an arginine conserved in the sixteen amino acid insertion module of Escherichia coli methionyl-tRNA formyltransferase with determinants for formylation in the initiator tRNA. Biochemistry 37:15925-15932. [DOI] [PubMed] [Google Scholar]
- 19.Sette, M., R. Spurio, P. van Tilborg, C. O. Gualerz, and R. Boelens. 1999. Identification of the ribosome binding sites of translation initiation factor IF3 by multidimensional heteronuclear NMR spectroscopy. RNA 5:82-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soffentini, A., R. Lorenzetti, L. Gastaldo, J. H. Parlett, R. Spurio, A. La Teana, and K. Islam. 1994. Purification procedures for bacterial translational initiation factors IF2 and IF3. Protein Expr. Purif. 5:118-124. [DOI] [PubMed] [Google Scholar]
- 21.Tedin, K., I. Moll, S. Grill, A. Resch, C. O. Gualerz, and U. Blasi. 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 31:67-77. [DOI] [PubMed] [Google Scholar]
- 22.Van Etten, W. J., and G. R. Janssen. 1998. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol. Microbiol. 27:987-1001. [DOI] [PubMed] [Google Scholar]
- 23.van Wezel, G. P., J. White, M. J. Bibb, and P. W. Postma. 1997. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, disruption and transcriptional analysis. Mol. Gen. Genet. 254:604-608. [DOI] [PubMed] [Google Scholar]
- 24.Wu, C.-J., and G. R. Janssen. 1996. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5′ untranslated leader. Mol. Microbiol. 22:339-355. [DOI] [PubMed] [Google Scholar]
- 25.Wu, C.-J., and G. R. Janssen. 1997. Expression of a streptomycete leaderless mRNA encoding chloramphenicol acetyltransferase in Escherichia coli. J. Bacteriol. 179:6824-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]