Abstract
Smc5 and Smc6 proteins form a heterodimeric SMC (structural maintenance of chromosome) protein complex like SMC1-SMC3 cohesin and SMC2-SMC4 condensin, and they associate with non-SMC proteins Nse1 and Nse2 stably and Rad60 transiently. This multiprotein complex plays an essential role in maintaining chromosome integrity and repairing DNA double strand breaks (DSBs). This study characterizes a Schizosaccharomyces pombe mutant rad62-1, which is hypersensitive to methyl methanesulfonate (MMS) and synthetically lethal with rad2 (a feature of recombination mutants). rad62-1 is hypersensitive to UV and gamma rays, epistatic with rhp51, and defective in repair of DSBs. rad62 is essential for viability and genetically interacts with rad60, smc6, and brc1. Rad62 protein physically associates with the Smc5-6 complex. rad62-1 is synthetically lethal with mutations in the genes promoting recovery from stalled replication, such as rqh1, srs2, and mus81, and those involved in nucleotide excision repair like rad13 and rad16. These results suggest that Rad62, like Rad60, in conjunction with the Smc5-6 complex, plays an essential role in maintaining chromosome integrity and recovery from stalled replication by recombination.
Mutants in Schizosaccharomyces pombe rad2, Saccharomyces cerevisiae RAD27, and Escherichia coli polA, all of which are defective in processing Okazaki fragments, are synthetically lethal with mutations in recombination repair genes (8, 11, 23, 24, 35, 37, 40, 43, 44). In these mutants, double strand breaks (DSBs) are thought to be produced when replication forks encounter the single strand gaps or nicks remaining unsealed due to the inefficient processing of Okazaki fragments, so they require a highly efficient capacity to repair DSBs for survival. In an effort to identify novel genes involved in recombination repair, we isolated mutants that were hypersensitive to methyl methanesulfonate (MMS) and synthetically lethal with rad2Δ and cloned the genes by complementation of the MMS sensitivity. Genes identified in this way include rhp57 (44), rad60 (35), rad32, nbs1 (45), and fdh1, which encodes an F-box DNA helicase (our unpublished data). A recent genome-wide search for mutations synthetically lethal with rad27 in S. cerevisiae identified mutations in all of the well-studied recombination repair genes including RAD52, RAD50, MRE11, XRS2 RAD51, RAD55, RAD57, RAD54, SGS1, MUS81, and MMS4 (43). The screen also identified genes involved in checkpoint regulation (RAD9, RAD17, RAD24, and DDC1) and those related to regulation of chromatin structure (CAC2, ESC2, HST1, and HST3).
We report here a novel gene, rad62, which was identified by isolating mutants that were hypersensitive to MMS and synthetically lethal with rad2 mutation and cloning the gene that complemented the MMS sensitivity of such a mutant. rad62 mutants show very similar phenotypes to those of rad60 mutants (7, 35) and rad18 mutants (30, 47). They are hypersensitive to UV, MMS, and gamma rays, epistatic with rhp51 with respect to the damage sensitivity, required for chromosome integrity during replication, and essential for growth. The spr18 and rad18 genes encode proteins whose structures are characteristic of the SMC family of proteins involved in chromosome condensation (Smc2-Smc4) and sister chromatid cohesion (Smc1-Smc3) (13, 21, 41). Since RAD18 is a different gene in budding yeast, and the human homologs of rad18 and spr18 are named hSMC6 and hSMC5, respectively (41), to avoid confusion we have used the names smc6 and smc5, respectively.
SMC proteins share a common structure in that they contain two ATPase motifs composed of Walker A motifs at the N termini and Walker B motifs at the C termini, which are separated by long coiled-coil domains with a hinge region in the middle. The two coiled-coil domains interact and bring two ATPase motifs into close contact to form a globular structure (21). Eukaryotic SMC proteins including Smc5 and Smc6 form tight heterodimers (13). The condensin complex plays an essential role in folding extended chromosomes in an ATP-dependent manner during mitosis. The cohesin complex forms a ring structure which holds sister chromatids in close proximity after replication until they are separated at anaphase in mitosis (16). The Smc5-Smc6 (Smc5-6) complex is proposed to play a role in repairing DSBs by recombination by holding broken DNA together (41). In common with the other eukaryotic SMC complexes, the Smc5-6 heterodimer is associated with non-SMC protein subunits, Nse1 and Nse2, and interacts with another essential protein, Rad60 (7, 19, 33, 35). Like smc5 and smc6, rad60, nse1, and nse2 are essential for growth, and the hypomorphic mutants are sensitive to DNA damaging agents.
Analysis of the genetic interactions between rad60 and smc6 mutants suggests that the products of these genes play a role in a common pathway for repairing DSBs and maintaining chromosome integrity (35). A recent study demonstrated that substoichiometric amounts of Rad60 were associated with Smc5-6 (7). Chromosomes were fragmented when a temperature-sensitive rad60-1 mutant was grown at nonpermissive temperature, suggesting that the Rad60 protein is required to prevent chromosome breakage during replication (35). Like smc6 and rad60 mutants (35, 47), the rad62-1 mutant is defective in joining DSBs produced by ionizing radiation and rad62 is essential for growth. From the phenotypic similarities and analysis of genetic and physical interactions, we propose that Rad62 plays a role as a non-SMC component of the Smc5-6 complex in maintaining chromosome integrity and repairing DSBs.
MATERIALS AND METHODS
S. pombe media and methods.
Yeast cells were grown in complete medium (YES), minimal medium (EMM2), or sporulation medium (ME), and standard yeast genetic and molecular procedures were as described previously (34). Genetic interactions were assessed through standard mating techniques, followed by tetrad analysis on YES plates as described previously (35).
Strains and plasmids.
The strains and plasmids used in this study are shown in Tables 1 and 2. pUR19 is a multicopy plasmid which contains the ars1 and ura4 genes (3). pREP41 is a plasmid that contains a weak nmt promoter, which is inducible by thiamine depletion (39).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| MMPX1 | mat1PΔ17::LEU2 leu1-32 ura4-D18 | Laboratory stock |
| MMPX2 | mat1PΔ17::LEU2 rad62-1 leu1-32 ura4-D18 | This study |
| MMPX3 | smt-0 rhp51::ura4 leu1-32 ura4-D18 | Laboratory stock |
| MMPX4 | mat1PΔ17::LEU2 rad62-1 rhp51::ura4 leu1-32 ura4-D18 | This study |
| MMPX5 | smt-0 rhp57::his3ura4-D18 leu1-32 his3-D1arg3-D1 | Laboratory stock |
| MMPX6 | smt-0 rad62-1 leu1-32 ura4-D18 | This study |
| MMPX7 | smt-0 rad62-1 leu1-32 ura 4-D18 his3-D1 | This study |
| MMPX8 | mat1PΔ17::LEU2 swi5::his3 leu1-32 ura4-D18 | Laboratory stock |
| MMPX9 | smt-0 rad62-1 swi5::his3 leu1-32 ura4-D18 his3-D1 | This study |
| MMP1 | h+leu1-32 ura4-D18 | Laboratory stock |
| MMP2 | h+rad62-1 leu1-32 ura4-D18 | This study |
| MMP5 | h−rad2::ura4 leu1-32 ura4-D18 ade6-M210 | Laboratory stock |
| MMP6 | rad62-1 rad2::ura4 leu1-32 ura4-D18 | This study |
| MMP7 | h+rad62-1 leu1-32 ura4-D18 his3-D1 | This study |
| MMP8 | rad62::ura4 leu1-32 ura4-D18 ade6-his7-366 (carrying pGFP62) | This study |
| MMP9 | h−rad13::ura4 leu1-32 ura4-D18 his3-D1 | This study |
| MMP10 | h+rhp18::ura4 leu1-32 ura4-D18 his3-D1 | Laboratory stock |
| MMP11 | h−rad62-1 rad13::ura4 leu1-32 ura4-D18 his3-D1 | This study |
| MMP12 | h+rad62-1 rhp18::ura4 leu1-32 ura4-D18 his3-D1 | This study |
| MMP13 | h−rad16::ura4 leu1-32 ura4-D18 his3-D1 | This study |
| MMP15 | rad62-1 rad50::LEU2 leu1-32 ura4-D18 | This study |
| MMP16 | h−srs2::Kanrleu1-32 ura4-D18 his3-D1 | This study |
| MMP17 | rad62-1 mus81::kanMX | This study |
| MMP18 | smt-0 rad62-1 uvel::his3 leu1-32 ura4-D18 his3-D1 | This study |
| MMP20 | h−leu1-32 ura 4-D18 rad62-FLAG-His smc5-Myc Kanr | This study |
| MMP21 | h−leu1-32 ura 4-D18 rad62-FLAG-His Kanr | This study |
| MMP22 | h+leu1-32 ura 4-D18 smc5-Myc Kanr | I. Miyabe |
| MMP23 | h− leu1-32 ura4-D18 his3-D1 uve1::his3 | This study |
| MMP24 | leu1-32 ura4-D18 his3-D1 rhp57::his3 rad62-1 | This study |
| MMP30 | h−mus81::kanMX | This study |
| PS3 | ade6-M375 int::pUC8/ura4/ade6-L469 | 17 |
| MPS401 | rad62-1 ade6-M375 int::pUC8/ura4/ade6-L469 | This study |
| MPS501 | rhp51::his3ade6-M375 int::pUC8/ura4/ade6-L469 | This study |
| 298 | h−rad50::LEU2 leu1-32 ura4-D18 | 42 |
| TE767 | rqh1::ura4 | 27 |
| NB2554 | mus81::kanMX | 6 |
| NCYC1979 | h−rad18-X | NCYCa |
| MMPD1 | h+/h−leu1-32/leu1-32 ura4-D18/ura4-D18 his7-366/+ ade6-M210/ade6-M216 rad62::ura4/+ | This study |
NCYC, National Collection of Yeast Cultures.
TABLE 2.
Plasmids used in this study
| Plasmid | Insertiona | Plasmid used for derivation | Source or reference |
|---|---|---|---|
| pSLR4 | rad62 genomic | pUC118 | This study |
| pSLR4D | rad62::ura4 | pSLR4 | This study |
| pREP41 | ars1 LEU2 nmt1* | 31 | |
| pREP41X | ars1 LEU2 nmt1* | 12 | |
| pBRC1 | ars1 LEU2 nmt1* brc1 cDNA | pREP41X | M. O'Connell |
| pRAD62 | ars1 LEU2 nmt1* rad62 cDNA | pREP41 | This study |
| pRAD60 | ars1 LEU2 nmt1* rad60 cDNA | pREP41 | This study |
| pREP41EGFP-N | ars1 LEU2 nmt1* EGFP | pREP41X | 9 |
| pGFP62 | ars1 LEU2 nmt1* EGFP-rad62 cDNA | pREP41EGFP-N | This study |
*, nmt medium-strength promoter.
Gamma ray, MMS, and UV sensitivity tests.
Cells were grown to mid-log phase in YES medium, resuspended in water, and then irradiated with gamma rays from a 60Co source. After irradiation, appropriately diluted samples were plated on YES plates and incubated at 30°C for 4 to 7 days before colony counting. For test of UV sensitivity, appropriately diluted samples were plated on YES plates, irradiated with the indicated dose of UV, and incubated at 30°C for 4 to 7 days. For the test of MMS sensitivity, 3 μl of sequential 10-fold dilutions of stationary-phase cells were spotted on EMM plates with the indicated MMS concentration.
Cloning of the rad62 gene.
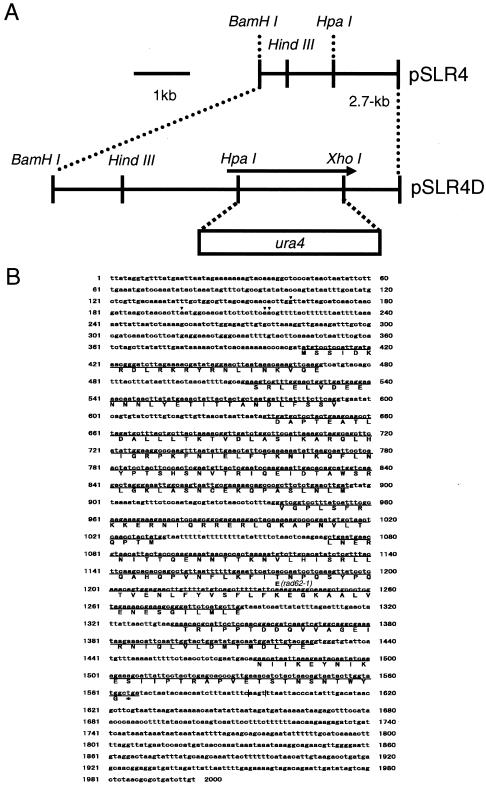
rad62-1 mutant cells were transformed with an S. pombe genomic DNA library (3), and MMS-resistant transformants were selected on plates containing 0.004% MMS (44). The plasmids that conferred MMS resistance to the mutant were isolated, and the region responsible for the recovery of the repair activity was identified (Fig. 1A) and sequenced.
FIG. 1.
Physical map of the rad62 genomic region and construction of a rad62 deletion mutant. (A) A restriction map of the rad62 region is shown at the top. The plasmid containing the rad62 deletion was constructed by replacing the HpaI-XhoI fragment in rad62 with the ura4 gene, as shown at the bottom. (B) Nucleotide sequence of the rad62 gene and predicted amino acid sequence of the Rad62 protein. The exons are underlined. Arrows show the transcription initiation sites. The poly(A) sites are shown by vertical lines. The amino acid alteration from Lys to Glu at position 223 caused by a mutation at position 1239 in rad62-1 mutant is shown.
Determination of the rad62 coding region.
The rad62 cDNA was amplified by PCR with an S. pombe cDNA library constructed on the pGAD GH vector (Clontech) as a template and analyzed as follows. Analysis of the 5′ region with primers GAD5-1 (5′-TATTCGATGATGAAGATACC-3′), corresponding to an upstream flanking sequence of the vector, and V1R1 (5′-ATAGTAGGTTGAGTTAGCAC-3′), corresponding to a middle region of a putative rad62 exon, revealed three introns, and analysis of the 3′ region with primers V1F2 (5′-GTCCTTTATCATTTCGCAAG-3′), corresponding to a region in the same putative rad62 exon, and T7 (5′-AATACGACTCACTATAG-3′), corresponding to a downstream flanking sequence of the vector, revealed an additional three introns, as shown in Fig. 1B. A 5′ rapid amplification of cDNA ends (5′ RACE) experiment to identify the transcription initiation site was carried out by using the FirstChoice RLM-RACE kit (Ambion) and the V1R1 primer. Nucleotide sequence analysis of the six clones obtained indicated that there were at least four initiation sites corresponding to the nucleotide numbers 162, 197, 216, and 217 (Fig. 1B). A 3′ RACE experiment was carried out by using the LA PCR kit, version 2 (TAKARA). DNA sequence analysis of the two clones indicated the poly(A) sites at nucleotide numbers 1592 and 1596, respectively.
Based on the above results, the rad62 open reading frame encoding 300 amino acids was identified (Fig. 1B). The cDNA for the rad62 coding region was amplified with primers S4NB (5′-GCGGATCCCATATGTCCTCCATTGATAAAC-3′), corresponding to the region for translational initiation, and S4CB (5′-GCGGATCCTCAGCCATACCAAGTATTAC-3′), corresponding to the region for translational termination by using the LA PCR kit, version 2. The resulting reverse transcription-PCR product was digested with BamHI and ligated to the BamHI site of pUC118. Nucleotide sequence analysis of the cDNA confirmed the existence of six introns.
Construction and characterization of rad62 deletion strain.
A rad62 deletion mutant was constructed essentially as described previously (15). pSLR4, a derivative of pUC118 carrying the 2.7-kb BamHI-BamHI region containing the rad62 gene (Fig. 1A), was used to generate a plasmid construct for gene disruption. The 0.9-kb HpaI-XhoI region of the rad62 gene on the plasmid was removed, and the staggered ends were converted to blunt ends by filling them in with E. coli DNA polymerase I. The ends of the 1.8-kb HindIII fragment containing the ura4 gene from pREP2 (31) were converted to blunt ends, and the fragment was ligated with the plasmid to generate pSLR4D (Fig. 1A). pSLR4D was linearized by BamHI digestion and transformed into the diploid S. pombe strain MPD1. The Ura+ transformed colonies were isolated and checked for the rad62 deletion by genomic Southern analysis. They were sporulated and subjected to tetrad analysis. For cytological observation, the diploid cells were incubated on ME at 26°C for 2 days for sporulation. The spores were treated with 0.5% glusulase, washed in water three times, and then spread onto an EMM plate lacking uracil. After 2 to 3 days, germinated spores of the rad62::ura4+ strain were collected and washed in water three times. The cells were then fixed in 70% ethanol and stained with 1.5 μg of 4, 6-diamidino-2-phenylindole (DAPI)/ml. The stained cells were observed under an epifluorescent microscope.
PFGE.
To examine the ability of the rad62-1 mutant to repair DSBs, the cells were irradiated with gamma rays at 600 Gy and the repair of DSBs was followed by pulsed-field gel electrophoresis (PFGE) as described previously (35), with slight modifications in the PFGE conditions: voltage gradient, 2 V/cm; pulse time, 30 min; angle, 120°; time, 48 h; temperature, 16°C.
Measurement of mitotic homologous recombination frequency.
The mitotic homologous recombination of rad62-1 was measured by the efficiency of integration of a linear DNA carrying the leu1+ gene into the leu1-32 locus on the chromosome as described previously (44).
Spontaneous mitotic intrachromosomal recombination frequencies of rad62-1, rhp51Δ, and wild-type cells were measured by using the strains PS3, MPS401, and MPS501, containing a nontandem direct repeat of ade6− heteroalleles flanking ura4+ as described previously (17, 38).
Construction of epitope-tagged genes.
For immunoprecipitation experiments and protein complex purification, an epitope-tagged rad62 gene generated as described previously (27a by the PCR-based method to place a FLAG-His epitope at the C terminus of the protein and mark the allele with the kanMx6 gene (27a). The rad62-1 strain containing the tagged rad62 gene was not sensitive to MMS and grew as normally as the wild-type strain, indicating that the tagged gene is functional.
Immunoprecipitation.
The cells carrying the epitope-tagged gene were grown in YES medium. Mid-log-phase cells from a 50-ml culture were collected, resuspended in 500 μl of buffer A (10 mM HEPES [pH 8.0], 0.1% NP-40, 25 mM KCl, 1 mM phenylmethylsulfonyl fluoride) containing protease inhibitor cocktail, and lysed by acid-washed glass beads. The lysate was clarified by centrifugation (12,000 × g, 15 min). Ten microliters of protein G-Sepharose 4FF (Amersham Pharmacia Biotech) prewashed in buffer A was added to adsorb nonspecific binding proteins to protein G. The suspension was incubated for 1 h at 4°C, and the supernatant was collected by centrifugation. Anti-Myc antibody (PL14; MBL) with protein G-Sepharose 4FF or anti-FLAG M2 affinity gel (Sigma) was added to the supernatant, and the suspensions were rocked for 1 h at 4°C. Immunoprecipitates were washed three times with buffer A, resuspended in 40 μl of 5% sodium dodecyl sulfate (SDS), and incubated for 10 min at 37°C. After centrifugation, the supernatants were mixed with 10 μl of SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, boiled for 10 min, separated by SDS-PAGE, and analyzed by Western blotting with the ECL Advance Western blotting detection kit (Amersham Bioscience).
Nucleotide sequence accession number.
Nucleotide sequence data, including the exon and intron information, have been deposited with the DDBJ under accession number AB158548.
RESULTS
Isolation of rad62-1 mutant defective in repairing DSBs and cloning of the gene that complemented the repair deficiency.
rad62-1 is one of the seven mutants previously isolated as sensitive to MMS and synthetically lethal with the rad2 mutation (44). The original mutant was repeatedly backcrossed with the wild-type strain, and the clones sensitive to MMS were selected. A genomic DNA fragment that conferred the MMS resistance to the mutant was isolated from an S. pombe genomic DNA library. The original plasmid carried a 4.1-kb genomic DNA insert, whose physical map is shown in Fig. 1A. The region that complemented the MMS sensitivity was subcloned on a shuttle vector, and the DNA sequence of the 2.8-kb BamHI region was determined. The sequenced region matches a region contained on the cosmid c20F10 (EMBL accession number AL021747).
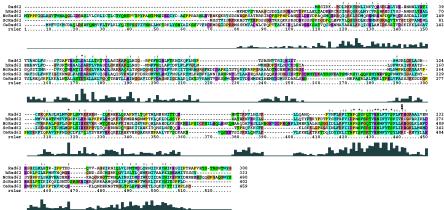
The rad62 coding region was analyzed by the 5′ and 3′ RACE and reverse transcription-PCR methods, and the existence of seven exons in the rad62 gene was shown by sequence analysis of the cDNA (Fig. 1B). The gene is unusual in having at least as many as four transcription initiation sites and such a long untranslated upstream exon. The open reading frame of rad62 was determined from these analyses and is shown in Fig. 1B. It was different from the one described in the DNA database as SPBC20F10.04c. The gene encodes a protein with 300 amino acids with no apparent functional sequence motifs, as analyzed by using databases (http://motif.genome.ad.jp/). Database search revealed that the Rad62 homologs exist ubiquitously throughout eukaryotes, as shown in Fig. 2. The nucleotide sequence covering the rad62 region of the rad62-1 allele and its flanking genomic regions were amplified by PCR, and the nucleotide sequence was analyzed directly. A single transition mutation from A to G was found at the first base of codon 223, altering the AAA codon for lysine to a GAA codon for glutamic acid. Codon 223 is likely to be important for function, as the corresponding sites are conserved as positive residues, including Lys and Arg, and the mutation changing Lys to an acidic residue, Glu, caused the functional defect in the rad62-1 mutant.
FIG. 2.
Sequence alignment of Rad62 proteins from various organisms. Multiple-sequence alignment was performed with CLUSTAL X, version 1.81 (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/). hRad62, human Rad62 (accession number AAH27612.1); NcRad62, Neurospora crassa Rad62 (accession number E1136561.1); ScRad62, S. cerevisiae Rad62 (Qri2) (accession number CAA55925.1); CeRad62, Caenorhabditis elegans Rad62 (accession number CAB09113.1). The arrow shows the position of the rad62-1 mutation. The levels of sequence homology are shown by the heights of the blocks at the bottom.
The rad62-1 mutant is defective in repairing DNA damage and epistatic with rhp51.
The sensitivities of the rad62-1 mutant to DNA-damaging agents, UV and gamma rays, were examined by counting survival fractions after irradiation at various doses (Fig. 3). The mutant showed two phases of response to UV irradiation, a highly sensitive phase at low doses up to 25 J/m2 and a relatively resistant phase at higher doses than that. The rhp51 gene, which is a homolog of S. cerevisiae RAD51 and E. coli recA, plays a major role in repairing DSBs by homologous recombination and is required for the UVER repair pathway, which is unique to fission yeast and some fungi (29, 32). The rad62-1 mutant was more sensitive to low doses (up to 50 J) of UV than the rhp51Δ mutant, but it was less sensitive at higher doses. rhp51Δ partially suppresses the UV sensitivity of rad62-1 at the lower doses, and the rad62-1 rhp51Δ double mutant showed a similar UV survival curve to that of the rhp51Δ single mutant. This result suggests that, at low doses of UV, the Rhp51 protein converts DNA damage to a toxic recombination intermediate in the rad62-1 mutant. The rad62-1 mutant is also hypersensitive to gamma rays, but it is less sensitive than the rhp51Δ mutant at all doses tested. The double mutant showed the same sensitivity as the rhp51Δ single mutant. These results suggest that rhp51 is epistatic to rad62 with respect to the DNA repair function. rhp57Δ is also epistatic to rad62-1 (data not shown). rhp51 is epistatic to rhp57, and both of them are involved in recombination repair (25, 44). rad60-1 and smc6-X (rad18-X) mutants are hypersensitive to UV and gamma rays and show a similar epistasis with rhp51 (30, 35).
FIG. 3.
The rad62-1 mutant is defective in DNA repair. (A and B) Sensitivities of rad62-1 mutant to UV and gamma rays. The cells at mid-logarithmic phase were irradiated with UV or gamma rays at the indicated doses, and the relative plating efficiencies were determined. Wild-type strain, MMP1 (♦); rad62-1 strain, MMP2 (▪); rhp51Δ strain, MMPX3 (▴); rad62-1 rhp51Δ strain, MMPX4 (•). The data represent the averages of the results from three experiments. (C) The rad62-1 mutant is defective in repairing DNA DSBs. Genomic DNA of wild-type and rad62-1 strains was analyzed by PFGE. Samples were taken at the indicated times (in hours) after irradiation by gamma rays at 500 Gy (+) or before irradiation (−).
Since rad62 belongs to the same epistasis group as the recombination repair genes rhp51 and rhp57 and the rad62-1 mutant showed phenotypes very similar to those of the rad60-1 and smc6-74 (rad18-74) mutants (35, 47), we examined whether the mutant was also defective in repairing DSBs. DSBs were induced by irradiation at 500 Gy, and DNA isolated at various times after irradiation was analyzed by PFGE. As shown in Fig. 3B, fragmented chromosomes were mostly rejoined in wild-type cells by incubation for 3 h while they remained unjoined fragments in the rad62-1 cells even after incubation for 6 h. The results strongly support the hypothesis that Rad62, Rad60, Smc5, and Smc6 function in the same pathway with recombination proteins such as Rhp51 and Rhp57 for repairing damaged DNA.
The synthetic lethality of rad62-1 with rad2Δ was examined by constructing a diploid strain by mating the two mutant strains rad62-1 (MMP7) and rad2Δ (MMP5) and analyzing the resultant tetrads. The double mutants formed extremely small colonies, and when they were streaked on a YES plate, they showed little sign of growth, confirming that the two mutations are synthetically lethal (data not shown).
rad62-1 is not defective in Rhp51-dependent genetic recombination.
Since the efficiency of homology-dependent genetic recombination is reduced in rhp51 and rhp57 mutants, we examined the ability of the rad62-1 mutant to carry out the recombination. The efficiency of integrating the leu1+ gene into the leu1-32 locus of the rad62-1 mutant (MMPX6) was not reduced, whereas it was reduced threefold (34%) in the rhp57Δ mutant (MMPX5). This shows that the rad62-1 mutant is not defective in recombination during mitosis, as examined by the integration of linear homologous DNA into the chromosome. rad60-1 was also not defective in such recombination (our unpublished data).
Next we examined spontaneous mitotic intrachromosomal recombination frequencies in the rad62-1 mutant containing a nontandem direct repeat of ade6− heteroalleles flanking ura4+, and intrachromosomal mitotic recombination was assayed by the formation of Ade+ recombinants. Two main classes of Ade6+ recombinants could be distinguished: Ade+ Ura− deletion type recombinants and Ade+ Ura+ conversion type recombinants.
The frequency of conversion type of recombination was much reduced in the rhp51Δ mutant (0.16 ± 0.07 per viable 104 cells) compared to wild-type cells (1.58 ± 0.30), but it was not reduced in rad62-1 cells (1.42 ± 0.72 per 104 viable cells). This result suggests that the rad62-1 mutant is not defective in Rhp51-dependent spontaneous intrachromosomal recombination such as the gene conversion type of recombination. On the other hand, deletion type recombination was significantly increased in the rhp51Δ mutant (9.68 ± 1.44 per 104 viable cells) compared to wild-type cells (1.29 ± 0.85 per 104 viable cells and it was not affected in the rad62-1 mutant (1.30 ± 0.59 per 104 viable cells). Therefore, the rad62-1 mutant is similar to the wild-type strain and different from rhp51 and rhp57 in regard to the recombination phenotypes examined by these methods.
rad62 is essential for growth.
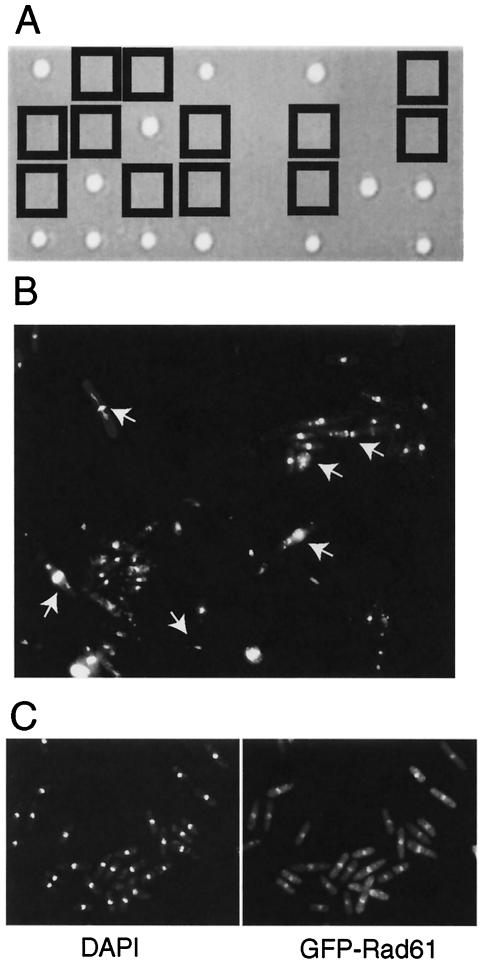
To study the phenotype of the rad62 null mutant, 70% of the rad62 coding region of one of the chromosomes in a diploid strain was replaced with the DNA fragment containing the ura4 gene by homologous recombination. The heterozygous diploid cells (MMPD1) were sporulated and subjected to tetrad analysis. Thirty asci were dissected and allowed to germinate on YES medium at 30°C, and only two viable segregants arose from each of the 30 asci (Fig. 4A). All 60 segregants required uracil for growth, indicating that these segregants did not carry the rad62::ura4+ allele and that the rad62+ gene is essential for growth. Microcolonies consisting of a few 10s of elongated cells were formed from the rad62::ura4+ segregants. The terminal morphology of the mutant cells grown for 2 days on a minimal plate lacking uracil was examined under an epifluorescent microscope after staining with DAPI (Fig. 4B). They were elongated, and the nuclei were mostly abnormal, sometimes extended, shrunk, fragmented, bisected by a septum (cut phenotype), or biased in position. The terminal morphology was very similar to that of rad60 and smc5 (spr18) deletion cells (13, 35).
FIG. 4.
Rad62 is an essential nuclear protein. (A) Tetrad analysis of rad62 heterozygous diploids. The diploid strain heterozygous for the rad62 deletion mutation was sporulated and subjected to tetrad analysis. The segregants from the dissected spores were grown on a YES plate at 30°C for 5 days. The rad62Δ segregants are boxed. (B) Terminal morphology of rad62Δ cells. The diploid strain MMPD1 was sporulated and germinated on an EMM2 plate lacking uracil. The rad62Δ cells grown for 2 days were fixed with 70% ethanol, stained with DAPI (1 μg/ml) and calcofluor white (20 μg/ml), and photographed. The abnormal nuclei are shown by arrows. (C) Nuclear localization of Rad62 protein. Cells of the rad62Δ (MMP8) carrying a plasmid for expression of GFP-Rad62 fusion protein, pGFP62, were fixed, stained with DAPI (1 μg/ml), and observed under an epifluorescence microscope. Fluorescence images of DAPI (left) and GFP-Rad62 (right) are shown.
The localization of Rad62 protein in the cell was examined by expressing Rad62 protein tagged with enhanced green fluorescent protein (EGFP) at its N terminus in the rad62Δ strain (MMP8). The cells were fixed, DNA stained with DAPI, and observed under an epifluorescent microscope (Fig. 4C). Rad62 colocalized with DNA, suggesting that it localizes to the nucleus.
rad62 genetically interacts with rad60, smc6, and brc1.
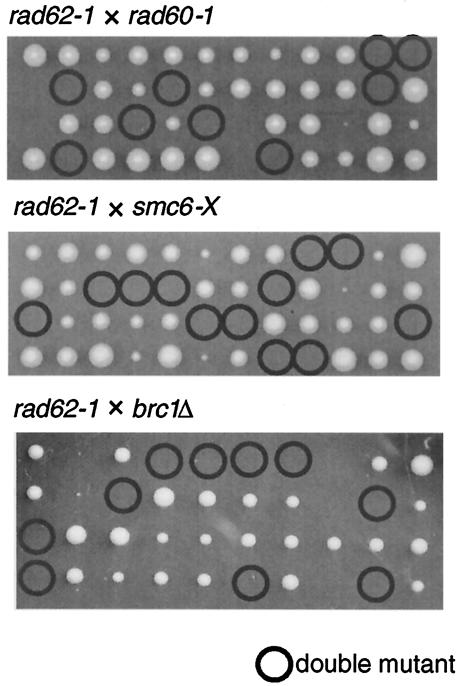
Since the phenotypes of rad62 mutants were very similar to those of rad60 and smc6 mutants, we suspected that these genes may be involved in a common pathway for maintaining chromosome integrity and repairing DNA damage by recombination. The brc1 gene was isolated as an allele-specific multicopy suppressor of the smc6-74 (rad18-74) mutation, and the deletion mutant, which is viable, was synthetically lethal with smc6 mutants, indicating close functional interaction with smc6 (47). The genetic interactions among the above genes were examined by constructing double mutants between rad62-1, rad60-1, smc6-X (rad18-X), and brc1. The mutants were crossed between each other, and the spores were subjected to tetrad analysis at 26°C. The double mutants combining any of the above mutations were inviable (Fig. 5).
FIG. 5.
rad62-1 is synthetically lethal with rad60-1, smc6-X and brc1Δ. rad62-1 was crossed with rad60-1, smc6-X or brc1Δ, and spores were subjected to tetrad analysis at 26°C. The double mutants are circled. Some single mutants formed small colonies, but they showed normal growth when restreaked on plates.
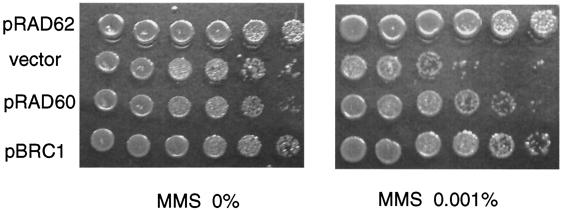
The rad60 gene overexpressed from a multicopy plasmid partially suppressed the MMS hypersensitivity of the smc6-X (rad18-X) mutant (35). We examined whether overexpression of the rad60 or brc1 gene suppressed the MMS hypersensitivity of rad62-1. The pBRC1 plasmid carrying brc1+ under the thiamine-repressible promoter suppressed the MMS and UV sensitivities of the rad62-1 mutant either under repressed or derepressed conditions (Fig. 6). The pRAD60 plasmid carrying rad60+ under the weaker nmt1 promoter suppressed the MMS and UV sensitivities of the rad62-1 mutant under derepressed conditions (Fig. 6). Furthermore, overexpression of Brc1 partly suppressed the MMS and UV sensitivity and raised the permissive temperature of the rad60-1 mutant (data not shown). The results of the genetic interactions revealed here and by previous works (35, 47) are summarized in Tables 3 and 4. These results strongly suggest that Rad62 is functionally related to Rad60, Smc5, Smc6, and Brc1 and that these proteins collaborate in repairing DNA damage and maintaining chromosome integrity during replication.
FIG. 6.
Suppression of MMS sensitivity of rad62-1 by overexpression of Rad60 or Brc1. rad62-1 cells (MMP7) carrying the pRAD62 plasmid, a vector plasmid, pREP41, the pRAD60 plasmid, or the pBRC1 plasmid were grown to saturation on EMM with required supplements. Serial 10-fold dilutions were spotted on YES plates with or without MMS. The plates were incubated at 30°C for 3 days.
TABLE 3.
Synthetic lethality of double mutations
| Gene | Lethality upon double mutation with:
|
||||
|---|---|---|---|---|---|
| rad62-1 | rad60-1 | brclΔ | smc6-74 | smc6-X | |
| rad62-1 | Lethal | Lethal | NDc | Lethal | |
| rad60-1 | Lethal | ND | Lethalb | ||
| brc1Δ | Lethala | Lethala | |||
| smc6-74 | |||||
| smc6-X | |||||
Verkade et al. (47).
Morishita et al. (35).
ND, not determined.
TABLE 4.
Suppression of UV or MMS sensitivity by overexpression of Smc5-6 related proteins
| Protein | Result with overexpression of:
|
||||
|---|---|---|---|---|---|
| rad62-1 | rad60-1 | brclΔ | smc6-74 | smc6-X | |
| Rad62 | No effect | NDfg | ND | No effecte | |
| Rad60 | Suppressedc | NDf | ND | Suppressedbe | |
| Brc1 | Suppressedc | Suppressedc | Suppressedad | Suppressedad | |
| Smc6 | No effect | No effect | NDf | ||
Verkade et al. (47).
Morishita et al. (35).
UV and MMS sensitivity.
UV sensitivity.
MMS sensitivity.
brc1Δ is not sensitive to UV.
ND, not determined.
Rad62 physically interacts with the Smc5-6 complex.
To identify proteins associated with Rad62, we purified a complex containing FLAG-His-tagged Rad62. MMP20 cells carrying the FLAG-His-tagged rad62 gene and the control cells were grown, and the extracts were prepared. The protein complex containing the tagged Rad62 was purified and analyzed by SDS-PAGE. In addition to a band corresponding to the tagged Rad62 protein (38 kDa), a 120-kDa protein band was specifically identified in the preparation from the MMP20 cells (data not shown). Since multiple genetic interactions of Rad62 with the Smc5-6 complex were observed in this work and the size of Smc5 and Smc6 proteins are about 120 kDa, we suspected that the 120-kDa protein found in the Rad62 complex was either Smc5, Smc6, or the doublet of them.
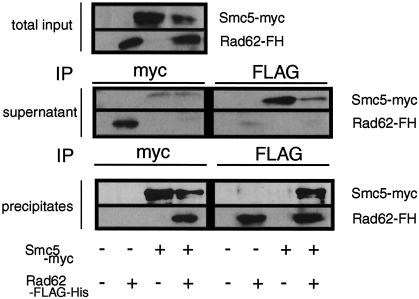
To examine whether Rad62 was included in the Smc5-6 complex, we performed coimmunoprecipitation experiments with FLAG-His-tagged Rad62 and Myc-tagged Smc5 expressed from their cognate promoters. From the cells expressing FLAG-His-tagged Rad62 and Myc-tagged Smc5, the tagged Rad62 was coimmunoprecipitated with the tagged Smc5 by the anti-Myc antibody and the tagged Smc5 was coimmunoprecipitated with the tagged Rd62 by the anti-FLAG M2 antibody (Fig. 7). As a negative control experiment, Rad62 was not immunoprecipitated by the anti-Myc antibody from the cells expressing FLAG-His-tagged Rad62 and untagged Smc5. The tagged Smc5 was not immunoprecipitated by the anti-FLAG M2 antibody from the cells expressing Myc-tagged Smc5 and untagged Rad62. From the comparison of the amounts of Rad62 and Smc5 in the extracts, supernatants, and immunoprecipitates, we conclude that large majorities of Rad62 and Smc5 were physically associated with each other (Fig. 7). These results suggest that Rad62 forms a protein complex with Smc5 in vivo.
FIG. 7.
Physical association of Rad62 with Smc5. The association of Rad62 and Smc5 in vivo was examined by coimmunoprecipitation of Rad62-FLAG-His (Rad62-FH) and Smc5-Myc. Cells expressing both tagged proteins (MMP20) or singly tagged protein (MMP21 or MMP22) were prepared and immunoprecipitated with anti-Myc or anti-FLAG. The immune complexes were separated by SDS-PAGE and immunoblotted with antibodies against Myc or FLAG. Sample volumes were adjusted to make a direct comparison of the relative amounts of Rad62-FH and Smc5-Myc in the extracts, supernatants, and precipitates possible.
Relationship of rad62-1 with genes involved in recovery from replication block.
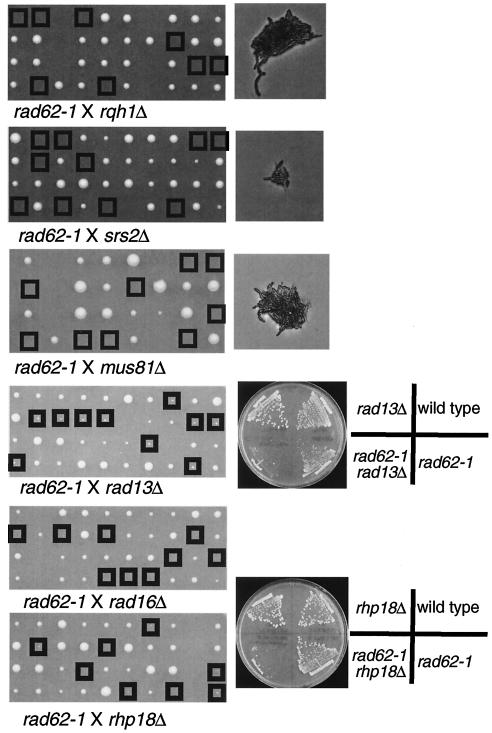
It occurred to us that one of the functions of rad62+ may be to provide a pathway for a resolution of the replication block by recombination. To test this hypothesis, we examined the synthetic lethality of the rad62-1 mutation with other mutations in the genes involved in repairing DNA damage and resolving the replication fork block one way or another (Fig. 8). The mutant became lethal when combined with mutations in rqh1+, encoding a RecQ DNA helicase homolog involved in recombination repair (36); srs2+, whose budding yeast homolog encodes a DNA helicase proposed to remove Rad51 recombination protein from toxic recombination intermediates (28, 46); or mus81, encoding a structure-specific endonuclease proposed to resolve recombination intermediates and rescue stalled replication forks (5, 6, 22). It showed a severe growth defect when combined with a mutation in rhp18+, which encodes a subunit of the ubiquitin ligase that regulates postreplication repair (48). It is also synthetically lethal or nearly so when combined with mutations in the genes involved in nucleotide excision repair, rad16+ or rad13+, encoding an excision endonuclease (29) (Fig. 8). Since these genes play roles in repairing various types of DNA damage, the only common feature caused by the mutations will be a block to replication, and together, these data suggest that the function defective in rad62-1 becomes essential for viability when replication blocks are not resolved during replication.
FIG. 8.
Synthetic growth defect of rad62-1 with rqh1Δ, srs2Δ, mus81Δ, rad13Δ, rad16Δ, and rhp18Δ. rad62-1 was crossed with rqh1Δ, srs2Δ, mus81Δ, rad13Δ, rad16Δ, or rhp18Δ, and spores were subjected to tetrad analysis at 30°C. The double mutants are boxed. Small colonies were formed from the double mutants, rad62-1 rad13Δ and rad62-1 rhp18Δ, and they were tested for growth by streaking on plates. Some single mutants formed small colonies like the double mutants on the figure, but they showed normal growth when restreaked on plates.
DISCUSSION
In this study, we identified another novel essential gene, rad62, whose function is very similar to the one previously reported for rad60 (35). Both of them are essential for viability, required to repair DSBs, and epistatic with recombination genes rhp51 and rhp57 with respect to their DNA repair function. The terminal morphologies of the deletion mutant cells were very similar; cells are elongated with abnormal nuclear structures, diffuse, shrunk, fragmented, or cut, where the nuclear material is bisected by a septum. These phenotypes are also shared by smc6 (rad18) mutants (13, 30, 47) and the recently identified nse1 and nse2 mutants (19, 33). S. cerevisiae nse1 mutants showed similar phenotypes (14). Smc6, like other SMC proteins, forms a heterodimer with its partner Smc5 (13), and they form a tight complex with Nse1 and Nse2 (33). Only a very small proportion of Rad60 (0.5 to 2.0%) was detected to be associated with Smc5-6 (7). We have demonstrated in this work that rad62 shows very intimate genetic interactions with rad60, smc6, and brc1. These results strongly suggest that Rad62 collaborates with the Smc5-6 complex for maintenance of chromosome integrity and repairing DSBs. Indeed, we demonstrated that the majority of Rad62 is associated with Smc5 in vivo, and thus, it is a subunit of the Smc5-6 complex (Fig. 7). We could not detect the association of Rad60 with Smc6 by similar experiments (unpublished data). Therefore, Rad60 association with the Smc5-6 complex is different from the association of the other subunits, as previously reported (7). Although, Rad62 was shown to associate with Smc5, it remains to be shown whether the interaction is direct or through other subunits of the complex. According to a recent paper that identified proteins copurifying with Smc5 and Smc6 in S. cerevisiae, the Smc5-6 proteins form two distinct complexes, one with Qri2 (Rad62 homolog), Nse1, Mms21 (Nse2 homolog), and an uncharacterized protein, YDR288W, and another with two uncharacterized proteins, YML023C and Kre29 (20). The homologs of the components of the first complex have been characterized for S. pombe, and physical interactions among Smc5, Smc6, Nse1, and Nse2 have been demonstrated (13, 19, 33). The phenotypes of the mutants were very similar. We recently identified a homolog of YDP288W in S. pombe named nse3 by a two-hybrid experiment with rad62 as a bait, and the mutants show similar phenotypes with those of the mutants of the smc5-6-related genes (H. Morikawa, I. Miyabe, T. Morishita, and H. Shinagawa, unpublished data). Although a fraction of S. pombe Rad60 was found in association with the Smc5-6 complex (7), its homolog in S. cerevisiae, Esc2, was not found in either of the Smc5-6 complexes. The ESC2 gene is not essential for viability and is involved in the silencing of genes in silent mating type loci and telomere regions (10). Since none of the non-SMC components of the latter complex have been studied, whether or not the two subcomplexes are involved in the same function is an open question.
We made an extensive analysis of the genetic interactions of rad62 with other genes involved in resolving the replication fork block in various ways (Fig. 8 and Table 1). The rad62-1 mutant was synthetically lethal with mutations in the rqh1, srs2, mus81, and rhp18 genes, all of which are required for repairing DNA damage or bypassing blocks to replication (26). To our surprise, it was also synthetically lethal with mutations in rad16 and nearly so with mutations in rad13, which encodes nucleases required for making incisions at the sites flanking large DNA adducts, such as pyrimidine dimers in nucleotide excision repair (29). These results are consistent with Rad62 being required for viability when replication is blocked by spontaneous damage in the absence of the functions that repair the damage or override the block by translesion DNA synthesis. The rad62 gene seems to possess at least two discrete functions, one essential for growth and the other for repair. The latter function is epistatic with rhp51, which plays a dominant role in homologous recombination and recombination repair. The rhp51 mutation could not suppress the lethality of a rad62 deletion mutant. So lethality is not due to the accumulation of recombination intermediates. Both the rad62-1 and rad60-1 mutants were not defective in homology-dependent recombination, as examined by integration of homologous linear DNA into the chromosome and by formation of spontaneous intrachromosomal conversion type recombinants. Preliminary results suggest that rad60-1 is not defective in meiotic recombination (data not shown). However, rad62 and rad60 belong to the same epistasis group with rhp51 and rhp57 with respect to repair function. The apparent discrepancies could be resolved if we assume that they function for recombination repair only at the replication fork but are not required for recombination in nonreplicating regions of chromosome. However, we cannot rule out a role for rad60 and rad62 in genetic recombination, since they are essential genes and these hypomorphic alleles may not give a strong change in recombination rates.
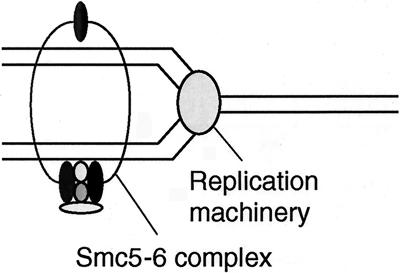
The Smc5-Smc6-Nse1-Nse2-Rad62 complex together with Rad60 may be a specific SMC complex functioning only at the replication fork (Fig. 9). Human Smc6 localizes to the nuclear region during the interphase and is excluded from condensed mitotic chromosomes (41). The Smc5-6 complex may provide a scaffold for the recombination reaction by holding the newly replicated sister chromatids in close proximity so that DSBs produced by replication block can be efficiently repaired by homologous recombination. The essential role of this SMC complex may be to maintain proper structure of the chromosome at the fork during replication. Spontaneous fragmentation of chromosomes and the diffuse nuclear structure observed in rad60-1 cells grown at the nonpermissive temperature may be caused by the defect of this function. The fact that the rhp51Δ mutation partially suppresses UV sensitivity at lower doses than the rad62-1 mutant (Fig. 3B) may suggest that Rhp51 bound to the UV-damaged fork may be toxic rather than beneficial when the newly replicated chromatids are not properly held. Since Rad60 seems transiently associated with Smc5-6 in contrast to other non-SMC components, which are more tightly associated, and is phosphorylated in response to a replication block in a manner dependent on Cds1 kinase (7), Rad60 may regulate the function of the Smc5-6 complex in response to the replication block. Further experiments are required to prove that the Smc5-6 complex functions at the replication fork.
FIG. 9.
Model of the Smc5-6 complex for maintenance of chromosome integrity and repair of DSBs at the replication fork. The Smc5-6 complex holds replicated chromatids in close proximity at the replication fork. Smc5 and Smc6 proteins form a heterodimeric tight complex similar to other SMC protein complexes like condensin and cohesin. Nse1, Nse2, and Rad62 associate tightly with the complex. Rad60 and Brc1 may transiently associate with the complex.
We examined the epistatic relationship with other repair and recombination genes, and the results are summarized in Table 5. Interestingly the UV sensitivity of the rad62-1 mutant was additive with rad50Δ and swi5Δ. rad50 has been proposed to play a role in promoting recombination between sister chromatids in collaboration with the cohesin complex (18). Swi5 has dual roles, one in mating type switching and the other in recombination repair in a novel pathway dependent on rhp51 but independent of rhp55-57 (1). Since the swi5Δ rhp57Δ double mutants are additive in UV sensitivity and equally as sensitive as rhp51Δ, the recombination repair pathway involving the Smc5-6 complex function is likely to be the same as the one involving the rhp55-57 functions. The additive UV sensitivity in the rad62-1 uve1Δ double mutant indicates that rad62 may function in a pathway different from the UV nucleotide excision repair pathway (32).
TABLE 5.
Effects of repair mutations on UV sensitivity of rad62-1 mutant
| Mutationa | UV sensitivity |
|---|---|
| rhp51Δ | Epistatic |
| rhp57Δ | Epistatic |
| rad50Δ | Additive |
| swi5Δ | Additive |
| uve1Δ | Additive |
Strains used are shown in Table 1.
It should be noted that cohesion- and condensin-related genes like rad21/scc1 (4, 21) and cnd2 (2) are also involved in repairing DNA damage. The cnd2-1 mutant is sensitive to hydroxyurea, and cnd2 is required for activation of the checkpoint kinase Cds1 (2). Rad60 is phosphorylated in a manner dependent on the cds1 function upon replication block (7), and both rad60-1 and rad62-1 are sensitive to hydroxyurea and defective in maintaining checkpoint arrest (our unpublished data) in a similar manner to that of smc6-74 and nse1 mutants (19, 47). Therefore, the integrity of the Smc5-6 complex seems to be essential for the maintenance of the checkpoint. The complex may be monitoring the completion of DNA repair, as suggested recently (19), and premature resumption of mitosis in these mutants may cause disintegration of the chromosomes, resulting in stretched or bisected nuclear structures. The essential role of the condensin complex in mitosis is well documented, but the involvement of the condensin complex in DNA repair in the interphase has been only recently demonstrated by the analysis of the phenotypes of the cdn2-1 mutant (2). Conversely, defects in S-phase function in the Smc5-6 complex affect mitosis. rad60, rad62, and smc6 mutations cause abnormal nuclear segregation. Recent studies on various SMC complexes have revealed interacting roles of the SMC complexes in the functions related to the DNA damage response, and the importance of the Smc5-6 complex in these processes is just beginning to be unveiled.
Acknowledgments
We are grateful to Matthew J. O'Connell, Paul Russell, Tamar Enoch, Juerg Kohli, Izumi Miyabe, Yufuko Akamatsu, Masaru Ueno, and Yasuhiro Tsutsui for strains and plasmids and to Johanne M. Murray for critically reading the manuscript. We thank Takashi Hishida for useful suggestions and Toshiji Ikeda for gamma irradiation.
This work is supported by the Human Frontier Science Program (A.M.C. and H.S.) and Grants-in-Aid for Scientific Research of Priority Areas from Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.S.
REFERENCES
- 1.Akamatsu, Y., D. Dziadkowiec, M. Ikeguchi, H. Shinagawa, and H. Iwasaki. 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100:15770-15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aono, N., T. Sutani, T. Tomonaga, S. Mochida, and M. Yanagida. 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197-202. [DOI] [PubMed] [Google Scholar]
- 3.Barbet, N., W. J. Muriel, and A. M. Carr. 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114:59-66. [DOI] [PubMed] [Google Scholar]
- 4.Birkenbihl, R. P., and S. Subramani. 1992. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20:6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates III, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 6.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boddy, M. N., P. Shanahan, W. H. McDonald, A. Lopez-Girona, E. Noguchi, I. J. Yates, and P. Russell. 2003. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 23:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, Y., and T. Kogoma. 1995. The mechanism of recA polA lethality: suppression by RecA-independent recombination repair activated by the lexA(Def) mutation in Escherichia coli. Genetics 139:1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221:59-68. [DOI] [PubMed] [Google Scholar]
- 10.Cuperus, G., and D. Shore. 2002. Restoration of silencing in Saccharomyces cerevisiae by tethering of a novel Sir2-interacting protein, Esc8. Genetics 162:633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fousteri, M. I., and A. R. Lehmann. 2000. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujioka, Y., Y. Kimata, K. Nomaguchi, K. Watanabe, and K. Kohno. 2002. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J. Biol. Chem. 277:21585-21591. [DOI] [PubMed] [Google Scholar]
- 15.Grimm, C., J. Kohli, J. Murray, and K. Maundrell. 1988. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet. 215:81-86. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, S., C. H. Haering, and K. Nasmyth. 2003. Chromosomal cohesin forms a ring. Cell 112:765-777. [DOI] [PubMed] [Google Scholar]
- 17.Gysler-Junker, A., Z. Bodi, and J. Kohli. 1991. Isolation and characterization of Schizosaccharomyces pombe mutants affected in mitotic recombination. Genetics 128:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartsuiker, E., E. Vaessen, A. M. Carr, and J. Kohli. 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20:6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey, S. H., D. M. Sheedy, A. R. Cuddihy, and M. J. O'Connell. 2004. Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol. Biol. Cell 24:662-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 21.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399-414. [DOI] [PubMed] [Google Scholar]
- 22.Hollingsworth, N. M., and S. J. Brill. 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishioka, K., A. Fukuoh, H. Iwasaki, A. Nakata, and H. Shinagawa. 1998. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells 3:209-220. [DOI] [PubMed] [Google Scholar]
- 24.Ishioka, K., H. Iwasaki, and H. Shinagawa. 1997. Roles of the recG gene product of Escherichia coli in recombination repair: effects of the delta recG mutation on cell division and chromosome partition. Genes Genet. Syst. 72:91-99. [DOI] [PubMed] [Google Scholar]
- 25.Jang, Y. K., Y. H. Jin, Y. S. Shim, M. J. Kim, E. J. Yoo, I. S. Choi, J. S. Lee, R. H. Seong, S. H. Hong, and S. D. Park. 1996. Identification of the DNA damage-responsive elements of the rhp51+ gene, a recA and RAD51 homolog from the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 251:167-175. [DOI] [PubMed] [Google Scholar]
- 26.Kai, M., and T. S. Wang. 2003. Checkpoint responses to replication stalling: inducing tolerance and preventing mutagenesis. Mutat. Res. 532:59-73. [DOI] [PubMed] [Google Scholar]
- 27.Kostrub, C. F., F. al-Khodairy, H. Ghazizadeh, A. M. Carr, and T. Enoch. 1997. Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol. Gen. Genet. 254:389-399. [DOI] [PubMed] [Google Scholar]
- 27a.Krawchuk, M. D., and W. P. Wahls. 1999. High-efficiency gene targeting in Schizosaccharcmyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15:1419-1427. [DOI] [PMC free article] [PubMed]
- 28.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 29.Lehmann, A. R. 1996. Molecular biology of DNA repair in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 363:147-161. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann, A. R., M. Walicka, D. J. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr. 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15:7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 32.McCready, S. J., F. Osman, and A. Yasui. 2000. Repair of UV damage in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 451:197-210. [DOI] [PubMed] [Google Scholar]
- 33.McDonald, W. H., Y. Pavlova, J. R. Yates III, and M. N. Boddy. 2003. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 278:45460-45467. [DOI] [PubMed] [Google Scholar]
- 34.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 35.Morishita, T., Y. Tsutsui, H. Iwasaki, and H. Shinagawa. 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22:3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray, J. M., M. Tavassoli, R. al-Harithy, K. S. Sheldrick, A. R. Lehmann, A. M. Carr, and F. Z. Watts. 1994. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol. Cell. Biol. 14:4878-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osman, F., M. Adriance, and S. McCready. 2000. The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 38:113-125. [DOI] [PubMed] [Google Scholar]
- 39.Osman, F., E. A. Fortunato, and S. Subramani. 1996. Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics 142:341-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor, E. M., J. S. Moghraby, J. H. Lees, B. Smit, P. B. Moens, and A. R. Lehmann. 2001. Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell 12:1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita, K., A. Matsuura, T. Caspari, A. M. Carr, Y. Akamatsu, H. Iwasaki, K. Mizuno, K. Ohta, M. Uritani, T. Ushimaru, K. Yoshinaga, and M. Ueno. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 23:5186-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 44.Tsutsui, Y., T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa. 2000. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics 154:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueno, M., T. Nakazaki, Y. Akamatsu, K. Watanabe, K. Tomita, H. D. Lindsay, H. Shinagawa, and H. Iwasaki. 2003. Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol. Cell. Biol. 23:6553-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Komen, S., M. S. Reddy, L. Krejci, H. Klein, and P. Sung. 2003. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 278:44331-44337. [DOI] [PubMed] [Google Scholar]
- 47.Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr, and M. J. O'Connell. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10:2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verkade, H. M., T. Teli, L. V. Laursen, J. M. Murray, and M. J. O'Connell. 2001. A homologue of the Rad18 postreplication repair gene is required for DNA damage responses throughout the fission yeast cell cycle. Mol. Genet. Genomics 265:993-1003. [DOI] [PubMed] [Google Scholar]