Abstract
Current hypotheses suggest the Mre11 nuclease activity could be directly involved in double-strand break (DSB) resection in the presence of a large number of DSBs or limited to processing abnormal DNA ends. To distinguish between these possibilities, we used two methods to create large numbers of DSBs in Saccharomyces cerevisiae chromosomes, without introducing other substrates for the Mre11 nuclease. Multiple DSBs were created either by expressing the HO endonuclease in strains containing several HO cut sites embedded within randomly dispersed Ty1 elements or by phleomycin treatment. Analysis of resection by single-strand DNA formation in these systems showed no difference between strains containing MRE11 or the mre11-D56N nuclease defective allele, suggesting that the Mre11 nuclease is not involved in the extensive 5′ to 3′ resection of DSBs. We postulate that the ionizing radiation (IR) sensitivity of mre11 nuclease-defective mutants results from the accumulation of IR-induced DNA damage that is normally processed by the Mre11 nuclease. We also report that the processivity of 5′ to 3′ DSB resection and the yield of repaired products are affected by the number of DSBs in a dose-dependent manner. Finally, we show that the exonuclease Exo1 is involved in the processivity of 5′ to 3′ resection of an HO-induced DSB at the MAT locus.
The first step in the repair of a DNA double-strand break (DSB) by homologous recombination (HR) is the 5′ to 3′ degradation of DNA ends (58, 70). The resulting 3′ single-stranded DNA (ssDNA) tails are substrates for Rad51 binding to promote pairing and strand exchange with a homologous duplex (59). In Escherichia coli, the RecBCD and RecJ nucleases are responsible for the formation of 3′ single-strand tails by the RecBCD and RecF pathways of HR, respectively (2). In eukaryotes, several observations suggest a nonessential role of the Mre11-Rad50-Xrs2 complex (MRX) in the 5′ to 3′ resection of DSB ends, but the other enzyme(s) responsible for this process remains unknown.
The MRE11, RAD50, and XRS2 genes were identified in screens for meiotic recombination deficiency or sensitivity to ionizing radiation (IR) (59). The corresponding null mutants exhibit the same phenotypes with respect to DNA metabolism, namely, defects in HR (1, 26), nonhomologous end joining (NHEJ) (43), telomere maintenance (9, 34, 48), and the intra-S-phase checkpoint (16, 27, 51). The three proteins form a high-affinity complex where two Mre11 molecules bridge together two Rad50 molecules with one Xrs2 molecule (11, 67). MRE11 and RAD50 encode evolutionarily conserved multidomain proteins homologous to E. coli SbcD and SbcC, respectively (20, 55). The Xrs2 sequence diverged more rapidly, and NBS1 is the human counterpart, forming a complex with the human MRE11 and RAD50 proteins (10, 20). The structure of Rad50 is similar to that of the structural maintenance of chromosomes family of proteins involved in chromosome condensation and sister chromatid cohesion (55, 56). Five phosphodiesterase motifs are present in the N-terminal region of Mre11, and these are required for 3′ to 5′ exonuclease activity on ssDNA and double-stranded DNA substrates, as well as structure-specific ssDNA endonuclease activity (25, 44, 64, 67).
Three observations suggest a role for the MRX complex in the 5′ to 3′ resection of DSBs. First, specific alleles of RAD50 (rad50S) and nuclease-defective alleles of MRE11 (mre11-nd) confer a defect in processing meiosis-specific DSBs (1, 25, 44, 66, 67). Second, the 5′ to 3′ resection at the MAT locus after HO endonuclease cleavage is delayed in mre11, rad50, and xrs2 null strains, but not abolished (31, 66). Third, strains containing the Mre11 nuclease-defective allele mre11-D56N or mre11-H125N are sensitive to IR and methyl methanesulfonate (MMS), but to a much lesser extent than an mre11 null strain. Although the polarity of the exonuclease activity observed in vitro is opposite to that predicted to function in DSB resection, the ssDNA endonuclease activity of Mre11 could function with an unwinding activity to degrade the 5′ ends at break sites (44, 64). Together, these data suggest that the MRX complex could act indirectly in resection by recruiting the proper nuclease to the DSB, or could be directly involved, and could function with at least one other partially redundant enzyme.
The existence of a nuclease different from Mre11 that is capable of degrading the 5′ ends of clean DSBs is implied by the observation that a functional 3′ ssDNA tail capable of strand invasion to promote mating type switching is still formed in both mre11 null and mre11-nd strains after a single HO-induced DSB (36, 44, 66). However, mre11-nd diploids accumulate unprocessed DSBs during meiosis, suggesting a defect in the removal of Spo11 from 5′ ends. In addition, mre11-H125N and mre11-D56N strains are sensitive to IR (44), suggesting a defect in processing chemically heterogeneous DNA ends that are generated by gamma radiolysis (30, 50, 68, 69). Based on these phenotypes, as well as the biochemical properties of Mre11 and SbcCD, we envision two possibilities concerning the role of the Mre11 nuclease activity in the processing of DSB ends. The first one is that Mre11 is not required for the extensive 5′ to 3′ resection of DSB ends, but only for the removal of chemical adducts bound to the ends. Thus resection would depend completely on another undetermined nuclease. The second possibility is that the nuclease activity of Mre11 is involved in the 5′ to 3′ resection process but that another nuclease, which can fully complement the absence of the Mre11 nuclease when only one DSB is present, would become limiting when additional DSBs are made. These two hypotheses could explain the mild sensitivity of the mre11-D56N mutant to IR, the first one because of the presence of blocked DSB ends and the second one because of the large number of DSBs generated.
To distinguish between these two possibilities, we examined 5′ to 3′ resection in a set of strains containing between 1 and 10 “clean” DSBs made by the HO endonuclease. The first model predicts no defect in resection in the mre11-D56N strain, while the second predicts a defect in resection when several breaks are present. As described, we found no defect in 5′ to 3′ resection of HO-induced DSBs in the mre11-D56N strain, regardless of the number of breaks, consistent with the view that the Mre11 nuclease is not involved in the resection of clean DSBs. Surprisingly, we found that the stability of DNA ends was markedly altered in strains containing multiple breaks and that this was independent of the Mre11 nuclease or the ability to undergo strand invasion.
MATERIALS AND METHODS
Media, growth conditions, and genetic methods.
All the media, growth conditions, and genetic methods are as described previously (44). When used, phleomycin (Zeocin; Invitrogen) was directly added to the growth medium and the cultures were protected from the light with aluminum foil.
Yeast strains.
All strains are isogenic and in the W303 background (leu2-3,112 trp1-1 ura3-1 can1-100 ade2-1 his3-11,15) with the corrected RAD5 allele (Table 1). To replace the chromosomal MRE11 allele with mre11-D56N, the integrating plasmid containing the mre11-D56N allele (pSM444) was linearized with SphI and used to transform strains LSY678, LSY679, LSY1259, and LSY1264. The resulting uracil-positive (Ura+) transformants were patched onto rich medium and then replica plated onto medium containing 5-fluoroorotic acid (5-FOA) to select for “pop out” events. The presence of the mre11-D56N allele in the resulting Ura− cells was assessed by determining IR sensitivity at 90 krad (44). To create rad52 strains, the plasmid pSM20 (D. Schild, personal communication) was digested with BamHI and transformed into strains LSY1259, LSY1264, and LSY1285. The presence of a disrupted rad52 allele in the resulting Leu+ transformants was assessed by determining sensitivity to IR.
TABLE 1.
Yeast strains
| Strain | Genotypea | Origin or reference |
|---|---|---|
| W1588-4C (LSY678) | MATa | R. Rothstein |
| W1588-4A (LSY679) | MATα | R. Rothstein |
| LSY723 | MATarad52::LEU2 met17::ADE2 | 5 |
| LSY779 | MATamre11::LEU2 | 45 |
| LSY894-2B | MATα met17-s ADE2 exo1::HIS3 | 60 |
| LSY894-4C | MATamet17-s ADE2 exo1::HIS3 | 60 |
| LSY1032 | MATamre11-D56N | Transformation of W1588-4C with pSM444 |
| LSY1159 | MATa 2 Ty1-HOcs-HIS3 | pBL001 retrotransposition in W1588-4C |
| LSY1164 | MATa 3 Ty1-HOcs-HIS3 | pBL001 retrotransposition in LSY1159 |
| LSY1168 | MATa 4 Ty1-HOcs-HIS3 | pBL001 retrotransposition in W1588-4Cb |
| LSY1170 | MATa 1 Ty1-HOcs-HIS3 | pBL001 retrotransposition in W1588-4C |
| LSY1173 | MATa 3 Ty1-HOcs-HIS3 | pBL001 retrotransposition in LSY1170 |
| LSY1216 | MATa 4 Ty1-HOcs-HIS3 | pBL001 retrotransposition in LSY1164 |
| LSY1220 | MATα 4 Ty1-HOcs-HIS3 | LSY1216 × W1588-4A spore |
| LSY1223 | MATa 6 Ty1-HOcs-HIS3 | pBL001 retrotransposition in LSY1173 |
| LSY1228 | MATa 6 Ty1-HOcs-HIS3 | pBL001 retrotransposition in LSY1168 |
| LSY1240 | MATα 8 Ty1-HOcs-HIS3 | LSY1228 × LSY1220 spore |
| LSY1248 | MATa 10 Ty1-HOcs-HIS3 | pBL001 retrotransposition into LSY1223 |
| LSY1259 | MATα 10 Ty1-HOcs-HIS3 | pBL001 retrotransposition in LSY1240 |
| LSY1264 | MATα 9 Ty1-HOcs-HIS3 | LSY1248 × LSY1240 spore |
| LSY1285 | MATα 10 Ty1-HOcs-HIS3 mre11-D56N | Derived from LSY1259 by transformation with pSM444 |
| LSY1299 | MATα 9 Ty1-HOcs-HIS3 mre11-D56N | Derived from LSY1264 by transformation with pSM444 |
| LSY1477 | MATα 10 Ty1-HOcs-HIS3 rad52::LEU2 | Derived from LSY1259 by transformation with pSM20 |
| LSY1481 | MATα 9 Ty1-HOcs-HIS3 rad52::LEU2 | Derived from LSY1264 by transformation with pSM20 |
| LSY1483 | MATα 10 Ty1-HOcs-HIS3 mre11-D56N rad52::LEU2 | Derived from LSY1285 by transformation with pSM20 |
All the strains are of the W303 genotype (leu2-3,112 trp1-1 ura3-1 can1-100 ade2-1 his3-11,15 RAD5); only the mating type and differences from this genotype are shown.
Three rounds of retrotransposition were performed before analysis.
Plasmid construction.
The relevant genetic elements of the plasmid BLR96 (15) are the Saccharomyces cerevisiae URA3 gene for selection and an active modified Ty1 element from S. cerevisiae (Fig. 1). This Ty1 is under the transcriptional control of the GAL1 promoter and contains the HIS3 gene inserted after ORFA and ORFB with the opposite transcriptional orientation. HIS3 is interrupted by an artificial intron (AI) whose splicing sites are in the same transcriptional orientation as that of Ty1. pBL001 was constructed by inserting the complementary oligonucleotides 5′CGTTTTTCTTTTAGTTTCAGCTTTCCGCAACAGTATAATTTTATAAACCCTGGTTTTGGTTTATGCATT and 5′CGAATGCATAAACCAAAACCAGGGTTTATAAAATTATACTGTTGCGGAAAGCTGAAACTAAAAGAAAAA into BLR96 at the unique ClaI site located near the stop codon of HIS3. The oligonucleotides were designed to contain 60 bp identical to the HO cut site sequence near HMRa1 (coordinates 294350 to 294409 on the chromosome III sequence), as well as an NsiI site used to confirm the presence of the insertion. The correct sequence of the HO cut site in pBL001 was confirmed by DNA sequence analysis.
FIG. 1.
Physical map of pBL001 containing the HOcs-marked Ty1 derived from the Ty1his3 AI (15). Striped arrowheads indicate Ty1 LTRs and the direction of Ty1 transcription. Open box, HIS3 gene (arrow, direction of its transcription); solid box, 104-bp AI (in antisense orientation relative to HIS3); vertical bars, relevant restriction sites (P, PvuII; N, NdeI; HO, HO cut site).
Retrotransposition of the HOcs-HIS3 Ty1 element.
pBL001 was transformed into the reference strains LSY678 and LSY679. Individual transformants were selected on synthetic complete medium minus uracil at 30°C and then streaked out for single colonies on galactose-containing synthetic complete medium minus His and incubated at 18 to 23°C. Ty1 retrotransposition was induced by the presence of galactose at low temperature and identified by the appearance of His+ colonies, indicative of the splicing of the HIS3 AI during transcription of the artificial Ty1. His+ colonies were patched onto yeast extract-peptone-dextrose (YPD) to allow the loss of pBL001 and then replica plated onto 5-FOA plates to select for its loss. Cells that grew on 5-FOA-containing medium were streaked out for single colonies onto YPD, and the presence of newly retrotransposed copies of the HIS3-marked Ty1 element was determined by DNA analysis. Genomic DNA was digested with PvuII, which has a unique cleavage site in the HIS3-marked Ty1, and the number of PvuII fragments containing HIS3 was determined by Southern analysis using a fragment of HIS3 as a probe (the 443-bp HindIII-SspI fragment from pRS403). His+ colonies analyzed contained between one and three marked Ty1 elements. To obtain strains with more marked Ty1s, the same procedure was repeated as many times as needed. The difference is that the starting cells were already His+, so the newly retrotransposed marked Ty1s could be detected only by physical analysis. Marked Ty1-containing strains were also crossed, and the spore clones obtained from tetrad dissection were analyzed by Southern blotting in order to optimize the number of these elements per strain. Strains containing between 1 and 10 marked Ty1s were generated this way. Randomness of the insertions throughout the genome was confirmed by pulsed-field gel electrophoresis (PFGE; CHEF-DR II; Bio-Rad) followed by Southern hybridization with the same HIS3 probe, as described above. Marked Ty1s on different chromosomes were detected, and quantification of the signals was in agreement with the number of insertions deduced from the PvuII analysis. The resulting strains did not exhibit obvious growth defects or sensitivity to IR (data not shown).
Physical analysis of DSB formation and resection.
Cells were transformed to Trp+ with plasmid pFH800 (47). Three hundred milliliters of cells was grown in raffinose-containing synthetic complete medium minus Trp to an optical density between 0.2 and 0.8, and 30 ml of 20% (wt/vol) galactose was added to the 270 ml of cells remaining after the removal of 30 ml of the culture for the 0-min time point. Thirty milliliters of cells was harvested by centrifugation every 30 min and washed with 1 ml of water, and the resulting pellet was frozen with liquid nitrogen. One hour after addition of galactose, the cultures were harvested by centrifugation, washed with water, and resuspended in an equal volume of rich medium containing glucose. DNA was extracted and digested with NdeI or PvuII for analysis of HO cut site-marked Ty1 elements or with BamHI and StyI for analysis of mating type switching. The resulting DNA fragments were separated by electrophoresis through 0.8% neutral or alkaline agarose gels and then transferred to nylon membranes. Membranes were hybridized with a PCR fragment generated by amplification of a 405-bp sequence distal to the HO cut site (coordinates 201173 to 201577 on chromosome III) or with HIS3. Hybridization signals were normalized to the LEU2 hybridization signal, obtained after reprobing the blots with a LEU2 fragment generated by PCR (coordinates 91371 to 92243 on chromosome III).
RESULTS
Construction and characterization of strains containing multiple HO cut sites.
To distinguish between the possibilities that (i) the Mre11 nuclease is required only to remove end adducts and (ii) it is involved in 5′ to 3′ resection but another redundant nuclease is limiting when multiple DSBs are made, we designed a system to measure 5′ to 3′ resection in the presence of one or more DSBs with identical ends. Our strategy was to generate a set of strains containing a range of HO cut sites in addition to the endogenous HO cut site present at the MAT locus. A 60-bp sequence containing the recognition site for HO (HOcs) was first cloned into a HIS3-marked Ty1 element regulated by the galactose-inducible GAL1 promoter (15) and then spread randomly throughout the genome by several cycles of retrotransposition (Table 1 and Fig. 1). After each round of transposition, individual clones were analyzed by Southern blotting to determine the copy number of the marked Ty1 element. Crosses between independently derived clones were also performed to optimize the number and random distribution of the marked Ty1 elements. We monitored the overall cleavage efficiency of the experimentally induced HO cut sites by Southern hybridization with HIS3 to probe genomic DNA cut with NdeI, which liberates an NdeI fragment internal to the HIS3-marked Ty1 encompassing the HOcs (Fig. 2). Cleavage efficiencies were calculated by determining the decrease of the intensity of the uncut band and were found to be about 40% for each strain after 1 h of HO induction (Fig. 2). To determine if cleavage by HO was proficient at all the marked Ty1 elements, we probed PvuII-digested genomic DNA with HIS3. The HIS3-marked Ty1 has a unique PvuII site, and the sizes of the fragments produced vary according to the distance between the site of the insertion and the closest genomic PvuII site. After 1 h of HO induction in a strain containing 10 marked Tys (LSY1259), the number of DNA fragments that hybridized to the HIS3 probe increased (Fig. 3), indicating that at least 6 of the 10 HO cut sites could be cut. Because two of the marked Tys are on PvuII fragments of the same size, we are unable to determine whether both are cut; two of the cut fragments comigrate with two other parental bands and thus cannot be accurately quantified, and we also cannot distinguish between the parental and cut PvuII fragments for the marked Ty present on the largest hybridizing fragment. The efficiency of cleavage was between 20 and 60% for individual HO cut sites. It is unlikely that the 60-bp sequence containing the HO cut site that we used was not long enough to allow complete cleavage by HO. Diede and Gottschling (19) obtained more-efficient cleavage with a 30-bp HO cut site, and Parket et al. (49) found an efficiency of HO cleavage similar to that described here using a 130-bp HO cut site within a Ty1 element. It is more likely that the Ty1 chromatin structure is suboptimal for HO cleavage (6).
FIG. 2.
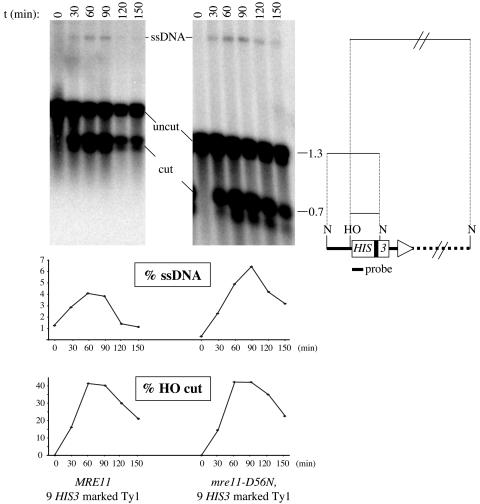
Single-stranded DNA intermediates are formed at the experimentally induced HO cut sites in MRE11 and mre11-D56N strains. DNA samples from MRE11 (LSY1264) (left) and mre11-D56N strains (LSY1299) (right) were prepared from cells undergoing a 60-min HO induction, digested with NdeI, electrophoresed under alkaline conditions, blotted onto a nylon membrane, and probed with a HIS3 fragment (see Materials and Methods). These two strains contain nine HIS3-marked Ty1s. Because splicing of the AI present in HIS3 is not an efficient process (15), some HOcs-HIS3 Ty1 elements still contain the 104-bp AI, as shown by the presence of two closely migrating bands at around 1.3 and 0.7-kb. t, time in minutes between the beginning of the HO induction and the collection of cells for DNA analysis; uncut, NdeI fragment from the HIS3-marked Ty1 containing the HO cleavage site; cut, HO-NdeI fragment overlapping with HIS3 from the HIS3-marked Ty1; ssDNA, ssDNA-containing fragment from the HO cleavage site of a particular HIS3-marked Ty1 and a downstream chromosomal NdeI cut site. (Bottom) Quantitation of the ssDNA intermediates and cut fragments for the two strains, expressed as percentages of their intensities relative to the sum of the intensities of all fragments at each time point. Symbols in the map to the right are as defined for Fig. 1.
FIG. 3.
In vivo HO cleavage of individual HO cut sites in a strain containing 10 HOcs-Ty1 elements (LSY1259). Left and right, 60- and 300-min HO induction, respectively. The arrows on the left start at the positions of the uncut parental PvuII fragments and end at the positions of the corresponding HO cut fragments (numbered 1 to 6 at the corresponding arrow). Any HO cut fragment is 1.6-kb smaller than its corresponding PvuII parental fragment. Stars indicate doublets, and the highest cut fragment comigrates with its uncut parental fragment (curved arrow). Two cut fragments comigrate with two of the smaller uncut parental fragments. As a result, only 6 cut fragments out of 10 are visible. Values are the percentages of the ratios of the intensities of the cut fragments to that of his3. t, time in minutes between the beginning of HO induction and the collection of cells for DNA analysis. Symbols in the map to the right are as defined for Fig. 1.
A DSB within a Ty1 element should be reparable by NHEJ or HR. HR could occur between the cut Ty1 and one of the approximately 30 other endogenous copies of Ty1 (33) or possibly another uncut copy of a HIS3-marked Ty1. HR could also occur by single-strand annealing between the two long terminal repeats (LTRs) of the cut Ty1, leading to the loss of the entire element except one of the LTRs. Repair by NHEJ does not require 5′ to 3′ resection of the ends of the DSBs, while HR does. Although Parket et al. (49) have shown that an HO-induced DSB in a Ty1 element induces repair by gene conversion or single-strand annealing between the LTRs, it was of primary importance to ensure that the HO-induced DSBs in the strains described here were recognized by the HR machinery. Rad52 is involved in all types of HR, but not in NHEJ. Unresected DSBs are not bound by Rad52 in vivo (24); therefore, Rad52-yellow fluorescent protein (YFP) focus formation is a marker for the repair of DSBs by HR (39). We reasoned that formation of Rad52-YFP foci in strains containing multiple HO-induced DSBs would indicate that the DSBs are processed and recognized by the HR system. Strains containing between one and nine HO cut sites were cotransformed with the HO-expressing plasmid and pWJ1344 expressing Rad52-YFP under the control of its native promoter. The Rad52-YFP foci formed in the resulting transformants were quantified after 2 h of HO induction in exponentially growing cells. A significant increase in the fraction of cells containing at least one Rad52-YFP focus and a 10-fold increase in the number of cells containing at least two distinct Rad52-YFP foci were observed in all the HOcs-HIS3 Ty1-containing strains, compared to strain LSY678, which contained only the HO cut site at the MAT locus (data not shown). We conclude that the HR machinery is recruited to some, but not necessarily all, of the HO-induced DSBs present in the marked Ty1 elements.
DSBs in Ty1 elements induce gross chromosomal rearrangements.
Two 60-min HO inductions were performed in a strain with 10 marked Ty1 elements (LSY1259), and individual cells were plated directly onto YPD medium. The karyotypes of resulting clones were analyzed by PFGE, followed by Southern hybridization with a HIS3 probe. Figure 4 shows three clones with modified karyotypes (lanes 2 to 4), as evident by an aberrant migration corresponding to either chromosome I or VI (lane 2) and chromosome XVI or XIII (lanes 3 and 4). (These pairs of chromosomes normally comigrate under the conditions used.) This aberrant migration reflects gross chromosomal rearrangements (GCRs) induced by cleavage at the experimentally introduced HO cut sites. At least 5 of the 54 HO-induced clones analyzed showed evidence of a GCR, while no GCR was observed in clones containing HOcs-HIS3 Ty1 elements that were not induced for HO. The formation of HO-induced translocation chromosomes has been reported previously (22, 28, 72). In the system described here, translocations could arise by NHEJ, HR, single-strand annealing, or possibly break-induced replication (18, 35). It is likely that the numbers presented underestimate the actual frequency of HO-induced GCRs because lethal events, such as the formation of acentric and dicentric chromosomes, would not be recovered.
FIG. 4.
Visualization of GCRs by PFGE in a strain containing 10 marked Ty1s (LSY1259) after HO induction. The cells underwent a 60-min HO induction and were then plated on the noninducing medium. Three resulting individual colonies were used to prepare the DNA plugs (lanes 2 to 4). Lane 1 corresponds to the starting strain prior to HO induction. (Left) Pulsed-field gel stained with SYBR Gold. Arrows, chromosomes with an abnormal mobility; numbers on the left, chromosome numbers. (Right) Pulsed-field gel blotted and probed with HIS3. Numbers on the left, numbers of HIS3-marked Ty1s in the corresponding band in LSY1259; *, signal from the endogenous his3 gene.
The nuclease activity of Mre11 is not required for 5′ to 3′ resection at multiple HO-induced DSBs.
HO was induced for 60 min in MRE11 (LSY1264) and mre11-D56N (LSY1299) strains, both of which contain nine HOcs-HIS3-marked Ty1 elements. To monitor ssDNA formation (resection) at the Ty1 HO cut sites, genomic DNA was digested with NdeI and the fragments were separated by alkaline agarose gel electrophoresis and then hybridized to a HIS3 probe (Fig. 2). At the 0-min time point, HIS3 hybridized to the endogenous HIS3 locus, as well as two NdeI fragments of slightly different sizes originating from HOcs-HIS3-marked Ty1s. The presence of the larger NdeI fragment revealed that some of these Ty1s did not lose their AI during retrotransposition. At the 30-min time point, two HO cut fragments appeared and were also separated on the basis of the size of the AI present in HIS3. An additional less abundant fragment appeared at this time point and was interpreted as being the result of 5′ to 3′ degradation through the NdeI site internal to HIS3 from a specific Ty1. Because NdeI cuts only duplex DNA and the 3′ ends at break sites are undegraded, 5′ to 3′resection through regions containing NdeI restriction sites can be monitored by observing the appearance of higher-molecular-weight DNA species by alkaline gel electrophoresis (58, 70). The intensity of the ssDNA intermediate peaked between the 60- and 90-min time points. The ssDNA profiles of the MRE11 and mre11-D56N strains were similar, indicating that the HO-induced DSBs are resected and, more importantly, that the nuclease activity of Mre11 is not required for 5′ to 3′ resection when multiple clean DSB ends are present. In this experiment, only one NdeI ssDNA fragment was visualized, suggesting that the ssDNA intermediate from one artificially introduced HO cut site was detectable. Because the HOcs-HIS3-marked Ty1 integrations are random, we had expected to see several other ssDNA species. Although we could have tested other restriction enzymes to identify those suitable to monitor ssDNA formation, the technical limitation is that some restriction fragments are likely to be undetectable due to their size. Thus, analysis of ssDNA formation at all of the artificially introduced HO cut sites was not straightforward. Assuming that the machinery involved in the processing of the DSB generated by HO at the MAT locus is the same as that for the other HO-induced DSBs (4), we focused on 5′ to 3′ resection at the well-characterized MAT locus (Fig. 5A).
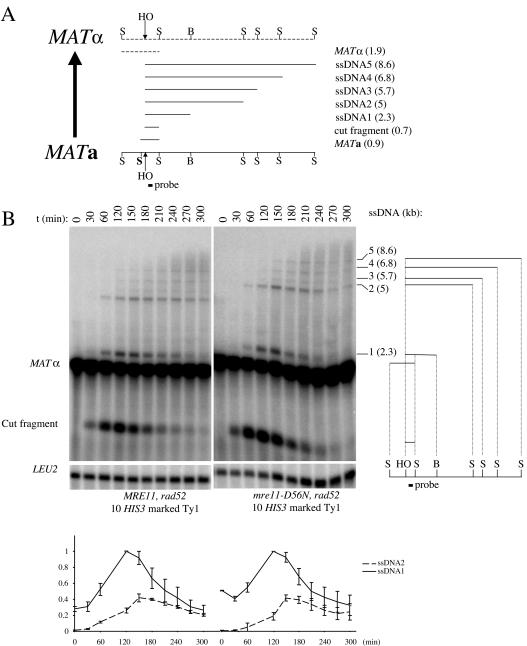
FIG. 5.
5′ to 3′ resection at the MAT locus is unaffected by mre11-D56N in the presence of additional DSBs. (A) Physical map of the MATa- MATα switching process and illustration of the DNA intermediates as observed by alkaline gel electrophoresis after a StyI-BamHI double digestion. The probe used reveals the 0.9- and 1.9-kb StyI fragments from the uncut MATa and MATα loci, respectively. When expressed, HO cuts the MAT locus to produce a smaller 0.7-kb HO-StyI cut fragment. As a result of the 5′ to 3′ resection of the right end of the HO break, some StyI and BamHI cut sites become single stranded and resistant to cleavage, generating high-molecular-weight ssDNA fragments when electrophoresed under alkaline conditions (ssDNA1 to -5). The sizes of the different DNA intermediates are indicated in parentheses. S, StyI; B, BamHI; HO, HO cut site. (B) A 60-min HO induction was performed in MRE11 (LSY1477) and mre11-D56N (LSY1483) strains, and ssDNA formation at MAT was analyzed by alkaline gel electrophoresis (see Materials and Methods). The sizes of the ssDNA species (ssDNA1 to -5) are indicated in parentheses. LEU2, hybridization control; t, time in minutes between the beginning of HO induction and the collection of cells for DNA analysis. (Bottom) Quantitation of the ssDNA intensities, normalized with the LEU2 signal and expressed as the fraction of the maximum intensity of ssDNA1 for each strain. Error bars correspond to the range of the data from two independent experiments.
All the following analyses of ssDNA formation were performed at the MAT locus. Because the ssDNA tails are relatively short, the initial analyses were performed in a rad52 mutant background, which was known to accumulate longer ssDNA tails due to the defect in strand invasion (70). ssDNA formation at the MAT locus in MRE11 (LSY1477) and mre11-D56N (LSY1483) strains, both of which contain a rad52 null allele and 10 HOcs-HIS3-marked Ty1s, was analyzed (Fig. 5B). At the 30-min time point for both strains, the HO cut fragment was present in addition to the parental fragment corresponding to the MATα allele. The shortest ssDNA intermediate was visible at 30 to 60 min and peaked at around 120 to 150 min. The second-longer ssDNA intermediate appeared at 60 min, and its intensity peaked at about 150 to 180 min. These two ssDNA intermediates were detectable until the 300-min time point. Finally, additional higher-molecular-weight ssDNA intermediates appeared at 120 min and were detectable until the 300-min time point. These discrete ssDNA intermediates were surrounded by a smeary signal, which we assume corresponds to degradation of the 3′ ends of ssDNA intermediates. Quantitation of the intensities of these intermediates is not presented because they are barely above the high background caused by the smeary signal in which they are embedded. The kinetics of appearance and disappearance of the ssDNA intermediates, as well as the amount of ssDNA observed in the mre11-D56N strain, are similar to those seen in the corresponding MRE11 strain. In conclusion, Mre11 nuclease activity is not required for the extensive 5′ to 3′ resection of DSBs in strains containing multiple DSBs. This is consistent with, and extends the conclusions made previously concerning, normal resection of a single DSB at the MAT locus in the mre11-H125N and mre11-3 strains (36, 44).
The presence of multiple DSBs in the yeast genome affects the kinetics of appearance and disappearance of the 3′-tailed ssDNA intermediates.
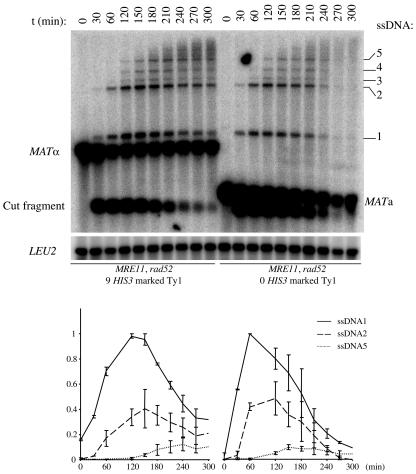
The ssDNA intermediates formed in the rad52 strain (LSY723) with only one HO break at the MAT locus were analyzed and compared to the profile observed in an isogenic strain (LSY1481) containing nine additional HO cut sites (Fig. 6). The same profile of ssDNA intermediates was observed in the two strains, but the kinetics of appearance and disappearance were different. The major difference comes from the longer persistence of the discrete ssDNA intermediates in the strain with nine additional HO cut sites, which were still detectable at 300 min, while none of them was clearly detectable at this time in the strain with one HOcs. This longer persistence of the ssDNA intermediates was also observed in the MRE11 rad52 and mre11-D56N rad52 strains, containing 10 additional HO cut sites (Fig. 5B), but is even more striking in the present experiment, where the intensities of ssDNA intermediates are greater. This longer persistence of the discrete ssDNA species could be due to a delay in the resection process and/or a stabilization of the 3′ ends of the break fragments. Starting from the 270-min time point, ssDNA intermediates were visualized only as a smear of high molecular weight in the strain with one HOcs, indicative of very long resection. Although this smear was also observed in the strain with 10 HO cut sites, other discrete ssDNA species were still visible after 270 min, especially ssDNA1, -2, and -5. A second difference comes from a delay of at least 30 min in the peaks of the intensities of the first two partially ssDNA intermediates of the strain with 10 HOcs sequences compared to the strain with one HOcs. The intensities of these two ssDNA species peaked at the 150-min time point in the strain with 10 HO cut sites and between the 60- and 120-min time points in the strain with 1 HOcs. In conclusion, the presence of multiple DSBs delays resection of the 5′ ends and stabilizes the 3′ ends of the DSBs.
FIG. 6.
Decrease in ssDNA turnover after multiple HO-induced DSBs are made. A 60-min HO induction was performed in rad52 strains containing nine (LSY1481; left) or zero (LSY723; right) HIS3-marked Ty1s. (Top) ssDNA formation at MAT was analyzed by alkaline gel electrophoresis (see Materials and Methods). LEU2, hybridization control; t, time in minutes between the beginning of HO induction and the collection of cells for DNA analysis. (Bottom) Quantitation of the ssDNA intensities, normalized with the LEU2 signal and expressed as the fraction of the maximum intensity of ssDNA1 for each strain. Error bars correspond to the range of the data from two independent experiments.
The nuclease activity of Mre11 is not required for the 5′ to 3′ resection at MAT when additional DSBs are made by phleomycin.
To investigate further the issue of altered kinetics of repair in the presence of extensive DNA damage, we used the radiomimetic agent phleomycin to induce DSBs. Two cultures each of the MRE11 strain (LSY678) and the corresponding mre11-D56N strain (LSY1032) were induced for HO expression for 60 min, and one of the two cultures of each genotype was simultaneously treated with phleomycin. Two independent experiments were performed, one using phleomycin at 100 μg/ml and one using it at 250 μg/ml. ssDNA formation at the MAT locus was analyzed every 30 min by alkaline gel electrophoresis and Southern blotting (Fig. 7). In the MRE11 strain, two ssDNA fragments were clearly visible at 60 min, with comparable intensities both in the presence and absence of phleomycin. In the absence of phleomycin, the intensities of these ssDNA species stayed close to their maximum between the 60- and 120-min time points. They then decreased progressively to become barely detectable by 270 min. In the presence of 250 μg of phleomycin/ml, the intensities of the smaller ssDNA intermediate increased significantly until 120 min but then plateaued until 270 min and then started to drop. The intensity of the second ssDNA intermediate increased until the 180-min time point and then plateaued until 300 min. Finally, we also noticed that the yield of switched product was decreased by at least a factor of two after the treatment with 250 μg of phleomycin/ml. For the corresponding mre11-D56N strain, the observations are almost completely superimposable with those made with the MRE11 strain. The same observations were made with 100 μg of phleomycin/ml, but with a less pronounced effect regarding the stability of the ssDNA species (Fig. 7) and the yield of switched product (data not shown). The accumulation of the ssDNA intermediates in the presence of multiple DSBs is not due solely to the delay in strand invasion because this phenomenon is also observed in rad52 strains treated with phleomycin, in which strand invasion does not occur (data not shown), or in rad52 strains with nine marked Ty1 elements (Fig. 6). The yield and kinetics of ssDNA intermediates were the same for MRE11 and mre11-D56N strains, similar to the results obtained with rad52 derivatives (Fig. 5), eliminating the possibility that another nuclease normally unable to process ends gains access in the absence of RAD52. As described previously (37), an mre11Δ strain showed reduced resection of an HO-induced DSB at the MAT locus, as evidenced by reduced accumulation of ssDNA1 and ssDNA2 in the absence of phleomycin (data not shown). The appearance of switched products was delayed, and the yield of switched products was less than that observed for the MRE11 and mre11-D56N strains. In the presence of phleomycin, there was an even more pronounced effect on the yield of switched product in the mre11Δ strain, compared with the MRE11strain, but there was little change in the yield of ssDNA intermediates (data not shown).
FIG. 7.
Accumulation of ssDNA intermediates at MAT after phleomycin treatment. (Top) A 60-min HO induction was performed in two equivalent cultures of MRE11 (LSY678; left two panels) and mre11-D56N (LSY1032; right two panels) strains. Phleomycin was added to one culture of each strain during the entire period of HO induction, at a final concentration of 250 μg/ml (163 μM). ssDNA formation at MAT was analyzed by alkaline gel electrophoresis (see Materials and Methods). t, time in minutes between the beginning of HO induction and the harvest of the corresponding cells. Values for time zero for LSY678 with no phleomycin are not plotted because of partial DNA degradation. (Bottom) Quantitation of the ssDNA intensities, normalized with the LEU2 signal and expressed as the fraction of the maximum intensity of ssDNA1 for each strain. Error bars correspond to the range of the data from two independent experiments for mre11-D56N and the standard deviations of three independent experiments for MRE11.
In parallel, the killing effect of phleomycin was estimated by plating dilutions of the cultures at different time points. There was a 30-fold reduction in survival of both the MRE11 and mre11-D56N strains when treated with 100 μg of phleomycin/ml for 60 min, and about a 1,000-fold reduction in survival when the strains were exposed to 250 μg of phleomycin/ml (data not shown). The equivalence of the phleomycin sensitivities of these two strains was also confirmed by a halo assay using several different concentrations of phleomycin (data not shown). Thus, the Mre11 nuclease activity is not required for the removal of the phosphoglycolate residues present at the 3′ ends of the DSBs generated by phleomycin (53).
The extensive resection of DSBs is partially dependent on EXO1.
The results presented above indicate that extensive 5′ to 3′ resection of DSBs occurs in the absence of the Mre11 nuclease. An obvious candidate for a redundant activity is Exo1, which was identified biochemically by its ability to generate 3′ single-strand tails from linear duplex DNA (23, 61). Previous studies showed no defect in mating type switching in exo1 mutants, but ssDNA intermediates were not analyzed (45, 65). We observed a slight delay in the appearance of ssDNA intermediates at the HO cut site at the MAT locus in the exo1 mutant, but the most striking observation was that the intermediate corresponding to ssDNA2 was fourfold less abundant than in the wild-type strain (Fig. 8), indicating a delayed and reduced resection in the exo1 strain.
FIG. 8.
Decreased resection of the HO-induced DSB at the MAT locus in exo1 mutants. Two 60-min HO inductions were performed in exo1 strains (LSY894-2B and LSY894-4C), and the formation of ssDNA intermediates generated was detected by alkaline gel electrophoresis. The amounts of ssDNA1 and ssDNA2 shown are expressed as the fractions of the maximum intensity of ssDNA1 for each strain. Error bars correspond to the range of the data from two independent experiments for exo1 and the standard deviations of three independent experiments for EXO1.
DISCUSSION
The Mre11 nuclease is not required to resect DSBs made by HO endonuclease.
The Mre11 complex is thought to function in the 5′ to 3′ resection of DSBs either directly via the Mre11 nuclease activity or indirectly by the recruitment of a dedicated nuclease. Previous studies have shown a requirement for the Mre11 nuclease in the processing of meiosis-specific DSBs and for a subset of IR-induced DNA damage, but not in resection of a single HO-induced DSB at the MAT locus (36, 44). One interpretation of these results is that a nuclease redundant with Mre11 is limiting in the presence of multiple DSBs. To test this hypothesis, we investigated the influence of multiple DSBs on the 5′ to 3′ resection of the DSB made by HO endonuclease at the MAT locus in MRE11 and Mre11 nuclease-defective (mre11-D56N) strains. Strains with up to 10 HO cut sites dispersed throughout the genome were made by retrotransposition of a modified Ty1 element containing an HO cut site. HO endonuclease was transiently expressed in these strains, and ssDNA resection of the breaks was monitored by determining the resistance to digestion with restriction enzymes. Resection was unaffected in the mre11-D56N strain compared with the MRE11 strain, even in the presence of multiple DSBs, suggesting that the Mre11 nuclease is not required for extensive resection of clean DSBs.
Because of the limited number of integration events of the HOcs-Ty1 element and the inefficiency of HO cleavage, phleomycin was used to generate a larger number of DSBs. As no difference in 5′ to 3′ resection of the HO-induced break at the MAT locus between the MRE11 and mre11-D56N strains after phleomycin treatment was observed, we conclude that Mre11 nuclease activity is not essential for the 5′ to 3′ resection process, and therefore that the IR sensitivity of the mre11-D56N mutant is not due to a defect in resecting numerous DSBs. This extends previous observations showing that the kinetics of mating-type switching are unaffected in mre11-nd mutants (36, 44). To explain the hypersensitivity to IR of the mre11-nd mutants, we propose that the Mre11 nuclease processes some types of potentially lethal DNA ends that specifically arise after IR treatment. It is also formally possible that the stability of the MRX complexes is slightly reduced in mre11-nd mutants, resulting in reduced efficiency of some other nuclease-independent functions of the complex.
Substrates for the Mre11 nuclease.
The mre11-D56N mutant does not exhibit hypersensitivity to phleomycin, whereas an mre11 null mutant does (data not shown), suggesting that the 3′ DNA ends bound to a phosphoglycolate group generated by phleomycin (53) can be processed by a mechanism that does not require Mre11 nuclease activity. IR generates DSBs with more complex ends, with phosphate or phosphoglycolate groups bound to the 3′ DNA ends (30). Lesions to the bases or sugar residues surrounding the DSBs also occur and are referred to as multiply damaged sites (MDS) (68, 69). We postulate that MDS created by IR are substrates for the Mre11 nuclease.
Topoisomerases form transient covalent intermediates with DNA, which can be trapped by topoisomerase poisons. Tdp1 is specifically required to remove Top1 from the 3′ ends of the DNA when trapped by camptothecin (52). We tested the camptothecin sensitivity of the mre11-D56N mutant and found a 10-fold decrease in survival compared to that of a wild-type strain using 10 μg of camptothecin/ml (data not shown). This hypersensitivity to camptothecin suggests that the nuclease activity of Mre11 can act on some DNA ends that are blocked by a covalently bound protein. During meiosis, the topoisomerase II-like protein Spo11 creates DSBs (8, 32). In mre11-nd mutants, Spo11 remains covalently bound to the 5′ DNA ends and no 5′ to 3′ resection takes place (25, 44, 66, 67). This strongly suggests that the nuclease activity of Mre11 can remove covalently bound protein from the 5′ DNA ends. This hypothesis is reinforced by the study of Connelly et al. (12), who recently showed that the endonuclease activity of the SbcCD complex, homologous to the Rad50-Mre11 complex, plays a direct role in freeing covalently bound protein from DNA ends in vitro (14). Finally, note that the SbcCD, Rad50-Mre11, and MRX complexes cleave DNA hairpin structures in vitro (13, 63, 64) and are implicated in processing similar structures in vivo (17, 40, 54, 71).
Which nucleases are responsible for the 5′ to 3′ resection at DSBs?
The mre11Δ and mre11-D56N mutations confer different phenotypes with respect to DSB resection. The mre11Δ strain shows delayed and reduced accumulation of ssDNA intermediates (37), whereas the yield and kinetics of appearance of ssDNA intermediates in the mre11-D56N strains are indistinguishable from those for the wild type (Fig. 2, 5, and 7). These observations suggest that the MRX complex recruits a resection nuclease and that this nuclease is still recruited by the Mre11-D56N-Rad50-Xrs2 complex, or that binding of the MRX complex (or the Mre11-D56N complex) to DSBs provides the appropriate environment for resection. If the MRX complex does recruit a nuclease, it is unlikely to be Exo1 because the exo1 mutation alone confers a subtle defect in resection (Fig. 8) and is synergistic with mre11Δ for IR and methyl methanesulfonate sensitivity (38, 45, 65). Physical analysis of DSB repair in the exo1 mre11Δ double mutant is complicated by the elevated cell death in this strain; about 70% of the cells in log-phase culture are inviable (38, 45). However, the synthetic interaction between exo1 and mre11Δ and the partial suppression of IR sensitivity, MMS sensitivity, and delayed mating type switching of mre11Δ mutants due to EXO1 in high copy number are consistent with a role for Exo1 in processing DSBs in vivo (36, 38, 45, 65). Furthermore, the processivity of the resection of meiotic DSBs and of uncapped telomeres is impaired in exo1 mutants (29, 42, 65). The synergism between exo1 and mre11 for IR sensitivity is also observed for mre11-nd alleles (mre11-D56N and mre11-H125N), but the exo1 mre11-nd double mutants have normal growth rates, and the kinetics of mating type switching are indistinguishable from those for the wild type (45). If the Mre11 nuclease was involved in processing DSBs and was fully redundant with Exo1, the double mutant would be predicted to exhibit a stronger defect in DSB repair. EXO1 present in high copy number does not suppress the IR sensitivity of mre11-nd strains, consistent with the hypothesis that the Mre11 nuclease is uniquely required to process MDS created by radiolysis and that clean ends are efficiently processed by other nucleases, including Exo1 (45).
Exo1 possesses structural motifs characteristic of a large family of 5′ to 3′ nucleases, including the S. cerevisiae Rad2, Rad27, Din7, and Yen1 proteins. We observed shorter 3′ ssDNA tails at the HO cut site in exo1 null mutants (Fig. 8), but not in a rad27 mutant (data not shown). A strain containing din7, rad2, exo1, and yen1 mutations had the same defect in ssDNA formation as the exo1 single mutant, indicating that the other nucleases (or putative nucleases) in this family are not redundant with Exo1. Certain mutant combinations within the RAD2 family and with mre11 have not been tested because of the synthetic growth defects of the double mutants; these include exo1 rad27, exo1 mre11, and mre11 rad27 (44, 45, 62). A defect in 5′ to 3′ resection is expected to lead to a hypersensitivity to IR, and exhaustive screens to look for single viable mutants hypersensitive to IR have been performed, but none of the mutants identified in these screens appeared to be good candidates for involvement in the 5′ to 3′ resection process (7). Therefore, it seems likely that the 5′ to 3′ resection of DNA ends is performed by several partially redundant players, Exo1 being one of them, or, alternatively, that an essential protein could be responsible for 5 to 3′ resection.
Recently, Aylon and Kupiec (3) showed that 5′ to 3′ resection of an HO break is delayed in a rad24 mutant. Rad24 is thought to associate with Rfc2-5 to form an alternative clamp loader protein complex, and the homologous complex from humans binds preferentially to 5′ recessed DNA ends, suggesting a specific role in the HR pathway (21). No nuclease activity has been demonstrated for the Rad24 complex, suggesting that it acts indirectly in DSB resection. Interestingly, we also observed a delayed and protracted resection after phleomycin treatment, suggesting that Rad24 may become limiting in the cell in the presence of numerous DSBs.
Comparison of phleomycin treatment and the action of Spo11.
Neale et al. (46) showed that end resection of a DSB created by the naturally occurring VMA1-derived endonuclease (VDE) during meiosis is affected by the levels of Spo11-induced DSBs. A decrease in the number of Spo11-induced DSBs resulted in an increase in the lengths of 3′ ssDNA tails at the VDE-induced DSB, consistent with the increase in gene conversion tract length observed at a meiotically induced HO break in a spo11 null mutant (41). One possible explanation for this observation is that meiotic DSBs compete for the resection machinery and consequently the rate of 5′ to 3′ resection of meiotic DSBs would be directly affected by the number of DSBs. Because Spo11-induced breaks require Mre11 for processing, the delay could be in part due to titration of the Mre11 complex. Another possibility is that Spo11 or Spo11-induced DSBs trans-regulate the resection of meiotic DSBs.
In this study, we have addressed the influence of the number of DSBs on the 5′ to 3′ resection of an HO-induced DSB in mitotic cells. We found a slight delay in the formation of ssDNA intermediates, indicative of reduced processivity of resection in the presence of additional HO cuts or phleomycin. We favor the hypothesis that this is due to partial saturation of the resection apparatus. Alternatively it could result from a partial saturation of the end-joining machinery, leaving more substrates available for resection, resulting in an increase in the ssDNA signal; however, this does not explain the delay or increased stability of these intermediates. The effect of multiple DSBs on resection in mitotic cells is much less than that observed during meiosis (46). If the same players are responsible for resection of meiotic DSBs, this would suggest that the meiotic resection process is regulated mostly in trans by Spo11 or Spo11-induced DSBs.
Finally, we also observed a partial saturation of the strand invasion machinery after phleomycin treatment because less repaired product was formed (Fig. 7). This saturation may be due to the limited quantity of Rad51 in mitotic cells, as suggested by Sugawara et al. (57), or to limiting amounts of other factors that act with Rad51 to promote strand invasion. Saturation of the strand invasion machinery cannot be the only cause for the longer half-lives of ssDNA species observed when multiple DSBs are present because they are also observed in a rad52 background where no strand invasion can take place (Fig. 6 and our unpublished results).
Acknowledgments
We are grateful to members of the Symington laboratory for stimulating discussions and L. Langston and W. K. Holloman for critical reading of the manuscript. We thank D. Schild, D. Garfinkel, J. Nickoloff, R. Rothstein, and M. Lisby for gifts of plasmids and strains. We thank M. Lisby for help with microscopy and R. Reid for assistance with PFGE. Camptothecin was a gift from R. Reid.
This research was supported by Public Health Service grant GM41784 from the National Institutes of Health and a postdoctoral fellowship from l'ARC (B.L.).
REFERENCES
- 1.Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419-436. [DOI] [PubMed] [Google Scholar]
- 2.Amundsen, S. K., and G. R. Smith. 2003. Interchangeable parts of the Escherichia coli recombination machinery. Cell 112:741-744. [DOI] [PubMed] [Google Scholar]
- 3.Aylon, Y., and M. Kupiec. 2003. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol. Cell. Biol. 23:6585-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aylon, Y., B. Liefshitz, G. Bitan-Banin, and M. Kupiec. 2003. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23:1403-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartsch, S., L. E. Kang, and L. S. Symington. 2000. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Aroya, S., P. A. Mieczkowski, T. D. Petes, and M. Kupiec. 2004. The compact chromatin structure of a Ty repeated sequence suppresses recombination hotspot activity in Saccharomyces cerevisiae. Mol. Cell 15:221-231. [DOI] [PubMed] [Google Scholar]
- 7.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 8.Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414-417. [DOI] [PubMed] [Google Scholar]
- 9.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 11.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 12.Connelly, J. C., E. S. de Leau, and D. R. Leach. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair 2:795-807. [DOI] [PubMed] [Google Scholar]
- 13.Connelly, J. C., L. A. Kirkham, and D. R. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 95:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connelly, J. C., and D. R. Leach. 2004. Repair of DNA covalently linked to protein. Mol. Cell 13:307-316. [DOI] [PubMed] [Google Scholar]
- 15.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darlow, J. M., and D. R. Leach. 1998. Evidence for two preferred hairpin folding patterns in d(CGG).d(CCG) repeat tracts in vivo. J. Mol. Biol. 275:17-23. [DOI] [PubMed] [Google Scholar]
- 18.Davis, A. P., and L. S. Symington. 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 20.Dolganov, G. M., R. S. Maser, A. Novikov, L. Tosto, S. Chong, D. A. Bressan, and J. H. Petrini. 1996. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol. Cell. Biol. 16:4832-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellison, V., and B. Stillman. 2003. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 1:E33. [Online.] http://biology.plosjournals.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fasullo, M., P. Giallanza, Z. Dong, C. Cera, and T. Bennett. 2001. Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics 158:959-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiorentini, P., K. N. Huang, D. X. Tishkoff, R. D. Kolodner, and L. S. Symington. 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 17:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank-Vaillant, M., and S. Marcand. 2002. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell 10:1189-1199. [DOI] [PubMed] [Google Scholar]
- 25.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Game, J. C., and R. K. Mortimer. 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24:281-292. [DOI] [PubMed] [Google Scholar]
- 27.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 28.Haber, J. E., and W. Y. Leung. 1996. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc. Natl. Acad. Sci. USA 93:13949-13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hackett, J. A., and C. W. Greider. 2003. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 23:8450-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henner, W. D., L. O. Rodriguez, S. M. Hecht, and W. A. Haseltine. 1983. Gamma ray induced deoxyribonucleic acid strand breaks. 3′ glycolate termini. J. Biol. Chem. 258:711-713. [PubMed] [Google Scholar]
- 31.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375-384. [DOI] [PubMed] [Google Scholar]
- 33.Kim, J. M., S. Vanguri, J. D. Boeke, A. Gabriel, and D. F. Voytas. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8:464-478. [DOI] [PubMed] [Google Scholar]
- 34.Kironmai, K. M., and K. Muniyappa. 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2:443-455. [DOI] [PubMed] [Google Scholar]
- 35.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, S. E., D. A. Bressan, J. H. Petrini, and J. E. Haber. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair 1:27-40. [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 38.Lewis, L. K., G. Karthikeyan, J. W. Westmoreland, and M. A. Resnick. 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160:49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lisby, M., R. Rothstein, and U. H. Mortensen. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98:8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 41.Malkova, A., F. Klein, W. Y. Leung, and J. E. Haber. 2000. HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc. Natl. Acad. Sci. USA 97:14500-14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neale, M. J., M. Ramachandran, E. Trelles-Sticken, H. Scherthan, and A. S. Goldman. 2002. Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Mol. Cell 9:835-846. [DOI] [PubMed] [Google Scholar]
- 47.Nickoloff, J. A., E. Y. Chen, and F. Heffron. 1986. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl. Acad. Sci. USA 83:7831-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 49.Parket, A., O. Inbar, and M. Kupiec. 1995. Recombination of Ty elements in yeast can be induced by a double-strand break. Genetics 140:67-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pastwa, E., R. D. Neumann, K. Mezhevaya, and T. A. Winters. 2003. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiat. Res. 159:251-261. [DOI] [PubMed] [Google Scholar]
- 51.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 52.Pouliot, J. J., K. C. Yao, C. A. Robertson, and H. A. Nash. 1999. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286:552-555. [DOI] [PubMed] [Google Scholar]
- 53.Povirk, L. F., Y. H. Han, and R. J. Steighner. 1989. Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5′ extensions. Biochemistry 28:5808-5814. [DOI] [PubMed] [Google Scholar]
- 54.Rattray, A. J., C. B. McGill, B. K. Shafer, and J. N. Strathern. 2001. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharples, G. J., and D. R. Leach. 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17:1215-1217. [DOI] [PubMed] [Google Scholar]
- 56.Strunnikov, A. V. 1998. SMC proteins and chromosome structure. Trends Cell Biol. 8:454-459. [DOI] [PubMed] [Google Scholar]
- 57.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 58.Sun, H., D. Treco, and J. W. Szostak. 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64:1155-1161. [DOI] [PubMed] [Google Scholar]
- 59.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Symington, L. S., L. E. Kang, and S. Moreau. 2000. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 28:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szankasi, P., and G. R. Smith. 1992. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 267:3014-3023. [PubMed] [Google Scholar]
- 62.Tishkoff, D. X., A. L. Boerger, P. Bertrand, N. Filosi, G. M. Gaida, M. F. Kane, and R. D. Kolodner. 1997. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA 94:7487-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trujillo, K. M., D. H. Roh, L. Chen, S. Van Komen, A. Tomkinson, and P. Sung. 2003. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J. Biol. Chem. 278:48957-48964. [DOI] [PubMed] [Google Scholar]
- 64.Trujillo, K. M., and P. Sung. 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276:35458-35464. [DOI] [PubMed] [Google Scholar]
- 65.Tsubouchi, H., and H. Ogawa. 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol. Cell 11:2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsubouchi, H., and H. Ogawa. 1998. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 18:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 68.Ward, J. F. 2000. Complexity of damage produced by ionizing radiation. Cold Spring Harbor Symp. Quant. Biol. 65:377-382. [DOI] [PubMed] [Google Scholar]
- 69.Ward, J. F. 1981. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat. Res. 86:185-195. [PubMed] [Google Scholar]
- 70.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu, J., K. Marshall, M. Yamaguchi, J. E. Haber, and C. F. Weil. 2004. Microhomology-dependent end joining and repair of transposon-induced DNA hairpins by host factors in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:1351-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, X., and A. Gabriel. 2004. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics 166:741-751. [DOI] [PMC free article] [PubMed] [Google Scholar]