Abstract
Accumulation of misfolded proteins in the endoplasmic reticulum (ER) induces a highly conserved homeostatic response in all eukaryotic cells, termed the unfolded-protein response (UPR). Here we describe the characterization of stanniocalcin 2 (STC2), a mammalian homologue of a calcium- and phosphate-regulating hormone first identified in fish, as a novel target of the UPR. Expression of STC2 gene is rapidly upregulated in cultured cells after exposure to tunicamycin and thapsigargin, by ATF4 after activation of the ER-resident kinase PERK. In addition, STC2 expression is also activated in neuronal cells by oxidative stress and hypoxia but not by several cellular stresses unrelated to the UPR. In contrast, expression of another homologue, STC1, is only upregulated by hypoxia independent of PERK or ATF4 expression. In vivo studies revealed that rat cortical neurons rapidly upregulate STC2 after transient middle cerebral artery occlusion. Finally, siRNA-mediated inhibition of STC2 expression renders N2a neuroblastoma cells and HeLa cells significantly more vulnerable to apoptotic cell death after treatment with thapsigargin, and overexpression of STC2 attenuated thapsigargin-induced cell death. Consequently, induced STC2 expression is an essential feature of survival component of the UPR.
The endoplasmic reticulum (ER) provides a specialized environment for folding and maturation of transmembrane and luminal proteins. Physiological, pathological, and experimental conditions that perturb ER function cause accumulation of misfolded proteins within the ER and, as a result of ER stress, activate intracellular signaling pathways collectively known as the unfolded protein response (UPR) (12, 38). Three ER-resident type-I transmembrane proteins—namely, IRE1 (α and β), PERK, and ATF6—function as proximal stress sensors and share a common pathway of activation (38). The lumenal stress-sensing domain of each of these proteins is normally bound to BiP/GRP78 (an ER protein chaperone), which keeps them in an inactive form. BiP/GRP78 dissociates from IRE, PERK, and ATF6 when the levels of unfolded proteins exceed a certain threshold under conditions of ER stress. Release from BiP/GRP78 leads to oligomerization, transautophosphorylation, and activation of IRE1 and PERK, whereas ATF6 translocates to the Golgi apparatus (38, 55, 62). IRE1 is a type I membrane protein with cytoplasmic serine/threonine kinase and endoribonuclease domains. The endoribonuclease activity of activated IRE1 splices a small intron from the XBP-1 mRNA, allowing the synthesis and accumulation of a larger XBP-1 protein, which functions as a transcription factor (4, 56, 69). PERK is a serine/threonine kinase that, when activated, phosphorylates eukaryotic translation initiation factor 2α (eIF2α), causing transient translational attenuation in an attempt to alleviate ER overload (16). The transmembrane ATF6 undergoes proteolytic processing upon reaching the Golgi, and the released cytoplasmic transcription factor domain translocates to the nucleus, where it activates transcription of a set of genes, including XBP-1 (19, 34, 69). Accumulation of phosphorylated eIF2α allows for enhanced translation of the transcription factor ATF4, which has an important role in activating a transcriptional cascade involving transcription factors ATF3 and CHOP (15, 17, 27).
Expression profiling has identified several classes of UPR-related genes in yeast and mammals, which coordinate protein folding and maturation, secretory trafficking, amino acid metabolism, protein translation, mitochondrial function, ER-associated degradation of misfolded proteins, etc. (5, 47, 60). Thus, in concert with translational attenuation, UPR induces the expression of genes that function specifically in mitigating ER stress and enhancing cell survival. Nevertheless, cells subjected to ER stress also upregulate the expression of the proapoptotic transcription factor CHOP, stimulate proapoptotic JNK signaling pathway, and activate caspase-12 (45, 46, 62). Thus, apoptosis ensues if the burden of misfolded protein accumulation cannot be relieved by the UPR.
UPR is activated in cultured cells by exposure to xenotoxic agents that affect ER homeostasis, such as tunicamycin, thapsigargin, Ca2+ ionophores, and reducing agents. However, it is becoming clear that certain cells and tissue specialized in protein secretion upregulate aspects of the UPR under physiological conditions in vivo. For example, IRE1 and XBP-1 are activated during the terminal differentiation of B cells to antibody-secreting plasma cells (51). Another example is the case of insulin-producing cells of the islets of Langerhans in pancreas, where PERK appears to modulate protein biosynthesis under physiological conditions (14). On the other hand, mounting evidence also implicates UPR in the development of diabetes and in the pathogenesis of neurodegenerative diseases such as Alzheimer's disease, polyglutamine disease, and Parkinson's disease (11, 13, 49, 57). In addition, several studies have also shown that UPR is rapidly activated in ischemic neurons, suggesting close association between ER dysfunction and ischemic neuronal damage (25, 32, 48).
Here, we describe the gene encoding stanniocalcin 2 (STC2) as a novel target of the mammalian UPR. STC2 shares limited sequence similarity with an antihypercalcemic hormone first discovered in fish (3, 65). Fish STC is secreted by corpuscles of Stannius, a small endocrine gland found on the ventral surface of the kidney. Two widely expressed STC-related proteins, STC1 and STC2, have been identified in mammals, but their physiological functions have not been clearly elucidated (8, 9, 21, 22). As in fish, elevated Ca2+ levels induce STC1 expression and stimulate phosphate uptake in the neural crest-derived Paju cell line, suggesting functional conservation for mammalian STC1 in mineral metabolism (33, 70). Interestingly, STC1 expression is upregulated in neurons after ischemic insult in human and mouse brain, and overexpression of STC1 increased cell viability in Paju cells when exposed to thapsigargin or hypoxia (70). There has been only limited information available on STC2 expression and function. We show here that STC2 expression is induced in cultured cells by ER stress agents, hypoxia, and oxidative stress, downstream of PERK activation. In addition, rat cortical neurons rapidly upregulate STC2 expression after cerebral ischemia. Short interfering RNA (siRNA)-mediated attenuation of STC2 expression during the UPR renders mouse N2a neuroblastoma cells and HeLa cells extremely vulnerable to cell death elicited by thapsigargin, whereas overexpression of STC2 protected cells against thapsigargin-induced apoptotic cell death. Thus, STC2 carries out distinct function in mammals as a critical survival component of the UPR.
MATERIALS AND METHODS
Cell culture and stress induction.
Mouse N2a neuroblastoma cells were cultured in 1:1 Dulbecco modified Eagle medium-OptiMEM (Gibco) supplemented with 5% fetal bovine serum. Wild-type (K1) and S2P-deficienct (clone M19) Chinese Hamster ovary (CHO) cells were cultured as described previously (18). All other cells were maintained in Dulbecco modified Eagle medium (Gibco) containing 10% fetal bovine serum. Stress-inducing agents were purchased from Sigma unless noted otherwise. To induce ER stress, cells were treated with tunicamycin (2 μg/ml), thapsigargin (300 nM), dithiothreitol (0.5 mM), methyl methanesulfonate (1 mM), or 300 μM hydrogen peroxide (H2O2). To induce hypoxia, cells were placed in an anaerobic jar with BBL GasPak Plus system (Becton Dickinson Microbiology Systems). In some experiments, cells were also exposed to high extracellular Ca2+ (5.4 mM), high osmolarity (0.3 M NaCl), short-wavelength UV light (40 J/m2), staurosporine (STS; 500 nM) (EMD Biosciences), or heat shock (42°C, 1 h).
Microarray hybridization studies.
Total RNA was isolated from HEK293 cells treated with tunicamycin (2 μg/ml) or thapsigargin (300 nM) for 5 or 10 h by using the TRIzol reagent (Invitrogen) and purified by using an RNeasy minikit (Qiagen). Microarray analyses were performed essentially as described by Lockhart et al. (40). HG_U95Av1 Affymetrix arrays containing oligonucleotide probes representing 11,879 human genes were used in our studies. Each sample was profiled in duplicate, with cRNA prepared separately from total RNA, and all chips were scaled to a target intensity of 200. Filters used to select UPR genes were as follows: a minimum twofold change in expression levels, a minimum mean expression value of 100 for at least one of the two groups compared, a “present” score required for at least half of the chips compared, and an “increase” or “decrease” call.
cDNAs and antibodies.
Mouse STC2 cDNA was amplified by PCR with the primer pair 5′-GTCGGTACCACCATGTGTGCGGAGCGGCTG-3′ and 5′-GACGGATCCTCACCTCCGGATGTCGGAATACTCAGAC-3′ by using N2a cell cDNA as the template and then cloned into pBluescript. After sequence verification, the cDNA was subcloned into pGEX-4T and pAG3Zeo vectors for expression in bacteria and mammalian cells, respectively. Myc epitope-tagged IRE1β and PERK expression plasmids were kind gifts from David Ron. ATF6 plasmid was kindly provided by Jian Ma. Polyclonal STC2 antisera were raised in rabbits against a glutathione S-transferase-mouse STC2 fusion protein expressed in bacteria. Two anti-STC2 antisera (STC260 and STC261) were characterized to be specific for STC2 by Western blotting, immunoprecipitation, and immunohistochemical analyses. Other antibodies were purchased from commercial sources as follows: anti-KDEL (Stressgen); CHOP and anti-myc 9E10 (Santa Cruz Biotechnology); β-actin, Golgi58K protein, and AP1/AP2 (Sigma); syntaxin 6 (BD Transduction Laboratories); and cytochrome c oxidase IV (COX-IV; Molecular Probes).
Analysis of mRNA levels.
Total RNA was prepared from cells subject to various stresses as described above, and 1-μg aliquots were reverse transcribed into cDNA by using random hexamers. Aliquots corresponding to 1/25 of the resulting cDNA were incubated in PCR with the following primer pairs: BiP (sense [5′-CTGGGTACATTTGATCTGACTGG-3′] and antisense [5′-GCATCCTGGTGGCTTTCCAGCCATTC-3′]), CHOP (sense [5′-GTCCAGCTGGGAGCTGGAAG-3′] and antisense [5′-CTGGTCAGGCGCTCGATTTCC-3′]), GADD45 (sense [5′-GAGCAGAAGACCGAAAGGATG-3′] and antisense [5′-CTTCAGTGCAATTTGGTTCAG-3′]), Mif 1 (sense [5′-GTCCAGAGGACCAGAGGTTA-3′] and antisense [5′-GTATCTCTTGTGCACTTGGTG-3′]), NUCB2 (sense [5′-GGAAGAGACTGATGGATTGGA-3′] and antisense [5′-CATCCTCTTCATTTTTAGGGTCA-3′ for human or 5′-CACTTTCTCCAACTCTCTTGTGAA-3′ for mouse]), ATP2A2 (sense [5′-ATGAGCAAGATGTTTGTGAAGG-3′] and antisense [5′-CAGTGGGTTGTCATGAGTGG-3′]), human CGRP2 (sense [5′-AGCCCTGCTGACACCTAGAG-3′] and antisense [5′-CTCCAAGGCATTTACCCAAA-3′]), mouse CGRP2 (sense [5′-CCGTGTGTGTGTGCTCTTCT-3′] and antisense [5′-GGTCTAGGCTGCTCTCCAAA-3′]), ATF6 (sense [5′-GCCTTTATTGCTTCCAGCAG-3′] and antisense [5′-TGAGACAGCAAAACCGTCTG-3′]), human STC2 (sense [5′-GGGAATGCTACCTCAAGCAC-3′] and antisense [5′-CACAGGTCAGCAGCAAGTTC-3′]), mouse STC2 (sense [5′-AGGAGAACGTCGGTGTGATT-3′] and antisense [5′-CTGTTCACACTGAGCCTGGA-3′]), STC1 (sense [5′-CCCAATCACTTCTCCAACAGA-3′] and antisense [5′-GAAGAGGCTGGCCATGTTAG-3′] for human or 5′-GAAGAGGCTGGCCATGTTG-3′ for mouse), and β-actin (sense [5′-GGCTACAGCTTCACCACCAC-3′] and antisense 5′-GTCAGGCAGCTCGTAGCTCT-3′ for human or 5′-TCTCCAGGGAGGAAGAGGAT-3′ for mouse). For the negative control, reverse transcription-PCR (RT-PCR) was performed in the absence of reverse transcriptase. The PCR products were separated by electrophoresis through 5 or 8% polyacrylamide gels, stained with ethidium bromide, and visualized by using Typhoon 8600 variable mode imager (Amersham Biosciences). The validity of β-actin as an internal control gene was confirmed by no significant changes in its expression under our experimental conditions. For quantitative analysis of mRNA levels, amplification and detection of STC2 and β-actin mRNA were performed in the same reaction with the iCycle iQ Multi-Color Real Time PCR detection system (Bio-Rad) with specific primers (above) and the following fluorescent oligonucleotides (Integrated DNA Technologies): STC2 (5′-/FAM/TTCAAGGATCTCCTGCTGC/BHQ-1/-3′ for mouse and 5′-/FAM/CAGAGACAGCCTGATGGAGA/BHQ-1/-3′ for human) and β-actin (5′-/HEX/GGAAATCGTGCGTGACAT/BHQ-1/-3′). STC2 signals were normalized to β-actin signals. RNA was isolated from three independent experiments, assayed by real-time quantitative PCR, and results are expressed as means ± the standard errors (SE). Statistically significant differences compared to basal levels were determined by one-way analysis of variance with Fisher protected least-squares difference test by using Statview 5.0 software.
Protein analysis and subcellular fractionation.
Cells were lysed in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, 0.5% sodium deoxycholate, 0.25% sodium dodecyl sulfate, 5 mM EDTA, and a mixture of protease inhibitors (Sigma). Lysates were briefly sonicated and fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. To immunoprecipitate secreted STC2, aliquots of conditioned medium were incubated overnight with STC2 antiserum, followed by binding to protein A-agarose beads (Pierce). For subcellular fractionation, cell pellet from two 100-mm dishes were homogenized in 0.5 ml of ice-cold sucrose buffer (0.25 M Sucrose, 10 mM Tris-HCl [pH 7.4], 1 mM magnesium acetate, and a mixture of protease inhibitors) by 10 passages through a prechilled ball bearing homogenizer with 16-μm clearance. Homogenates were centrifuged at 800 × g for 10 min at 4°C, and postnuclear supernatants were loaded on sucrose step gradients consisted of (from top to bottom) 0.25 M (1 ml), 0.5 M (2 ml), 0.8 M (2 ml), 1.16 M (2.5 ml), 1.3 M (2.5 ml), and 2 M (1.5 ml) sucrose in 10 mM Tris-HCl (pH 7.4)-1 mM magnesium acetate. The gradients were centrifuged at 39,000 rpm for 2.5 h at 4°C in an SW41Ti rotor (Beckman Instruments), and 1-ml fractions were collected from the top by using an Autodensiflow fractionator (Labconco). Aliquots of each fraction (60 μl) were analyzed by Western blotting with STC2, anti-KDEL, and syntaxin 6 antibodies.
Mitochondria were isolated by using a differential centrifugation method (58). Cells from one 100-mm dish were incubated for 5 min in 700 μl of ice-cold hypotonic buffer (10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl [pH 7.4]) and lysed by 10 passes through a prechilled ball bearing homogenizer with 12-μm clearance. Then, 233 μl of 4× mannitol-sucrose buffer (840 mM mannitol, 280 mM sucrose, 20 mM Tris-HCl [pH 7.4], 4 mM EDTA) was added, and the lysates were spun at 830 × g for 10 min at 4°C to remove nuclei. Supernatants were then briefly sonicated (30 s at 2 W by using GE130 Ultrasonic Processor with a microtip) to disperse large membrane fragments, and mitochondria were sedimented by centrifugation at 10,000 × g for 20 min at 4°C. Aliquots of each fraction (60 μl) were analyzed by Western blotting with STC2, anti-KDEL, and COX-IV antibodies.
Immunofluorescence labeling.
COS cells grown on coated coverslips were transfected with pAG3Zeo-STC2 plasmid. At 48 h after transfection, cells were fixed in 4% paraformaldehyde at room temperature for 10 min and then permeabilized in 0.2% Triton X-100 for 5 min. After nonspecific binding was blocked, coverslips were incubated with polyclonal STC2 antiserum (1:500) and monoclonal organelle marker antibodies diluted in phosphate-buffered saline (PBS) containing 0.2% Tween 20 and 3% bovine serum albumin (anti-KDEL, 1:250; Golgi58K, 1:1,000; AP-1/AP-2, 1:200). To label mitochondria, cells were incubated with 50 nM Mitotracker red (Molecular Probes) for 30 min before fixing. After three washes, coverslips were incubated with fluorescein isothiocyanate-conjugated anti-rabbit and Cy3-conjugated anti-mouse secondary antibodies and then mounted. Immunofluorescence staining was examined by confocal microscopy (LSM 5 Pascal; Carl Zeiss) by using a ×63 oil immersion objective lens, and images were processed by using Metamorph software (Universal Imaging Corp.).
MCA occlusion studies.
All animal procedures were done according to the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the guidelines endorsed by the Canadian Council for Animal Care. Middle cerebral artery (MCA) occlusion was induced by using the intraluminal filament technique (24, 41) as modified below. Adult male Sprague-Dawley rats were anesthetized in a chamber by using 5% isoflurane in 30% oxygen-70% nitrous oxide. A surgical depth of anesthesia was maintained throughout the operation, with 2.5% isoflurane delivered through a nose mask. Under the operating microscope, the left common carotid artery (CCA) was exposed through a midline cervical incision and gently separated from the adjacent nerves. Microvascular clips were placed around the left CCA, the external carotid artery, and the internal carotid artery (ICA) to prevent bleeding during the insertion of the filament. A heat-blunted nylon filament (0.45-mm rounded tip) was inserted into the ICA lumen through a puncture made in the CCA and was advanced 1.8 to 2.1 cm, or until resistance was felt, to occlude the blood flow to the MCA territory. The filament was held in place by tightening the suture around the CCA. Body temperature was monitored and maintained at 37°C by using a heating pad connected to a rectal probe (Harvard Homeothermic blanket control unit). After 90 or 120 min of MCA occlusion, the filament was removed, the CCA was permanently ligated with a 3-0 silk suture, and the neck incision was closed with a 5-0 silk suture. Supplementary 0.9% NaCl solution was administered to the animals, along with rectal Tylenol. The animals were given free access to food and water between procedures and allowed to survive for 3.5, 6, 12, 24, or 48 h (n = at least 4 for each time point). At the end of the recirculation period, the rats were fixed by perfusion with PBS containing 4% paraformaldehyde. The brains were subsequently removed, and immunohistochemistry was performed on 16-μm sections essentially as described previously (23-25) with anti-STC2 antisera or the respective preimmune serum (1:1,000). For mRNA analysis, ischemic and contralateral brain tissue were dissected, and total RNA was isolated by using the acid guanidinium thiocyanate-chloroform-phenol extraction technique as previously described (25). RT-PCRs were performed as described above.
siRNA inhibition, STC2 overexpression, and cell viability assays.
To suppress STC2 expression in N2a cells, complementary oligonucleotides corresponding to nucleotides 153 to 171 (GGAGATCCAGCACTGTTTG) of mouse STC2 cDNA separated by a nine-nucleotide noncomplementary spacer (TTCAAGAGA) were synthesized and cloned into pSUPER plasmid (2). N2a cells were cotransfected with 1.5 μg of pSUPER or pSUPER-STC2 plasmids, along with 0.5 μg of pEYFP-C1 (Clontech). After 48 h, cells were treated or not treated with thapsigargin (150 nM), tunicamycin (2 μg/ml), H2O2 (100 μM), or STS (250 nM) for 16 h, and stained with ethidium homodimer 1 (EthD-1) (Molecular Probes). Fluorescent images of three random fields were acquired by using Zeiss Axioplan2 inverted microscope with a ×2.5 objective lens, and yellow fluorescent protein (YFP)-positive EthD-1-negative live cells were counted by using MetaMorph software. The data from four independent experiments were statistically examined by paired Student t test expressed as means ± the SE.
Synthetic siRNA duplexes containing the sequences 5′-CAGCGGAGAUCCAGCACUGdTdT-3′ (sense) and 5′-CAGUGCUGGAUCUCCGCUGdTdT-3′ (antisense) (Dharmacon) were used to suppress STC2 expression in HeLa cells. Mismatch STC2 siRNA (6 bp changes in the STC2 siRNA), with no significant sequence homology to mammalian genes by NCBI BLAST search, was used as the control in these experiments (mismatch STC2 siRNA: 5′-CUGCAGAGCUCGACCACUCdTdT-3′, sense; 5′-GAGUGGUCGAGCUCUGCAGdTdT-3′, antisense). HeLa cells were transfected in 12-well plates with 0.1 nmol of each siRNA by using Oligofectamine (Invitrogen). After 24 h, cells were treated or not treated with thapsigargin (500 nM) or tunicamycin (2 μg/ml) for 40 h. In some experiments, cells were treated with STS (250 nM) for 9 h. After treatment, cells were stained with 1.5 μg of Hoechst 33342 (Molecular Probes)/ml and a 1:40 dilution of Annexin-V-Alexa 568 (Molecular Probes). Fluorescent images of three random fields per coverslip were acquired on Zeiss Axioplan2 microscope with a ×5 objective lens, and morphologically distinct apoptotic cells with condensed nuclei or Annexin-V positive cells were counted by using MetaMorph software. The data from three independent experiments were statistically examined by paired t test and are expressed as means ± the SE. For overexpression studies, HeLa cells were transfected in 12-well plates with 0.6 μg of STC2 expression plasmid by using Lipofectamine (Invitrogen). After 24 h, cells were treated or not treated with thapsigargin (500 nM) or tunicamycin (2 μg/ml) for 40 h and then stained with Hoechst 33342 and Annexin-V-Alexa 568, and apoptotic cell death was estimated as described above.
RESULTS
Microarray profiling of novel UPR-related gene expression.
We sought to identify novel genes associated with the UPR in human embryonic kidney 293 cells by using high-density oligonucleotide microarray hybridization strategy (Affymetrix human HG_U95Av1 microarrays), which allows simultaneous analysis of 11,879 gene transcripts. To induce the UPR, HEK293 cells were exposed for 5 or 10 h to either tunicamycin, an inhibitor of N-linked glycosylation, or thapsigargin, an irreversible inhibitor of sarco(endo)plasmic reticulum Ca2+-ATPase. These studies revealed a total of 127 genes whose expression is upregulated at least twofold at either time point after exposure to tunicamycin and/or thapsigargin. The list of upregulated genes includes well-characterized UPR markers such as BiP/GRP78 (7.3-fold), CHOP (35-fold), and Mif 1/Herp (11.2-fold) and other genes that have been reported by previous microarray investigations of the UPR (17, 47, 50). Functional categorization of the UPR targets uncovered four genes that are known to regulate cellular Ca2+ homeostasis: the sarco(endo)plasmic reticulum Ca2+-ATPase 2b isoform (SERCA2b; ATP2A2), nucleobindin 2 (NUCB2), calcitonin gene-related peptide 2 (CGRP2), and STC2 (Table 1) genes. We initially focused on these genes because alterations in Ca2+ homeostasis is known to be tightly associated with UPR and directly involved in multiple models of apoptosis and neurodegenerative disorders (1, 43, 61).
TABLE 1.
UPR-mediated upregulation of genes involved in cellular Ca2+ homeostasisa
| Gene | Name | GenBank no. | Fold induction
|
|||
|---|---|---|---|---|---|---|
| Tm
|
Tg
|
|||||
| 5 h | 10 h | 5 h | 10 h | |||
| NUCB2 | Nucleobindin 2 | X76732 | 0.2 | 3.1 | 1.5 | 2.2 |
| ATP2A2 | Sarco(endo)plasmic reticulum Ca2+-ATPase 2 | M23114 | 1.2 | 2 | 2.2 | 1.9 |
| CGRP2 | Calcitonin gene-related peptide 2 | X02404 | 1.2 | 2.2 | 1.2 | 1.6 |
| STC2 | Stanniocalcin 2 | AF098462 | 6.3 | 10.4 | 4.6 | 8.4 |
HEK293 cells were treated with 2 μg of tunicamycin (Tm) per ml or 300 nM thapsigargin (Tg) for 5 or 10 h. RNA was prepared and assayed in duplicate by hybridization to Affymetrix HG_U95A microarrays and then analyzed as described in Materials and Methods. Induction is the average ratio of the hybridization signals in treated cells versus nontreated controls.
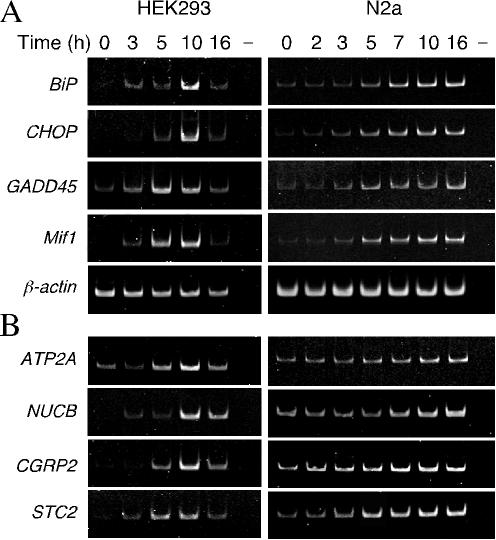
To validate the microarray results, we determined the temporal profile of UPR-mediated gene expression of select genes in HEK293 and N2a cells after treatment with tunicamycin. First, we examined time-dependent upregulation of a few well-characterized UPR-induced genes. As expected, expression of BiP, CHOP, GADD45, and Mif 1 increased from 3 to 5 h onward after exposure to tunicamycin in both cell types (Fig. 1A). Next, we assessed the upregulation of four UPR-related genes involved in regulating Ca2+ homeostasis. As shown in Fig. 1B, the expression of genes regulating Ca2+ homeostasis was increased in HEK293 and N2a cells during UPR beginning 3 to 5 h after exposure to tunicamycin. SERCA2b Ca2+-ATPases are intracellular pumps located in the sarcoplasmic reticulum or ER (42), and NUCB (also referred as Calnuc) is an abundant Golgi resident protein and the major Ca2+-binding protein in Golgi (37). Both proteins are well known to be involved in the maintenance of cytoplasmic Ca2+ concentrations, and UPR induction of ATP2A2 expression has been characterized (6, 50). CGRP is produced by alternative processing of the RNA transcribed from the calcitonin gene, which is involved in plasma Ca2+ regulation and skeletal maintenance, and is also an important regulator of vascular tone and blood flow (28). STC2 is a glycoprotein hormone thought to be associated with Ca2+ and phosphate metabolism (21). STC2 was listed among the genes upregulated by exposure to tunicamycin in SH-SY5Y neuroblastoma cells in a recent report (50), but STC2 expression was not examined in any detail. Therefore, the remainder of the present study focuses on the association of STC2 with the UPR.
FIG. 1.
Expression of UPR target genes in HEK293 and N2a cells treated with tunicamycin. HEK293 and N2a cells were treated with 2 μg of tunicamycin/ml for the indicated periods. Total RNA isolated from the cells was subjected to RT-PCR with specific primers, and products were analyzed on polyacrylamide gels. (A) The levels of established UPR target genes: BiP, CHOP, GADD45, and Mif 1. β-actin mRNA served as the control. For the negative control (lanes −), RT-PCR was performed in the absence of reverse transcriptase. (B) Upregulation of UPR genes that regulate Ca2+ homeostasis: ATP2A2, NUCB2, CGRP2, and STC2.
Characterization of STC2 expression.
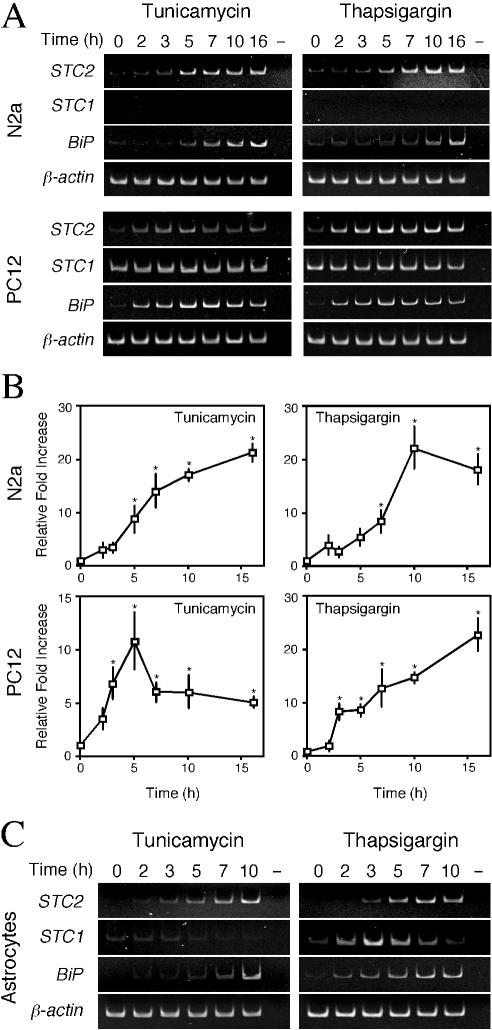
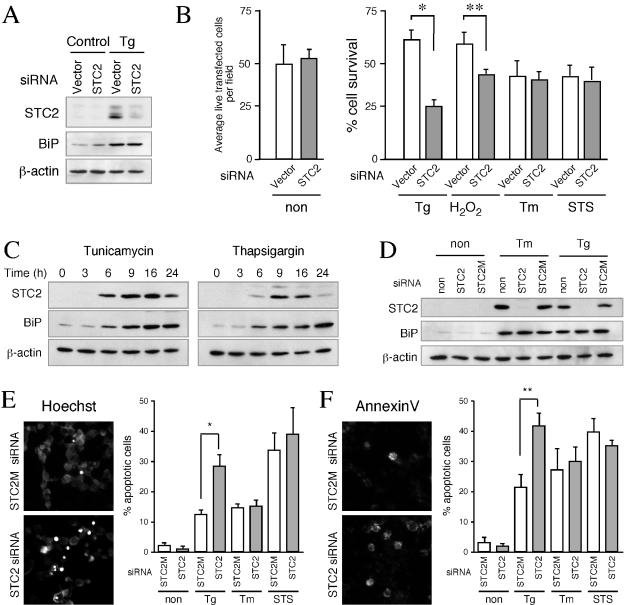
First, we analyzed the temporal expression pattern of STC2 in N2a neuroblastoma, rat PC12 pheochromocytoma, and primary rat astrocytes treated with tunicamycin or thapsigargin. The expression of STC2 mRNA was clearly increased upon ER stress in these neuronal cells; expression of BiP, a well-established UPR marker, served as a positive control for ER stress-mediated upregulation of gene expression (Fig. 2). We also examined the steady-state levels of STC1, the homologue of STC2. Unlike STC2, STC1 expression was not upregulated by ER stress in any of the cell types examined, except for a small and transient increase in primary rat astrocytes exposed to thapsigargin. Real-time quantitative PCR analyses revealed that UPR-mediated increases in STC2 mRNA were 16- to 25-fold and 10- to 12-fold during the time periods tested in N2a and PC12 cells, respectively (Fig. 2B). Due to the considerably low basal expression level, UPR-mediated expression of STC2 reached 400- and 900-fold in astrocytes treated with thapsigargin and tunicamycin, respectively (Fig. 2C).
FIG. 2.
STC2 is upregulated by the UPR. (A) N2a and PC12 cells were treated with 2 μg of tunicamycin/ml or 300 nM thapsigargin for indicated periods. Total RNA was subject to RT-PCR analyses with STC1 and STC2 primers. UPR was assessed by the induction of BiP mRNA. (B) Quantitative real-time PCR. Steady-state levels of STC2 and β-actin mRNAs were simultaneously quantified in one reaction by using specific primer pairs and fluorescently labeled internal primers. STC2 values were normalized to β-actin mRNA signal and plotted as the fold increase compared to no treatment control. The data represent means ± the SE from three independent experiments. ✽, P < 0.02 relative to time zero. (C) UPR-mediated induction of STC1 and STC2 expression in primary rat astrocytes.
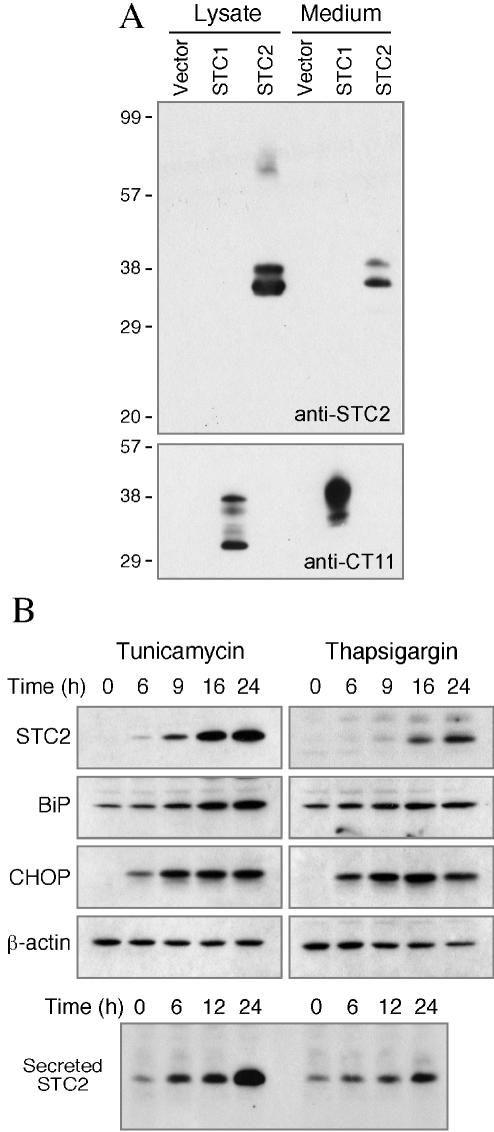
Next, we analyzed STC2 protein levels in N2a cells subject to ER stress. For these studies, we generated two polyclonal antisera against full-length STC2. Immunoblot analysis of transfected cell lysates confirmed that each STC2 antiserum uniquely reacts with STC2 and does not cross-react with transfected STC1 (Fig. 3A). STC2 protein was present at nearly undetectable levels in lysates of untreated N2a cells. After exposure to tunicamycin or thapsigargin, there was a time-dependent increase in the levels of STC2 (Fig. 3B). Whereas there was only a small increase in the levels of BiP, markedly induced expression of CHOP was observed with both treatments. STC2 amino acid sequence deduced from cDNA reveals an N-terminal hydrophobic stretch that can serve as a signal sequence for import into the ER lumen, and it has been reported that STC2 is secreted from HT1080 fibrosarcoma cells (26). Therefore, we sought to determine whether STC2 is secreted from N2a cells under conditions of ER stress. Western blot analysis of media conditioned by untreated N2a cells revealed detectable levels of STC2. The levels of STC2 in the media increased markedly after exposure of cells to ER stress agents (Fig. 3B). Together, these findings suggest that STC2 mRNA expression is upregulated, and STC2 protein is readily secreted during ER stress.
FIG. 3.
UPR-mediated expression and secretion of STC2. (A) Specificity of STC2 antiserum. N2a cells were transfected with empty vector or plasmids encoding C-terminally tagged STC1 or untagged STC2. Aliquots of detergent lysates and conditioned media were analyzed by immunoblotting with STC2 antiserum or CT11 (tag) antibody. Note that STC2 antibody does not cross-react with STC1. (B) N2a cells were treated with tunicamycin or thapsigargin for the indicated periods, and detergent lysates were analyzed by blotting with STC2, BiP, CHOP, and β-actin antibodies. Secreted STC2 were immunoprecipitated with STC2 antiserum and then analyzed by immunoblotting with the same antibody. Note the UPR-mediated induction and secretion of STC2.
Subcellular localization of STC2.
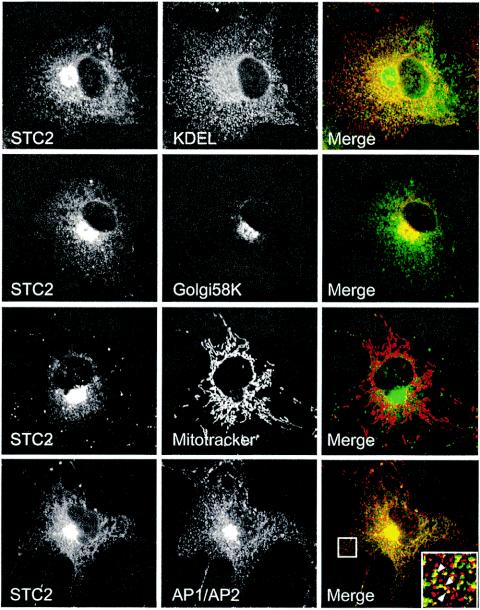
In addition to reports that STC1 is secreted from cultured from HT1080 fibrosarcoma and Madin-Darby canine kidney cells (26, 54), a recent study reported that >90% of cellular STC1 localized to the mitochondria of the kidneys, liver, spleen, and lungs (44). To determine the subcellular distribution of STC2, we first performed immunofluorescence localization studies. In transfected COS cells, the majority of STC2 colocalized with BiP and GRP94 (anti-KDEL staining) and 58K protein, which are markers of the ER and Golgi apparatus (Fig. 4). STC2 immunofluorescence can also be seen in small vesicles that colocalized with AP1/AP2 clathrin-associated adaptor protein complexes, which are markers for TGN-derived secretory and plasma membrane-derived endocytic vesicles. However, we found no overlap between STC2 and mitochondria. Similar results were observed in mouse N2a cells transfected with STC2 (data not shown).
FIG. 4.
Immunofluorescence localization of STC2 to the ER and Golgi apparatus. COS cells transfected with STC2 were fixed with 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and double labeled with STC2 antiserum (green) and monoclonal antibodies (red) to KDEL (marker of ER), 58K (Golgi apparatus), and AP1/AP2 (TGN-derived secretory vesicles and plasma-membrane derived endocytic vesicles). Mitochondria were labeled by incubating cells with MitoTracker red. Images were acquired on a laser scanning confocal microscope. Note the extensive colocalization of STC2 with the ER, the Golgi/TGN, and the presence of STC2 in some AP1/AP2-positive small vesicles.
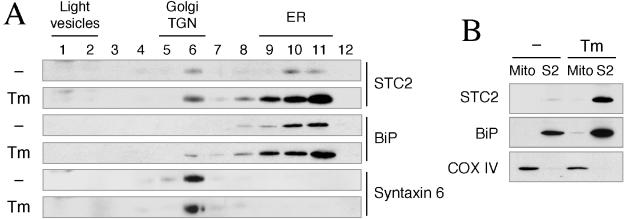
Subcellular localization of endogenous STC2 in ER and Golgi compartments was also confirmed by sucrose density fractionation of N2a cells. In these experiments we compared the distribution of STC2 in untreated cells and cells exposed to tunicamycin. Consistent with the immunofluorescence data, endogenous STC2 was predominantly found in subcellular fractions enriched in ER- and Golgi-resident proteins (BiP and syntaxin 6, respectively) that serve as organelle markers (Fig. 5A). This pattern of distribution remained unchanged in N2a cells subject to ER stress, indicating that STC localization is not altered by the UPR. These results are consistent with the residence and trafficking of STC2 through the secretory pathway under normal conditions and during induction of the UPR. We also examined potential localization of endogenous STC2 in mitochondria by subcellular fractionation. In these experiments we isolated mitochondrial fractions enriched in COX-IV. Consistent with the immunofluorescence localization studies, we found that in control and tunicamycin-treated N2a cells, STC2 was not present in mitochondria but cofractionated with the ER marker BiP (Fig. 5B).
FIG. 5.
Subcellular fractionation of endogenous STC2. (A) Sucrose density gradient analysis. Subconfluent dishes of untreated (−) and tunicamycin treated (Tm) N2a cells were lysed by using a ball-bearing homogenizer and fractionated by velocity sedimentation. Aliquots of fractions were analyzed by immunoblotting with STC2, anti-KDEL (ER marker), and syntaxin 6 (Golgi/TGN marker) antibodies. Note that endogenous STC2 is predominantly localized in ER and Golgi/TGN fractions. (B) Mitochondrial fractionation. Untreated (lane −) and tunicamycin-treated (lane Tm) N2a cells were lysed in a hypotonic buffer and separated into mitochondrial and supernatant fractions (containing the cytosol and other organelles) by differential centrifugation. Equal amounts of mitochondrial pellet (Mito) and supernatant fraction (S2) proteins were analyzed by immunoblotting with antibodies to STC2, BiP (ER marker), and COX-IV (mitochondrial marker). Note that endogenous STC2 is present in the S2 fraction but not in the mitochondrial fraction.
Selective upregulation of STC2 expression by ER stress-inducing agents.
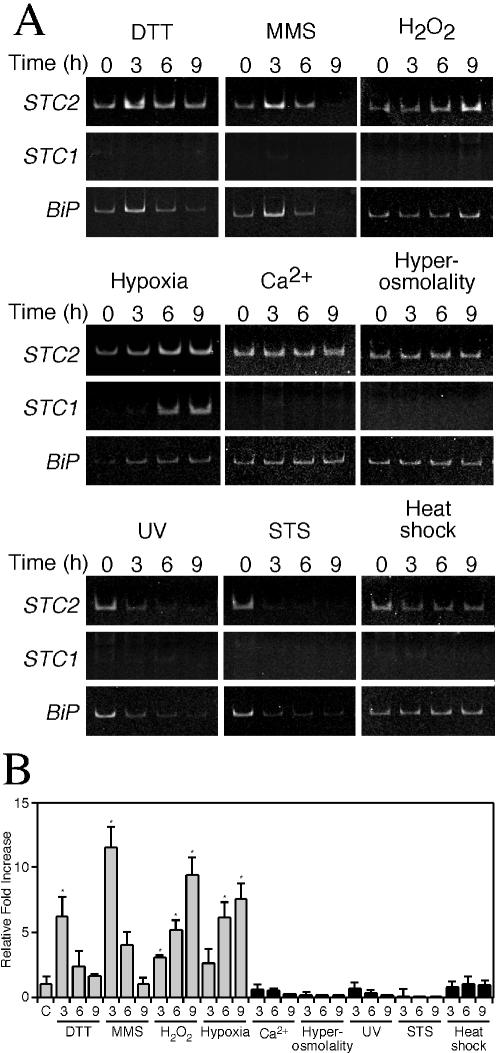
Next, we set out to determine whether upregulation of STC2 by tunicamycin and thapsigargin reflects intimate association with the induction of the UPR or is part of a general cellular response to diverse stress stimuli. First, we subjected N2a cells to additional treatments that are known to effectively induce the UPR. Exposure of cells to the reducing agent, dithiothreitol (0.5 mM) or the DNA alkylating agent, methyl methanesulfonate (1 mM) resulted in upregulation of STC2 mRNA (Fig. 6). Consistent with a recent report that oxidative stress induces aspects of UPR (20), cells subject to oxidative stress by treatment with H2O2 (300 μM) upregulated STC2 and BiP mRNA. Moreover, exposure of cells to hypoxia, a treatment known to activate PERK, phosphorylation of eIF2α, and the UPR (30), also induced expression of STC2 mRNA (Fig. 6). In contrast, we observed no change or reduction in STC2 mRNA levels in cells subject to diverse stress treatments with no association to the UPR, including high extracellular Ca2+ (5.4 mM), high osmolarity (0.3 M NaCl), short-wavelength UV light exposure (40 J/m2), apoptosis-inducible agent, staurosporine (500 nM), and heat shock (42°C, 1 h) (Fig. 6A). Interestingly, unlike STC2, STC1 expression was selectively induced by exposure of cells to hypoxia and not by other treatments that successfully induced the UPR or by unrelated stress conditions (Fig. 6A). Collectively, these results suggest that upregulation of STC2 expression is tightly associated with the induction of the UPR, whereas STC1 expression is limited to hypoxia.
FIG. 6.
Selective induction of STC2 expression by ER and oxidative stress agents. N2a cells were left untreated or were treated with the various stress agents for the indicated periods. (A) Levels of STC2, STC1, and BiP mRNA were evaluated by RT-PCR with specific primers. (B) Quantitative analysis of STC2 induction by real-time PCR normalized to β-actin mRNA. The graph shows fold increase in STC2 mRNA compared to untreated controls (C). Values represent means ± the SE (✽, P < 0.03) of three independent experiments, each assayed in triplicate.
Induction of STC2 through PERK-ATF4-mediated pathway.
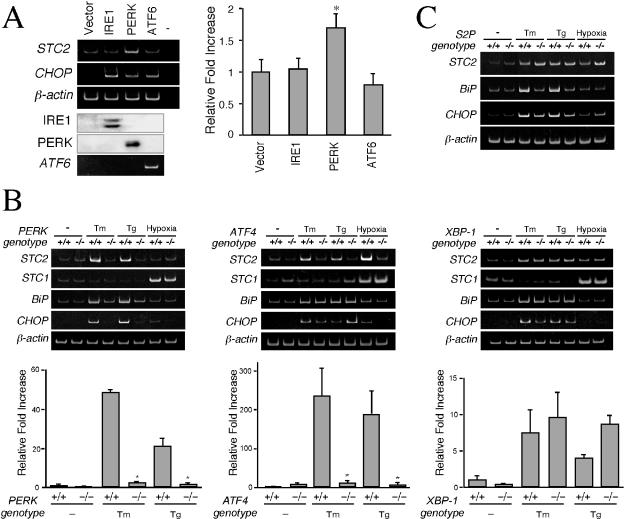
Three major ER-resident stress sensors—IRE1, PERK, and ATF6—are activated during induction of the UPR. The signaling from these transducers merges in the nucleus to coordinately activate transcription of UPR target genes (38, 52). To identify the upstream transducer that activates STC2 expression, we tested levels of STC2 mRNA in HEK293 cells overexpressing IRE1, PERK, and ATF6. As shown Fig. 7A, only cells transfected with PERK increased STC2 mRNA, whereas CHOP mRNA was upregulated by expression of any of the three early transducers. To confirm the role of PERK in STC2 expression during the UPR, we examined mouse embryonic fibroblasts with homozygous deletion of PERK alleles (16). Unlike wild-type cells, PERK−/− fibroblasts failed to strongly upregulate STC2 expression upon treatment with tunicamycin and thapsigargin (Fig. 7B). Together, these results suggest that stress within the ER results in enhanced STC2 expression through the PERK-mediated pathway.
FIG. 7.
STC2 expression is induced through PERK-dependent pathway. (A) HEK293 cells were transfected with empty vector or indicated expression plasmids, and total RNA was isolated 48 h after transfection. IRE and PERK expression was analyzed by Western blotting with 9E10 antibody. The levels of STC2, CHOP, β-actin, and ATF6 mRNA were evaluated by RT-PCR. For negative control (lane −), RT-PCR was performed in the absence of reverse transcriptase. STC2 expression was assessed by quantitative real-time PCR with specific primers, normalized to β-actin mRNA signal, and plotted as the increase compared to cells transfected with empty vector. The data represent means ± the SE from three independent experiments. ✽, P < 0.05 (relative to cells transfected with empty vector). Note the increase in STC2 mRNA level in cells transfected with PERK cDNA. (B) Wild-type and PERK−/−, ATF4−/−, or XBP-1−/− mouse embryonic fibroblasts were treated with 2 μg of tunicamycin/ml, 300 nM thapsigargin, or hypoxic conditions for 8 h. STC2 mRNA level were analyzed by RT-PCR and quantitative real-time PCR. The data represent means ± the SE from three independent experiments. ✽, P < 0.01 (relative to wild-type cells). Note the marked attenuation in ER stress-induced expression of STC2 mRNA in PERK−/− and ATF4−/− fibroblasts. (C) Wild-type and S2P-deficient CHO cells were treated cell with 2 μg of tunicamycin/ml or 300 nM thapsigargin for 8 h. Levels of STC2, STC1, BiP, and β-actin mRNA were evaluated by RT-PCR. STC2 mRNA is induced by ER stress in S2P-deficient CHO cells.
During activation of the UPR, PERK causes transient translational arrest by phosphorylation of eIF2α and facilitates enhanced translation of the transcription factor ATF4 (15, 17, 27). To determine whether activation of PERK alters STC2 expression via ATF4, we performed a set of experiments in ATF4−/− fibroblasts. Exposure of ATF4−/− fibroblasts to tunicamycin or thapsigargin fails to upregulate STC2 expression, providing a mechanistic insight into STC2 expression by ER stress downstream of PERK activation (Fig. 7B). To further confirm these findings, we examined STC2 expression in cells lacking the active transcription factors XBP-1 and ATF6. We found that XBP-1−/− and wild-type control fibroblasts upregulate STC2 expression to comparable levels upon ER stress (Fig. 7B). Next, we tested the requirement for ATF6 in STC2 induction by using M19 mutant CHO cells, which fail to process ATF6 due to a deficiency in Site-2 protease (34, 68). M19 cells induce STC2 expression after ER stress to levels similar to that of parental CHO cells, whereas as reported previously, we observe attenuated induction of BiP expression in M19 cells (Fig. 7C). These results are in agreement with a major role for ATF4 in STC2 upregulation during the UPR independent of XBP-1 and ATF6.
Since STC1 expression is not associated with the UPR, we reasoned it is highly unlikely that PERK-ATF4 pathway contributes to STC1 induction during hypoxia. Consistent with this notion, STC1 expression was upregulated by hypoxia in PERK−/− and ATF4−/− fibroblasts (Fig. 7B). Similarly, XBP-1 expression was also not required for hypoxia-mediated induction of STC1 expression.
STC2 expression in ischemic rat brain.
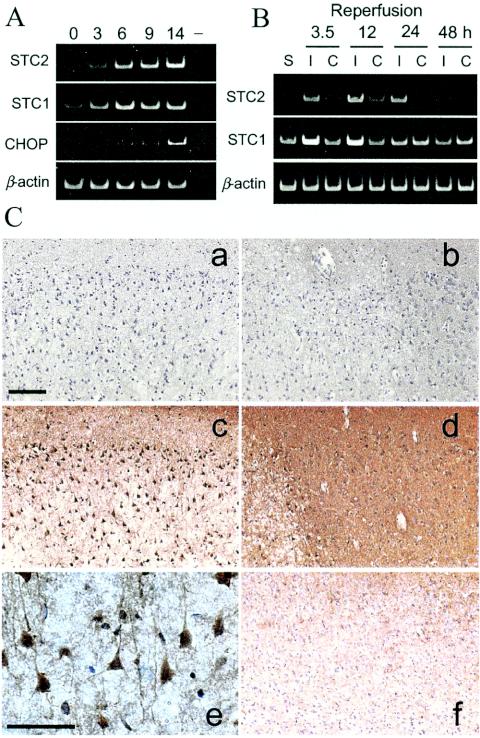
For in vivo analysis of induced STC2 expression we chose to focus on the rat MCA occlusion model because it is known that PERK is activated in the brain after transient cerebral ischemia (31, 32). RT-PCR analysis of cortex excised from the ischemic side shows marked induction of STC2 expression in ischemic cortex after 3.5 to 24 h of reperfusion after 90 min of MCA occlusion (Fig. 8B). Expression of STC2 was extremely low in the contralateral cortex or sham-operated animals. Consistent with a previous report (70) STC1 mRNA is also detectable at higher levels in the ischemic cortex compared to contralateral cortex.
FIG. 8.
Induced expression of STC2 in ischemic rat brain. (A) Rat PC12 cells were subject to hypoxic conditions for the indicated periods. Levels of STC2, STC1, BiP, and β-actin mRNA were evaluated by RT-PCR with specific primers. Note the coordinated induction of STC2 and STC1 expression by hypoxia. (B) Rats were subject to transient MCA occlusion, and total RNA was extracted from cortex after reperfusion for the indicated periods. STC2, STC1, and β-actin mRNA levels were examined by RT-PCR. Lanes: S, sham-operated control; I, ischemic side cortex; C, contralateral side cortex. (C) Immunohistochemical analysis of STC2 expression in ischemic rats after 12 h of reperfusion. Sections were stained with preimmune serum (a and b) or STC2 antibody (c to f). Subpanels: a and c, ischemic core; b and d, penumbra; e, higher magnification of STC2 staining in ischemic cortex; f, contralateral cortex. Scale bar, 100 μm. Note the overall increase in STC2 immunoreactivity in the penumbra compared to the contralateral cortex and the intense staining of vulnerable neurons within the ischemic core.
We performed immunohistochemical analysis to determine where STC2 expression is induced after cerebral ischemia. Increased STC2 immunoreactivity was observed at 6 h reperfusion after 120 min MCA occlusion and persisted after 12, 24, and 48 h of reperfusion after MCA occlusion. Representative STC2 staining in ischemic side frontoparietal motor cortex at 12 h of reperfusion is shown in Fig. 8Cc and d. We observed strong STC2 immunoreactivity within the ischemic core and generally higher diffuse staining in the penumbra on the ischemic cortex compared to contralateral side (Fig. 8Cf). Moreover, the distinctive morphology and distribution of STC2-positive cells within the ischemic core are consistent with cortical neurons (Fig. 8Ce). Based on these results, we conclude that STC2 is a target of the UPR in vulnerable neurons during cerebral ischemia and reperfusion.
STC2 contributes to cell survival during thapsigargin- and H2O2-induced stress.
As mentioned above, PERK is one of the most proximal ER stress sensors that is activated by accumulation of misfolded proteins within the ER. Upon activation, PERK rapidly phosphorylates eIF2α and causes translational arrest, which is a protective measure as it attenuates ER overload. However, the accumulation of phosphorylated eIF2α leads to selective translation of ATF4, a transcription factor that upregulates the expression of ER chaperones, XBP-1, and also the proapoptotic transcription factor CHOP. Thus, PERK activation regulates both prosurvival and proapoptotic signaling during ER stress (12, 38). Since STC2 upregulation was mediated by PERK/ATF4 pathway, we sought to determine whether STC2 contributes to cytoprotection or facilitates apoptotic signaling. To address this issue, we decided to attenuate STC2 expression by using siRNA strategy (2) and assess the extent of stress-induced cell death in neuronal cells. In these experiments, mouse N2a neuroblastoma cells were transfected with pSUPER vector or pSUPER STC2 siRNA plasmid, along with small amounts of a plasmid encoding YFP. After cells were exposed to ER and oxidative stresses, we stained the cells with EthD-1 (dead cells stain red); YFP-positive EthD-negative cells were counted as surviving cells. In parallel, we performed Western blots to monitor the efficiency of siRNA inhibition of STC2 upregulation. Transient transfection of STC2 siRNA plasmid effectively attenuated induced expression of STC2 after treatment with thapsigargin. Expression of STC2 siRNA did not affect upregulation of BiP in cells exposed to thapsigargin (Fig. 9A). As shown in Fig. 9B, there was no difference in the number of average YFP-positive EthD-negative cells per field when cultures were transfected with either empty vector or STC2 siRNA plasmid, suggesting that expression of siRNA per se did not adversely affect cell survival. Approximately 60% of N2a cells transfected with empty vector survived when cultures were treated with thapsigargin or H2O2. On the other hand, only 30 and 40% of STC2 siRNA-treated cells survived exposure to thapsigargin and H2O2, respectively, indicating that STC2 siRNA-transfected cells were significantly more vulnerable to cell death (P < 0.03 [versus control cells]) (Fig. 9B). In contrast, no differences were observed among the percentages of dead cells in control or STC2 siRNA-transfected cells after exposure to tunicamycin or staurosporine. These results suggest that survival adaptation of cells to thapsigargin-induced ER stress and oxidative stress requires upregulation of STC2 expression.
FIG. 9.
siRNA inhibition of STC2 induction accentuates cell death by thapsigargin and oxidative stress. (A) N2a cells were cotransfected with empty pSUPER or pSUPER-STC2 siRNA plasmid, along with pEYFP plasmid. Two days after transfection, cells were treated or not treated with 150 nM thapsigargin (Tg), 100 μM H2O2, 2 μg of tunicamycin/ml (Tm), or 250 nM STS for 16 h. Western blot analysis of STC2, BiP, and β-actin levels. Note the marked reduction in STC2 levels in STC2 siRNA transfected cells. (B) Transfected N2a cells grown on coverslips were washed in PBS and stained with EthD-1 to stain dead cells. Fluorescence images were acquired, and live green cells devoid of EthD-1staining were counted by using MetaMorph software. On the left, the average number of live transfected cells per field in untreated cultures (non) were plotted. On the right, a graph shows the percentage of cell survival (means ± the SE) in cultures treated with various agents (above), normalized to untreated controls in each case, calculated from four independent experiments. ✽, P < 0.01; ✽✽, P < 0.03. (C) HeLa cells were treated with tunicamycin or thapsigargin for indicated periods and detergent lysates were analyzed by blotting with STC2, BiP, and β-actin antibodies. (D) HeLa cells were transfected with STC2 or mismatch STC2 (STC2 M) synthetic siRNA duplexes. One day after transfection, cells were treated or not treated with 500 nM thapsigargin (Tg), 2 μg of tunicamycin/ml (Tm), or 250 nM STS for 16 h, and the levels of STC2, BiP, and β-actin were analyzed by Western blotting. (E) HeLa cells grown on coverslips were transfected and treated with stress agents for 40 h (Tg and Tm treatment) or staurosporine for 9 h (STS). Coverslips were stained with Hoechst 33342 to visualize dead cells with condensed nuclei. Fluorescence images were acquired, and apoptotic cells were counted by using MetaMorph software. The graph shows percentage of cell death expressed as means ± the SE for three independent experiments. The data were analyzed by using a paired t test to compare the differences between STC2 M and STC2 siRNA-transfected cells. ✽, P < 0.05. (F) siRNA transfected HeLa cells were also stained with Annexin-V-Alexa 568 and annexin-V positive cells were quantified as described above. ✽✽, P < 0.01.
To confirm these findings, we performed a set of studies in HeLa cells by inhibiting STC2 expression with synthetic siRNA duplexes. HeLa cells also upregulate STC2 expression in a time-dependent manner after exposure to tunicamycin or thapsigargin (Fig. 9C). Western blot analyses showed that transfection with STC2 siRNA successfully suppressed ER-stress associated upregulation of STC2 expression compared to cells exposed only to the transfection reagents (no siRNA). Moreover, a mutant siRNA (STC2 M, which differs by 6 bp from STC2 siRNA) did not interfere with UPR induction of STC2 expression (Fig. 9D). In order to determine whether suppression of STC2 expression during the UPR leads to apoptotic cell death, we stained siRNA-transfected HeLa cells with Hoechst 33342 and Annexin-V-Alexa 568. There was minimal apoptotic staining of cells transfected with either STC2 or STC2 M siRNA in the absence of ER stress. After treatment with thapsigargin, we observed ca. 10% (with Hoechst staining) and 20% (with Annexin-V staining) of cell death in mismatch control STC2 M siRNA transfected cultures. In contrast, ca. 30% (with Hoechst staining) and 40% (with Annexin-V staining) cell death was observed in STC2 siRNA-transfected cultures exposed to thapsigargin. As was expected from N2a siRNA studies, the extent of cell death induced by tunicamycin or staurosporine treatment was not significantly different between STC2 and the mismatch STC2 M siRNA. Thus, STC2 expression seems to be necessary for cell survival under conditions in which thapsigargin but not tunicamycin was used as the ER stress agent.
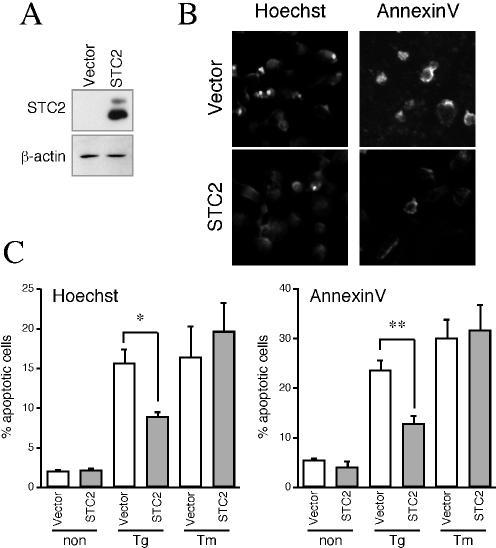
In addition to the STC2 knock-down experiments described above, we performed a set of studies to determine whether overexpression of STC2 will offer protection against ER stress-induced cell death. To this end, we overexpressed STC2 by transient transfection in HeLa cells (Fig. 10A). Transfected cells not exposed to ER stressors exhibited only minimal cell death, and there was no significant difference between transfection with control vector or STC2 expression plasmid. However, upon treatment with thapsigargin, there were significantly fewer dead cells staining positive with Hoechst or Annexin-V in STC2 transfected cultures compared to cultures transfected with control vector (Fig. 10B and C). On the other hand, no significant difference was observed in the percentage of cell death between the two groups when cells were exposed to tunicamycin. Thus, overexpression of STC2 significantly increased the resistance to apoptotic death after treatment with thapsigargin but not after treatment with tunicamycin (Fig. 10).
FIG. 10.
Overexpression of STC2 protects cells from thapsigargin-induced cell death. (A) HeLa cells were transfected with empty vector or STC2 expression plasmid. Expression of STC2 was analyzed by Western blotting. (B and C) One day after transfection, cells were treated or not treated with thapsigargin (Tg) or tunicamycin (Tm) and then processed as described in Fig. 9E and F to quantify cell death. The graph shows the percentage of cell death expressed as means ± SE for three independent experiments. ✽, P < 0.05; ✽✽, P < 0.01.
DISCUSSION
Mammalian cells activate UPR, a homeostatic response, when challenged with diverse set of stress conditions that affect ER function such as induced the accumulation of misfolded proteins and alterations in Ca2+ homeostasis in the ER. Two major components of this response, transcriptional induction of genes encoding protein chaperones and molecules associated with ER-associated degradation, are highly conserved from yeast to mammals (52). A third component, involving activation of PERK, leading to transient attenuation of protein translation, is a salient feature of the UPR in metazoans. Together, these pathways mount an adaptive response critical for cell recovery and survival from injury, as well as activate key steps mediating cell death in case the protein misfolding defect is not corrected (29, 38, 62). It is becoming increasingly clear that the balance between the survival and cell death responses of the UPR is crucial to maintaining normalcy under certain physiological and pathological settings, including acute and chronic brain diseases (11, 49, 57). Characterization of proximal ER stress sensors in the past few years, together with large-scale gene profiling of UPR, have fueled the discovery of additional sets of UPR targets and revealed the complexity of the mammalian UPR (17, 47, 50, 52, 60). In the present study, we investigated the gene expression profile during UPR in HEK293 cells by using microarray technique in order to identify novel UPR targets. We observed UPR-related upregulation of 127 genes (ca. 1% of the genes profiled) in cells treated with tunicamycin or thapsigargin. This outcome is fairly consistent with earlier large-scale gene profiling studies of the UPR in mouse and human cells (17, 50, 47). Consistent with previous reports, we observed upregulation of classes of genes encoding proteins involved in translation, protein folding and posttranslational modifications, ER-associated degradation, vesicular transport, glucose metabolism, transcriptional regulation, etc. Since the vast majority of the UPR-related genes identified in our study have been linked to ER stress previously or during the course of our investigation, we turned our attention to genes with known function in regulating Ca2+ homeostasis.
In HEK293 and mouse N2a neuroblastoma cells, exposure to tunicamycin or thapsigargin upregulates four genes involved in regulating Ca2+ homeostasis (Table 1 and Fig. 1); three of these genes, namely, NUCB2, CGRP2, and STC2, are novel targets of the UPR. During the course of our investigation, NUCB2 and STC2 were also found to be induced by tunicamycin treatment in SH-SY5Y neuroblastoma cells (6, 50). Previous studies have identified ATP2A2, which encodes a SERCA2b isoform, as a gene inducible by ER stress (6, 47, 57). SERCA2b is expressed ubiquitously and plays a critical role in Ca2+ signaling and homeostasis in all cells by translocating Ca2+ from the cytosol to the ER lumen. Genetic mutations in ATP2A2 cause an autosomal-dominant skin disorder termed Darier-White disease and are associated with an increased prevalence of neuropsychiatric disorders, including bipolar disorders, schizophrenia, epilepsy, and mental retardation (OMIM no. 124200 [http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=124200]). NUCB2 encodes a ubiquitous protein with two EF-hand motifs, termed nucleobindin 2 (also referred to as Calnuc), which is the major Ca2+-binding protein in the Golgi apparatus (36). Nucleobindin 2 is thought to establish an agonist-mobilizable Ca2+ store within the Golgi lumen (37). CGRP2 encodes a 127-amino-acid protein that gets processed into 37-amino-acid CGRP-II, which is nearly identical to neuropeptide CGRP-I (with three amino acid differences in human). CGRP-II is expressed in central and peripheral nervous system pituitary, the thyroid, and medullary thyroid carcinoma. Biological functions of CGRPs include vasodilation, nociception, glucose and calcium metabolism, etc. (63). STC2 encodes a secreted phosphoprotein related to calcium- and phosphate-regulating glycoprotein hormone first described in fish (21).
The function of STC in fish is to prevent hypercalcemia by targeting gill (reduces Ca2+ influx), gut (inhibits Ca2+ reabsorption), and kidney (increased PO4 reabsorption) (59, 64). Of the two mammalian STC homologues, STC1 shares greater sequence similarity with fish STC. In contrast to the restricted expression documented for fish STC, mouse STC1 is widely expressed in brain and several peripheral tissues, including the kidney, prostate, thyroid gland, and ovary (21). Nevertheless, STC function appears to be retained during evolution, because, as in fish, mammalian STC1 also stimulates phosphate absorption in the kidney and intestine and regulates calcium homeostasis (53, 66). STC2 is also widely expressed in mammals, but its physiological role(s) has not been fully understood. Since STC2 shares much lower identity (∼35%) with STC1 and fish STC, it is possible that STC2 is functionally divergent from STC1 and fish STC (21). This notion is supported by the differential activation of STC2 expression by ER stress agents in neuronal and nonneuronal cells (Fig. 2 and 4). Interestingly, expression of both mammalian STC appear to correlate with the levels of estrogen receptor, and STC2 expression is induced by estrogen, indicating a link between STC expression and human cancer (7).
Despite the differential UPR induction of STC genes, the available evidence suggests that both STC1 and STC2 share cytoprotective functions. Previously, it was reported that overexpression of STC1 in Paju cells increased the viability of cells exposed to thapsigargin (70). The siRNA and overexpression studies reported here indicate that STC2 function is required for cell survival during exposure to thapsigargin and oxidative stress. Thapsigargin, an inhibitor of Ca2+ ATPase in the ER, rapidly mobilizes the intracellular calcium store and leads to increased levels of intracellular cytosolic Ca2+. Similarly, oxidative stress also elevates cytosolic Ca2+ levels by stimulating Ca2+ influx from the extracellular environment and ER and by inhibiting ER Ca2+ ATPase (10). Given the negative regulation of Ca2+ influx by fish STC and mammalian STC1 (53), it is very likely that STC2 also functions in a similar manner by regulating Ca2+ homeostasis in cells subject to ER stress, thus promoting cell survival. Finally, although STC2 expression is induced by treatment of cells with tunicamycin, siRNA and overexpression experiments showed no correlation between STC2 expression and cell viability in the presence of tunicamycin, suggesting that protein N glycosylation of STC2 or other protein(s) is critical for cytoprotective properties of STC2 during ER stress.
The manner in which STC2 expression is induced indicates that STC2 function is specifically required during the cellular response to challenge with ER stress agents, oxidative stress, and hypoxia (Fig. 6). We did not observe changes in STC2 expression when cells were exposed to several other xenotoxic stresses that would indicate a more generalized response to cytotoxicity. Hence, it is not surprising that STC2 is a downstream target of signaling activated by the proximal ER stress-sensing transmembrane protein kinase PERK and its downstream transcription factor, ATF4. Recent evidence suggests that activation of PERK pathway is not unique to ER stress response. For example, Koumenis et al. (30) reported that PERK is hyperphosphorylated upon hypoxic stress and is required for adaptation and survival of cells exposed to hypoxia. Recently, Harding et al. (17) demonstrated that PERK−/− cells show impaired expression of genes involved in antioxidative stress response and accumulate endogenous peroxides during ER stress. Thus, the cellular adaptive response to ER stress and oxidative stress share a common mechanism involving the early activation of PERK pathway. Based on these findings, we speculate that STC2 function may be associated with the ability of cells to adapt to not only UPR but also oxidative stress and hypoxia. Indeed, we find rapid induction of STC2 expression in cortical neurons reacting to ischemia within 3.5 h of reperfusion after transient MCA occlusion. Cerebral ischemia is a well-known pathophysiological condition in which aspects of UPR are activated (25, 32, 48). Strong STC2 immunoreactivity is observed in the ischemic cortex 6 h after reperfusion and persisted even after 48 h of reperfusion (Fig. 8). Our in vitro findings, together with marked induction of STC2 in neurons within the ischemic core and penumbra, suggest that STC2 upregulation might have a nonredundant physiological role in the survival of ischemic neuronal cells.
Interestingly, induced STC1 expression appears to be limited to hypoxia and not associated with ER stress or other xenotoxic stresses examined in the present study. The only exception we noted was thapsigargin-induced transient upregulation of STC1 in primary cortical astrocytes (Fig. 2 and 6). Thus, STC1 may have a restricted role during hypoxia compared to a more general role of STC2 during UPR and hypoxia. While the present study was being prepared, Leonard et al. (35) reported STC2 as one of the genes induced by hypoxia in a kidney cell line and dependent on hypoxia-inducible factor 1 (HIF-1). Furthermore, it was observed that STC1 is among the genes induced by vascular endothelial growth factor, a HIF-1-dependent gene (39, 67). Our results from analysis of mouse embryonic fibroblasts are consistent with both of these findings. In agreement with a central role of HIF, hypoxia-mediated upregulation of STC1 in fibroblasts occurs under conditions when hallmarks of the UPR (BiP and CHOP) are not activated and in the absence of PERK, ATF4, or XBP-1 expression (Fig. 7B).
Recent evidence suggests that >90% of STC1 is concentrated in the mitochondria of the kidney, liver, spleen, and lung. It was proposed that STC1 is internalized from the plasma membrane by binding to an unidentified receptor, and internalized STC1 is shuttled to the mitochondria (44). Our confocal microscopy analyses and subcellular fractionation studies reveal that STC2 is localized mainly in the ER and Golgi apparatus, with no overlap with mitochondrial markers. Purified mitochondrial fractions were also devoid of STC2 (Fig. 5). One possibility that might explain this apparent discrepancy is that cultured cells used in our study do not express high levels of the putative STC receptor(s). The molecular details of STC function in neuronal cells still need to be elucidated. In the future it will be important to determine whether STC1 and STC2 have different subcellular sites of action in regulating cellular mineral metabolism under physiological conditions and under conditions of stress.
Acknowledgments
This study was supported by grants from the National Institutes of Health and the Brain Research Foundation (G.T.) and from the Sankyo Life Science Foundation (D.I.).
We are grateful to David Ron (Skirball Institute, New York University) for providing IRE1β and PERK cDNAs and PERK−/− fibroblasts, Linda M. Hendershot (St. Jude Children's Research Hospital, Memphis, Tenn.) for providing ATF4−/− fibroblasts, Laurie H. Glimcher (Harvard School of Public Health, Boston, Mass.) for providing XBP-1−/− fibroblasts, and T. Y. Chang (Dartmouth Medical School, Hanover, N.H.) for providing the S2P-deficient CHO cells. We thank Manuel F. Utset (University of Chicago) for discussions and advice on histology.
REFERENCES
- 1.Berridge, M. J. 2002. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium 32:235-249. [DOI] [PubMed] [Google Scholar]
- 2.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 3.Butkus, A., P. J. Roche, R. T. Fernley, J. Haralambidis, J. D. Penschow, G. B. Ryan, J. F. Trahair, G. W. Tregear, and J. P. Coghlan. 1987. Purification and cloning of a corpuscles of Stannius protein from Anguilla australis. Mol. Cell Endocrinol. 54:123-133. [DOI] [PubMed] [Google Scholar]
- 4.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 5.Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P. O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of Saccharomyces cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5:729-735. [DOI] [PubMed] [Google Scholar]
- 6.Caspersen, C., P. S. Pedersen, and M. Treiman. 2000. The sarco/endoplasmic reticulum calcium-ATPase 2b is an endoplasmic reticulum stress-inducible protein. J. Biol. Chem. 275:22363-22372. [DOI] [PubMed] [Google Scholar]
- 7.Chang, A. C., D. A. Jellinek, and R. R. Reddel. 2003. Mammalian stanniocalcins and cancer. Endocrinol. Relat. Cancer 10:359-373. [DOI] [PubMed] [Google Scholar]
- 8.Chang, A. C., and R. R. Reddel. 1998. Identification of a second stanniocalcin cDNA in mouse and human: stanniocalcin 2. Mol. Cell Endocrinol. 141:95-99. [DOI] [PubMed] [Google Scholar]
- 9.DiMattia, G. E., R. Varghese, and G. F. Wagner. 1998. Molecular cloning and characterization of stanniocalcin-related protein. Mol. Cell Endocrinol. 146:137-140. [DOI] [PubMed] [Google Scholar]
- 10.Ermak, G., and K. J. Davies. 2002. Calcium and oxidative stress: from cell signaling to cell death. Mol. Immunol. 38:713-721. [DOI] [PubMed] [Google Scholar]
- 11.Forman, M. S., V. M. Lee, and J. Q. Trojanowski. 2003. ′Unfolding' pathways in neurodegenerative disease. Trends Neurosci. 26:407-410. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 13.Harding, H. P., and D. Ron. 2002. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes 51(Suppl. 3):S455-S461. [DOI] [PubMed] [Google Scholar]
- 14.Harding, H. P., H. Zeng, Y. Zhang, R. Jungries, P. Chung, H. Plesken, D. D. Sabatini, and D. Ron. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7:1153-1163. [DOI] [PubMed] [Google Scholar]
- 15.Harding, H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5:897-904. [DOI] [PubMed] [Google Scholar]
- 16.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 17.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 18.Hasan, M. T., C. C. Chang, and T. Y. Chang. 1994. Somatic cell genetic and biochemical characterization of cell lines resulting from human genomic DNA transfections of Chinese hamster ovary cell mutants defective in sterol-dependent activation of sterol synthesis and LDL receptor expression. Somat. Cell Mol. Genet. 20:183-194. [DOI] [PubMed] [Google Scholar]
- 19.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtz, W. A., and K. L. O'Malley. 2003. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J. Biol. Chem. 278:19367-19377. [DOI] [PubMed] [Google Scholar]
- 21.Ishibashi, K., and M. Imai. 2002. Prospect of a stanniocalcin endocrine/paracrine system in mammals. Am. J. Physiol. Renal Physiol. 282:F367-F375. [DOI] [PubMed] [Google Scholar]
- 22.Ishibashi, K., K. Miyamoto, Y. Taketani, K. Morita, E. Takeda, S. Sasaki, and M. Imai. 1998. Molecular cloning of a second human stanniocalcin homologue (STC2). Biochem. Biophys. Res. Commun. 250:252-258. [DOI] [PubMed] [Google Scholar]
- 23.Ito, D., Y. Imai, K. Ohsawa, K. Nakajima, Y. Fukuuchi, and S. Kohsaka. 1998. Microglia-specific localization of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 57:1-9. [DOI] [PubMed] [Google Scholar]
- 24.Ito, D., K. Tanaka, S. Suzuki, T. Dembo, and Y. Fukuuchi. 2001. Enhanced expression of Iba1, ionized calcium-binding adapter molecule 1, after transient focal cerebral ischemia in rat brain. Stroke 32:1208-1215. [DOI] [PubMed] [Google Scholar]
- 25.Ito, D., K. Tanaka, S. Suzuki, T. Dembo, A. Kosakai, and Y. Fukuuchi. 2001. Upregulation of the Ire1-mediated signaling molecule, Bip, in ischemic rat brain. Neuroreport 12:4023-4028. [DOI] [PubMed] [Google Scholar]
- 26.Jellinek, D. A., A. C. Chang, M. R. Larsen, X. Wang, P. J. Robinson, and R. R. Reddel. 2000. Stanniocalcin 1 and 2 are secreted as phosphoproteins from human fibrosarcoma cells. Biochem. J. 350(Pt. 2):453-461. [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Lu, T. Hai, H. P. Harding, X. Wang, D. Ron, D. R. Cavener, and R. C. Wek. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24:1365-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonas, V., C. R. Lin, E. Kawashima, D. Semon, L. W. Swanson, J. J. Mermod, R. M. Evans, and M. G. Rosenfeld. 1985. Alternative RNA processing events in human calcitonin/calcitonin gene-related peptide gene expression. Proc. Natl. Acad. Sci. USA 82:1994-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 30.Koumenis, C., C. Naczki, M. Koritzinsky, S. Rastani, A. Diehl, N. Sonenberg, A. Koromilas, and B. G. Wouters. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 22:7405-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, R., S. Azam, J. M. Sullivan, C. Owen, D. R. Cavener, P. Zhang, D. Ron, H. P. Harding, J. J. Chen, A. Han, B. C. White, G. S. Krause, and D. J. DeGracia. 2001. Brain ischemia and reperfusion activates the eukaryotic initiation factor 2alpha kinase, PERK. J. Neurochem. 77:1418-1421. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, R., G. S. Krause, H. Yoshida, K. Mori, and D. J. DeGracia. 2003. Dysfunction of the unfolded protein response during global brain ischemia and reperfusion. J. Cereb. Blood Flow Metab. 23:462-471. [DOI] [PubMed] [Google Scholar]
- 33.Lafeber, F. P., R. G. Hanssen, Y. M. Choy, G. Flik, M. P. Herrmann-Erlee, P. K. Pang, and S. E. Bonga. 1988. Identification of hypocalcin (teleocalcin) isolated from trout Stannius corpuscles. Gen. Comp. Endocrinol. 69:19-30. [DOI] [PubMed] [Google Scholar]
- 34.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonard, M. O., D. C. Cottell, C. Godson, H. R. Brady, and C. T. Taylor. 2003. The role of HIF-1 alpha in transcriptional regulation of the proximal tubular epithelial cell response to hypoxia. J. Biol. Chem. 278:40296-40304. [DOI] [PubMed] [Google Scholar]
- 36.Lin, P., H. Le-Niculescu, R. Hofmeister, J. M. McCaffery, M. Jin, H. Hennemann, T. McQuistan, L. De Vries, and M. G. Farquhar. 1998. The mammalian calcium-binding protein, nucleobindin (Calnuc), is a Golgi resident protein. J. Cell Biol. 141:1515-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, P., Y. Yao, R. Hofmeister, R. Y. Tsien, and M. G. Farquhar. 1999. Overexpression of CALNUC (nucleobindin) increases agonist and thapsigargin releasable Ca2+ storage in the Golgi. J. Cell Biol. 145:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, C. Y., and R. J. Kaufman. 2003. The unfolded protein response. J. Cell Sci. 116:1861-1862. [DOI] [PubMed] [Google Scholar]
- 39.Liu, D., H. Jia, D. I. Holmes, A. Stannard, and I. Zachary. 2003. Vascular endothelial growth factor-regulated gene expression in endothelial cells: KDR-mediated induction of Egr3 and the related nuclear receptors Nur77, Nurr1, and Nor1. Arterioscler. Thromb. Vasc. Biol. 23:2002-2007. [DOI] [PubMed] [Google Scholar]
- 40.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 41.Longa, E. Z., P. R. Weinstein, S. Carlson, and R. Cummins. 1989. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke 20:84-91. [DOI] [PubMed] [Google Scholar]
- 42.MacLennan, D. H., C. J. Brandl, B. Korczak, and N. M. Green. 1985. Amino-acid sequence of a Ca2+ + Mg2+-dependent ATPase from rabbit muscle sarcoplasmic reticulum, deduced from its complementary DNA sequence. Nature 316:696-700. [DOI] [PubMed] [Google Scholar]
- 43.Mattson, M. P., F. M. LaFerla, S. L. Chan, M. A. Leissring, P. N. Shepel, and J. D. Geiger. 2000. Calcium signaling in the ER: its role in neuronal plasticity and neurodegenerative disorders. Trends Neurosci. 23:222-229. [DOI] [PubMed] [Google Scholar]
- 44.McCudden, C. R., K. A. James, C. Hasilo, and G. F. Wagner. 2002. Characterization of mammalian stanniocalcin receptors. Mitochondrial targeting of ligand and receptor for regulation of cellular metabolism. J. Biol. Chem. 277:45249-45258. [DOI] [PubMed] [Google Scholar]
- 45.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 46.Nishitoh, H., A. Matsuzawa, K. Tobiume, K. Saegusa, K. Takeda, K. Inoue, S. Hori, A. Kakizuka, and H. Ichijo. 2002. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16:1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okada, T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366:585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paschen, W., C. Aufenberg, S. Hotop, and T. Mengesdorf. 2003. Transient cerebral ischemia activates processing of xbp1 messenger RNA indicative of endoplasmic reticulum stress. J. Cereb. Blood Flow Metab. 23:449-461. [DOI] [PubMed] [Google Scholar]
- 49.Paschen, W., and A. Frandsen. 2001. Endoplasmic reticulum dysfunction: a common denominator for cell injury in acute and degenerative diseases of the brain? J. Neurochem. 79:719-725. [DOI] [PubMed] [Google Scholar]
- 50.Reimertz, C., D. Kogel, A. Rami, T. Chittenden, and J. H. Prehn. 2003. Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J. Cell Biol. 162:587-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reimold, A. M., N. N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E. M. Gravallese, D. Friend, M. J. Grusby, F. Alt, and L. H. Glimcher. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412:300-307. [DOI] [PubMed] [Google Scholar]
- 52.Rutkowski, D. T., and R. J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20-28. [DOI] [PubMed] [Google Scholar]
- 53.Sheikh-Hamad, D., R. Bick, G. Y. Wu, B. M. Christensen, P. Razeghi, B. Poindexter, H. Taegtmeyer, A. Wamsley, R. Padda, M. Entman, S. Nielsen, and K. Youker. 2003. Stanniocalcin-1 is a naturally occurring L-channel inhibitor in cardiomyocytes: relevance to human heart failure. Am. J. Physiol. Heart Circ. Physiol. 285:H442-H448. [DOI] [PubMed] [Google Scholar]
- 54.Sheikh-Hamad, D., D. Rouse, and Y. Yang. 2000. Regulation of stanniocalcin in MDCK cells by hypertonicity and extracellular calcium. Am. J. Physiol. Renal Physiol. 278:F417-F424. [DOI] [PubMed] [Google Scholar]
- 55.Shen, J., X. Chen, L. Hendershot, and R. Prywes. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3:99-111. [DOI] [PubMed] [Google Scholar]
- 56.Shen, X., R. E. Ellis, K. Lee, C. Y. Liu, K. Yang, A. Solomon, H. Yoshida, R. Morimoto, D. M. Kurnit, K. Mori, and R. J. Kaufman. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for Caenorhabditis elegans development. Cell 107:893-903. [DOI] [PubMed] [Google Scholar]
- 57.Soto, C. 2003. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4:49-60. [DOI] [PubMed] [Google Scholar]
- 58.Spector, D. L., R. D. Goldman, and L. A. Leinwand. 1998. Culture and biochemical analysis of cells, p. 41.1-41.4. In Cells: a laboratory manual, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Sundell, K., B. T. Bjornsson, H. Itoh, and H. Kawauchi. 1992. Chum salmon (Oncorhynchus keta) stanniocalcin inhibits in vitro intestinal calcium uptake in Atlantic cod (Gadus morhua). J. Comp. Physiol. B 162:489-495. [DOI] [PubMed] [Google Scholar]
- 60.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 61.Treiman, M. 2002. Regulation of the endoplasmic reticulum calcium storage during the unfolded protein response-significance in tissue ischemia? Trends Cardiovasc. Med. 12:57-62. [DOI] [PubMed] [Google Scholar]
- 62.Urano, F., A. Bertolotti, and D. Ron. 2000. IRE1 and efferent signaling from the endoplasmic reticulum. J. Cell Sci. 113(Pt. 21):3697-3702. [DOI] [PubMed] [Google Scholar]
- 63.van Rossum, D., U. K. Hanisch, and R. Quirion. 1997. Neuroanatomical localization, pharmacological characterization and functions of CGRP, related peptides, and their receptors. Neurosci. Biobehav. Rev. 21:649-678. [DOI] [PubMed] [Google Scholar]
- 64.Wagner, G. F. 1994. The molecular biology of the corpuscles of stannius and regulation of stanniocalcin gene expression, p. 273-306. In C. Hew (ed.), Fish physiology, vol. 13. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 65.Wagner, G. F., M. Hampong, C. M. Park, and D. H. Copp. 1986. Purification, characterization, and bioassay of teleocalcin, a glycoprotein from salmon corpuscles of Stannius. Gen. Comp. Endocrinol. 63:481-491. [DOI] [PubMed] [Google Scholar]
- 66.Wagner, G. F., B. L. Vozzolo, E. Jaworski, M. Haddad, R. L. Kline, H. S. Olsen, C. A. Rosen, M. B. Davidson, and J. L. Renfro. 1997. Human stanniocalcin inhibits renal phosphate excretion in the rat. J. Bone Miner. Res. 12:165-171. [DOI] [PubMed] [Google Scholar]
- 67.Wary, K. K., G. D. Thakker, J. O. Humtsoe, and J. Yang. 2003. Analysis of VEGF-responsive genes involved in the activation of endothelial cells. Mol. Cancer 2:25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 69.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, K., P. J. Lindsberg, T. Tatlisumak, M. Kaste, H. S. Olsen, and L. C. Andersson. 2000. Stanniocalcin: a molecular guard of neurons during cerebral ischemia. Proc. Natl. Acad. Sci. USA 97:3637-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]