Abstract
The human transcription factor CA150 modulates human immunodeficiency virus type 1 gene transcription and contains numerous signaling elements, including six FF domains. Repeated FF domains are present in several transcription and splicing factors and can recognize phosphoserine motifs in the C-terminal domain (CTD) of RNA polymerase II (RNAPII). Using mass spectrometry, we identify a number of nuclear binding partners for the CA150 FF domains and demonstrate a direct interaction between CA150 and Tat-SF1, a protein involved in the coupling of splicing and transcription. CA150 FF domains recognize multiple sites within the Tat-SF1 protein conforming to the consensus motif (D/E)2/5-F/W/Y-(D/E)2/5. Individual FF domains are capable of interacting with Tat-SF1 peptide ligands in an equivalent and noncooperative manner, with affinities ranging from 150 to 500 μM. Repeated FF domains therefore appear to bind their targets through multiple weak interactions with motifs comprised of negatively charged residues flanking aromatic amino acids. The RNAPII CTD represents a consensus FF domain-binding site, contingent on generation of the requisite negative charges by phosphorylation of serines 2 and 5. We propose that CA150, through the dual recognition of acidic motifs in proteins such as Tat-SF1 and the phosphorylated CTD, could mediate the recruitment of transcription and splicing factors to actively transcribing RNAPII.
Dynamic associations between molecular complexes involved in pre-mRNA processing events are important for sustaining efficient transcription in the nucleus. Protein modules such as WW and FF domains participate in these complexes by regulating specific protein-protein interactions. The potential consequences of these associations range from changes in the rates of transcriptional elongation to splice site recognition and capping of pre-mRNA. The human transcription factor CA150 is a regulator of RNA polymerase II (RNAPII) activity that contains numerous elements characteristic of signaling proteins, including three WW and six FF domains (10, 52).
Overexpression of CA150 reduces the ability of the human immunodeficiency virus type 1 (HIV-1) transactivator (Tat) to mediate production of transcripts from the long terminal repeat (LTR) promoter (53). This function requires core promoter elements such as an intact TATA box, suggesting a role for CA150 in HIV-1 gene regulation, and is dependent on both the WW and FF domains of CA150, which can associate with the pre-mRNA splicing factor SF1 and RNAPII, respectively (11, 21). Such interactions may bridge splicing complexes to actively transcribing RNAPII. Although there is excellent evidence for transcriptional coupling to the spliceosome (32, 34, 35, 44), there have been few examples of direct interactions between known elongation and splicing factors; thus, the ability of CA150 to engage both the transcriptional and splicing machinery is intriguing. Furthermore, the domains of CA150 orthologs from Caenorhabditis elegans to humans are highly conserved, arguing that they control biologically important protein interactions.
While WW domains have been extensively characterized (51), the functions of the more recently described FF domain (6) are not as well understood. FF domains are usually found in repeats of 4 to 6 and are present in several nuclear proteins related to splicing and transcription, as well as the p190 RhoGAP family of GTPases. The solution structure of a single FF repeat from HYPA/FBP11 revealed three α-helices arranged in an orthogonal bundle with the N and C termini at opposite ends, a feature that could facilitate the linking of multiple functional repeats (1). FF domains from CA150, HYPA/FBP11, and the yeast splicing factor Prp40 associate with the C-terminal domain (CTD) of the large subunit of RNAPII in a phospho-dependent manner (1, 11, 36).
The mammalian CTD contains 52 tandem repeats of the heptad amino acid sequence Y1S2P3T4S5P6S7 and is alternatively phosphorylated on Ser5 in promoter regions and Ser2 during elongation (27, 28). Several cellular kinases have the capacity to phosphorylate CTD residues (3, 14, 30, 31, 33, 37, 45, 46), and the first FF domain of HYPA/FBP11 binds a CTD peptide phosphorylated on both serines with a dissociation constant of 50 μM (1). It is still unclear, however, exactly which serine residues must be modified to facilitate FF domain binding.
Here we present evidence that the FF domains of CA150 can associate with multiple transcription- and splicing-related proteins, notably Tat-specific factor 1 (Tat-SF1). Tat-SF1 is essential for Tat-activated transcription from the HIV-1 LTR promoter and recruits U small nuclear ribonucleoproteins (snRNPs) to active sites of transcription (18, 60). Its yeast ortholog CUS2 plays an important role in splicing as well (59). CA150 FF domains can recognize multiple weak binding sites within Tat-SF1, and individual domains within the FF repeats show equivalent and noncooperative binding properties. We also identify a consensus FF domain binding motif which suggests that these domains can recognize both phosphorylated and nonphosphorylated targets.
MATERIALS AND METHODS
Plasmid constructs and antibodies.
Murine CA150 cDNA (IMAGE Clone; Open Biosystems, Huntsville, Ala.) was cloned into the expression vector pFLAG-CMV2 (Sigma, Oakville, Ontario, Canada). Tat-SF1 cDNA (NCBI accession no. BAB27483) was generated from a reverse transcription-PCR on murine testis poly(A) RNA and cloned into pFLAG-CMV2. Fragments of Tat-SF1 (residues 1 to 134, 1 to 216, 1 to 346, 266 to 561, or 347 to 561) were subcloned into the pFLAG-CMV2 vector. The Tat-SF1 EW to AA mutant was generated by using the QuikChange system (Stratagene, La Jolla, Calif.). Constructs expressing glutathione S-transferase (GST) fusion proteins of CA150 FF domains 1 (residues 639 to 694), 2 (residues 705 to 761), 3 (residues 772 to 828), 5 to 6 (residues 934 to 1059), 1 to 3 (residues 639 to 828), and 1 to 6 (residues 639 to 1059) were generated by subcloning fragments of CA150 cDNA into the pGEX-4TK vector (Amersham Pharmacia Biotech, Piscataway, N.J.). Expression constructs containing yellow fluorescent protein (YFP)-tagged CA150 (residues 1 to 1079), CA150 N terminus (residues 1 to 632), or CA150 C terminus (residues 598 to 1079) were subcloned into pEYFP-C1 (Clontech/BD Biosciences, Mississauga, Ontario, Canada). His-tagged Tat-SF1 was generated by subcloning Tat-SF1 cDNA into pET-15b (Novagen, Madison, Wis.). The YEWDLD oligomer was constructed by using overlapping primers, where the product was ligated into pET-15b directly or into a pGEX-4TK vector carrying FF domains 1 to 3.
Mouse monoclonal anti-Flag and anti-Tat-SF1 were from Sigma and Transduction Laboratories (BD Biosciences). Rabbit polyclonal anti-RNAPII and horseradish peroxidase-conjugated anti-GST antibodies were from Santa Cruz Biotechnology (Santa Cruz, Calif.), as was goat polyclonal anti-CA150. Rabbit polyclonal anti-GST was a gift from Sigal Gelkop (Ben Gurion University, Beer-Sheva, Israel). Texas Red-conjugated anti-mouse antibody and green fluorescent protein (GFP)-conjugated anti-goat antibody were purchased from Molecular Probes (Eugene, Oreg.), and rabbit polyclonal anti-GFP was purchased from Abcam (Cambridge, Mass.).
Cell culture and immunofluorescence.
Human embryonic kidney 293 (HEK 293) cells were maintained in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal calf serum and antibiotics. For exogenous expression of proteins, cells were transiently transfected with cDNAs by the calcium phosphate precipitation method (13).
For immunofluorescence, cells grown on glass coverslips were washed with phosphate-buffered saline and fixed with 4% paraformaldehyde. Samples were permeabilized with 0.25% Triton X-100 and blocked with 3% bovine serum albumin. Staining with anti-Tat-SF1 was detected with Texas Red-conjugated anti-mouse immunoglobulin (Ig), and CA150 was detected with GFP-conjugated anti-goat Ig. All samples were mounted with p-phenylenediamine to retard photobleaching. Microscopy was carried out with a Leica (Heerbrugg, Switzerland) DMIRE2 inverted microscope equipped with fluorescence and transmitted light optics and Openlab software (Quorum Technologies, Guelph, Ontario, Canada).
The images in Fig. 2B were obtained on live cells by using an Olympus 1X-70 inverted microscope equipped with fluorescent optics and Deltavision Deconvolution Microscopy software (Applied Precision, Bratislava, Slovakia).
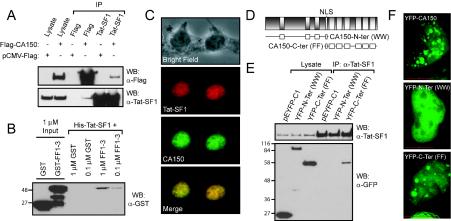
FIG. 2.
CA150 and Tat-SF1 colocalize in cells and interact both in vivo and in vitro in an FF domain-dependent manner. (A) CA150 and Tat-SF1 coimmunoprecipitate. HEK 293 cells were transfected with plasmid expressing N-terminal Flag-tagged CA150 or with empty vector. CA150 and Tat-SF1 were immunoprecipitated with antibodies against either Flag or endogenous Tat-SF1. Associated proteins were subsequently detected by immunoblotting. Whole-cell lysates are shown as controls. (B) Recombinant Tat-SF1 and CA150 FF domains interact in vitro. GST alone (0.1 or 1 μM) or GST-tagged FF domains 1 to 3 were mixed with His-tagged Tat-SF1 immobilized on beads. The 1 μM inputs were loaded in the first two lanes on the left. Immunoblotting with antibodies against GST allowed detection of bound proteins. Recombinant proteins containing the CA150 FF domains were retained on the beads, whereas GST alone was not. (C) CA150 and Tat-SF1 colocalize in the nucleus. HEK 293 cells were fixed, permeabilized, and stained with antibodies against endogenous CA150 and Tat-SF1. The proteins colocalized to punctate nuclear structures resembling speckles. (D) Representation of CA150 N-terminal (WW domain) and CA150 C-terminal (FF domain) fragments used for examining the localization and Tat-SF1 binding properties of CA150. Constructs overlap at the nuclear localization sequence (NLS). (E) CA150 FF domains are coimmunoprecipitated with Tat-SF1 while the WW domain-containing N terminus is not. HEK 293 cells were transfected with pEYFP-C1 vector alone or YFP-tagged fragments encompassing the N or C terminus of CA150 (as in panel D). Tat-SF1 immunoprecipitations were probed with antibodies against GFP to identify associated proteins. (F) CA150 localization to nuclear speckles is dependent on its FF domains. Plasmids expressing YFP-tagged full-length, C-terminal, or N-terminal fragments of CA150 were transfected into HEK 293 cells. Images of single nuclei in live cells, taken by deconvolution microscopy, show localization of full-length CA150 or the C-terminal FF domains to nuclear speckles but not the WW domains. Red bar, 10 μm.
Purification of recombinant proteins and mixing experiments.
GST fusion proteins containing FF domains were expressed in Escherichia coli BL21 cells (Amersham Pharmacia Biotech). Cells were lysed in PLC buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EDTA, 100 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of aprotinin ml−1, 10 μg of leupeptin ml−1, and 1 mM dithiothreitol) and sonicated. The lysate was cleared by centrifugation and incubated with glutathione beads (Amersham Pharmacia Biotech) at 4°C for 1 to 2 h. His-tagged Tat-SF1 or YEWDLD oligomers were purified on TALON metal affinity resin (Clontech, BD Biosciences) with buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 1 mM PMSF, 10 μg of aprotinin ml−1, and 10 μg of leupeptin ml−1. Mixing experiments with GST-tagged FF domains and His-tagged proteins were conducted in TCL buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 2 mM EDTA, 1 mM sodium vanadate, 1 mM PMSF, 10 μg of aprotinin ml−1, and 10 μg of leupeptin ml−1) plus 1 mM dithiothreitol.
Immunoprecipitation, GST pull downs, and Western blotting.
Transfected HEK 293 cells were lysed in NP-40 buffer (50 mM Tris-HCl [pH 7.4], 75 mM NaCl, 10% glycerol, 0.5% NP-40, 2 mM EDTA, 1 mM sodium vanadate, 1 mM PMSF, 10 μg of aprotinin ml−1, and 10 μg of leupeptin ml−1). Proteins were immunoprecipitated for 1 to 6 h at 4°C and washed multiple times with NP-40 buffer. For GST pull downs, HEK 293 cells lysed with TCL buffer were incubated with glutathione beads carrying recombinant GST fusion proteins for 1 to 2 h at 4°C. All bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Schleicher and Schuell Bioscience, Keene, N.H.). Membranes were blocked in Tris-buffered saline containing 5% skim milk and immunoblotted. Primary antibodies were detected with anti-mouse Ig or anti-rabbit Ig antibodies conjugated to horseradish peroxidase followed by treatment with enhanced chemiluminescence (Pierce, Rockford, Ill.).
Identification of interacting proteins by mass spectrometry.
HEK 293 cells were lysed in TCL buffer and cleared by ultracentrifugation. The lysate was passed through a 0.45-μm-pore-size filter (Pall Corporation, Ann Arbor, Mich.) and precleared with glutathione beads before incubation at 4°C for 2 h with 6 μg of recombinant GST fusion proteins. After separation by SDS-PAGE, proteins were visualized with colloidal Coomassie (GelCode Blue Stain Reagent; Pierce). Bands were excised, reduced, alkylated, and digested with trypsin by using a ProGEST tryptic digestion robot (Genomic Solutions, Ann Arbor, Mich.). Mass spectrometry analysis was performed on a Finnigan LCQ Deca XP ion trap (Thermo Finnigan, Mississauga, Ontario, Canada) and by database searching with the Sonar MS/MS search engine (ProteoMetrics; Genomic Solutions).
Peptide spot array synthesis.
Peptide arrays were produced according to the spot synthesis method (19). Acid-hardened cellulose membranes prederivatized with a polyethylene glycol spacer (Intavis, Cologne, Germany) and standard 9-fluorenylmethoxy carbonyl (Fmoc) chemistry were used for synthesis. Fmoc-protected and -activated amino acids (Intavis) were spotted in high-density 24-by-18 arrays on 130- by 90-mm membranes with an AbiMed (Langfield, Germany) ASP422 robot. Membranes were blocked overnight in 10% skim milk, incubated with 1 μM purified GST fusion proteins in Tris-buffered saline with Tween for 2 h at 4°C, washed three times, and probed with rabbit polyclonal anti-GST antibody. Primary antibodies were detected by horseradish peroxidase-conjugated anti-rabbit antibody followed by enhanced chemiluminescence (Pierce).
Peptide synthesis and fluorescent polarization.
Peptides were synthesized on an AbiMed 431 synthesizer with the standard Fastmoc protocol. The authenticity of the product was confirmed by matrix-assisted laser desorption ionization mass spectrometry and amino acid analysis. Equilibrium binding constant determination was carried out on a Beacon fluorescence polarization system (Pan Vera, Wis.) equipped with a 100-μl sample chamber. Fluorescein-labeled probes were prepared through the reaction of carboxy-terminally labeled peptides with 5- (and −6)-carboxyfluorescein succinimidyl ester (Molecular Probes). Binding studies were conducted with 5 nM fluorescein-labeled probes dissolved in phosphate-buffered saline containing 100 μg of bovine serum albumin ml−1 and 1 mM dithiothreitol. Reaction mixtures were allowed to equilibrate for 30 to 60 min at room temperature. All fluorescent polarization measurements were conducted at 22°C.
RESULTS
CA150 FF domains pull-down transcription and splicing-related nuclear proteins.
The FF domains of CA150 have been shown to associate with the CTD of RNAPII in a phospho-dependent manner (11). In support of this observation, we consistently found that recombinant proteins containing multiple CA150 FF repeats (see Fig. 8C) or single domains bound a fraction of RNAPII present in cell lysates. To identify additional proteins that interact with FF domains, we used a mass spectrometry-based analysis of CA150-binding proteins. Two overlapping regions of murine CA150 were used as bait, encompassing either all six FF domains or the fifth and sixth domains alone (Fig. 1A). Beads carrying recombinant GST-tagged FF domains or GST control were incubated with HEK 293 cell lysate and washed extensively. Bound proteins were separated by SDS-PAGE and stained with colloidal Coomassie (Fig. 1B). Proteins that associated specifically with FF domains (lanes 4 and 5) were excised and identified by tandem mass spectrometry (liquid chromatography/mass spectrometry/mass spectrometry). The majority of these potential FF domain-binding partners are nuclear proteins known or thought to be involved in transcription or splicing (Table 1). Many contain long regions rich in acidic residues or serines and protein domains predicted to possess nucleic acid binding properties.
FIG. 8.
Binding to mock-phosphorylated RNAPII CTD is consistent with the defined FF domain interaction motif. (A) Heptad amino acid sequence repeated 52 times in mammalian RNAPII CTD. Phosphorylation of serine residues at positions 2 and 5 plays an important role in transcriptional progression. (B) A requirement for two negatively charged residues and Tyr for CTD peptides binding to CA150 FF domains. Peptide arrays were synthesized with CTD repeats containing Asp to mimic pSer and incubated with recombinant GST-tagged FF1 to 3. Asp (in boldface type) was used at either the Ser2 or Ser5 position alone (peptides 1 and 2), both positions together (peptide 3), or Ser2 with Thr4 (peptide 4). The first peptide on each membrane contains the wild-type sequence (listed horizontally across the top). Below each residue are peptides incorporating alanine substitutions. (C) Only CTD peptides with two negative charges are able to compete out CA150 FF domain binding to RNAPII in pull-down assays. Whole-cell lysate from HEK 293 cells was mixed with recombinant GST-tagged FF1 to 3 bound to glutathione beads. One millimolar synthetic peptides corresponding to two repeats of the RNAPII CTD were added. Peptides either contained two wild-type repeats, had Asp substitutions at the Ser2 or Ser5 positions, or had both together. Proteins retained on beads after washing were separated by SDS-PAGE, and the associated RNAPII was detected with specific antibody. α-RNA Pol II, anti-RNAPII; WB, Western blotting.
FIG. 1.
Identification of cellular proteins binding to FF domains of CA150. (A) Domain architecture of CA150, which includes three WW domains at its N terminus followed by six FF domains. FF1-6 and FF5-6 represent segments of the protein used to identify FF domain binding partners. (B) Recombinant proteins encompassing either the FF1-6 or FF5-6 CA150 domain were used to precipitate cellular proteins from HEK 293 whole-cell lysate. Interacting proteins were separated by SDS-PAGE and detected by staining with colloidal Coomassie. The addition of lysate to GST alone (lane 1) or GST-FF fusion proteins minus lysate (lanes 2 to 3) served as controls. Protein bands identified as uniquely binding to FF domains were excised and subsequently identified by mass spectrometry.
TABLE 1.
Nuclear proteins identified by mass spectrometry as interacting with the FF domains of CA150
| Protein | Locus identification | Domain(s) (n) |
|---|---|---|
| ATBF1 | 463 | ZnF/C2H2 (23), homeobox (4) |
| +BR140a | 7862 | ZnF/RBZ, PHD (2), BROMO, PWWP |
| +BTF | 9774 | |
| +CFIM | 11052 | RRM |
| DDX1 | 1653 | SPRY, helicase (2, DEXD, HELIC) |
| DDX3 | 1654 | Helicase (2, DEXD, HELIC) |
| DDX36 | 170506 | Helicase (2, DEXD, HELIC), HA2 |
| DDX9 | 1660 | DSRM (2), helicase (2, DEXD, HELIC) |
| DNAPK | 5591 | FAT, PI3K, FATc |
| FBP2 | 8570 | KH (4) |
| hnRNPK | 3190 | KH (3) |
| +hnRNPU | 3192 | SAP, SPRY |
| HRPT2 | 79577 | Cdc73-like |
| KARP-1 | 10545 | |
| MSI1 | 4440 | RRM (2) |
| NONO | 4841 | RRM (2) |
| +NOPP140 | 9221 | LisH, SRP40-like |
| +Nucleolin | 4691 | RRM (4) |
| +p150TSP | 9646 | TPR (9) |
| PARP-1 | 142 | BRCT |
| +PD2 | 54623 | Paf-1 like |
| +PNN | 5411 | |
| PSF | 6421 | RRM (2) |
| PRP8 | 10594 | JAB/MPN |
| +S164 | 58517 | PWI, RRM |
| SAP130 | 79595 | |
| +SC35 | 6427 | RRM |
| +SRm300 | 23524 | AT hook |
| +SRP55 | 6431 | RRM (2) |
| +SRP75 | 6429 | RRM (2) |
| +TAT-SF1 | 27336 | RRM (2) |
| +TCOF1 | 6949 | LisH |
| +TRAP150 | 9967 | |
| TRAP95 | 10025 | WD40 (repeats) |
| +UBF-1 | 7343 | HMG (6) |
| +ZNF265 | 9406 | ZnF/RBZ |
A + indicates proteins containing regions rich in serine or acidic residues.
CA150 colocalizes and interacts in vivo and in vitro with Tat-SF1 via its FF domains.
To explore CA150 FF domain-mediated interactions, we focused on one of the identified binding partners, Tat-SF1. CA150 regulates mRNA transcription from the HIV-1 LTR promoter. Likewise, Tat-SF1 and its associated kinase, P-TEFb, are specifically required for Tat activation of HIV transcription in vitro (18, 41, 60, 61). Recruitment of RNAPII containing a functional CTD to Tat-mediated transcription complexes is also a requirement for copying from the LTR promoter (40). It therefore seemed possible that an interaction between the CA150 FF domains and Tat-SF1 could support the formation of elongation-efficient complexes.
The ability of CA150 FF domains to bind Tat-SF1 was insensitive to RNase treatment in vitro, which suggested a direct protein-protein interaction. To pursue this association, HEK 293 cells were transfected with murine CA150 tagged with an N-terminal Flag epitope. Upon immunoprecipitation with anti-Flag or anti-Tat-SF1 antibodies, we observed coprecipitation of CA150 with endogenous Tat-SF1 (Fig. 2A). This provides evidence that CA150 and Tat-SF1 do interact, potentially in a higher-order complex containing RNAPII and other transcription or splicing factors such as those identified as in vitro FF domain-binding partners.
To examine whether purified Tat-SF1 binds directly to CA150 FF domains, recombinant full-length Tat-SF1 protein containing an N-terminal histidine (His) tag was purified, bound to metal affinity beads, and incubated with either purified GST-tagged FF domains 1 to 3 or GST alone (Fig. 2B). FF domains were specifically precipitated by Tat-SF1 from solutions containing either 1 or 0.1 μM recombinant FF domains (lanes 5 to 6), indicating that Tat-SF1 can associate directly with the CA150 FF domains in vitro.
To test whether they could interact in vivo, we examined HEK 293 cells that endogenously express both proteins. Immunofluorescence staining of CA150 and Tat-SF1 revealed that these proteins colocalize in a heterogeneous speckle pattern within the nucleus (Fig. 2C), consistent with the possibility that CA150 and Tat-SF1 do interact in cells. We further examined the determinants of CA150 localization and Tat-SF1 binding by using YFP-tagged fragments encompassing either the N- or C-terminal region containing the WW or FF domains, respectively (Fig. 2D). These fragments overlap by only 35 amino acids in a region containing a putative nuclear localization sequence. Expression of these constructs in HEK 293 cells and immunoprecipitation with anti-Tat-SF1 antibodies revealed coprecipitation of the FF domain-containing C-terminal region of CA150 but not the N-terminal fragment (Fig. 2E). This is consistent with the idea that CA150 interacts through its FF domains with Tat-SF1 in vivo. Using deconvolution microscopy, we also examined the localization of these protein fragments and determined that the C-terminal FF domain-containing region consistently localized to speckle structures, whereas the N-terminal region encompassing the WW domains was observed uniformly throughout the nucleus (Fig. 2F). These data suggest that FF domains are specifically involved in targeting CA150 to nuclear speckles, where interactions with Tat-SF1 would occur.
FF domains of CA150 bind Tat-SF1 peptide motifs containing acidic and aromatic residues.
To delineate the regions of Tat-SF1 recognized by FF domains, we synthesized a spot peptide array spanning residues 1 to 561 of murine Tat-SF1 with 12-mer peptides and a moving window of five amino acids. The membrane was incubated with 1 μM recombinant GST-tagged CA150 FF domains 1 to 3, and bound protein was detected with anti-GST antibodies (Fig. 3A). Multiple Tat-SF1 peptides interacted with the FF domains in this assay (Fig. 3B). These sequences encompassed 10 nonoverlapping regions of the protein and therefore appeared to represent multiple distinct sites for FF domain recognition. The interacting peptides were all rich in the acidic amino acids Asp and Glu, suggesting that this may be a common feature of FF domain-binding sites.
FIG. 3.
The FF domains of CA150 bind to multiple motifs of Tat-SF1 rich in aromatic and acidic residues. (A) Recombinant GST-tagged CA150 FF domains 1 to 3 bind to several peptide sequences derived from Tat-SF1. Overlapping 12-mer peptides spanning residues 1 to 561 of murine Tat-SF1 were synthesized on a peptide spot array. The resulting membrane was incubated with either the FF domains or GST alone as a control (spots circled in grey). Immunoblotting with antibodies against GST identified interacting peptides. (B) Complete sequences of peptides identified as interacting with FF domains 1 to 3. Sequences with overlapping regions, seen as adjacent spots, are grouped together with brackets and numbered accordingly. Acidic residues Asp or Glu are shown in boldface type. Peptides binding to GST alone or the anti-GST antibody were omitted. (C) FF domain binding to Tat-SF1-derived peptides is dependent on interactions with aromatic and acidic residues. Four peptides able to bind FF domains were selected for mutational analysis. Each residue in a given sequence was systematically mutated to alanine on a spot array. The wild-type sequence is shown above the membrane, with spots below each amino acid representing the incorporation of alanine at that position. Blots were probed with GST-tagged FF domains 1 to 3.
To address this possibility, peptide arrays incorporating mutations to Ala at every position from four of these Tat-SF1 peptides were synthesized and probed with FF domains 1 to 3 (Fig. 3C). Although peptides containing the wild-type Tat-SF1 sequences bound the CA150 FF domains as anticipated, a number of the Ala substitutions abolished detectable peptide binding to the domains. The acidic residues Asp and Glu, as well as aromatic residues Phe, Trp, and Tyr, were identified as the major requirements for binding to FF domains. Aromatic residues, together with negatively charged Asp or Glu, are therefore characteristic of CA150 FF domain-binding motifs in Tat-SF1.
FF domains bind to multiple (D/E)2/5-F/W/Y-(D/E)2/5 motifs with low affinity.
To expand the definition of an FF domain ligand, spot arrays were synthesized in which all 20 natural amino acids were substituted at each position of specific Tat-SF1 peptide sequences. The membranes were probed with FF domains 1 to 3 (Fig. 4A). Asp and Glu were accepted at the majority of positions in every peptide sequence, as were Phe, Trp, and Tyr, confirming that these amino acids are important for peptide interactions with FF domains. Each sequence contains several acidic residues that potentially contribute to binding, yet only in SGWFHVEEDRNT are these contiguous. The involvement of negatively charged amino acids in FF domain binding is most obvious in this peptide, since only acidic residues are tolerated in this core region.
FIG. 4.
FF domains bind preferentially to the consensus sequence (D/E)2/5-F/W/Y-(D/E)2/5. (A) All 20 natural amino acids were substituted at each position of three representative Tat-SF1 peptide ligands with a spot array. Sequences of the wild-type Tat-SF1 peptides are indicated vertically at the left of each array. Amino acid substitutions are listed horizontally across the top. Membranes were incubated with GST-tagged CA150 FF domains 1 to 3, and bound protein was detected with antibodies against GST. A strong selection for acidic amino acids, aromatic residues, and against Lys can be seen. (B) Consensus FF domain binding sequence. Tat-SF1 motifs recognized by FF domains 1 to 3 of CA150 (Fig. 3) are listed. Sequences conforming to the parameters for FF domain binding identified in panel A are underlined. Aromatic residues of Tyr, Phe, or Trp (shown in black boxes) are found in 8 of 10 motifs but may be replaced with the smaller leucine, valine, or isoleucine (shown in gray boxes). Either N- or C-terminal to the hydrophobic residues are two or more of the acidic amino acids Glu or Asp (shown in boldface type). In 7 of 10 sequences, a basic Lys or Arg residue is also observed in close proximity to the binding motif (shown in italics). (C) Potential FF domain binding sites in full-length murine Tat-SF1 (residues 1 to 757) conforming to the (D/E)2/5-F/W/Y-(D/E)2/5 consensus motif. Shown are the number of interaction motifs identified by peptide arrays as well as other potential sites found in Tat-SF1 that may influence FF domain binding, such as those created or enhanced by 19 putative DNAPK or casein kinase II phosphorylation sites (*).
A preference for Phe, Trp, or Tyr was observed at all positions naturally occupied by these aromatic residues, although substitution with acidic amino acids at these sites also allowed significant binding. This was likely because each peptide possesses multiple aromatic residues that would compensate for the loss of any one such amino acid. In contrast to the requirement for acidic and aromatic residues, Lys was excluded at any position. Binding to PDDTPYEWDLDK, the only Tat-SF1 peptide sequence analyzed which contained a naturally occurring Lys, was significantly enhanced by mutation of this residue to any other amino acid. Selection against Arg was not as strong.
Comparison of wild-type Tat-SF1 peptide sequences found to bind FF domains was used to establish a consensus domain-binding motif in conjunction with the amino acid scans. Eight of the 10 FF binding motifs of Tat-SF1 have aromatic residues surrounded by multiple acidic amino acids (Fig. 4B). This suggested a consensus binding motif of (D/E)2/5-F/W/Y-(D/E)2/5, where two of the five residues either preceding or following an aromatic residue are negatively charged Glu or Asp. In some cases, substitution of F/W/Y with the smaller hydrophobic residues Leu or Val may be tolerated.
While 10 Tat-SF1 sites able to bind FF domains in vitro were identified by spot arrays, an additional 16 possible FF-binding motifs are present in the sequence of full-length murine Tat-SF1 (Fig. 4C). These consist of sites that interacted with a negative control on the spot scan, were not present in the arrays (residues 562 to 757), or could potentially be generated by serine-threonine phosphorylation to create negatively charged motifs. This latter possibility is plausible, as Tat-SF1 is known to be phosphorylated on serine (60). Serine phosphorylation could also enhance the affinity of several of the identified FF domain-binding motifs.
To determine FF domain affinities for Tat-SF1 peptide ligands, we quantified binding of recombinant FF domains 1 to 3 to synthetic peptides in vitro by fluorescence polarization. Fluorescently labeled peptides representing three of the sequences from Tat-SF1 had modest affinities for FF domains. The SGWFHVEEDRNT motif bound with a dissociation constant of 61 μM (Fig. 5), whereas the other two peptides bound more weakly. For the additional two sequences, we tested whether substitutions based on the results of the amino acid scans could increase their affinities. A Lys mutation to Ala in PDDTPYEWDLDA (Fig. 4A, top panel) conferred a 207 μM affinity, whereas the addition of two Tyr residues and incorporation of a Gly and Ser in YGNDEFDEGLYS (Fig. 4A, bottom panel) gave an affinity of 170 μM. Hill coefficients of 1 indicated that there was no cooperative binding between individual domains, signifying that a single domain-peptide interaction does not increase the affinity of adjacent domains for these peptide ligands. To further examine the FF domain-binding motifs, Tat-SF1 peptides with only the core consensus motif were synthesized (YEWDLD and WFHVEED). These peptides bound to FF domains 1 to 3 with slightly tighter affinity than the extended sequences (132 and 48 μM, respectively). Finally, we found that FF domains interacted with a minimal motif of WDD with a Kd of 128 μM and a Hill coefficient of approximately 1, indicating that one aromatic residue followed by two negatively charged aspartic acids is sufficient to confer a single-site, low-affinity FF domain interaction. These data are consistent with the notion that CA150 FF domain interactions with Tat-SF1 could occur due to the avidity of multiple FF domains for multiple low-affinity binding sites. These sites are composed of the (D/E)2/5-F/W/Y-(D/E)2/5 motif but may have residues that impair affinity, such as basic residues within or adjacent to the core motif or small hydrophobic amino acids in place of F/W/Y.
FIG. 5.
FF domains bind with low affinity to several Tat-SF1-derived peptides. (A, B, and C) CA150 FF domain binding to Tat-SF1 peptides as measured by fluorescence polarization. Recombinant protein containing FF domains 1 to 3 was incubated with fluorescently labeled peptides representing optimal Tat-SF1 binding motifs. Peptide sequences are indicated at the top left. Mutations increasing affinities to panels A and C were made according to peptide spot analysis. Wild-type Tat-SF1 residues are shown below the sequence (black boxes) where mutations were employed. Michaelis-Menten (left), Scatchard (inset), and Hill (right) plots for peptide interactions with FF domains are shown. (D) Summary of FF domain peptide binding data displaying observed Kd and Hill coefficient values. In addition to peptides represented in the top panels, binding results for fluorescently labeled peptides corresponding to minimal FF domain binding motifs are also shown.
Individual CA150 FF domains are capable of associating with target motifs.
To examine whether individual FF domains from CA150 can interact with Tat-SF1 peptide ligands, we performed binding experiments using only domain 1, 2, or 3 alone. Recombinant proteins containing the individual domains bound a Tat-SF1-derived peptide (containing the core motif WFHVEED) or a minimal FF domain consensus binding site (WDD) (Fig. 6). All three domains possessed an equivalent ability to bind either ligand and showed similar kinetics. Equilibrium constants for each interaction were threefold greater than those measured with domains 1 to 3 together, and Hill coefficients again indicated a single peptide-binding site within each domain. These data suggest that individual FF domains within the repeats act as distinct interaction modules, capable of low-affinity binding to (D/E)2/5-F/W/Y-(D/E)2/5 motifs.
FIG. 6.
Individual CA150 FF domains bind target motifs with similar kinetics and affinity. (A) Recombinant protein containing GST-tagged FF domain 2 of CA150 interacts with Tat-SF1 peptide as measured by fluorescent polarization. The Tat-SF1 motif WFHVEED (identified as an FF domain ligand by spot arrays) was fluorescently labeled and mixed with increasing concentrations of FF2. Michaelis-Menten (top), Scatchard (inset), and Hill (bottom) plots are shown. (B) Summary of kinetic data obtained from individual CA150 FF domains interacting with peptides. In addition to FF2, interaction data for the first and third domains binding to Tat-SF1 peptide WFHVEED were gathered. Similarly, the affinity of individual domains for a minimal FF domain motif-peptide (WDD) was also measured. Equilibrium constants (Kd) for the interactions are listed.
Oligomerized binding motifs interact with FF domains in vitro and compete for domain binding to Tat-SF1.
These data suggest that CA150 may bind its partners through multiple weak FF domain interactions. To test this model, we sought to mimic a physiological FF domain ligand by oligomerizing the minimal YEWDLD motif from Tat-SF1, which by itself binds only weakly to a single domain (Kd of ∼450 μM) and is not able to precipitate the CA150 FF domains when immobilized on streptavidin beads. A construct containing an N-terminal His tag fused to five YEWDLD repeats (Fig. 7A) was examined for its ability to precipitate GST-tagged FF domains 1 to 3 or GST alone in a mixing experiment (Fig. 7B). The CA150 FF domains were able to interact with the multimeric motifs while GST alone could not. Indeed, the amount of FF domains precipitated by 5× YEWDLD beads was similar to that pulled down by beads carrying an equivalent quantity of full-length Tat-SF1 (Fig. 2B). This indicates that a minimal YEWDLD sequence is sufficient to mediate a stable interaction with multiple FF domains when oligomerized in a fashion similar to CA150 FF domain-binding sites in Tat-SF1. To determine whether this multimeric motif could outcompete the FF domain interaction with full-length Tat-SF1, we generated a construct containing the 5× YEWDLD oligomer fused to the C terminus of FF domains 1 to 3. The oligomeric YEWDLD motifs were able to successfully compete for the FF domain interaction with Tat-SF1 (Fig. 7C). Interestingly, the addition of a peptide containing only a single YEWDLD motif to a pull down with FF domains 1 to 3 was not able to inhibit binding to full-length Tat-SF1, even at peptide concentrations up to 2 mM (data not shown). Therefore, though the YEWDLD sequence is sufficient to generate an interaction with FF domains, this weak association requires multiple copies of the target motif to mediate stable binding.
FIG. 7.
FF domain repeats interact with multimeric ligand. (A) Sequence of recombinant His-tagged protein containing five copies of the FF domain binding motif YEWDLD, each separated by a five-residue linker (GAGAG). (B) Recombinant YEWDLD oligomer and CA150 FF domains interact in vitro. Constructs expressing the His-tagged YEWDLD motifs were expressed in bacteria. Either 1 or 5 μM GST alone or GST-tagged FF domains 1 to 3 were incubated with the immobilized, multimeric binding sites. One micromolar inputs were loaded in the first two lanes on the left. Immunoblotting with antibodies against GST allowed detection of bound proteins. (C) Oligomerized motifs can compete for Tat-SF1 interaction. Beads carrying either GST alone or recombinant GST-tagged proteins containing FF domains 1 to 3 with or without the tethered binding motifs were mixed with HEK 293 whole-cell lysate. Interacting protein was detected with antibodies against endogenous Tat-SF1. α-Tat-SF1, anti-Tat-SF1. (D) Domain architecture of Tat-SF1, which has two RRM domains. Several regions of the protein, including parts of the N terminus, RRM domain 1 or 2, and C terminus were used to map the interaction with the CA150 FF domains. A point mutation abolishing binding to the FF domain target site YEWDLD (YEAALD) was also made. Fragments are shown at the bottom, with open boxes denoting RRM domains. The number of potential FF domain binding motifs in each segment is shown in parentheses. (E) The FF domains of CA150 are able to associate with several regions of Tat-SF1. Flag-tagged fragments of Tat-SF1 were transfected into HEK 293 cells, and expression was monitored by immunoblotting with anti-Flag (α-Flag) antibodies (top panel). Lysate was incubated with recombinant GST-tagged FF1 to 3 bound to glutathione beads. Proteins retained after washing were detected by immunoblotting (bottom panel). WB, Western blotting.
The observed binding of CA150 FF domains to multiple peptides dispersed throughout the Tat-SF1 sequence, coupled with the binding properties of the oligomerized motif, suggested that FF domains can interact with distinct regions of the Tat-SF1 protein. To confirm this, N-terminal Flag-tagged fusions containing distinct regions of Tat-SF1 were expressed in HEK 293 cells (Fig. 7D). These polypeptides spanned either the N-terminal, central RRM domain, or C-terminal region of Tat-SF1. The expression of each construct was normalized by Western blotting (Fig. 7E, top), and immobilized GST-tagged CA150 FF domains 1 to 3 or GST alone was incubated with lysate containing the Flag-tagged fusion proteins. While none of the Tat-SF1 fragments could interact with GST (data not shown), all of the expressed proteins were consistently pulled down with FF domains apart from the N-terminal region (Fig. 7E, bottom), which interacted only weakly. The extent of binding correlated roughly with the number of predicted FF domain-binding motifs in each fragment (Fig. 7D). The result of this experiment argues that the FF domains of CA150 can bind with multiple sites in Tat-SF1 and that the quantity of target motifs relates to the strength of the interaction.
FF domain-peptide interactions are consistent with binding to an RNAPII CTD phosphorylated on serines 2 and 5.
Although recruitment of several proteins to the CTD requires phosphorylation of either serine 2 or 5, no known CTD-interacting proteins have an absolute requirement for phosphorylation of both sites. Binding of the nuclear matrix protein SCAF8 to RNAPII appears to favor a doubly phosphorylated CTD, but this interaction is not well understood (42). An individual FF domain from HYPA/FBP11 was shown to bind the CTD heptad amino acid sequence (Fig. 8A) containing pSer at both the 2 and 5 positions with a binding constant of 50 μM (1). As our consensus binding motif suggested that two negatively charged residues are needed to generate an FF domain ligand, we investigated whether this was also the case for binding to the CTD. To address this, we synthesized spot peptide arrays containing 12-mer CTD peptides in which Asp was incorporated at potential Ser or Thr phosphorylation sites, owing to the inefficient synthesis of peptides containing multiple pSer residues; these arrays were then probed with CA150 FF domains 1 to 3 (Fig. 8B). While no binding was observed to peptides with Asp at only the second or fifth positions, those with substitutions for both Ser residues or Ser2 in conjunction with Thr4 associated with the CA150 FF domains. Ala substitutions of specific amino acids within each of these peptides resulted in an almost complete loss of binding, notably, substitution of Tyr1 and Asp at position 2 or 5 (acting as pSer mimics) interfered with FF domain-binding.
To pursue the notion that two negatively charged amino acids in a CTD repeat are preferred for effective binding to FF domains, we examined the capacity of these CTD peptide ligands to compete for the binding of CA150 FF domains to RNAPII. Synthetic peptides corresponding to two repeats of the CTD heptad amino acid sequence were prepared in an unmodified form or with Asp at position 2, 5, or both together to mimic pSer. These peptides (1 mM) were mixed with HEK 293 cell lysate and GST-tagged FF domains 1 to 3 on beads. Bound RNAPII was detected with antibodies to the endogenous protein (Fig. 8C). While the addition of peptides with no Asp or those with substitutions at the second or fifth positions alone did not significantly lower FF domain binding to RNAPII, peptides with Asp at both positions inhibited this interaction. The joint inclusion of peptides charged at either the second or fifth position separately had no effect as well. This result argues that two negative charges within the CTD motif enhance FF domain binding and that peptides with acidic residues at the Ser2 or Ser5 position alone have no, or significantly reduced, individual or cooperative effects on CA150 FF domain binding to RNAPII. These data, coupled with the CTD binding studies on HYPA/FBP11, indicate that FF domain interactions with RNAPII occur at sites that conform to the consensus binding motif identified by peptide arrays and could represent a CTD interaction requiring phosphorylation of Ser 2 and 5 together.
DISCUSSION
We have explored the mechanisms by which FF repeats recognize peptide ligands and have identified a number of nuclear proteins that directly or indirectly associate with the FF domains of CA150. Many of these FF domain-binding proteins contain serine- or acid-rich sequences reminiscent of the RNAPII phospho-CTD, and several are involved in transcription or splicing. The RNA binding protein Tat-SF1, for example, appears important in coupling these two processes. Like CA150, immunodepletion of Tat-SF1 from nuclear extracts abolishes Tat transactivation from the HIV-1 LTR promoter (52, 60), and an interaction between these two proteins may therefore be biologically relevant. In addition to being a general transcription factor, Tat-SF1 has also been linked to the spliceosome and the assembly of splicing-competent U2 snRNP in vivo (18). Since WW domains from CA150, HYPA/FBP11, and Prp40 interact with the pre-mRNA splicing factor SF1/mBBP and components of the U1 snRNP (4, 5, 21, 25), these FF domain-containing proteins could bridge transcription- and splicing-related molecules.
The identities of several FF domain-binding partners are consistent with a potential ability to link transcription and splicing. The CA150 FF domains isolated UBF-1, BR140, and the PD2/HRPT2 complex, which are implicated in the regulation of transcription from RNAPII promoters and represent connections to initiation and elongation (23, 50, 55). Likewise, the identification of Prp8, PSF, and p54nrb/NonO as FF domain-binding proteins suggests an interaction with the spliceosome, especially since purified spliceosome fractions contain CA150 (7, 17, 22, 39, 49). Among other in vitro binding partners for the CA150 FF domains, Btf and FBP2 have roles in the transcriptional control of apoptosis, another process to which CA150 has been linked (26, 48, 58), and the RS proteins SRm300, SC35, SRP55, and SRP75 are involved in alternative splicing (8, 9, 20, 29, 47, 57).
Consistent with the functional relevance of this proteomic analysis, several of the FF domain-interacting proteins, such as SC35, PSF, and Prp8, localize to nuclear speckles, as observed for CA150 and Tat-SF1. Nuclear speckles are rich in pre-mRNA processing components and contain hyperphosphorylated RNAPII (16) along with fractions of HIV-1 Tat and PTEF-b (24, 43). The observation that the CA150 FF domains, when targeted to the nucleus, localize to speckles correlates with their binding to transcriptional regulators that associate with these nuclear structures.
Analysis of the interaction between the CA150 FF domains and their binding motifs in Tat-SF1 has suggested a model in which an avidity for multiple weak sites can result in stable binding between two protein partners. We identified multiple FF domain-binding sites in Tat-SF1 that yielded a consensus binding motif of (D/E)2/5-F/W/Y-(D/E)2/5. Although several of the proteins listed in Table 1 may not interact directly with the CA150 FF domains, many of them contain multiple copies of this target sequence, with Tat-SF1 possessing the largest number of potential FF domain binding sites. The average number of motifs present in these proteins is well above that calculated for a randomly generated list of protein sequences and would be further enhanced with the inclusion of potential Ser phosphorylation sites.
Full amino acid scans on several peptides showed that incorporation of Asp or Glu at most positions in each peptide resulted in an increased affinity for CA150 FF domains 1 to 3, and the same held true for Phe, Trp, and Tyr. Spacing constraints between the hydrophobic and negatively charged residues are not stringent, yet there is a definite requirement for at least two acidic residues in these peptide ligands. There are a number of conserved basic residues within the six CA150 FF domains and on the surface of the first HYPA/FBP11 FF domain (1). Each of these domains possesses an overall pKa of approximately 9.5, and it is possible that the large number of basic residues (22% on average) could drive a broad selection for acidic residues in the ligand. Interestingly, among the family of FF domains, only those from the p190 RhoGAPs do not possess these conserved basic charges and instead have a pKa of 5. This implies that these domains could have a divergent mechanism of ligand recognition.
An interesting aspect of several FF domain-binding sites on Tat-SF1 is their close proximity to disfavored Lys residues, yielding sites that are suboptimal. This is reminiscent of interactions between yeast Sic1 and the WD40 domain-containing F-box protein Cdc4 (38), in which a single Cdc4 binding site exists for 9 suboptimal phosphorecognition motifs in Sic1. The situation with FF domains, however, is likely to be different due to their regular arrangement in repeated arrays. Whereas the WD40 domain of Cdc4 is able to bind to optimal phosphopeptides with comparatively high affinity (Kd of ∼1 μM), it seems likely that FF domain binding, even to the most favorable ligands, is relatively weak. This may explain the requirement for multiple FF domains, which would provide a sufficiently high avidity for an interaction to occur with multiple weak target sites.
Binding data for the CA150 FF domains indicates that individual domains possess roughly equal affinities for Tat-SF1 peptides or for a minimal WDD motif. Hill coefficients also indicated that each domain has only one recognition site and that there does not appear to be any cooperative effect on domain binding, whereby interactions at one site would increase the affinity of adjacent binding events. Instead, these data suggest that each FF domain within a repeated structure possesses an independent binding activity, conferring an ability to engage multiple targets through weak interactions. This is in contrast to repeated motifs such as TPR or WD40 repeats, which pack together to form tertiary structures, creating a composite binding surface for a polypeptide ligand (2, 15). The ubiquitin-interacting motifs of the yeast endocytic adaptor protein Vps27 may represent a binding mode analogous to FF domains (54). These 15-residue motifs fold autonomously into single α-helices capable of noncooperative interactions with monoubiquitin at modest affinities (Kd of 117 to 277 μM), resulting in a stable interaction with ubiquitinated proteins.
The regulation of FF domain interactions is an intriguing issue. The FF domain was originally identified as a pSer-binding module. However, in contrast to other interaction domains that recognize phosphorylated Ser/Thr or Tyr sites and absolutely require a phosphoamino acid for binding, CA150 FF domains can recognize peptides with either pSer or acidic Glu/Asp residues and therefore have a strong selection for negative charges (in the context of an aromatic residue) rather than a strict dependence on phosphorylation. These data suggest two potential modes by which FF domains can recognize ligands. We observed interactions with several Tat-SF1-derived peptides containing acidic residues flanking an aromatic amino acid and obtained evidence that stable binding requires interaction of multiple CA150 FF domains with multimeric aromatic acidic motifs. CA150 FF domains also recognize the RNAPII CTD, but in this case, the requisite negative charges must be generated by phosphorylation of the Ser2 and Ser5 residues, and the interaction is therefore phospho dependent. Of note, we observe no binding of the CA150 FF domains to unphosphorylated CTD peptides by fluorescence polarization and only slightly greater affinities for CTD peptides phosphorylated on Ser2. Peptides phosphorylated on Ser5 bind FF domains with a slightly higher affinity but still more weakly than those which are phosphorylated at both sites, consistent with the possibility that two negatively charged residues are important for stabilizing the interaction (M. J. Smith, unpublished data). The phospho-CTD has 52 potential FF domain binding sites, and a doubly phosphorylated CTD motif was shown to bind a single HYPA/FBP11 FF domain with a Kd of 50 μM (1). This is stronger than the interaction observed between unphosphorylated and acidic Tat-SF1 peptides and single CA150 FF domains and comparable to the described binding of a single WW domain from the prolyl isomerase Ess1/Pin1 to differentially phosphorylated CTD peptides (56). These results, combined with our peptide array and competition data, suggest that an RNAPII CTD containing doubly phosphorylated motifs is potentially a preferred binding partner for FF domains compared with Tat-SF1.
These observations raise the speculation that CA150 FF domains may switch between different binding partners, depending on their phosphorylation status. For example, CA150 FF domains could bind partners such as Tat-SF1 until RNAPII progresses from an initiation mode to a processive complex capable of elongation. This is when it is most likely that both Ser2 and Ser5 would be phosphorylated within individual CTD repeats, creating binding sites for FF domains. FF domain interactions with these phosphorylated sites may therefore allow the recruitment of elongation and splicing factors, such as Tat-SF1, to active sites of transcription as they are released from CA150 due to its stronger affinity for the phospho-CTD. Additionally, weak interactions between the RNAPII CTD and WW domains (which are regularly found in conjunction with FF domains) may also stabilize the complex, as several WW domains have been shown to bind differentially phosphorylated forms of the CTD (12, 56).
We have described the biochemical characteristics of an interaction between the FF domains of the mammalian elongation factor CA150 and the transcription and splicing factor Tat-SF1. It will be of interest to explore the consequences of this interaction for the transcription of mammalian genes or for those of pathogens like HIV-1. FF domains comprise a class of repeated interaction domains, with novel and versatile binding properties.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR), the National Cancer Institute of Canada (NCIC), and the Ontario Research and Development Challenge Fund (ORDCF). T.P. is a Distinguished Scientist of the CIHR. M.J.S. is a recipient of an NSERC Postgraduate Scholarship.
REFERENCES
- 1.Allen, M., A. Friedler, O. Schon, and M. Bycroft. 2002. The structure of an FF domain from human HYPA/FBP11. J. Mol. Biol. 323:411-416. [DOI] [PubMed] [Google Scholar]
- 2.Andrade, M. A., C. Perez-Iratxeta, and C. P. Ponting. 2001. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134:117-131. [DOI] [PubMed] [Google Scholar]
- 3.Baskaran, R., M. E. Dahmus, and J. Y. Wang. 1993. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc. Natl. Acad. Sci. USA 90:11167-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedford, M. T., D. C. Chan, and P. Leder. 1997. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 16:2376-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedford, M. T., R. Reed, and P. Leder. 1998. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc. Natl. Acad. Sci. USA 95:10602-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedford, M. T., and P. Leder. 1999. The FF domain: a novel motif that often accompanies WW domains. Trends Biochem. Sci. 24:264-265. [DOI] [PubMed] [Google Scholar]
- 7.Beggs, J. D., S. Teigelkamp, and A. J. Newman. 1995. The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J. Cell Sci. Suppl. 19:101-105. [DOI] [PubMed] [Google Scholar]
- 8.Blencowe, B. J., R. Issner, J. A. Nickerson, and P. A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blencowe, B. J., G. Bauren, A. G. Eldridge, R. Issner, J. A. Nickerson, E. Rosonina, and P. A. Sharp. 2000. The SRm160/300 splicing coactivator subunits. RNA 6:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohne, J., S. E. Cole, C. Sune, B. R. Lindman, V. D. Ko, T. F. Vogt, and M. A. Garcia-Blanco. 2000. Expression analysis and mapping of the mouse and human transcriptional regulator CA150. Mamm. Genome 11:930-933. [DOI] [PubMed] [Google Scholar]
- 11.Carty, S. M., A. C. Goldstrohm, C. Sune, M. A. Garcia-Blanco, and A. L. Greenleaf. 2000. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc. Natl. Acad. Sci. USA 97:9015-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, A., S. Cheang, X. Espanel, and M. Sudol. 2000. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:20562-20571. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cisek, L. J., and J. L. Corden. 1989. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature 339:679-684. [DOI] [PubMed] [Google Scholar]
- 15.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 16.Dirks, R. W., and S. Snaar. 1999. Dynamics of RNA polymerase II localization during the cell cycle. Histochem. Cell Biol. 111:405-410. [DOI] [PubMed] [Google Scholar]
- 17.Emili, A., M. Shales, S. McCracken, W. Xie, P. W. Tucker, R. Kobayashi, B. J. Blencowe, and C. J. Ingles. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8:1102-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong, Y. W., and Q. Zhou. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414:929-933. [DOI] [PubMed] [Google Scholar]
- 19.Frank, R. 1992. Spot-synthesis: an easy technique for positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217-9232. [Google Scholar]
- 20.Fu, X. D., A. Mayeda, T. Maniatis, and A. R. Krainer. 1992. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc. Natl. Acad. Sci. USA 89:11224-11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstrohm, A. C., T. R. Albrecht, C. Sune, M. T. Bedford, and M. A. Garcia-Blanco. 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21:7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gozani, O., J. G. Patton, and R. Reed. 1994. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13:3356-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grueneberg, D. A., L. Pablo, K. Q. Hu, P. August, Z. Weng, and J. Papkoff. 2003. A functional screen in human cells identifies UBF2 as an RNA polymerase II transcription factor that enhances the beta-catenin signaling pathway. Mol. Cell. Biol. 23:3936-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann, C. H., and M. A. Mancini. 2001. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 114:1491-1503. [DOI] [PubMed] [Google Scholar]
- 25.Kao, H. Y., and P. G. Siliciano. 1996. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol. Cell. Biol. 16:960-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasof, G. M., L. Goyal, and E. White. 1999. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol. Cell. Biol. 19:4390-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 28.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaire, R., A. Winne, M. Sarkissian, and R. Lafyatis. 1999. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur. J. Immunol. 29:823-837. [DOI] [PubMed] [Google Scholar]
- 30.Liao, S. M., J. Zhang, D. A. Jeffery, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. van Vuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 31.Lu, H., L. Zawel, L. Fisher, J. M. Egly, and D. Reinberg. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641-645. [DOI] [PubMed] [Google Scholar]
- 32.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 33.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 34.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 35.Misteli, T., and D. L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3:697-705. [DOI] [PubMed] [Google Scholar]
- 36.Morris, D. P., and A. L. Greenleaf. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:39935-39943. [DOI] [PubMed] [Google Scholar]
- 37.Murray, S., R. Udupa, S. Yao, G. Hartzog, and G. Prelich. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 21:4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 39.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46-50. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto, H., C. T. Sheline, J. L. Corden, K. A. Jones, and B. M. Peterlin. 1996. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. USA 93:11575-11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parada, C. A., and R. G. Roeder. 1999. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 18:3688-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patturajan, M., X. Wei, R. Berezney, and J. L. Corden. 1998. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 18:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pendergrast, P. S., C. Wang, N. Hernandez, and S. Huang. 2002. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol. Biol. Cell. 13:915-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 45.Rickert, P., J. L. Corden, and E. Lees. 1999. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18:1093-1102. [DOI] [PubMed] [Google Scholar]
- 46.Rossignol, M., I. Kolb-Cheynel, and J. M. Egly. 1997. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 16:1628-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Screaton, G. R., J. F. Caceres, A. Mayeda, M. V. Bell, M. Plebanski, D. G. Jackson, J. I. Bell, and A. R. Krainer. 1995. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 14:4336-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seok, H., J. Cho, M. Cheon, and I. S. Park. 2002. Biochemical characterization of apoptotic cleavage of KH-type splicing regulatory protein (KSRP)/far upstream element-binding protein 2 (FBP2). Protein Pept. Lett. 9:511-519. [DOI] [PubMed] [Google Scholar]
- 49.Shav-Tal, Y., and D. Zipori. 2002. PSF and p54(nrb)/NonO-multi-functional nuclear proteins. FEBS Lett. 531:109-114. [DOI] [PubMed] [Google Scholar]
- 50.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudol, M., and T. Hunter. 2000. New wrinkles for an old domain. Cell 103:1001-1004. [DOI] [PubMed] [Google Scholar]
- 52.Sune, C., T. Hayashi, Y. Liu, W. S. Lane, R. A. Young, and M. A. Garcia-Blanco. 1997. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 17:6029-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sune, C., and M. A. Garcia-Blanco. 1999. Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol. Cell. Biol. 19:4719-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson, K. A., R. S. Kang, S. D. Stamenova, L. Hicke, and I. Radhakrishnan. 2003. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 22:4597-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson, K. A., B. Wang, W. S. Argraves, F. G. Giancotti, D. P. Schranck, and E. Ruoslahti. 1994. BR140, a novel zinc-finger protein with homology to the TAF250 subunit of TFIID. Biochem. Biophys. Res. Commun. 198:1143-1152. [DOI] [PubMed] [Google Scholar]
- 56.Verdecia, M. A., M. E. Bowman, K. P. Lu, T. Hunter, and J. P. Noel. 2000. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 7:639-643. [DOI] [PubMed] [Google Scholar]
- 57.Wang, J., and J. L. Manley. 1995. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1:335-346. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, K. C., A. L. Cheng, S. E. Chuang, H. C. Hsu, and I. J. Su. 2000. Retinoic acid-induced apoptotic pathway in T-cell lymphoma: identification of four groups of genes with differential biological functions. Exp. Hematol. 28:1441-1450. [DOI] [PubMed] [Google Scholar]
- 59.Yan, D., R. Perriman, H. Igel, K. J. Howe, M. Neville, and M. Ares, Jr. 1998. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol. 18:5000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]