Abstract
Significant knowledge about glucocorticoid signaling has accumulated, yet many aspects remain unknown. We aimed to discover novel factors involved in glucocorticoid receptor regulation that do not necessarily require direct receptor interaction. We achieved this by using a functional genetic screen: a stable cell line which cannot survive hormone treatment was engineered, randomly mutated, and selected in the presence of glucocorticoid. A hormone-resistant clone was analyzed by two-dimensional gel electrophoresis. Differentially expressed proteins were identified and tested as candidates for regulation of the glucocorticoid receptor. An unexpected candidate, cofilin 1, inhibited receptor activity. Cofilin is known to promote actin depolymerization and filament severing. Several experiments suggest that this feature of cofilin is involved in its inhibitory action. Both its actin depolymerization activity and its inhibitory action on the receptor are dependent on its phosphorylation state. Treatment of cells with a cytoskeleton-disrupting agent decreased receptor activity, as did overexpression of actin, particularly a mutant actin that does not polymerize. In addition, overexpression of cofilin and actin as well as chemical cytoskeleton disruption changed the subcellular receptor distribution and upregulated c-Jun, which could constitute the inhibitory mechanism of cofilin. In summary, cofilin represents a novel factor that can cause glucocorticoid resistance.
The glucocorticoid receptor (GR) belongs to the superfamily of ligand-modulated transcription factors (5) which regulate gene transcription by activation as well as by repression (3). GR is activated upon binding of glucocorticoids (GCs) in various tissues and is involved in many physiological and developmental processes (11, 51). GC resistance plays a central role in a wide variety of diseases (9).
Intense research over the last decades revealed many mechanistic details of GR signaling. In the absence of hormone, GR is bound in the cytosol by a complex termed the foldosome, which consists of chaperones and cochaperones that are required for the high hormone binding affinity of the receptor (22). After binding of the hormone, the receptor presumably dissociates from the foldosome and translocates to the nucleus, reportedly along cytoskeletal tracks (42). GR activates or represses transcription by binding to its cognate DNA elements (GC-responsive elements or negative GC-responsive elements, respectively) or decreases transcription by interaction with other transcription factors such as AP-1 or NF-κB (3, 25). These actions are modulated by an array of coactivators and corepressors which regulate the structure of chromatin and the recruitment of the basal transcriptional machinery (44).
Many of the regulatory factors of GR have been identified initially by screens which are based on physical interaction of the respective factor with GR, e.g., by coimmunoprecipitation or yeast two-hybrid systems (8, 14, 18, 20, 21, 55, 57). The aim of this study was to carry out a functional screen for factors which influence GR function by using GC-resistant cells. Some functional screens with different designs had already been performed before. One carried out with Saccharomyces cerevisiae resulted in the identification of two ligand effect modulators, LEM3 and LEM4 (47). Three studies employed different techniques to create GC-resistant cell lines, but no specific factors were identified or characterized as regulators of GR (15, 16, 43). Another screen resulted in the identification of GR mutations that cause reduced hormone binding (28).
We set out to establish a screen in a human cell line that has the potential to reveal not only functional mutations in GR but also factors that regulate GR, with or without direct physical interaction. We adapted a screen which has been very successful in identifying regulatory factors for alpha interferon (33, 40), gamma interferon (33), and interleukin-1 (29) signal transduction. In principle, we used herpes virus thymidine kinase (TK) driven by the GC-responsive mouse mammary tumor virus (MMTV) promoter as the selection marker. MMTV promoter-driven green fluorescent protein (GFP) and puromycin N-acetyltransferase (PAC; mediates puromycin resistance) served as additional control markers. These markers were stably integrated into HeLa cells which express GR endogenously. After random mutagenesis and negative selection against TK expression, GC-unresponsive cells could be isolated. We used a proteomic approach, i.e., two-dimensional (2-D) gel electrophoresis and mass spectrometry, to compare the parental cell line with one of the mutant cell lines. Several differentially expressed proteins could be detected on the gel, and four of them could be identified as reticulocalbin, thioredoxin, hsp27, and cofilin 1. To find out whether one of these candidates indeed regulates GR, we isolated and cloned the corresponding genes for further analysis. Thioredoxin has been reported as GR regulatory protein under oxidative stress (31), but we found no effect under normal conditions. The candidates hsp27 and reticulocalbin did not affect GR either, whereas cofilin 1 turned out to be an inhibitor of GR. Cofilin is an actin-depolymerizing protein and is thus involved in the organization of the cytoskeleton, but it has never been associated before with the function of GR or any other nuclear receptor. Based on several approaches, we show strong evidence that the actin depolymerization activity is involved in the mechanism of GR inhibition. First, the GR inhibitory activity of cofilin depends on its phosphorylation status, like its actin depolymerization function. Second, overexpression of actin reduces GR activity, in particular, overexpression of a mutant which is reported not to polymerize. Third, the cytoskeleton-disrupting drug cytochalasin B also impaired GR function. Intriguingly, all of these treatments that sever actin filaments also changed the cytoplasmic and nuclear distribution of the receptor. Moreover, they led to the overexpression of the established GR inhibitor c-Jun, offering an explanation for the inhibitory action of cofilin.
MATERIALS AND METHODS
Plasmids.
Vectors pCMV-βGal, MTV-luc, pRK7GR, flag-GR, GFP-GR, and pRK5MCS have been described previously (56). The vectors coll-Luc (collagenase promoter region from −573 to +63 linked to luciferase) and NF-κB-Luc (6 NF-κB sites linked to luciferase) were a kind gift from Stefanie Heck. pGEXGR encoding the glutathione S-transferase (GST)-tagged GR was a kind gift from Dietmar Spengler. For cloning of MMTV-GFP, the MMTV promoter was amplified from the MTV-luc plasmid (19), with a 5′ AseI site and a 3′ NheI site. From the pEGFP-C1 vector (Clontech), the cytomegalovirus (CMV) promoter was cut out with AseI and NheI and exchanged with the amplified MMTV promoter. For MMTV-TK, the TK sequence was cut out of plasmid pPNT (a kind gift from W. Wurst) with HincII and HindIII and inserted into MMTV-GFP cut with EcoR47III and HindIII. MMTV-puroR was constructed by amplification of the MMTV promoter from MTV-luc with a 5′ HindIII site and a 3′ PstI site and insertion downstream of the CMV promoter of the vector pRK5MCS. For pRK5cof, the cofilin cDNA was amplified from total RNA with 5′-TCC CTC GAG ACA TGG CCT CCG GTG TGG TCC GGA-3′ for the 5′ end and 5′-TCC TCA CAA AGG CTT GCC CTC C-3′ for the 3′ end, adding an XhoI site and a BamHI site at the respective ends for insertion into pRK5MCS or pEGFP-C1 (Clontech). For pRK5thio, cDNA was amplified from total RNA with 5′-TCC CTC GAG ATG GTG AAG CAG ATC GAG AG-3′ and 5′-TCC GGA TCC TTA GAC TAA TTC ATT AAT GGT GG-3′ and cloned into pRK5MCS like cofilin. For pRK5actin, cDNA was amplified from total RNA with 5′-TCC CTC GAG ATG GAT GAT GAT ATC GCC GC-3′ and 5′-TCC GGA CTA GAA GCA TTT GCG GTG GA-3′ and cloned into pRK5MCS like cofilin. Mutant and flag-tagged forms of cofilin and actin were cloned analogously.
Cell culture, stable transfection, and cell fusion.
All cell lines were kept in Dulbecco's modified Eagle medium completed with 10% normal or charcoal-stripped fetal calf serum. For creating the parental clone, cells were transfected by using ExGen (Fermentas) according to the manufacturer's instructions with 2 μg of each construct. One day after transfection, G418 (1,000 μg/ml) was added to the medium. After 16 days, single colonies could be isolated. For the cell line stably expressing cofilin, cells were transfected as described above except that 6 μg of pRK5cof was used and selected in puromycin (5 μg/ml). For cell fusion, mut1 and HeLa MTV-luc (containing a puromycin resistance cassette) were seeded together in 60-mm-diameter dishes. At 80% confluence, medium was removed and 1 ml of 50% polyethylene glycol solution (Hybri Max; Sigma) was added for 1 min followed by 4 washes with 0.3, 0.6, 1.2, and 2.4 ml of medium for 5 min. Cells were left to recover overnight, and then selection was carried out first with 3 μg of puromycin/ml for 4 days and then with 1,000 μg of G418/ml for 7 days.
Mutagenesis and selection.
Parental1 cells were grown in 150-mm2 flasks and treated with 1 to 5 μg of ICR191/ml or 50 to 200 μg of ethylnitrosourea (ENU)/ml for 3 and 24 h, respectively. Cells were washed twice with phosphate-buffered saline (PBS). Conditions were chosen so that between 70 and 90% of the cells were killed. After recovery, the cells were subjected to another 2 to 5 rounds of mutagenesis. For selection, cells were seeded a 1 cell/well in 96-well plates and grown for at least 7 days. Ganciclovir (10 μg/ml) and 1 μM dexamethasone (dex) were added to the medium. Medium without hormone and antibiotic was added every 3 to 4 days for 24 h. Cells were selected for 2 to 3 weeks and then tested for hormone resistance.
MTT assay.
Cells were seeded at 1,000 cells per well in 96-well plates and treated with 1 μM dex or ethanol combined with ganciclovir (0, 10, 20, 30, 50, or 100 μg/ml) or puromycin (0, 1, 2, 3, 5, or 10 μg/ml) for 7 or 3 days, respectively. MTT assays were performed as described previously (1).
RT-PCR.
Total RNA was prepared from 105 to 107 cells by using Trizol reagent (Gibco-BRL) according to the manufacturer's instructions. The reverse transcriptase (RT) reaction was carried out with murine leukemia virus RT (NEB). Two microliters of the RT reaction was used for PCR. PCR conditions were 94°C for 45 s, 55°C for 45 s, and 72°C for 2 min for 24 cycles. The following primers were used: MT forward, ATG GAT CCC AAC TGC TCC TGC G, and reverse, AGG GCT GTC CCA ACA TCA GGC; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, ACC ACA GTC CAT GCC ATC AC, and reverse, TCC ACC ACC CTG TTG CTG TA. PCR products were separated on a 2% agarose gel and stained with ethidium bromide. The bands corresponding to metallothionein I (MT-I) (250 bp) and GAPDH (490 bp) were quantified by using a gel imaging system (Image Station 440CF and 1-D Image Analysis software; Kodak, Rochester, N.Y.). The intensities of the MT-I bands were normalized to the GAPDH band intensities.
Transient transfection, luciferase, and β-galactosidase assays.
HeLa cells (2 × 107) or HEK cells (1 × 107) were resuspended in 400 μl of electroporation buffer (50 mM K2HPO4, 20 mM potassium acetate, 25 mM MgSO4 [pH 7.35]). Three micrograms of reporter plasmid MTV-Luc or 5 μg of coll-luc or 6× NF-κB-luc, 2.5 μg of β-galactosidase expression vector pCMVβ-Gal (Stratagene), 10 ng of pRK7GR (HEK cells only), and various amounts of pRK5thio, pRK5cof, pRK5actin, pRK5G13R, or empty expression vector were added as indicated. Transfection was performed with an electroporation system (Genepulser II; Bio-Rad) at 350 V and 700 μF. Electroporated cells were replated and cultured for 16 to 48 h in fresh medium containing 5% steroid-free fetal calf serum and vehicle, hormone, or phorbol myristate acetate. For treatment with cytochalasin B, only HEK cells were used, as the overall transcriptional activity in HeLa cells was heavily affected by this drug. After transfection, cells were left to recover overnight, incubated with 50 μg of cytochalasin B/ml for 1 h, and stimulated with 1 μM dex for another 24 h in the continuous presence of cytochalasin B.
Luciferase and β-galactosidase assays were performed as described before (17). All experiments were carried out in duplicates.
Immunoprecipitation and Western blotting.
Immunoprecipitation and immunoblotting were performed as described previously (56). The following primary antibodies and dilutions were used: anti-GR H-300, 1:2,000; anti-GFP B-2, 1:1,000; anti-p-cofilin (Ser3)-R, 1:2,000; anti-hsp90 H-114, 1:2,000; anti-actin I-19, 1:1,000 (all Santa Cruz); anti-cofilin 1, 1:1,000 (Cytoskeleton, Inc., Denver, Colo.); anti-c-Jun, 1:500 (Transduction Laboratories).
GST pull-down assays.
GST-GR was expressed in DH5α and purified by using glutathione-Sepharose (Amersham) according to the manufacturer's instructions. In vitro-translated [35S]cofilin or [35S]luciferase as a negative control was added to the immobilized GR overnight, and the eluates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were exposed to an X-ray film overnight.
2-D gel electrophoresis and mass spectrometry.
Cells (5 × 107) were lysed in urea-thiourea buffer {2 M thiourea, 7 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 0.2% Biolyte [Bio-Rad], 100 mM dithiothreitol, bromophenol blue} containing Complete protease inhibitor cocktail (1:25; Roche), 1 mM phenylmethylsulfonyl fluoride, and 1 μg of pepstatin/ml for 1 h on ice and centrifuged at 13,000 rpm (Biofuge Fresco centrifuge; Heraeus) and 4°C for 1 h. The protein concentration was determined with the Amersham Plus One 2-D Quant kit. Separation in the first dimension was performed in a PROTEAN isoelectric focusing (IEF) cell (Bio-Rad) with 300 μg of total protein on 17-cm ReadyStrip IPG strips (pH 4 to 7; Bio-Rad) according to the manufacturer's instructions with active rehydration and IEF at 10,000 V until 60,000 V · h. The second-dimension separation was carried out by SDS-PAGE according to the Laemmli method on 12% acrylamide gels of 20.5-cm height. Gels were stained with colloidal Coomassie brilliant blue G (Sigma) and compared with PDQuest 2-D gel analysis software (Bio-Rad). Spots of interest were excised, and proteins were cleaved with Promega sequencing grade trypsin. Peptides were separated by high-performance liquid chromatography on a nano column (RP-C18, 75-μm inner diameter by 10 cm) and analyzed by online tandem mass spectrometry with an ion trap (LCQ Deca Xplus; Thermo Electron, San José, Calif.). The spectra were subsequently searched against a nonredundant FASTA protein database.
Fluorescence analysis.
HeLa cells were seeded on glass plates covered with 0.1% gelatin in a six-well plate (∼2 × 105 cells/well) in steroid-free medium without phenol red. After 1 day, they were transfected by using ExGen (Fermentas) according to the manufacturer's instructions. GFP-GR (0.25 μg) and 0.75 μg of pRK5cof, pRK5actin, pRK5G3R, or empty vector were used for each well. For the investigation of mut1 and c1, only 1 μg of GFP-GR was transfected. At 24 h after transfection, cells were treated with hormone for different times or with cytochalasin B for 1 h and then fixed with 4% paraformaldehyde (PFA). After 20 min, cells were washed twice with 1× Tris-buffered saline-Tween and embedded in ProTaqs Mount Fluor (Biocyc GmbH & CoKG, Luckenwalde, Germany). Cells were analyzed with a fluorescence microscope (Axioplan 2 imaging; Zeiss, Jena, Germany). To score the cytoplasmic and nuclear distribution of the fluorescing receptors, more than 100 cells were evaluated according to a method adapted from the literature (13). A value of 0 was assigned to a cell with a balanced distribution, +1 was assigned for a cell with enhanced nuclear fluorescence (−1 for a cell with enhanced cytoplasmic fluorescence), and +2 was assigned for a cell with exclusively nuclear fluorescence (−2 for a cell with exclusively cytoplasmic fluorescence). Alternatively, the pixel density of the cytoplasm and nucleus in confocal microscope pictures (LSM 510 META NLO; Carl Zeiss) was determined by using the program Image J (Wayne Rasband, National Institutes of Health, Bethesda, Md.), and the cytoplasm to nucleus ratio was calculated. This time-consuming method was applied only to selected conditions to validate our approach by visual inspection described above.
For staining of the actin cytoskeleton, cells were transfected with 0.75 μg of enhanced GFP (EGFP)-tagged cofilin or empty vector, fixed with 4% PFA, and permeabilized by treatment with cold acetone and a 10-min incubation with 0.1% Triton X-100 in PBS. Actin was stained with 50 μg of tetramethyl rhodamine isothiocyanate-coupled phalloidin (Sigma)/ml in PBS containing 0.1% Triton X-100 for 40 min and visualized with a confocal laser microscope (LSM 510 META NLO, Carl Zeiss).
RESULTS
Creation of GC-resistant cell line.
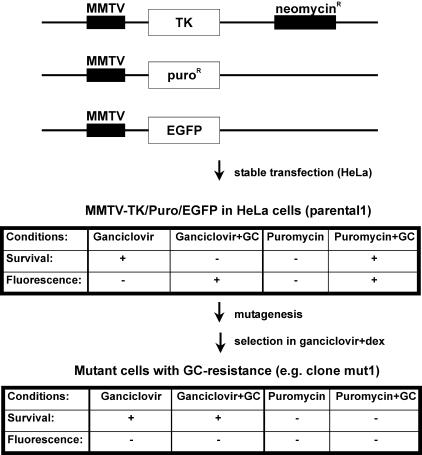
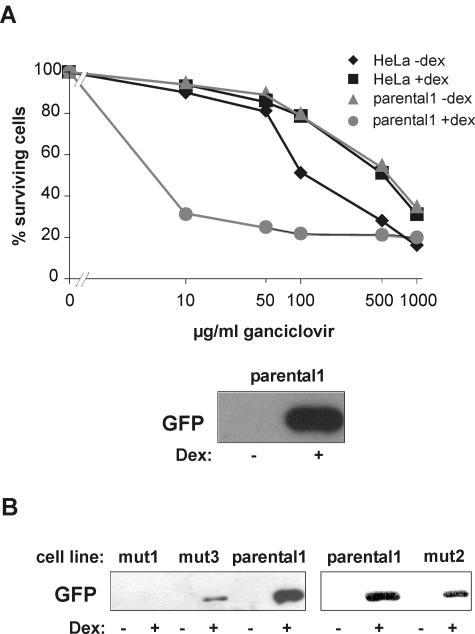
The concept was to develop a cellular screen that allows for the selection of factors which are functionally required for GR signaling. Thus, we set out to engineer a cell line for which activation of GR signal transduction by GCs is lethal. After random mutagenesis of this cell line, selection in GC-containing medium should produce only mutants with a dysfunction in GR signaling. We adapted a method which was previously used for the analysis of the interleukin-1 signaling pathway (29). As a selection marker, we used herpes virus TK under the control of the GC-responsive MMTV promoter. TK converts ganciclovir into a cytotoxic nucleoside analogue. Cells which have integrated the MMTV-TK construct can, therefore, not survive treatment with ganciclovir and hormone (Fig. 1). As additional markers, we used EGFP and PAC, both under the control of the MMTV promoter (Fig. 1). HeLa cells were transfected with the three constructs, and after selection, one clone (referred to as parental1) was isolated in which the three genes were well inducible upon treatment with 1 μM dex (Fig. 2A and data not shown). Parental1 was then used for the screen.
FIG. 1.
Outline of the functional genetic screen to create GC-resistant cells. The constructs MMTV-TK, MMTV-Puro, and MMTV-EGFP were transfected into HeLa cells, and the cells were cultivated in G418. Stable integration of the three plasmids was verified in the isolated clone parental1. For mutagenesis and drug selection, cells were subjected to three to five rounds of ICR191 or ENU treatment and selected in medium containing 10 μg of ganciclovir/ml and 1 μM dex. The tables show the characteristics of parental1 and the mutant cells. +, positive; −, negative.
FIG. 2.
Characterization of parental1 and mutant cell lines. (A) Inducibility of TK and EGFP in parental1 cells. TK expression was determined with an MTT assay. Cells were cultivated with (+) or without (−) 1 μM dex and with different concentration of ganciclovir. The MTT assay was performed after 7 days. The inducibility of EGFP was evaluated by Western blotting of whole-cell extracts treated with 1 μM dex or ethanol. A representative blot is shown. (B) Inducibility of EGFP in mutant cell lines. Representative Western blots are shown.
For mutagenesis, cells were subjected to several rounds of treatment with either ICR191 or ENU. ICR induces frameshift mutations (49), whereas ENU leads to point mutations (46). The mutated cells were then selected in medium containing 1 μM dex and 10 μg of ganciclovir/ml. It has been reported that prolonged treatment with GCs can lead to desensitization of the MMTV promoter (27). Therefore, we carefully tested different dexamethasone incubation protocols and found that short interruption of hormone exposure ensures continuing sensitivity of the cells to ganciclovir (data not shown). The outcome of mutagenesis and selection is summarized in Table 1. Three clones (termed mut1, mut2, and mut3) could not only survive the selection but also showed reduced induction of EGFP (Fig. 2B), PAC, and TK activity in an MTT assay (data not shown). These clones were then further analyzed. We report here the results for clone mut1, which emerged after mutagenesis with the frameshift-inducing agent ICR191.
TABLE 1.
List of clones isolated after mutagenesis and selection
| Clone | Mutagenic agent | Characteristic | Differentially expressed proteins |
|---|---|---|---|
| 1.3/1 | ICR191 | Mutation in TK gene or promoter | |
| 1.3/2 (mut1) | ICR191 | Mutation in trans | Thioredoxin (1), cofilin (2), hsp27 (3), reticulocalbin (4)a |
| 1.3/3 | ICR191 | Mutation in TK gene or promoter | |
| 1.3/4 | ICR191 | False positiveb | |
| 1.3/5 | ICR191 | False positive | |
| 1.3/6 | ICR191 | False positive | |
| 1.3/7 | ICR191 | False positive | |
| 1.4/1 | ICR191 | Mutation in TK gene or promoter | |
| 2.3 | ICR191 | False positive | |
| 3.3 | ICR191 | False positive | |
| 5.3/2 (mut2) | ICR191 | Mutation in trans or GR gene | Under investigation |
| ENU5 (mut3) | ENU | Mutation in trans or GR gene | Under investigation |
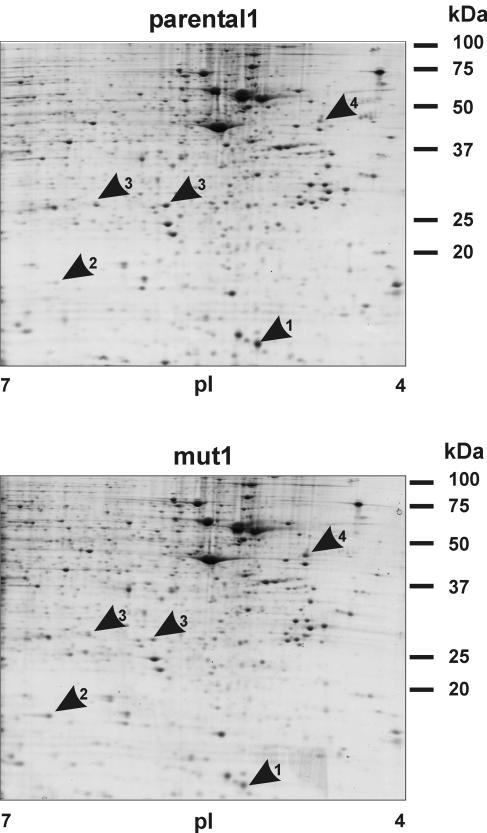
Numbers in parentheses refer to spots on the 2-D gels in Fig. 4.
False positive refers to clones that survived selection but did not show GC resistance, i.e., hormone was still able to induce the reporters.
Dominant phenotype in clone mut1 leads to reduced GR transactivation.
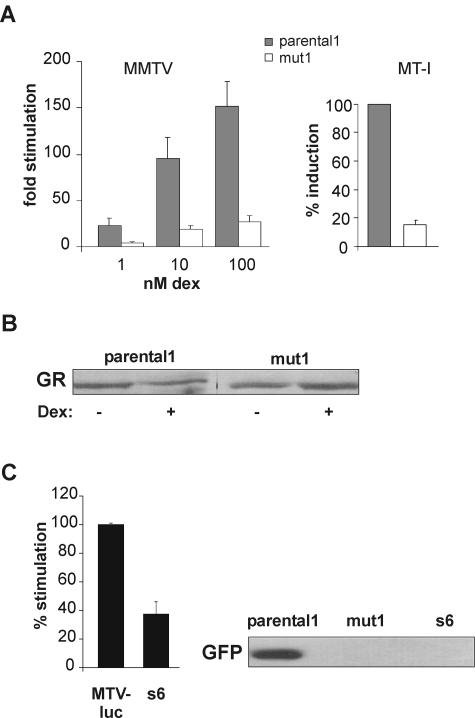
To further corroborate and characterize the observed GC insensitivity in mut1 cells, transient reporter assays with MMTV-driven luciferase were employed. As shown in Fig. 3A (left panel), luciferase activity was reduced approximately fivefold in mut1 cells compared to parental1 cells. This inhibition was independent of the hormone concentration. To test the possibility that factors specific for the MMTV promoter are affected in mut1 cells, we investigated the transcription of endogenous MT-I, another GC-inducible gene (26). The mRNA levels of MT-I were determined by RT-PCR. The induction of MT-I was reduced six- to sevenfold in mut1 cells (Fig. 3A, right panel). This demonstrates that the resistance is independent of the promoter and affects endogenous as well as transiently transfected DNA templates.
FIG. 3.
Analysis of the cell line mut1. (A) Left panel, transient luciferase assays in mut1 cells compared to parental1 cells. Cells were transiently transfected with MTV-Luc and pCMVβ-Gal to control for the transfection efficiency. Increasing concentrations of dex were applied for 16 h. The means of stimulation relative to luciferase activity in the absence of hormone + standard errors of the means of the results from five independent experiments are displayed. Right panel, inducibility of MT-I mRNA in the two cell lines. Total RNA was prepared from cells stimulated with 1 μM dex for 24 h, and RT-PCR was performed. Induction in parental1 cells is set to 100%. Means + standard errors of the means of the results from three independent experiments are shown. (B) GR levels in parental1 and mut1 cells. Representative Western blots of total cell extracts with (+) or without (−) hormone are displayed. (C) Analysis of s6, a fusion of mut1 and MTV-luc cells. Left panel, luciferase activity from the endogenous MMTV-Luc in s6 and in MTV-luc cells. Stimulation in MTV-Luc cells was set to 100%; mean values + standard errors of the means of the results from four independent experiments are displayed. Right panel, representative Western blot of GFP levels in dex-treated parental1, mut1, and s6 cells.
To exclude the possibility that the decrease in GR activity is due to a mutation in the GR gene (mutation in cis), GR levels were compared between parental1 and mut1 cells on a Western blot (Fig. 3B). No difference was detected. Furthermore, the GR cDNA from mut1 cells was amplified and completely sequenced and no mutation was found. To determine whether the mutation causing hormone insensitivity is dominant or recessive, mut1 cells were fused with a HeLa cell line which carries the MTV-luc construct in its genome. The fused cell lines were then tested for the induction of luciferase activity and GFP expression after hormone treatment. Three fusion clones were analyzed, and the results with one of the clones, s6, are shown in Fig. 3C. The luciferase activity was significantly reduced in s6 cells compared to the stable MTV-luc cells, and no GFP induction was detectable. The same results were found with the two other clones, indicating that the mutation which leads to GC resistance in mut1 is dominant. Taken together, we have established a method to generate GC-resistant cells. One of the obtained cell lines, mut1, has a GR-relevant dominant mutation which is not in cis. Thus, its resistance is caused by a mutation of some factor(s) important for GR signaling.
Identification of candidates for GR regulation.
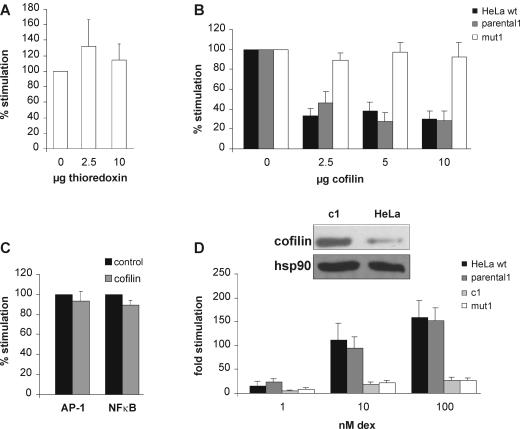
We expected that the cell line mut1 contains several mutations and that only one or some of them caused GC resistance. Our concept was to first uncover differences between the mutated and parental cell lines and then test these candidates individually in a reporter gene system. To evaluate the difference between the mut1 clone and the parental1 cell line, we chose a proteomic approach. Compared to microarray analysis, this technology frequently results in fewer candidates but has the power to reveal differences in posttranslational modifications that may be relevant for protein function. Total cell extracts of the two cell lines were compared by 2-D gel electrophoresis (Fig. 4). A total of 10 proteins were expressed with at least a twofold difference between the two lines. Five spots could be identified by tandem mass spectrometry analysis (Fig. 4; Table 1) (identification of the other five failed for technical reasons). The most interesting one at this point was thioredoxin, as it has been shown to be involved in the regulation of GR. As a cellular reducing catalyst, it restores GR function under oxidative conditions (31). Since thioredoxin may also regulate GR under conditions other than oxidative stress, we tested whether increasing the levels of thioredoxin in mut1 enhances the hormone response. Exogenous thioredoxin was expressed in mut1 cells, and luciferase activity was measured in a reporter assay. As shown in Fig. 5A, increasing the thioredoxin levels did not significantly change GR transactivation. We also tested the other candidates in reporter gene assays. Whereas hsp27 and reticulocalbin displayed no effect on GR function (data not shown), cofilin 1 reduced GR activity, as detailed in the following paragraphs.
FIG. 4.
2-D gel analysis of mut1 and parental1. Total protein was separated in the first dimension by IEF and in the second dimension by SDS-PAGE. Gels were stained with colloidal Coomassie brilliant blue. Several differentially expressed proteins were detected by visual inspection and confirmed by using PDQuest (Bio-Rad). The spots were excised and analyzed by tandem mass spectroscopy. Arrows mark thioredoxin (1), cofilin (2), 2 forms of hsp27 (3), and reticulocalbin (4).
FIG. 5.
Cofilin, but not thioredoxin, affects GR signaling. (A) Transient luciferase assay with coexpressed thioredoxin. Mut1 cells were transfected with MTV-Luc, pCMVβ-Gal, and increasing amounts of pRK5thio and stimulated with 1 μM dex for 24 h. Inducibility with empty vector was set to 100%. Data represent the means + standard errors of the means of the results from three independent experiments. (B) Transient luciferase assay with coexpression of cofilin. HeLa, parental1, and mut1 cells were transfected with MTV-Luc, pCMVβ-Gal, and increasing amounts of pRK5cof and stimulated with 1 μM dex for 24 h. Inducibility with empty vector was set to 100%. Data represent the means + standard errors of the means of the results from five independent experiments. (C) Transient reporter assay on AP-1- and NF-κB-responsive promoters. HeLa cells were transfected with either coll-luciferase or 6× NF-κB-luciferase, pCMVβ-Gal, and increasing amounts of pRK5cof and stimulated with 80 ng of phorbol myristate acetate/ml for 16 h. Inducibility with empty vector was set to 100%. Data represent the means + standard errors of the means of the results from three independent experiments. (D) Upper panel, representative Western blots of cofilin in c1 and HeLa extracts with an antibody which recognizes all forms of cofilin or hsp90 as a control. Lower panel, transient luciferase reporter assay with HeLa, parental1, c1, and mut1 cells performed as described for panel B.
Cofilin 1 acts as inhibitor of GR without direct interaction.
Cofilin is an 18-kDa protein and is involved in actin depolymerization. Its distribution, function, and regulation are well investigated, but it has never been described to be involved in signaling of GR or any other nuclear receptor. The intensity of the spot identified as cofilin was higher in mut1 than in the parental cell line. We therefore hypothesized that increasing the levels of cofilin in wild-type cells would interfere with GR signaling. Using reporter gene assays, we found that ectopic expression of cofilin in HeLa wild-type and parental1 cells as well as in HEK cells decreased GR-mediated transactivation (Fig. 5B and data not shown). In the less-hormone-responsive mut1 cells, additional expression of cofilin did not further reduce GR transactivation. To test whether cofilin is a general inhibitor of transcription factors, we investigated the activity of two other factors, AP-1 and NF-κB, in a reporter assay (Fig. 5C). Neither of them was affected by coexpression of cofilin.
To corroborate the inhibitory function of cofilin on GR, pRK5cof was stably integrated into HeLa cells, resulting in elevated levels of total cofilin (cell line c1) (Fig. 5D, top panel). In this cell line, the GR response to hormone was reduced about fivefold (Fig. 5D, bottom panel), independent of the hormone concentrations used.
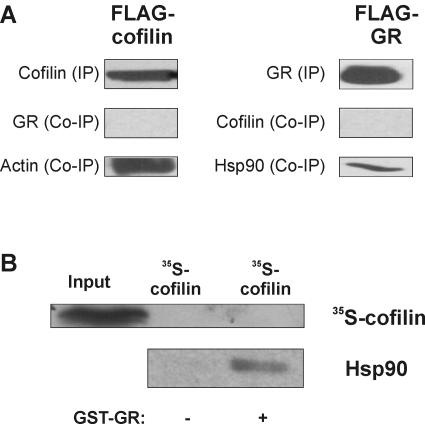
To determine whether cofilin has to directly interact with GR for its inhibitory action, coimmunoprecipitation was carried out with either flag-tagged GR or cofilin in the presence or absence of dex. GR was not coprecipitated with cofilin, whereas the positive control, actin, was readily detected as a binding partner (Fig. 6A, left panels). Conversely, cofilin was not precipitated together with GR, whereas hsp90 was (Fig. 6A, right panels). To confirm this finding by another precipitation method, GST pull-down assays were performed with GST-tagged GR and in vitro-translated 35S-labeled cofilin. As shown in Fig. 6B, 35S-cofilin was not bound to GR, whereas hsp90 was. We conclude from these experiments that cofilin acts as a specific inhibitor of GR transactivation. As the inhibition is independent of the hormone concentration, cofilin is not likely to affect the affinity of the receptor. Our experiments indicate that the inhibitory mechanism does not involve stable direct interaction between GR and cofilin. We cannot exclude the possibility that there is a weak and/or transient interaction (i.e., of different quality than our controls GR-hsp90 and cofilin-actin interactions) which is not detectable by either coimmunoprecipitation or GST pull-down assay.
FIG. 6.
The GR inhibitory effect of cofilin does not require physical interaction. (A) Coimmunoprecipitation of GR and cofilin. HEK cells were transfected with 10 μg of cofilin-flag or flag-GR. After precipitation, blots were probed with antibodies directed against cofilin, GR, actin, and hsp90. Representative blots are shown. (B) GST pull-down assay with GR and cofilin. GST-GR was expressed in bacteria and immobilized to glutathione-Sepharose beads. In vitro-translated 35S-cofilin was added to the beads, and eluates were tested for the presence of cofilin and hsp90 as a positive control. +, present; −, absent.
Dephosphorylation of cofilin 1, but not its nuclear presence, is essential for the inhibitory action on GR.
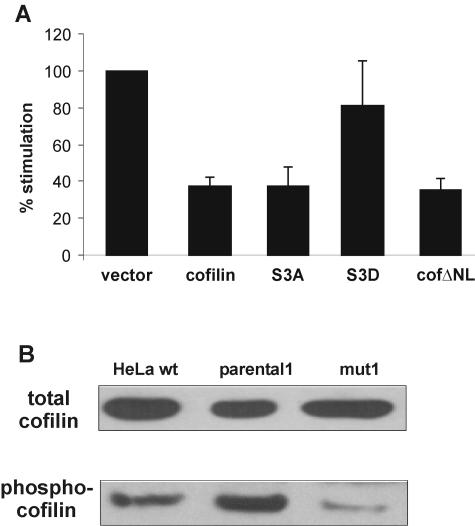
Cofilin 1 can be phosphorylated at the serine-3 residue, and this phosphorylation leads to inhibition of cofilin 1 as an actin depolymerization factor (36). If serine-3 is replaced with an alanine (S3A), the mutant is constitutively active. If, on the other hand, the negatively charged phosphate is mimicked by an aspartate (S3D), the mutant is completely inactive (36). To evaluate the relevance of the serine-3 phosphorylation status for the inhibitory activity of cofilin on GR, we used the mutants S3A and S3D in transient luciferase assays. If the dephosphorylated form is the active one, S3A should still inhibit GR, whereas the S3D mutant that mimics the effect of the phosphorylated form is likely to have lost its inhibitory potential. Indeed, we observed that S3A inhibits GR transactivation to an extent similar to that of wild-type cofilin, whereas S3D has lost this ability (Fig. 7A).
FIG. 7.
The inhibitory activity of cofilin on GR requires its dephosphorylation but not its nuclear presence. (A) Luciferase reporter assays with wild-type (wt) and mutant forms of cofilin. HeLa wild-type cells were transfected with MTV-Luc, pCMVβ-Gal, and 10 μg of pRK5cof, pRK5S3A, pRK5S3D, pRK5cofΔNL, or empty vector and incubated in the absence or presence of 1 μM dex for 48 h. The stimulation of cells transfected with vector control was set to 100%. Data are shown as means + standard errors of the means of the results from five independent experiments. (B) Comparison of different cofilin forms in wild-type, parental1, and mut1 cells. Representative Western blots with an antibody detecting all forms of cofilin (upper panel) or only cofilin phosphorylated at serine 3 (lower panel) are shown.
Phosphorylation of cofilin not only inhibits its function on actin filaments but may also prevent its translocation to the nucleus because the abolition of the phosphorylation site in S3A leads to preferential nuclear localization (37). Based on this, our findings with the cofilin mutants can have at least two explanations. First, the actin depolymerization function of cofilin must be intact for its inhibitory action on GR. Second, it can only inhibit GR function in the nucleus. In fact, severing the actin filaments in the nucleus may change the dynamics of the association of GR with the nuclear matrix, which has been postulated to be important for GR activity (50). To investigate this second possibility, a mutant, cofΔNL, was created which lacks the putative nuclear localization sequence KKRKK at positions 30 to 34 (23). The cellular distribution of this mutant was assessed with a GFP-tagged form and was confirmed to be mostly cytosolic (data not shown). cofΔNL was as efficient in inhibiting GR transactivation as wild-type cofilin (Fig. 7A).
The targeted mutations presented in Fig. 7A demonstrated the relevance of serine-3 phosphorylation for GR inhibition by cofilin. Therefore, it was of interest to determine which form of cofilin is overexpressed in mut1. We used an antibody recognizing all forms of cofilin and one specific for phospho-cofilin on Western blots (Fig. 7B). The levels of total cofilin showed no significant difference between mut1, parental1, and wild-type HeLa cells (top panel). However, the levels of the phosphorylated form are lower in mut1 than in parental1 and wild-type cells (bottom panel). We cannot directly compare the levels of dephosphorylated cofilin, as there are no specific antibodies available. However, we conclude from the comparison of the two panels that mut1 cells contain more dephosphorylated cofilin. Therefore, the cofilin spot in the 2-D gel may represent this form and the phosphorylated one was not detected. Alternatively, the spots represent the most abundant form of cofilin in each cell line, i.e., the phosphorylated cofilin in parental1 and the dephosphorylated cofilin in mut1. In support of this, we noted that the cofilin spot of mut1 was slightly shifted to basic pH values. These findings suggest that dephosphorylated cofilin may also inhibit GR in mut1 cells.
In conclusion, the inhibitory effect on GR is mediated mainly, if not exclusively, by cytoplasmic cofilin. Furthermore, since dephosphorylation is not only necessary for actions of cofilin in actin depolymerization (36) but also for its inhibitory action on GR (Fig. 7A), we hypothesize that actin depolymerization is involved in the inhibitory mechanism of cofilin.
Cofilin 1 increases the nuclear fraction of GR prior to hormone exposure.
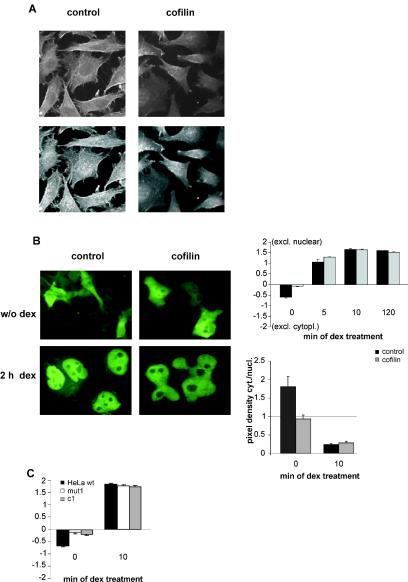
GR interacts with actin filaments via hsp90, and it was hypothesized that this leads to the anchoring of GR in the cytoplasm (34). Furthermore, it has been shown that efficient translocation of the activated GR is dependent on an intact cytoskeleton (13). Since we thus intended to examine the impact of cofilin on the subcellular distribution of GR, we first verified that overexpression of cofilin leads to impairment of the actin filaments in the cell (Fig. 8A). To test the influence of cofilin on GR distribution, cofilin or control vector was transfected into wild-type HeLa cells together with GFP-fused GR. The subcellular distribution was evaluated by visually classifying cells with a fluorescence microscope according to a method described previously (see Materials and Methods and reference 13). In the absence of hormone, cofilin increased the number of GFP-GR in the nucleus (Fig. 8B) while it did not change the distribution of GFP alone (data not shown). After 5 min of hormone exposure, there was still more GR in the nuclei of cells with additional cofilin; however, after 10 min, no difference was detectable any more. After 2 h, virtually all GR was nuclear in both cases.
FIG. 8.
Cofilin induces nuclear accumulation of GR in the absence of hormone. (A) Phalloidin staining of the actin cytoskeleton in cells transfected with empty vector (left panels) or pRK5cofilin (right panels). Representative sections (upper panels) or 3-D pictures (lower panels) are shown. (B) Determination of intracellular distribution of GR in cells overexpressing cofilin. HeLa cells were transfected with either vector control or pRK5cof together with GFP-tagged GR. At 24 h after transfection, cells were incubated with 100 nM dex or solvent (w/o dex) for the times indicated. Representative pictures are shown. The cytoplasmic and nuclear distribution of the fluorescing receptor was evaluated by scoring more than 100 cells (+2 denotes cells with exclusively [excl.] nuclear GR and −2 denotes cells with exclusively cytoplasmic [cytopl.] GR, upper diagram). Alternatively, the pixel densities of the cytoplasm (cyt.) and nucleus (nucl.) were determined in confocal microscope pictures, and the cytoplasm to nucleus ratio was calculated (lower diagram). Bars represent the mean values + standard errors of the means of the results from three independent experiments. (C) Determination of intracellular distribution of GR in mut1 and c1 cells. The analysis was performed as described for panel B, except that cells were transfected with GFP-GR alone.
We repeated the evaluation of GR distribution on a laser scanning microscope and compared the pixel densities in the cytosol and the nucleus in the absence of hormone and 10 min after hormone exposure. We could confirm the conclusions from our visual evaluation. In the absence of hormone, there was about a 1.8-fold-higher pixel density in the cytosol in the control cells without cofilin, whereas in the presence of cofilin, there was already more GR in the nucleus (Fig. 8B). Ten minutes after hormone exposure, the cytosolic/nuclear ratio of pixel densities was not significantly altered by the coexpression of cofilin. Therefore, the data from the laser scanning microscope fully confirmed the validity of our quantification by visual inspection.
It was important to test whether the stably cofilin-overexpressing cells c1 also display nuclear accumulation of GR in the absence of hormone. This was indeed the case (Fig. 8C). In addition, mut1 cells exhibited the same phenomenon (Fig. 8C). These results corroborate the conclusion that cofilin has no effect on the nuclear translocation of the activated GR but affects its subcellular distribution in the absence of hormone.
Disrupting actin filaments or increasing globular actin reduces GR function.
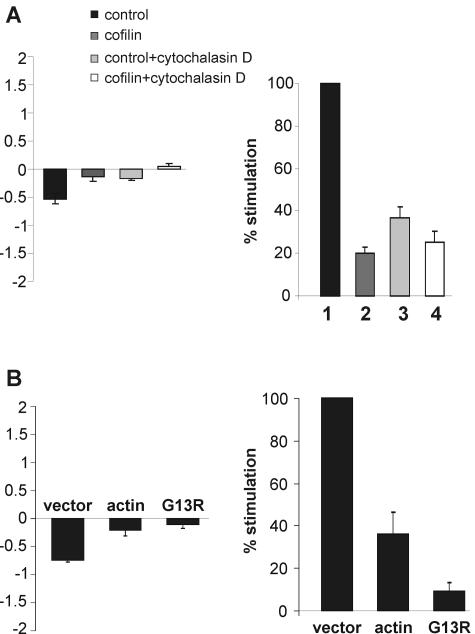
As we had gathered evidence that the inhibitory mechanism of cofilin requires actin depolymerization, we set out to tackle this idea more directly. We used the drug cytochalasin B to disrupt actin filaments and tested GR function under these conditions. We found that this treatment both reduces GR-dependent transcription, as shown previously (39), and causes nuclear accumulation of GR in the absence of hormone, similar to cofilin (Fig. 9A). Cytochalasin B also had additional effects on receptor trafficking, as we observed delayed GR transport to the nucleus after exposure to hormone (data not shown), as reported previously (13). This indicates a partly different mode of action on actin. Nevertheless, in the presence of cytochalasin B, cofilin had a much lesser effect on the nuclear translocation of GR and did not significantly alter GR transcription (Fig. 9A, compare columns 3 and 4). Thus, cytochalasin B apparently mimicked the effects of cofilin so that additional cofilin was less inhibitory for GR.
FIG. 9.
Disruption of the actin cytoskeleton or increase in G-actin reduces GR function. (A) Effect of cytochalasin B on intracellular distribution of GR (left panel) and transcriptional activity (right panel). For translocation assays, cells were transfected and evaluated as described for Fig. 8B, except that they were treated with cytochalasin B for 1 h before fixing. For luciferase assays, HEK cells were transfected with MTV-Luc, pCMVβ-Gal, pRK7GR, and pRK5cof, treated with cytochalasin B for 1 h, and stimulated with 1 μM dex for 24 h in the continued presence of cytochalasin B. Inducibility with empty vector was set to 100%. Data represent the means + standard errors of the means of the results for six independent experiments. (B) Effect of transient wild-type and mutant actin expression on intracellular distribution of GR (left panel) and transcriptional activity (right panel). For translocation assays, cells were transfected with pRK5actin, pRK5G13R, or empty vector and then treated and evaluated as described for Fig. 8B. For luciferase assays, HEK cells were transfected with MTV-Luc, pCMVβ-Gal, pRK7GR, and 5 μg of pRK5actin or pRK5G13R and stimulated with 1 μM dex for 48 h. Inducibility with empty vector was set to 100%. Data represent the means + standard errors of the means of the results for eight independent experiments.
Increased levels of G-actin have been shown to affect the transcription factor SRF (41). As increased levels of cofilin also increase G-actin (38), we tested whether overexpressing actin would have effects on GR similar to those of overexpressing cofilin. We observed that actin indeed enhances the nuclear fraction of GR in the absence of hormone, similar to cofilin (Fig. 9B, left panel). Furthermore, actin also reduced GR-dependent transcription (Fig. 9B, right panel). Intriguingly, a mutant form of actin (G13R) that does not polymerize (41) had a stronger effect on GR (Fig. 9B). These experiments are consistent with the idea that the effect of cofilin on actin depolymerization is relevant for its inhibitory mechanism.
Transient or stable overexpression of cofilin induces the GR inhibitor c-Jun.
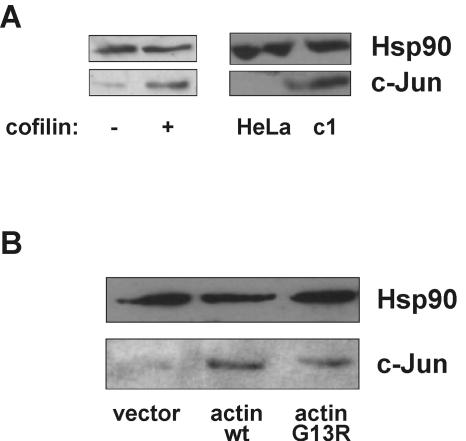
Chemical disruption of the cytoskeleton has been shown to induce expression of c-Jun (39). It is well described that c-Jun inhibits GR (24, 45, 58). Therefore, to further elucidate the mechanism of GR inhibition by cofilin, we tested whether its overexpression similarly activates this GR inhibitor. We found that both transient and stable overexpression of cofilin indeed induced expression of c-Jun (Fig. 10A). Moreover, overexpression of wild-type actin and its mutant G13R also induced c-Jun (Fig. 10B). Therefore, the mechanism of GR inhibition by cofilin likely includes induction of c-Jun, caused by an increased G-actin pool.
FIG. 10.
Disturbance of the actin cytoskeleton induces c-Jun. (A) Effect of increased cofilin on c-Jun. Left panel, HeLa cells were transfected with pRK5 cofilin (+) or vector control (−), and c-Jun levels were detected by Western blotting of total cell extracts. Right panel, HeLa and c1 cells were compared. Representative blots are shown. (B) Effect of increased actin or the G-actin mutant G13R on c-Jun. Lysates from the experiment described for Fig. 9B were analyzed for c-Jun protein levels by Western blotting. Representative blots are shown. wt, wild type.
DISCUSSION
The goal of this study was to set up a cellular mutational screen to identify novel factors relevant for GR function, with or without directly interacting with the receptor. We employed a combination of toxicity and reporter markers designed to be GC-responsive for mutagenesis and proteomic analysis. We applied relatively mild selection, as we reasoned that harsher conditions would suppress clones with mutations that lead to moderate impairment of GR. A mutation in GR itself, e.g., by frameshift mutations induced by ICR191, would in most cases lead to a complete loss of receptor function. Thus, we expected to increase the likelihood of detecting mutations of GR cofactors as opposed to mutations of GR itself.
The mutated cell line mut1 that we used to obtain candidates for GR regulators showed reduced activity of the MMTV-driven, stably integrated markers and of transiently transfected MMTV-driven luciferase and endogenous MT-I. Integrated and transiently transfected MMTV promoter templates differ in their chromosomal structure as well as in their transactivation by steroid receptors (4, 48). Since our cell mutant exhibited decreased GR-dependent activity with both kinds of templates, the mutation(s) is affecting a function of GR that acts independently of the chromosomal context.
2-D gel analysis comparing the mut1 cell line with its parental cell line yielded several differentially expressed proteins, four of which were identified by mass spectrometry as thioredoxin, hsp27, reticulocalbin, and cofilin 1. Thioredoxin, hsp27, and reticulocalbin were not confirmed as GR regulators. On the other hand, the unexpected candidate cofilin 1 inhibited GR. Stable and transient ectopic expression of cofilin reduced GR-dependent transcription in HeLa and HEK cells. Its transient expression did not affect transcription factors in general, as the activity of AP-1 and NF-κB in reporter assays was not changed.
The cause for the observed upregulation of cofilin in the mutated cell line is not known. The cDNA of cofilin was not mutated (data not shown). Therefore, the causal mutation may affect a regulator upstream of cofilin, e.g., a kinase or a phosphatase. In any case, we have established cofilin as a GR inhibitory protein, regardless of what the causal mutation in mut1 cells may be. Moreover, we cannot exclude the possibility that additional GR regulatory proteins are affected. The likelihood for multiple random mutations complicates the dissection of the actions of cofilin important for GR inhibition in mut1 cells. Therefore, it was mandatory to analyze the GR regulatory characteristics of cofilin in a cellular system (i.e., assays in common cell lines) that allows the introduction of targeted mutations.
Cofilin 1 is well characterized as an actin depolymerization factor (2, 10, 35). Together with actin depolymerization factor (ADF) and cofilin 2, it constitutes the evolutionarily conserved ADF/cofilin family (6) and is expressed abundantly in embryonic and adult tissue (53). ADF/cofilins are crucial for many cellular processes, such as cell motility, cell division, and membrane organization. Binding of cofilin to actin filaments alters the twist of the filament, thereby promoting filament severing and depolymerization (32).
To address the question of whether the GR inhibitory function of cofilin 1 requires its action on actin filaments, we first made use of the fact that the actin depolymerization function is inactivated by phosphorylation of cofilin at serine-3 (36). This phosphorylation generates a charge repulsion that inhibits actin binding. Thus, mutation of serine-3 to alanine creates a constitutively active form of cofilin, whereas a mutation of serine-3 to glutamate renders it constitutively inactive. Our observation that these cofilin mutants also exhibit the same pattern of constitutive activity and inactivity on GR, respectively, was the first strong indication of a mechanism whereby cofilin acts via the actin filaments, possibly by increasing G-actin, to influence GR. This is also in line with the decreased fraction of phosphorylated cofilin that we found in the mutated, less GC-responsive cell line. Moreover, an action of cofilin on GR that functions via the actin filaments of the cytoskeleton does not require direct association of cofilin with GR. Indeed, no stable association was detectable between GR and cofilin.
By use of the cytoskeleton-disrupting agent cytochalasin B, we corroborated the relevance of actin and actin filaments for GR activity and for the inhibitory action of cofilin. Cytochalasin B alone reduced GR function, as has been shown previously with Mueller glial cells of chicken embryo retina (39). In the presence of this drug, the ability of cofilin to impair GR was nearly abolished.
Overexpression of cofilin or treatment with cytochalasin B reduces F-actin and increases G-actin (2, 10, 35). To distinguish which of these changes causes inhibition of GR, we overexpressed actin and the mutant G13R, which does not polymerize (41). Overexpression of wild-type actin increases both F- and G-actin while overexpression of G13R actin specifically increases G-actin. Coexpression of each form inhibited GR; G13R was more efficient. Thus, it is the increased levels of G-actin that apparently initiate a process to impair GR. A similar observation has been reported for the transcription factor SRF (41).
Treatment of cells with cytochalasin B was shown to induce c-Jun, providing an explanation for the inhibition of GR (39). Thus, it is possible that the mechanism of cofilin for GR inhibition also involves c-Jun. Indeed, we observed induction of c-Jun after transient or stable overexpression of cofilin as well as after expression of wild-type actin and G13R actin. c-Jun was not detected in mut1 cells (data not shown). Therefore, the induction of c-Jun in these cells may be too small to be detectable. Alternatively, additional unknown mutations in these cells may contribute to GR regulation and/or obscure some effects of cofilin. This again underscores the necessity to analyze the behavior of cofilin in better definable systems. From all of our experiments with GR-inhibiting actin manipulations, we conclude that the increase of G-actin leads to induction of the established GR inhibitor c-Jun (24, 45, 58), representing an inhibitory mechanism of cofilin.
Intriguingly, we also observed nuclear accumulation of GR in the absence of hormone whenever we increased G-actin, i.e., by overexpression of cofilin and the two actin forms or treatment with cytochalasin B. A possible explanation for this is that the cytoskeleton may be required to keep the inactivated receptor in the cytoplasm, as suggested previously (34). Higher levels of G-actin may reduce the attachment of GR to F-actin so that more GR molecules reach the nucleus. Since we did not observe an effect of cofilin on the hormone-independent promoter activity, the additional GR in the nucleus is apparently not active. Since GR is normally folded in the cytosol to attain high hormone binding affinity, the conditions in the nucleus may not allow full hormone efficacy with these receptor molecules. On the other hand, there is a receptor recycling chaperone machinery also in the nucleus that maintains the hormone binding capacity of GR after steroid depletion (30). Nevertheless, the strict correlation of premature nuclear accumulation of GR with its inhibition strongly argues that this is part of the inhibitory mechanism of cofilin. We cannot yet exclude that it is an epiphenomenon, indicating a disturbance of GR regulation.
The inhibitory action of cofilin on GR may have physiological relevance. We were intrigued by the recently reported up-regulation of cofilin in male wild house mice selected for long attack latency (LAL) compared to mice with short attack latency (SAL) (12). SAL mice show highly aggressive behavior and an active coping style while LAL mice display the converse phenotype (52). LAL and SAL mice also differ in their hypothalamic-pituitary-adrenal system. The activity and hormone levels of the hypothalamic-pituitary-adrenal axis are balanced by a negative feedback loop which is mediated by GR (7). LAL mice show elevated corticosterone levels after forced swim stress compared to SAL mice (54). This could be explained by an impaired negative feedback, i.e., by decreased GR activity, which may be due to up-regulated cofilin. It will be interesting to further explore the physiological importance, possibly in diseases involving GC resistance, and reveal details of the mechanism of action of GR inhibition by cofilin.
Acknowledgments
We thank Paul Hill, Dale Milfay, and Isabel Birg for excellent technical assistance; Archana Jacob and Guiseppina Maccarrone for support in mass spectrometry analyses; and Gabriela Wochnik, Amalia Tsolakidou, Marilyn Tirard, and Dietmar Spengler for critical reading of the manuscript. We thank Peter Hutzler and E. Mannweiler (National Research Center for Environment and Health, Munich, Germany) for generous support with laser scanning microscopy.
J.R. was supported by a studentship from the German National Academic Foundation.
REFERENCES
- 1.Abel, A., G. Wochnik, J. Rüegg, A. Rouyer, F. Holsboer, and T. Rein. 2002. Activity of the glucocorticoid receptor in G2 and mitosis. Mol. Endocrinol. 16:1352-1366. [DOI] [PubMed] [Google Scholar]
- 2.Bamburg, J. R., and O. P. Wiggan. 2002. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 12:598-605. [DOI] [PubMed] [Google Scholar]
- 3.Beato, M., P. Herrlich, and G. Schütz. 1995. Steroid hormone receptors: many actors in search of a plot. Cell 83:851-857. [DOI] [PubMed] [Google Scholar]
- 4.Bonovich, M. T., H. J. List, S. Zhang, M. Danielsen, and A. T. Riegel. 1998. Identification of glucocorticoid receptor domains necessary for transcriptional activation of the mouse mammary tumor virus promoter integrated in the genome. Exp. Cell Res. 239:454-462. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann, A. O. 1994. Steroid hormone receptors: activators of gene transcription. J. Pediatr. Endocrinol. 7:275-282. [DOI] [PubMed] [Google Scholar]
- 6.Carlier, M. F. 1998. Control of actin dynamics. Curr. Opin. Cell Biol. 10:45-51. [DOI] [PubMed] [Google Scholar]
- 7.De Kloet, E. R., M. S. Oitzl, and M. Joels. 1993. Functional implications of brain corticosteroid receptor diversity. Cell. Mol. Neurobiol. 13:433-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Martino, M. U., N. Bhattachryya, S. Alesci, T. Ichijo, G. P. Chrousos, and T. Kino. 2004. The glucocorticoid receptor (GR) and the orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) interact with and mutually affect each other's transcriptional activities: implications for intermediary metabolism. Mol. Endocrinol. 18:820-833. [DOI] [PubMed] [Google Scholar]
- 9.DeRijk, R., and E. M. Sternberg. 1997. Corticosteroid resistance and disease. Ann. Med. 29:79-82. [DOI] [PubMed] [Google Scholar]
- 10.Dos Remedios, C., D. Chhabra, M. Kekic, I. V. Dedova, M. Tsubakihara, D. A. Berry, and N. J. Nosworthy. 2003. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 83:433-473. [DOI] [PubMed] [Google Scholar]
- 11.Evans-Storms, R. B., and J. A. Cidlowski. 1995. Regulation of apoptosis by steroid hormones. J. Steroid Biochem. Mol. Biol. 53:1-8. [DOI] [PubMed] [Google Scholar]
- 12.Feldker, D. E., N. A. Datson, A. H. Veenema, E. Meulmeester, E. R. De Kloet, and E. Vreugdenhil. 2003. Serial analysis of gene expression predicts structural differences in hippocampus of long attack latency and short attack latency mice. Eur. J. Neurosci. 17:379-387. [DOI] [PubMed] [Google Scholar]
- 13.Galigniana, M. D., J. L. Scruggs, J. Herrington, M. J. Welsh, C. Carter-Su, P. R. Housley, and W. B. Pratt. 1998. Heat shock protein 90-dependent (geldanamycin-inhibited) movement of the glucocorticoid receptor through the cytoplasm to the nucleus requires intact cytoskeleton. Mol. Endocrinol. 12:1903-1913. [DOI] [PubMed] [Google Scholar]
- 14.Göttlicher, M., S. Heck, V. Doucas, E. Wade, M. Kullmann, A. C. Cato, R. M. Evans, and P. Herrlich. 1996. Interaction of the Ubc9 human homologue with c-Jun and with the glucocorticoid receptor. Steroids 61:257-262. [DOI] [PubMed] [Google Scholar]
- 15.Grove, J. R., B. S. Dieckmann, T. A. Schroer, and G. M. Ringold. 1980. Isolation of glucocorticoid-unresponsive rat hepatoma cells by fluorescence-activated cell sorting. Cell 21:47-56. [DOI] [PubMed] [Google Scholar]
- 16.Harrison, R. W., S. S. Lippman, and R. VerHoeven. 1995. Selection of glucocorticoid-resistant mutations from an AtT-20 cell line containing a glucocorticoid-regulated selectable transgene. Biochem. Biophys. Res. Commun. 209:18-24. [DOI] [PubMed] [Google Scholar]
- 17.Herr, A., G. M. Wochnik, M. C. Rosenhagen, F. Holsboer, and T. Rein. 2000. Rifampicin is not an activator of the glucocorticoid receptor. Mol. Pharmacol. 57:732-737. [DOI] [PubMed] [Google Scholar]
- 18.Hittelman, A. B., D. Burakov, J. A. Iñiguez-Lluh, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenberg, S. M., and R. M. Evans. 1988. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell 55:899-906. [DOI] [PubMed] [Google Scholar]
- 20.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hulkko, S. M., H. Wakui, and J. Zilliacus. 2000. The pro-apoptotic protein death-associated protein 3 (DAP3) interacts with the glucocorticoid receptor and affects the receptor function. Biochem. J. 349:885-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchison, K. A., K. D. Dittmar, M. J. Czar, and W. B. Pratt. 1994. Proof that hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with hsp90. J. Biol. Chem. 269:5043-5049. [PubMed] [Google Scholar]
- 23.Iida, K., S. Matsumoto, and I. Yahara. 1992. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct. Funct. 17:39-46. [DOI] [PubMed] [Google Scholar]
- 24.Jonat, C., H. J. Rahmsdorf, K. K. Park, A. C. Cato, S. Gebel, H. Ponta, and P. Herrlich. 1990. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell 62:1189-1204. [DOI] [PubMed] [Google Scholar]
- 25.Karin, M. 1998. New twists in gene regulation by glucocorticoid receptor: is DNA binding dispensable? Cell 93:487-490. [DOI] [PubMed] [Google Scholar]
- 26.Karin, M., A. Haslinger, A. Heguy, T. Dietlin, and R. Imbra. 1987. Transcriptional control mechanisms which regulate the expression of human metallothionein genes. Experientia Suppl. 52:401-405. [DOI] [PubMed] [Google Scholar]
- 27.Lee, H. L., and T. K. Archer. 1998. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 17:1454-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S., K. A. Duncan, H. Chou, D. Chen, K. Kohli, C. F. Huang, and M. R. Stallcup. 1995. A somatic cell genetic method for identification of untargeted mutations in the glucocorticoid receptor that cause hormone binding deficiencies. Mol. Endocrinol. 9:826-837. [DOI] [PubMed] [Google Scholar]
- 29.Li, X., M. Commane, C. Burns, K. Vithalani, Z. Cao, and G. R. Stark. 1999. Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol. Cell. Biol. 19:4643-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, J., and D. B. DeFranco. 1999. Chromatin recycling of glucocorticoid receptors: implications for multiple roles of heat shock protein 90. Mol. Endocrinol. 13:355-365. [DOI] [PubMed] [Google Scholar]
- 31.Makino, Y., K. Okamoto, N. Yoshikawa, M. Aoshima, K. Hirota, J. Yodoi, K. Umesono, I. Makino, and H. Tanaka. 1996. Thioredoxin: a redox-regulating cellular cofactor for glucocorticoid hormone action. Cross talk between endocrine control of stress response and cellular antioxidant defense system. J. Clin. Investig. 98:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGough, A., B. Pope, W. Chiu, and A. Weeds. 1997. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 138:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKendry, R., J. John, D. Flavell, M. Muller, I. M. Kerr, and G. R. Stark. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. USA 88:11455-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyata, Y., and I. Yahara. 1991. Cytoplasmic 8 S glucocorticoid receptor binds to actin filaments through the 90-kDa heat shock protein moiety. J. Biol. Chem. 266:8779-8783. [PubMed] [Google Scholar]
- 35.Moon, A., and D. G. Drubin. 1995. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol. Biol. Cell 6:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriyama, K., K. Iida, and I. Yahara. 1996. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells 1:73-86. [DOI] [PubMed] [Google Scholar]
- 37.Nebl, G., S. C. Meuer, and Y. Samstag. 1996. Dephosphorylation of serine 3 regulates nuclear translocation of cofilin. J. Biol. Chem. 271:26276-26280. [DOI] [PubMed] [Google Scholar]
- 38.Nishida, E., S. Maekawa, and H. Sakai. 1984. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 23:5307-5313. [DOI] [PubMed] [Google Scholar]
- 39.Oren, A., A. Herschkovitz, I. Ben-Dror, V. Holdengreber, Y. Ben-Shaul, R. Seger, and L. Vardimon. 1999. The cytoskeletal network controls c-Jun expression and glucocorticoid receptor transcriptional activity in an antagonistic and cell-type-specific manner. Mol. Cell. Biol. 19:1742-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posern, G., A. Sotiropoulos, and R. Treisman. 2002. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 13:4167-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pratt, W. B., A. M. Silverstein, and M. D. Galigniana. 1999. A model for the cytoplasmic trafficking of signalling proteins involving the hsp90-binding immunophilins and p50cdc37. Cell. Signal. 11:839-851. [DOI] [PubMed] [Google Scholar]
- 43.Rabindran, S. K., M. Danielsen, G. L. Firestone, and M. R. Stallcup. 1987. Glucocorticoid-dependent maturation of viral proteins in mouse lymphoma cells: isolation of defective and hormone-independent cell variants. Somat. Cell Mol. Genet. 13:131-143. [DOI] [PubMed] [Google Scholar]
- 44.Robyr, D., A. P. Wolffe, and W. Wahli. 2000. Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol. Endocrinol. 14:329-347. [DOI] [PubMed] [Google Scholar]
- 45.Schule, R., P. Rangarajan, S. Kliewer, L. J. Ransone, J. Bolado, N. Yang, I. M. Verma, and R. M. Evans. 1990. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell 62:1217-1226. [DOI] [PubMed] [Google Scholar]
- 46.Shibuya, T., and K. Morimoto. 1993. A review of the genotoxicity of 1-ethyl-1-nitrosourea. Mutat. Res. 297:3-38. [DOI] [PubMed] [Google Scholar]
- 47.Sitcheran, R., R. Emter, A. Kralli, and K. R. Yamamoto. 2000. A genetic analysis of glucocorticoid receptor signaling. Identification and characterization of ligand-effect modulators in Saccharomyces cerevisiae. Genetics 156:963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, C. L., H. Htun, R. G. Wolford, and G. L. Hager. 1997. Differential activity of progesterone and glucocorticoid receptors on mouse mammary tumor virus templates differing in chromatin structure. J. Biol. Chem. 272:14227-14235. [DOI] [PubMed] [Google Scholar]
- 49.Taft, S. A., H. L. Liber, and T. R. Skopek. 1994. Mutational spectrum of ICR-191 at the hprt locus in human lymphoblastoid cells. Environ. Mol. Mutagen. 23:96-100. [DOI] [PubMed] [Google Scholar]
- 50.Tang, Y., and D. B. DeFranco. 1996. ATP-dependent release of glucocorticoid receptors from the nuclear matrix. Mol. Cell. Biol. 16:1989-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vacchio, M. S., J. D. Ashwell, and L. B. King. 1998. A positive role for thymus-derived steroids in formation of the T-cell repertoire. Ann. N. Y. Acad. Sci. 840:317-327. [DOI] [PubMed] [Google Scholar]
- 52.van Oortmerssen, G. A., and T. C. Bakker. 1981. Artificial selection for short and long attack latencies in wild Mus musculus domesticus. Behav. Genet. 11:115-126. [DOI] [PubMed] [Google Scholar]
- 53.Vartiainen, M. K., T. Mustonen, P. K. Mattila, P. J. Ojala, I. Thesleff, J. Partanen, and P. Lappalainen. 2002. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell 13:183-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veenema, A. H., O. C. Meijer, E. R. De Kloet, J. M. Koolhaas, and B. G. Bohus. 2003. Differences in basal and stress-induced HPA regulation of wild house mice selected for high and low aggression. Horm. Behav. 43:197-204. [DOI] [PubMed] [Google Scholar]
- 55.Wakui, H., A. P. Wright, J. Gustafsson, and J. Zilliacus. 1997. Interaction of the ligand-activated glucocorticoid receptor with the 14-3-3 eta protein. J. Biol. Chem. 272:8153-8156. [DOI] [PubMed] [Google Scholar]
- 56.Wochnik, G. M., J. C. Young, U. Schmidt, F. Holsboer, F. U. Hartl, and T. Rein. 2004. Inhibition of GR-mediated transcription by p23 requires interaction with Hsp90. FEBS Lett. 560:35-38. [DOI] [PubMed] [Google Scholar]
- 57.Yang, L., J. Guerrero, H. Hong, D. B. DeFranco, and M. R. Stallcup. 2000. Interaction of the tau2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol. Biol. Cell 11:2007-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang-Yen, H. F., J. C. Chambard, Y. L. Sun, T. Smeal, T. J. Schmidt, J. Drouin, and M. Karin. 1990. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell 62:1205-1215. [DOI] [PubMed] [Google Scholar]