Abstract
The MEK kinase TPL-2 (also known as Cot) is required for lipopolysaccharide (LPS) activation of the extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase cascade in macrophages and consequent upregulation of genes involved in innate immune responses. In resting cells, TPL-2 forms a stoichiometric complex with NF-κB1 p105, which negatively regulates its MEK kinase activity. Here, it is shown that lipopolysaccharide (LPS) stimulation of primary macrophages causes the release of both long and short forms of TPL-2 from p105 and that TPL-2 MEK kinase activity is restricted to this p105-free pool. Activation of TPL-2, MEK, and ERK by LPS is also demonstrated to require proteasome-mediated proteolysis. p105 is known to be proteolysed by the proteasome following stimulus-induced phosphorylation of two serines in its PEST region by the IκB kinase (IKK) complex. Expression of a p105 point mutant, which is not susceptible to signal-induced proteolysis, in RAW264.7 macrophages impairs LPS-induced release of TPL-2 from p105 and its subsequent activation of MEK. Furthermore, expression of wild-type but not mutant p105 reconstitutes LPS stimulation of MEK and ERK phosphorylation in primary NF-κB1-deficient macrophages. Consistently, pharmacological blockade of IKK inhibits LPS-induced release of TPL-2 from p105 and TPL-2 activation. These data show that IKK-induced p105 proteolysis is essential for LPS activation of TPL-2, thus revealing a novel function of IKK in the regulation of the ERK MAP kinase cascade.
Lipopolysaccharide (LPS) stimulation of Toll-like receptor 4 (TLR4) on macrophages leads to the induction of genes that function in the innate and adaptive immune responses to gram-negative bacterial infection (32). These include proinflammatory cytokines, chemokines, the major histocompatibility complex, and costimulatory molecules (23). LPS induction of these genes involves activation of NF-κB transcription factors and each of the major mitogen-activated protein (MAP) kinase subtypes (extracellular signal-regulated kinases 1 and 2 [ERK-1/2], Jun amino-terminal kinases, and p38) (32).
MAP kinases are phosphorylated and activated by MAP kinase kinases, which in turn are phosphorylated and activated by MAP kinase kinase kinases in conserved three-tiered kinase cascades (9). LPS activation of ERK-1/2 MAP kinases in macrophages requires the serine/threonine kinase TPL-2 (12) (also known as Cot [24]). TPL-2 functions as a MAP kinase kinase kinase which phosphorylates and activates the ERK-1/2 kinases, MEK-1/2 (28). LPS induction of tumor necrosis factor alpha (TNF-α) and cyclooxygenase 2 (COX-2) is dramatically reduced in TPL-2-deficient macrophages due to defective ERK-1/2 activation (12, 13). Consequently, TPL-2−/− mice are resistant to LPS/d-galactosamine-induced endotoxin shock (12). TPL-2 is also required for TNF-α and CD40 ligand to stimulate MEK-1/2 activation (14), suggesting an important role for TPL-2 in both innate and adaptive immune responses.
NF-κB dimers are retained in the cytoplasm of unstimulated cells through their interaction with a family of inhibitory proteins, termed IκBs (15). The IκB family includes NF-κB1 p105, which retains associated NF-κB dimers by virtue of its C-terminal ankyrin repeats. p105 is also constitutively processed by the proteasome to produce the NF-κB transcription factor, p50 (19). Genetic studies with mice have indicated that p105 is particularly important for the cytoplasmic retention of p50 homodimers (17). Following cellular stimulation with ligands, such as TNF-α, two serines in the p105 PEST region are rapidly phosphorylated by the IκB kinase (IKK) complex (20, 29). This creates a binding site for the ubiquitin E3 ligase, SCFβTrCP, which promotes p105 ubiquitination (16, 20, 26), leading predominantly to the complete degradation of p105 by the proteasome. Associated NF-κB (Rel) subunits are thereby released to translocate into the nucleus and modulate target gene expression.
Earlier studies from this laboratory demonstrated that the C-terminal half of NF-κB1 p105 forms a high-affinity, stoichiometric association with TPL-2 (3, 4). This interaction is required to maintain TPL-2 protein stability. Consequently, the steady-state levels of TPL-2 are very low in p105-deficient cells (3, 37), and LPS activation of MEK is severely reduced in bone marrow-derived macrophages (BMDMs) generated from NF-κB1−/− mice (37). Interaction of p105 with TPL-2 also negatively regulates its MEK kinase activity by preventing access to MEK (3, 37). In unstimulated BMDMs, therefore, TPL-2 MEK kinase activity is blocked (37), since all detectable TPL-2 is complexed with p105 (21). However, following LPS stimulation, TPL-2 MEK kinase activity increases, indicating that TPL-2 is released from p105 inhibition (37).
In the present study, the mechanism by which TPL-2 is activated after LPS stimulation of BMDMs was investigated. Evidence is presented that IKK-induced p105 proteolysis is required to generate a pool of p105-free TPL-2 in LPS-stimulated cells which phosphorylates and activates MEK. Consequently, LPS activation of ERK in macrophages is dependent on the activity of the IKK complex.
MATERIALS AND METHODS
cDNA constructs, antibodies, and recombinant proteins.
Hemagglutinin (HA) epitope-tagged wild-type NF-κB1 p105 (HA-p105), HA-p105S927A,S932A (HA-p105SSAA), Myc epitope-tagged wild-type TPL-2 (Myc-TPL-2), and kinase-inactive Myc-TPL-2D270A (Myc-TPL-2KD) cDNAs have been described previously (4, 20). For stable transfection of RAW264.7 cells, these cDNAs were subcloned in the pMX-1 vector (Ingenius). For retroviral infection of BMDMs, FLAG epitope-tagged wild-type p105 (FL-p105) and p105S927A,S932A (FL-p105SSAA) were generated by PCR, subcloned into a pMSCV-based vector, and verified by DNA sequencing.
The rabbit antibodies used to immunoprecipitate TPL-2 (70-mer) and NF-κB1 p105 (anti-p105N) have previously been described (20). For Western blotting, anti-p105C antibody was used to detect total p105 (29), and a previously described anti-phospho-S927-p105 antibody (29) was used to detect p105 phosphorylated on serine 927. A commercial anti-TPL-2 antibody (Santa Cruz) was used to detect TPL-2 on Western blots. Antibodies against MEK-1/2, phospho(S217/S221)-MEK-1/2 (activated MEK-1/2; phospho-MEK), p38, and phospho(T180/Y182)-p38 (activated p38; phospho-p38) were purchased from Cell Signaling Technology. Anti-ERK-1b was generously provided by Jeremy Tavare (University of Bristol, Bristol, United Kingdom), and anti-phospho(T185/Y187)-ERK (activated ERK; phospho-ERK) was purchased from Biosource. Antibodies used to immunoprecipitate and Western blot HA epitope-tagged proteins have been described previously (20). Tubulin (detected with TAT-1 anti-α-tubulin monoclonal antibody, kindly provided by Keith Gull, University of Manchester, United Kingdom) was used as a loading control protein on Western blots of total cell extracts. Glutathione S-transferase (GST)-MEK and GST-ERK proteins were both kindly provided by Richard Marais (CR-UK, London, United Kingdom).
Mouse strains and cell lines.
NF-κB1 knockout mice (30) (from Jackson Laboratories, Bar Harbor, Maine) and BALB/c mice were bred in a specific-pathogen-free environment at the National Institute for Medical Research (London, United Kingdom).
BMDMs from BALB/c mice were prepared as described previously (36). Briefly, bone marrow cells were plated in complete BMDM medium (RPMI 1640; Sigma) supplemented with 10% fetal bovine serum, antibiotics, and 20% L-cell conditioned medium. After 24 h, nonadherent cells were transferred to 90-mm dishes (Nunc; 6 × 106 cells). On day 4 of culture, the cells were further supplemented with complete BMDM medium. After a total of 7 days of culture, the adherent macrophages were harvested and plated for experiments. More than 95% of the resulting cell populations were positive for the macrophage marker F4/80, as judged by flow cytometric analysis (data not shown).
RAW264.7 cells were kindly provided by Lynn Williams (Kennedy Institute, London, United Kingdom) and maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum and antibiotics. For stable transfection, 5 × 107 RAW264.7 cells were transfected by electroporation with 10 μg of pMX-1 expression vector containing cDNA inserts encoding HA-p105, HA-p105SSAA, Myc-TPL-2, or MycTPL-2KD. As a control, cells were transfected with pMX-1 vector containing no insert (empty vector [EV]). Transfected cells were cultured for a further 48 h and cloned by limiting dilution under neomycin selection (complete Dulbecco's modified Eagle's medium plus 1 mg of G418/ml; Invitrogen). After 1 to 3 weeks, clones were expanded and tested for expression of transfected proteins by Western blotting for HA epitope tag or TPL-2. Positive clones were maintained in selection medium. Prior to experiments, the RAW264.7 cell lines were passaged once without G418. All experiments were performed with at least two independent clones and generated similar results.
Protein analysis.
BMDMs (3 × 106 cells) or RAW264.7 cells (4 × 106) were plated in 60-mm dishes (Nunc). After 18 h in culture, cells were stimulated with LPS (1 μg/ml; Salmonella enterica serovar Minnesota; Alexis Biochemicals) for the times shown or phorbol myristate acetate (PMA) (100 ng/ml; Sigma) for 7.5 min or left untreated. Where appropriate, cells were preincubated with the indicated concentrations of MG132 proteasome inhibitor (Biomol), BAY 11-7082 IKK inhibitor (Calbiochem), or dimethyl sulfoxide (DMSO) vehicle control for 30 min prior to stimulation. Cells were washed once in phosphate-buffered saline prior to lysis in 1% NP-40 containing buffer A (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 1 mM Na3VO4, 100 nM okadaic acid [Calbiochem], 2 mM Na4P2O7 plus a mixture of protease inhibitors [Roche Molecular Biochemicals]).
For experiments in which total p105 levels were quantified, washed BMDMs were extracted directly into sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Extracts were passed through a 23-gauge needle to shear DNA and boiled prior to SDS-PAGE. p105 levels were detected on Western blots by enhanced chemiluminescence (Amersham Biosciences) and quantified using a Fuji image reader (LAS-3000). p105 levels were normalized against α-tubulin.
Covalent coupling of antibodies to protein A-Sepharose (Amersham Biosciences), immunoprecipitation, and Western blotting of proteins were carried out as described previously (18). p105 was precleared from BMDM lysates by immunodepletion twice with anti-p105N antibody coupled to protein A-Sepharose. p105 was similarly immunodepleted from lysates of stably transfected RAW264.7 cells but with a mixture of anti-p105N and anti-HA (12CA5) antibodies coupled to protein A Sepharose. Complete depletion of p105 was confirmed by Western blotting. The percentage of TPL-2 released from p105 after LPS stimulation of BMDMs was determined by scanning Western blots using a Bio-Rad calibrated imaging densitometer (GS710). TPL-2 levels were normalized against α-tubulin.
Assay of TPL-2 phosphorylation and MEK kinase activity.
To analyze TPL-2 phosphorylation, LPS-stimulated BMDMs (8 × 106 cells) were lysed in kinase lysis buffer (buffer A containing 0.5% NP-40, 5 mM Na β-glycerophosphate, and 0.1% 2-mercaptoethanol,) and TPL-2 was immunoprecipitated for 4 h with anti-TPL-2 antibody coupled to protein A-Sepharose. Beads were washed twice in kinase lysis buffer and twice in protein phosphatase 2A (PP2A) buffer (Upstate) and then resuspended in 50 μl of PP2A buffer. PP2A (0.5 U; Upstate) was added to the appropriate tubes with or without the phosphatase inhibitors, NaF (1 mM) and okadaic acid (100 nM). Control tubes had no addition. Samples were incubated for 30 min at 30°C. Bound TPL-2 was eluted from beads by boiling in SDS-PAGE sample buffer and then subjected to Western blotting after 10%-acrylamide SDS-PAGE.
To assay TPL-2 MEK kinase activity, BMDMs (8 × 106 cells) or RAW264.7 cells (10 × 106 cells) were plated in 90-mm dishes (Nunc). After 18 h in culture, cells were stimulated with LPS for the indicated times and then extracted with kinase lysis buffer. Lysates were immunoprecipitated for 4 h with anti-TPL-2 antibody coupled to protein A-Sepharose. To analyze p105-free TPL-2, lysates were first immunodepleted twice for 2 h with p105N antibody or control immunoglobulin coupled to protein A-Sepharose. Subsequently, these cleared lysates were immunoprecipitated with anti-TPL-2 antibody, anti-p105N antibody, or control immunoglobulin (Ig). Specific antibody-coupled beads were washed four times in kinase lysis buffer and twice with kinase buffer (50 mM Tris [pH 7.5], 5 mM β-glycerophosphate, 0.1 mM sodium vanadate, 100 nM okadaic acid, 10 mM MgCl2, 0.1 mM EGTA, 0.03% Brj35, 0.1% 2-mercaptoethanol). The beads were resuspended in 25 μl of kinase buffer supplemented with 1 mM ATP, 6.5 μg of GST-MEK/ml, and 100 μg of GST-ERK/ml and incubated for 30 min at room temperature on a shaker. After beads were pelleted by centrifugation, 2 μl of the supernatant was added to 48 μl of kinase buffer containing 0.33 mg of myelin basic protein (MBP) (Sigma)/ml, 0.1 mM ATP, and 2.5 μCi of [γ-32P]ATP (Amersham Biosciences) and incubated at room temperature for 10 min. The assay was terminated by adding 50 μl of 2× SDS sample buffer, and labeled MBP was revealed by autoradiography after 12.5% acrylamide SDS-PAGE. Immunoprecipitated protein was eluted from the antibody-coupled beads with 0.2 M glycine (pH 2.5), resolved by 10% acrylamide SDS-PAGE, and subjected to Western blotting.
Retrovirus infection of BMDMs.
Amphoteric recombinant retrovirus were produced by transfecting wild-type FL-p105 or FL-p105SSAA, subcloned in the pMSCV-based vector, into the Plat-E packaging cell line (25). These cells were transfected using GeneJuice transfection reagent (Merck Biosciences) according to the manufacturer's instructions. Transfected cells were cultured at 37°C for 48 to 72 h, at which time culture supernatants were removed and filtered (pore size, 0.4 μm). Virus was concentrated 10-fold by centrifugation prior to BMDM infection.
BMDMs from NF-κB1−/− mice were prepared essentially as described above, with minor modifications. Bone marrow cells were plated in complete BMDM medium at 106 cells/well (culture volume, 2 ml) of a six-well plate (Sarstedt). Following 48 h of incubation, 200 μl of concentrated virus was added per well, and the plates were centrifuged at 2,000 × g for 1 h at room temperature. Each well was further supplemented with complete BMDM medium (2 ml) 2 and 48 h after infection. On day 7, cells were replated in fresh BMDM medium (1.5 × 106 cells/well; Nunc six-well plates). On the following day, BMDMs were stimulated with LPS (10 ng/ml) and analyzed as described above. Flow cytometric analysis confirmed that >95% of cells prepared in this way were F4/80 positive.
RESULTS
LPS stimulation of primary macrophages induces release of TPL-2 from p105.
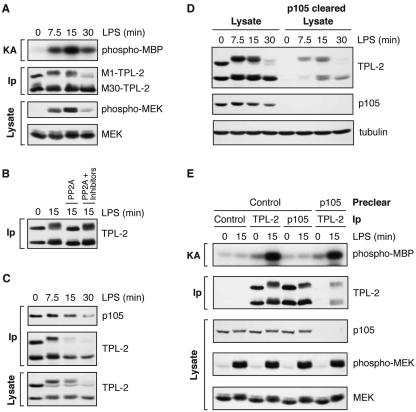
The MEK kinase activity of TPL-2 is inhibited by its interaction with NF-κB1 p105 (3, 37). In unstimulated BMDMs, the majority of TPL-2 is associated with p105 (Fig. 1D) (21), and consequently, TPL-2 MEK kinase activity cannot be detected in anti-TPL-2 immunoprecipitates (37) (Fig. 1A). However, LPS stimulation of BMDMs causes a marked increase in TPL-2 MEK kinase activity with kinetics similar to phosphorylation of endogenous MEK on its activation loop (Fig. 1A) (37). In the following sections, the physiological mechanism by which TPL-2 is released from p105 inhibition after LPS stimulation is investigated.
FIG. 1.
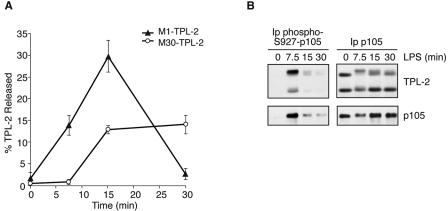
LPS stimulation induces a p105-free pool of TPL-2 which activates MEK. BMDMs (BALB/c) were stimulated with LPS for the indicated times. (A) TPL-2 was immunoprecipitated from cell lysates with anti-TPL-2 antibody, and its MEK kinase activity was determined by coupled MEK/ERK kinase assay. Labeled MBP substrate was visualized by autoradiography after SDS-PAGE, and the levels of immunoprecipitated TPL-2 were determined by Western blotting. Lysates were also subjected to Western blotting with an anti-phospho-(S217/S221)-MEK-1/2 antibody (phospho-MEK) to determine activation of endogenous MEK by LPS. (B) TPL-2 was immunoprecipitated from cell lysates. Beads were incubated with 0.5 U of PP2A, 0.5 U of PP2A plus the phosphatase inhibitors NaF and okadaic acid (PP2A + Inhibitors), or control buffer. Immunoprecipitated TPL-2 was revealed by Western blotting. (C) Cell lysates and anti-p105 immunoprecipitates were subjected to Western blotting. (D) Total cell lysates (lysates) and p105 cleared lysates were subjected to Western blotting for TPL-2 and p105. Equal protein loading was confirmed by probing for α-tubulin. (E) Cell lysates were immunoprecipitated with anti-TPL-2 antibody, anti-p105N antibody, or control Ig (control) after first immunodepleting with either anti-p105N antibody (p105) or control Ig (control) as indicated. Immunoprecipitates were assayed for associated MEK kinase activity and TPL-2 levels as for panel A. Cell lysates were subjected to Western blotting to confirm depletion of p105 and activation of MEK phosphorylation by LPS.
In initial experiments, the effect of LPS on TPL-2 was determined by Western blotting. Alternative translational initiation on a second methionine (at residue 30) results in the expression of two TPL-2 isoforms (1), M1-TPL-2 and M30-TPL-2 (Fig. 1A). Similar to published experiments (37), LPS induced proteolysis of the longer isoform, M1-TPL-2, at 30 min, whereas the shorter isoform, M30-TPL-2, was stable (Fig. 1A and C). Prior to inducing its degradation, LPS stimulation induced a marked decrease in mobility of essentially all of M1-TPL-2 in SDS-PAGE (Fig. 1A and C). In some experiments, a very small fraction of M30-TPL-2 also underwent an LPS-induced mobility shift. Treatment of anti-TPL-2 immunoprecipitates with PP2A demonstrated that these mobility shifts were due to phosphorylation of TPL-2 (Fig. 1B).
Western blotting of anti-p105N immunoprecipitates revealed a decrease in copurifying M1-TPL-2 after LPS stimulation (Fig. 1C). Previously this LPS-induced decrease in p105-associated TPL-2 had been suggested to result from the selective dissociation of M1-TPL-2 from p105 (37). However, this was not clear from the present analyses due to proteolysis of both M1-TPL-2 and p105 after LPS stimulation (Fig. 1A and C). Therefore, to determine conclusively whether LPS induces the dissociation of TPL-2 from its inhibitor p105, an assay was developed which could directly detect low levels of p105-free TPL-2. To do this, lysates of BMDMs were precleared of p105 by immunodepletion and then subjected to Western blotting for TPL-2. Similar to previous results (21), no p105-free TPL-2 was detected in unstimulated cells (Fig. 1D). However, LPS stimulation induced the appearance of both M1-TPL-2 and M30-TPL-2 in the p105-depleted lysate, peaking at 15 min, at which time TPL-2 MEK kinase activity was also maximal (Fig. 1A). By densitometric scanning of Western blots, it was determined that 21% (standard error of the mean [SEM], ±4.6%; n = 4) of total TPL-2 present in unstimulated cells was released from p105 after 15 min of LPS stimulation.
To establish whether activated TPL-2 resides in the p105-free pool, TPL-2 was immunoprecipitated from lysates of LPS-stimulated BMDMs which had been precleared with either anti-p105 antibody or control Ig. Similar levels of TPL-2 MEK kinase activity were isolated from p105-free and control lysates, despite the amount of TPL-2 immunoprecipitated from p105-free lysates being only a fraction of that isolated from control lysates (Fig. 1E). Consistent with published data (37), no MEK kinase activity was detected when p105 was isolated directly with anti-p105N antibody, although large amounts of TPL-2 were copurified (Fig. 1E).
Together these data show that LPS activation of TPL-2 in BMDMs involves the release of both M1 and M30 TPL-2 from p105 and that TPL-2 MEK kinase activity is restricted to this p105-free pool.
LPS activation of TPL-2 and MEK requires proteasome activity.
LPS stimulates p105 proteolysis by the proteasome in THP-1 monocytes and 70Z/3 pre-B cells (11, 16). Therefore, one possible mechanism by which LPS might promote the release of TPL-2 from p105 in BMDMs is to stimulate proteasome-mediated proteolysis of p105.
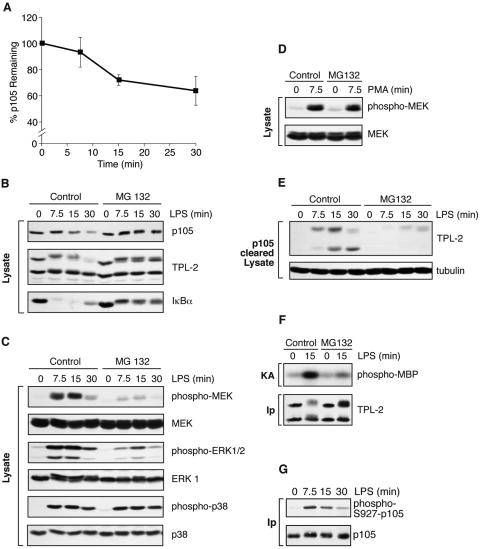
To determine whether LPS induces p105 proteolysis in BMDMs rapidly enough to account for the kinetics of TPL-2 release from p105 and TPL-2 activation, p105 levels were quantified by Western blotting in replicate experiments. To ensure that total p105 was assayed, cells were extracted directly into SDS-PAGE sample buffer. After 15 min of LPS stimulation, when LPS-induced release of TPL-2 from p105 and TPL-2 MEK kinase activity were maximal (Fig. 1A and D), p105 levels decreased by 28% (standard error of the mean, ±4%; n = 3; P = 0.02) compared with unstimulated control (Fig. 2A). Treatment of BMDMs with the proteasome inhibitor MG132 prior to stimulation blocked the LPS-induced decreases in the levels of both p105 and IκBα (Fig. 2B). Proteolysis of M1-TPL-2 induced by LPS stimulation was also inhibited by MG132 (Fig. 2B). These data indicate that LPS stimulation of BMDMs induces proteasome-mediated proteolysis of p105 with kinetics that correlate with TPL-2 release from p105 and TPL-2 activation.
FIG. 2.
Proteasome activity is required for LPS activation of TPL-2. (A) BMDMs were simulated with LPS for the indicated times. Total p105 protein levels in cell lysates were determined by Western blotting and quantified using a Fuji Image reader. Data are presented as means (± SEM; n = 3), normalized against α-tubulin. (B to E) BMDMs were preincubated with the proteasome inhibitor MG132 (40 μM) or DMSO vehicle control for 30 min and then stimulated with LPS or PMA for the indicated times. Total cell lysates were subjected to Western blotting for the indicated proteins. LPS activation of endogenous MEK, ERK, and p38 was monitored with the appropriate phospho-specific antibodies. (E) Cell lysates were immunodepleted of p105 prior to Western blotting for TPL-2 and α-tubulin. (F) BMDMs were treated with MG132 (40 μM) prior to stimulation with LPS for 15 min. TPL-2 was immunoprecipitated from total cell lysates and then assayed for MEK kinase activity as for Fig. 1A. (G) BMDMs, pretreated with 40 μM MG132, were simulated with LPS for the indicated times. p105 was immunoprecipitated from total cell lysates, subjected to Western blotting, and probed with a specific anti-phospho-peptide antibody to monitor p105 serine 927 phosphorylation.
Next, pharmacological blockade of the proteasome with MG132 was used to investigate initially whether p105 proteolysis might be an important step in the generation of p105-free active TPL-2 in BMDMs. Western blotting with phospho-antibodies revealed that MG132 inhibited LPS stimulation of MEK and ERK phosphorylation (Fig. 2C) in a dose-dependent fashion (supplemental figure). However, LPS activation of p38 phosphorylation (Fig. 2C) and phorbol ester stimulation of MEK phosphorylation (Fig. 2D), which are both independent of TPL-2 (12), were unaffected by MG132 treatment. Two other inhibitors of proteasome function, clasto-lactacystin β-lactone and ALLN, also inhibited LPS-induced phosphorylation of MEK and ERK in a dose-dependent fashion (supplemental figure). Each of the proteasome inhibitors blocked LPS stimulation of MEK and ERK phosphorylation, p105 proteolysis, and IκBα degradation with similar potency. ALLM, an ALLN congener that is not active on the proteasome, did not affect LPS induction of MEK and ERK phosphorylation. Similar to MG132, clasto-lactacystin β-lactone and ALLN had no effect on LPS stimulation of p38 phosphorylation.
Western blotting of p105-depleted lysates revealed that LPS-induced release of TPL-2 from p105 was prevented by MG132 treatment (Fig. 2E). Furthermore, MG132 was also found to significantly reduce LPS stimulation of TPL-2 MEK kinase activity detected in coupled MEK/ERK kinase assays (Fig. 2F). The experiments described in this section demonstrate that there is an essential proteasome-mediated proteolytic step in the TLR4 signaling pathway that activates the ERK MAP kinase cascade and suggest that this regulates the generation of the pool of p105-free TPL-2 which can phosphorylate MEK.
Signal-induced proteolysis of NF-κB1 p105 is essential for LPS activation of MEK phosphorylation.
Stimulation of p105 proteolysis by LPS is mediated via the IKK complex (16). TNF-α-induced proteolysis of p105 is induced by IKK phosphorylation of p105 serines 927 and 932 (20, 29). A phosphopeptide-specific antibody which recognizes p105 phosphorylated on serine 927 (29) was used to confirm that LPS-induced proteolysis of p105 also involves phosphorylation of the p105 PEST region. To do this, p105 was immunoprecipitated from lysates of BMDMs, which had been pretreated with MG132 prior to LPS stimulation, and subjected to Western blotting with an anti-phospho-S927-p105 antibody (29). LPS stimulation induced the rapid phosphorylation of p105 on serine 927 (Fig. 2G). Significantly, LPS stimulation induced maximal p105 serine 927 phosphorylation at 7.5 min. Thus, LPS-induced phosphorylation of p105 is very rapid and precedes MEK phosphorylation, which peaks at 15 min (Fig. 1A). In the following experiments, a genetic approach was taken to determine the importance of IKK-induced phosphorylation of p105 in LPS-induced TPL-2 activation.
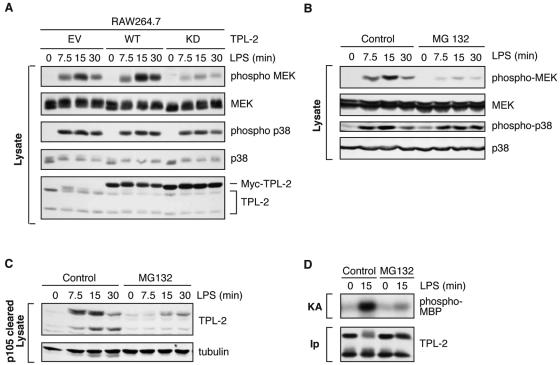
TPL-2 is the major MEK kinase activated by LPS in the murine macrophage cell line RAW264.7 (8). Stable expression of kinase-inactive TPL-2 in these cells blocked LPS induction of MEK phosphorylation, while induction of p38 phosphorylation was not affected (Fig. 3A). Thus, LPS activation of MEK is dependent on TPL-2 kinase activity in RAW264.7 cells. In addition, MG132 pretreatment of RAW264.7 cells inhibited LPS stimulation of MEK phosphorylation (Fig. 3B), TPL-2 release from p105 (Fig. 3C), and TPL-2 activation (Fig. 3D). Therefore, similar to primary macrophages, LPS activation of TPL-2 and MEK requires proteasome activity in RAW264.7 cells. In the following experiments, RAW264.7 cells were used as a model system to determine whether the inhibitory effect of MG132 reflects a requirement for signal-induced p105 proteolysis in LPS activation of TPL-2 and MEK.
FIG. 3.
LPS activation of MEK in RAW264.7 cells is dependent on TPL-2 and proteasome activity. (A) RAW 264.7 cells stably transfected with expression vectors encoding wild-type Myc-TPL-2 (WT), kinase-inactive Myc-TPL-2 (KD), or no insert control (empty vector [EV]) were stimulated with LPS, and cell lysates were subjected to Western blotting. LPS activation of endogenous MEK and p38 activation were assayed using phospho-specific antibodies. (B to D) RAW264.7 cells were preincubated with MG132 (40 μM) or DMSO vehicle control for 30 min and then stimulated with LPS for the times indicated. (B) Total cell lysates were subjected to Western blotting for the indicated proteins. LPS activation of MEK and p38 activation were monitored with the phospho-specific antibodies. (C) Cell lysates were immunodepleted of p105 and Western blotted for TPL-2 and α-tubulin. (D) TPL-2 was immunoprecipitated from total cell lysates and then assayed for MEK kinase activity as for Fig. 1A.
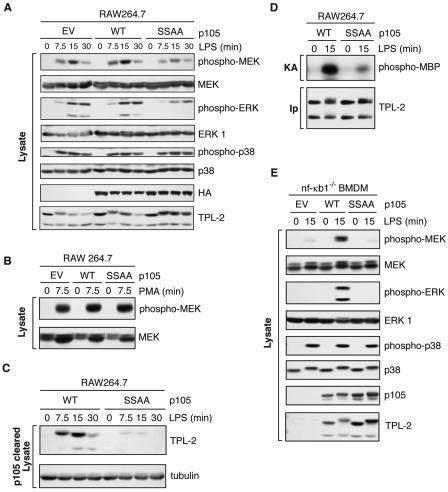
RAW264.7 cells were stably transfected with vectors encoding wild-type HA-p105 (HA-p105WT) or a mutant with inactivating mutations at the IKK phosphorylation sites, HA-p105SSAA (20, 29), which is not susceptible to signal-induced proteolysis. Clones were selected which expressed similar levels of transfected protein. LPS stimulation of MEK phosphorylation was very similar in cells transfected with HA-p105WT compared with empty vector control (Fig. 4A). However, LPS-induced MEK phosphorylation was significantly reduced (60% ± 10% reduction; n = 3; P = 0.01) in HA-p105SSAA-transfected cells from levels in HA-p105WT cells (Fig. 4A). LPS-induced p38 phosphorylation (Fig. 4A) was similar in the two cell lines (98% ± 12%; n = 3). There was also no difference in phorbol ester activation of MEK phosphorylation (Fig. 4B).
FIG. 4.
LPS activation of TPL-2 is dependent on signal-induced p105 proteolysis. (A to D) RAW 264.7 cells stably transfected with vectors encoding wild-type HA-p105 (WT), HA-p105S927A,S932A (SSAA), or with no insert (EV) were stimulated with LPS or PMA for the indicated times. (A and B) Total cell lysates were subjected to Western blotting, and phospho-specific antibodies were used to monitor MEK, ERK, and p38 activation. (C) Lysates were immunodepleted of p105 and subjected to Western blotting for TPL-2 and α-tubulin. (D) TPL-2 was immunoprecipitated from lysates of RAW264.7 cells expressing HA-p105 (WT) or HA-p105S927A,S932A (SSAA), and MEK kinase activity was determined by a coupled MEK/ERK kinase assay as for Fig. 1A. (E) BMDMs generated from NF-κB1−/− mice were infected with recombinant retroviruses encoding wild-type FL-p105 (WT), FL-p105S927A,S932A (SSAA), or with no insert (EV). Cells were stimulated with LPS (10 ng/ml) for 15 min or left untreated, and total cell lysates were subjected to Western blotting for the indicated proteins. Endogenous MEK, ERK, and p38 activation was assayed using phospho-specific antibodies.
Analysis of p105-depleted lysates indicated that LPS induction of TPL-2 release from p105 was markedly reduced by expression of HA-p105SSAA compared with results for HA-p105WT (Fig. 4C). Additionally, coupled MEK/ERK kinase assays demonstrated that LPS activation of TPL-2 MEK kinase activity was dramatically reduced in p105SSAA-transfected cells relative to that in HA-p105WT transfected cells (Fig. 4D). Thus, p105 proteolysis is required for LPS stimulation to induce efficiently release of TPL-2 from p105, TPL-2 MEK kinase activity, and MEK phosphorylation in RAW264.7 cells.
Expression of HA-p105SSAA in RAW264.7 cells reduced but did not eliminate LPS activation of the TPL-2/MEK/ERK pathway. This was presumably due to LPS-induced proteolysis of endogenous wild-type p105, which was expressed at approximately 20 to 40% of the level of transfected HA-p105SSAA in the clones analyzed (data not shown). LPS stimulation would therefore be expected to still liberate some TPL-2 from p105 in these cells due to signal-induced proteolysis of wild-type endogenous p105, resulting in the residual levels of induced MEK and ERK phosphorylation detected (Fig. 4A).
To determine whether there is an absolute requirement for signal-induced p105 proteolysis for LPS activation of the ERK MAP kinase cascade in primary macrophages, BMDMs were generated from NF-κB1−/− mice (30). These cells lack any p105 protein and are also deficient in TPL-2 due to its metabolic instability in the absence of p105 (3, 37). FL-p105WT and FL-p105SSAA were expressed in NF-κB1−/− BMDMs using recombinant retroviruses. Similar to reported results (37), LPS stimulation of NF-κB1−/− BMDMs infected with control EV retrovirus did not induce MEK and ERK phosphorylation due to TPL-2 deficiency (Fig. 4E). Expression of wild-type FL-p105 dramatically increased steady-state levels of TPL-2 and reconstituted the ability of LPS to stimulate both MEK and ERK phosphorylation, as expected (37). Steady-state levels of TPL-2 protein were increased to a similar degree by expression of FL-p105SSAA (Fig. 4E). However, LPS stimulation completely failed to induce MEK and ERK phosphorylation in cells expressing FL-p105SSAA, although LPS-induced p38 phosphorylation was similar to control EV (Fig. 4E). These data, which are consistent with the results of the RAW264.7 cell experiments, demonstrate that LPS-induced p105 proteolysis is an essential step in the TLR4 signaling pathway that activates the ERK MAP kinase pathway in primary macrophages.
IKK complex activity is required for LPS activation of TPL-2.
The experiments in the previous section demonstrated that expression of a p105 mutant that is insensitive to IKK-induced proteolysis (20) blocked LPS stimulation of the TPL-2/ERK signaling pathway in macrophages (Fig. 4). Since LPS stimulation of p105 proteolysis is mediated via the IKK complex (16), these data suggest that IKK activity is required for LPS activation of this MAP kinase pathway.
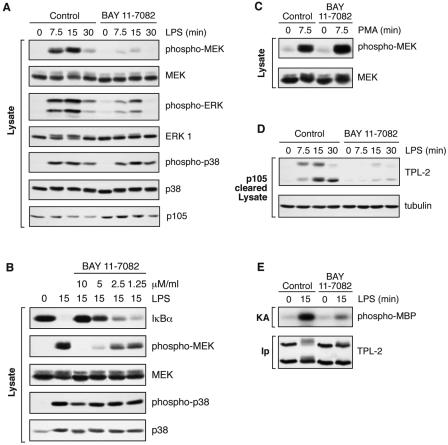
To investigate this possibility, BMDMs were pretreated with the IKK inhibitor BAY 11-7082 (27). Western blotting of cell lysates demonstrated that this inhibitor blocked LPS-induced p105 proteolysis, as expected (Fig. 5A, bottom panel). LPS induction of MEK and ERK phosphorylation was also dramatically reduced by BAY 11-7082 treatment (Fig. 5A). A titration experiment revealed that BAY 11-7082 inhibition of LPS-induced MEK phosphorylation occurred over a concentration range similar to that associated with its inhibitory effects on LPS-induced IκBα degradation (Fig. 5B). Since IκBα degradation is triggered by its direct phosphorylation by the IKK complex (19), these data are consistent with the effect of BAY 11-7082 on LPS-induced MEK phosphorylation being mediated by inhibition of the IKK complex. LPS stimulation of p38 phosphorylation (Fig. 5A) or phorbol ester stimulation of MEK phosphorylation (Fig. 5C) were unaffected by BAY 11-7082, confirming that its effect on LPS activation of MEK was specific.
FIG. 5.
IKK is required for LPS activation of TPL-2. BMDMs (BALB/c) were preincubated with the IKK inhibitor BAY11-7082 or DMSO vehicle control and stimulated with LPS or PMA for the indicted times. BAY11-7082 was used at a concentration of 7.5 μM unless otherwise indicated. (A to C) Total cell lysates were subjected to Western blotting for the indicated proteins. (D) Lysates were immunodepleted of p105 and subjected to Western blotting for TPL-2 and α-tubulin. (E) TPL-2 was immunoprecipitated from total cell lysates and assayed for MEK kinase activity in a coupled MEK/ERK kinase assay as for Fig. 1A.
As expected from its ability to inhibit IKK-triggered p105 proteolysis, BAY 11-7082 blocked both LPS induction of TPL-2 release from p105 (Fig. 5D) and TPL-2 MEK kinase activity (Fig. 5E). Control experiments demonstrated that BAY 11-7082 did not inhibit TPL-2 MEK kinase activity measured in a coupled kinase assay in vitro (data not shown). Treatment of BMDMs with another IKK2 inhibitor, BMS-345541 (5), also blocked LPS induction of MEK phosphorylation and TPL-2 release from p105, similar to results with BAY 11-7082 (data not shown). Thus, in primary macrophages, IKK activity is required for LPS stimulation of the TPL-2/ERK signaling pathway.
Preferential serine 927 phosphorylation of M1-TPL-2-associated p105.
Densitometric scanning of Western blots of p105-depleted lysates revealed that LPS stimulation more efficiently induced the release of M1-TPL-2 from p105 than M30-TPL-2 at early time points (Fig. 6A). With more-prolonged LPS stimulation, M1-TPL-2 levels were reduced due to proteolysis, whereas the amount of M30-TPL-2 reached a plateau. Since both TPL-2 isoforms associate with p105 at similar levels (Fig. 1A and 6B, right panels), these data suggest that the pool of p105 that is associated with M1-TPL-2 is more efficiently degraded than that which is associated with M30-TPL-2.
FIG. 6.
IKK preferentially phosphorylates p105 complexed with M1-TPL-2. BMDMs (BALB/c) were simulated with LPS for the indicated times. (A) Total cell lysates or cell lysates depleted of p105 were subjected to Western blotting for TPL-2. Bands were quantified by densitometry, and data are presented as a mean (± SEM; n = 4) of the fraction of M1- or M30-TPL-2 that is released from p105 normalized against α-tubulin levels. (B) Total p105 or p105 phosphorylated on S927 was immunoprecipitated from total cell lysates by using anti-p105N or anti-phospho-S927-p105 antibodies, respectively. Associated TPL-2 was detected by Western blotting after SDS-PAGE.
Signal-induced p105 proteolysis is triggered by IKK-mediated phosphorylation of the p105 PEST region on serines 927 and 932 (20, 29). It was therefore interesting to determine whether M1-TPL-2-associated p105 is differentially phosphorylated by the IKK complex relative to M30-TPL-2-associated p105. To do this, p105 phosphorylated on serine 927 was immunoprecipitated from lysates of LPS-stimulated BMDMs using a phospho-specific antibody (29) and subjected to Western blotting for associated TPL-2. M1-TPL2 coimmunoprecipitated with phospho-S927-p105 at much higher levels than M30-TPL-2 from LPS-stimulated cell lysates (Fig. 6B, left panels), although both TPL-2 isoforms copurified in approximately equal amounts with anti-p105N antibody (Fig. 6B, right panels). This suggests that M1-TPL-2-associated p105 is phosphorylated by IKK more strongly than M30-TPL-2-associated p105. Since IKK-induced phosphorylation of p105 induces its degradation (16, 20, 29), this differential phosphorylation provides an explanation for why M1-TPL-2 is released more efficiently from p105 than M30-TPL-2 in LPS-stimulated cells.
DISCUSSION
This study identifies a key step in the activation of the ERK MAP kinase cascade by LPS in macrophages. LPS stimulation activates the IKK complex to phosphorylate the p105 PEST region, which triggers p105 proteolysis by the proteasome. Consequently, TPL-2 is liberated from its inhibitor, p105 (3, 37). This p105-free active pool of TPL-2 then triggers activation of the MEK/ERK MAP kinase cascade.
It has previously been suggested that LPS stimulation of BMDMs induces the release of only M1-TPL-2 from p105 (37). However, using an assay which directly detects p105-free TPL-2 (Fig. 1D), it is evident that both M1-TPL-2 and M30-TPL-2 are actually released from p105 following LPS stimulation of BMDMs. Therefore, both TPL-2 isoforms may contribute to activation of MEK.
LPS stimulation of BMDMs or RAW264.7 macrophages induces a change in the mobility of the majority of M1-TPL-2 in SDS-PAGE that is due to phosphorylation (Fig. 1B and data not shown). A phosphorylation-induced mobility shift is also evident for the M30-TPL-2 band (Fig. 1B), but only a very small fraction is modified. Western blotting of anti-p105N immunoprecipitates suggests that M1-TPL-2 is phosphorylated when it is still bound to p105 (Fig. 1C). It has recently been reported that PP2A phosphatase treatment has no inhibitory effect on the MEK kinase activity detected in anti-TPL-2 immunoprecipitates from LPS-stimulated RAW264.7 cells (8). Thus, phosphorylation of TPL-2 may not regulate its catalytic activity. Rather, the correlation between the quantitative phosphorylation of M1-TPL-2 and its susceptibility to proteolysis after LPS stimulation (Fig. 1A and C) suggests that phosphorylation of M1-TPL-2 may play a role in promoting its proteolysis.
MG132 pretreatment of BMDMs blocked LPS-induced proteolysis of M1-TPL-2 (Fig. 2B), showing that this process requires proteasome activity. Analysis of p105-depleted lysates clearly indicates that the p105-free pool of TPL-2 is degraded (Fig. 1D). However, since only a fraction of total TPL-2 (approximately 20%) is released from p105, it is possible that M1-TPL-2 proteolysis can occur when it is still bound to p105. Interestingly, expression of HA-p105SSAA in RAW264.7 cells prevented LPS-induced degradation of M1-TPL-2 (Fig. 4A). Thus, p105 proteolysis is required for LPS-induced degradation of M1-TPL-2.
At early time points, M1-TPL-2 is preferentially released from p105 compared with M30-TPL-2. However, since M1-TPL-2 is degraded on prolonged LPS stimulation (30 min), only p105-free M30-TPL-2 accumulates at later time points (Fig. 6A). Kinetic experiments suggest that following release from p105, both TPL-2 isoforms may contribute to MEK phosphorylation which peaks at 15 min (Fig. 1A and D). However, since p105-free M30-TPL-2 persists after MEK phosphorylation has substantially declined (Fig. 1A and D), this isoform may have a lower specific activity for MEK than M1-TPL-2. In the future, it will be interesting to determine whether the kinetic dissimilarities in their LPS-induced release from p105 and degradation translate into functional differences between the TPL-2 isoforms.
The increased release of M1-TPL-2 from p105 at early time points relative to M30-TPL-2 (Fig. 6A) appears to occur as a consequence of preferential IKK-mediated phosphorylation of M1-TPL-2-associated p105 (Fig. 6B). This is expected to trigger more-pronounced proteolysis of this pool of p105 relative to M30-TPL-2-associated p105 (Fig. 5) (20, 29). These data raise the question of why associated TPL-2 affects IKK phosphorylation of the p105 PEST region. Both TPL-2 and the IKK complex bind to p105 via its death domain (2, 3). It is therefore possible that the M1-TPL-2/p105 complex binds to the IKK complex more strongly than the M30-TPL-2/p105 complex, facilitating increased IKK-mediated phosphorylation of M1-TPL-2-associated p105. An alternative possibility is that M1-TPL-2 and M30-TPL-2 differentially regulate activation of IKK associated with p105, since overexpressed TPL-2 activates the IKK complex and induces the proteolysis of cotransfected p105 (4, 22). However, this model is not supported by the absence of a global defect in LPS-induced NF-κB activation in TPL-2-deficient macrophages (12) and the fact that TPL-2 would be expected to be inactive when it is bound to p105 (37; this study). Further experiments will clearly be necessary to determine the mechanism underlying the preferential phosphorylation by IKK of the pool of p105 that is associated with M1-TPL-2.
The IKK-dependent mechanism of TPL-2 activation raises the question of the physiological advantage of linking LPS activation of ERK and NF-κB via p105. LPS upregulation of both TNF-α and COX-2 is blocked in TPL-2-deficient BMDMs due to defective activation of the ERK MAP kinase cascade (12, 13). The blockade in LPS induction of TNF-α is due to defective transport of TNF-α mRNA from the nucleus to the cytoplasm (12), whereas COX-2 upregulation is inhibited as a consequence of reduced CREB phosphorylation (13), a critical regulator of COX-2 transcription (6, 35). Transcriptional induction of both TNF-α (31, 33, 34) and COX-2 (7, 10) by LPS requires NF-κB binding to sites in their respective promoters. Thus, LPS upregulation of TNF-α and COX-2 production involves coordinated activation of ERK and NF-κB. The mechanism of TPL-2 activation, which is dependent on phosphorylation of p105 by the IKK complex, ensures that ERK is only activated simultaneously with NF-κB activation, facilitating activation of these genes.
In conclusion, this study demonstrates that LPS activation of TPL-2 in macrophages requires release from its inhibitor, NF-κB1 p105. TPL-2 release is shown to occur as a consequence of IKK-triggered p105 proteolysis by the proteasome. In future studies, it will be important to determine whether a similar mechanism to activate MEK is utilized by TNF-α and CD40 ligand, which also require TPL-2 MEK kinase to activate the ERK MAPK cascade (14).
Supplementary Material
Acknowledgments
We thank Alain Israel, Toschio Kitamura, and Phillip Tsichlis for reagents used in this study. We are also grateful to Lee Johnston and Hamish Allen for critical reading of the manuscript, to NIMR Biological Services, and to other members of the Ley laboratory for their support during the course of this work.
This study was supported by the United Kingdom Medical Research Council.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Aoki, M., F. Hamada, T. Sugimoto, S. Sumida, T. Akiyama, and K. Toyoshima. 1993. The human cot proto-oncogene encodes two protein serine/threonine kinases with different transforming activities by alternative initiation of translation. J. Biol. Chem. 268:22723-22732. [PubMed] [Google Scholar]
- 2.Beinke, S., M. P. Belich, and S. C. Ley. 2002. The death domain of NF-κB1 p105 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 277:24162-24168. [DOI] [PubMed] [Google Scholar]
- 3.Beinke, S., J. Deka, V. Lang, M. P. Belich, P. A. Walker, S. Howell, S. J. Smerdon, S. J. Gamblin, and S. C. Ley. 2003. NF-κB p105 negatively regulates TPL-2 MEK kinase activity. Mol. Cell. Biol. 23:4739-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belich, M. P., A. Salmeron, L. H. Johnston, and S. C. Ley. 1999. TPL-2 kinase regulates the proteolysis of the NF-κB inhibitory protein NF-κB1 p105. Nature 397:363-368. [DOI] [PubMed] [Google Scholar]
- 5.Burke, J. R., M. A. Pattoli, K. R. Gregor, P. J. Brassil, J. F. MacMaster, K. W. McIntyre, X. Yang, V. S. Iotzova, W. Clarke, J. Strnad, Y. Qiu, and F. C. Zusi. 2003. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J. Biol. Chem. 278:1450-1456. [DOI] [PubMed] [Google Scholar]
- 6.Caivano, M., and P. Cohen. 2000. Role of mitogen-activated protein kinase cascades in mediating lipopolysaccharide-stimulated induction of cyclooxygenase-2 and IL-1β in RAW264 macrophages. J. Immunol. 164:3018-3025. [DOI] [PubMed] [Google Scholar]
- 7.Caivano, M., B. Gorgoni, P. Cohen, and V. Poli. 2001. The induction of cyclooxygenase-2 mRNA in macrophages is biphasic and requires both CCAAT enhancer-binding protein β (C/EBP β) and C/EBPδ transcript. J. Biol. Chem. 276:48693-48701. [DOI] [PubMed] [Google Scholar]
- 8.Caivano, M., C. Rodriguez, P. Cohen, and S. Alemany. 2004. 15-deoxy-Δ-12,14-prostaglandin J2 regulates enodgenous Cot MAP kinase kinase kinase 1 activity induced by LPS. J. Biol. Chem. 278:52124-52130. [DOI] [PubMed] [Google Scholar]
- 9.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C.-C., Y.-T. Sun, J.-J. Chen, and Y.-J. Chang. 2001. Tumor necrosis factor-α-induced cyclooxygenase-2 expression via sequential activation of ceramide-dependent mitogen-activated protein kinases, and IκB kinase 1/2 in human alveolar epithelial cells. Mol. Pharmacol. 59:493-500. [DOI] [PubMed] [Google Scholar]
- 11.Donald, R., D. W. Ballard, and J. Hawiger. 1995. Proteolytic processing of NF-κB/IκB in human monocytes. J. Biol. Chem. 270:9-12. [DOI] [PubMed] [Google Scholar]
- 12.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J.-H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNFα induction by LPS is regulated post-transcriptionally via a TPL2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 13.Eliopoulos, A. G., C. D. Dumitru, C.-C. Wang, J. Cho, and P. N. Tsichlis. 2002. Induction of COX-2 by LPS in macrophages is regulated by TPL2-dependent CREB activation signals. EMBO J. 21:4831-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eliopoulos, A. G., C.-C. Wang, C. D. Dumitru, and P. N. Tsichlis. 2003. TPL-2 transduces CD40 and TNF signals that activate ERK and regulates IgE induction by CD40. EMBO J. 22:3855-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionary conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 16.Heissmeyer, V., D. Krappmann, E. N. Hatada, and C. Scheidereit. 2001. Shared pathways of IκB kinase-induced SCFβTrCP-mediated ubiquitination and degradation for the NF-κB precursor p105 and IκBa. Mol. Cell. Biol. 21:1024-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa, H., E. Claudio, D. Dambach, C. Raventos-Suarez, C. Ryan, and R. Bravo. 1998. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p) 105 precursor (NF-κB1) but expressing p50. J. Exp. Med. 187:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabouridis, P. S., A. I. Magee, and S. C. Ley. 1997. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J. 16:4983-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 20.Lang, V., J. Janzen, G. Z. Fischer, Y. Soneji, S. Beinke, A. Salmeron, H. Allen, R. T. Hay, Y. Ben-Neriah, and S. C. Ley. 2003. βTrCP-mediated proteolysis of NF-κB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol. Cell. Biol. 23:402-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang, V., A. Symons, S. J. Watton, J. Janzen, Y. Soneji, S. Beinke, S. Howell, and S. C. Ley. 2004. ABIN2 forms a ternary complex with TPL-2 and NF-κB1 p105 and is essential for TPL-2 protein stability. Mol. Cell. Biol. 24:5235-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, X., E. T. Cunningham, Y. Mu, R. Geleziunas, and W. C. Greene. 1999. The proto-oncogene Cot kinase participates in CD3/CD28 induction of NF-κB acting through the NF-κB-inducing kinase and IκB kinases. Immunity 10:271-280. [DOI] [PubMed] [Google Scholar]
- 23.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135-145. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi, J., T. Higashi, H. Mukai, T. Ohuchi, and T. Kakunaga. 1991. Structure and transforming potential of the human cot oncogene encoding a putative protein kinase. Mol. Cell. Biol. 11:4088-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063-1066. [DOI] [PubMed] [Google Scholar]
- 26.Orian, A., H. Gonen, B. Bercovich, I. Fajerman, E. Eytan, A. Israel, F. Mercurio, K. Iwai, A. L. Schwartz, and A. Ciechanover. 2000. SCFβTrCP ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 19:2580-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierce, J. W., R. Schoenleber, G. Jesmock, J. Best, S. A. Moore, T. Collins, and M. E. Gerritsen. 1997. Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effect in vivo. J. Biol. Chem. 272:21096-21103. [DOI] [PubMed] [Google Scholar]
- 28.Salmeron, A., T. B. Ahmad, G. W. Carlile, D. Pappin, R. P. Narsimhan, and S. C. Ley. 1996. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 15:817-826. [PMC free article] [PubMed] [Google Scholar]
- 29.Salmeron, A., J. Janzen, Y. Soneji, N. Bump, J. Kamens, H. Allen, and S. C. Ley. 2001. Direct phosphorylation of NF-κB p105 by the IκB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 276:22215-22222. [DOI] [PubMed] [Google Scholar]
- 30.Sha, W. C., H.-C. Liou, E. I. Tuomanen, and D. Baltimore. 1995. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell 80:321-330. [DOI] [PubMed] [Google Scholar]
- 31.Swantek, J. L., L. Christerson, and M. H. Cobb. 1999. Lipopolysaccaride-induced tumor necrosis factor-α promoter activity is inhibitor of nuclear factor-κB kinase dependent. J. Biol. Chem. 274:11667-11671. [DOI] [PubMed] [Google Scholar]
- 32.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 33.Udalova, I. A., J. C. Knight, V. Vidal, S. A. Nedospasov, and D. Kwiatkowski. 1998. Complex NF-κB interactions at the distal tumor necrosis factor promoter region in human monocytes. J. Biol. Chem. 273:21178-21186. [DOI] [PubMed] [Google Scholar]
- 34.Udalova, I. A., A. Richardson, A. Denys, C. Smith, H. Ackerman, B. Foxwell, and D. Kwiatkowski. 2000. Functional consequences of a polymorphism affecting NF-κB p50-p50 binding to the TNF promoter region. Mol. Cell. Biol. 20:9113-9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadleigh, D. J., S. T. Reddy, E. Kopp, G. Ghosh, and H. R. Herschman. 2000. Transcriptional activation of the cylooxygenase-2 gene in endotoxin-treated RAW 264.7 macrophages. J. Biol. Chem. 275:6259-6266. [DOI] [PubMed] [Google Scholar]
- 36.Warren, M. K., and S. N. Vogel. 1985. Bone marrow-derived macrophages: development and regulation of differentiation markers by colony-stimulating factor and interferons. J. Immunol. 134:982-989. [PubMed] [Google Scholar]
- 37.Waterfield, M. R., M. Zhang, L. P. Norman, and S.-C. Sun. 2003. NF-κB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the TPL-2 kinase. Mol. Cell 11:685-694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.