Abstract
A myriad of stimuli including proinflammatory cytokines, viruses, and chemical and mechanical insults activate a kinase complex composed of IκB kinase β (IKK-β), IKK-α, and IKK-γ/N, leading to changes in NF-κB-dependent gene expression. However, it is not clear how the NF-κB response is tailored to specific cellular insults. Signaling molecule that interacts with mouse pelle-like kinase (SIMPL) is a signaling component required for tumor necrosis factor alpha (TNF-α)-dependent but not interleukin-1-dependent NF-κB activation. Herein we demonstrate that nuclear localization of SIMPL is required for type I TNF receptor-induced NF-κB activity. SIMPL interacts with nuclear p65 in a TNF-α-dependent manner to promote endogenous NF-κB-dependent gene expression. The interaction between SIMPL and p65 enhances p65 transactivation activity. These data support a model in which TNF-α activation of NF-κB dependent-gene expression requires nuclear relocalization of p65 as well as nuclear relocalization of SIMPL, generating a TNF-α-specific induction of gene expression.
The innate immune response is an evolutionarily conserved process that plays a pivotal role in an organism's defense against pathogenic agents and is integral to the repair of damaged tissue. Recent data have implied that the innate immune response may direct the nature of the adaptive immune response (21). Moreover, dysregulation of the innate immune response contributes to the pathophysiological states associated with diabetes, atherosclerosis, and certain forms of cancer (for a review, see references 1 and 18). Changes in innate immune response gene expression are controlled by the transcription factor NF-κB. In vivo, the cytokine tumor necrosis factor alpha (TNF-α), necessary for the innate immune response, increases the expression of genes whose products facilitate recruitment, activation, and adherence of phagocytic cells. Analysis of fibroblasts derived from TNF receptor knockout animals has linked TNF-α activation of NF-κB-dependent gene expression to the type I TNF receptor (TNF RI) (11).
In most cells, under steady-state conditions, NF-κB is in an IκB-containing complex that shuttles between the cytoplasm and the nucleus (for a review, see reference 14). Current models predict that stimulation of cells activates a preformed complex containing the following: IκB kinase α (IKKα), IKKβ, and IKKγ/NEMO, a nonenzymatic protein that may function as a scaffold. Activated IKKα and IKKβ phosphorylate two key serine residues located amino terminal to the ankyrin repeat region in IκBα and IκBβ (3, 19, 20, 29, 30), targeting phospho-IκB to a protein complex containing ubiquitin-conjugating enzymes (2). Phosphorylated IκB is ubiquitinated on lysine residues near the phosphorylated residues, leading to removal via proteolysis of IκB from the NF-κB-containing complex. IκB degradation allows NF-κB nuclear translocation and allows NF-κB-dependent changes in gene transcription (activation and repression). These data predict a model in which TNF RI-dependent NF-κB activation requires IKKα/β activation. Consistent with this model, TNF-α-dependent NF-κB activation does not occur in fibroblasts derived from animals in which the IKKα and IKKβ genes have been deleted (15). Analysis of animals in which either the IKKα or the IKKβ gene is deleted revealed that TNF-α-dependent NF-κB activity is compromised more severely in IKKβ than with IKKα nulligenics (8, 14, 16-20, 24, 25). While nuclear translocation of NF-κB is clearly necessary, it is unclear whether it is sufficient for TNF-α-dependent activation of NF-κB-dependent gene expression.
In a previous report, we described the isolation of a novel protein, signaling molecule that interacts with pelle-like kinase (SIMPL), which we determined was required for TNF-α- and not interleukin 1 (IL-1)-dependent activation of NF-κB activity (28). SIMPL was originally identified in a yeast two-hybrid screen with IRAK-1; mass spectrometry and immunocomplexing assays confirmed the interaction between endogenous IRAK-1 and endogenous SIMPL (28). We demonstrate here that nuclear localization of SIMPL is required for TNF RI-induced NF-κB activity. SIMPL interacts with p65 in a TNF-α-dependent manner to enhance the transactivation activity of p65, leading to increased endogenous gene expression. These data taken together with our previous work allow us to describe a model in which TNF-α-dependent activation of endogenous NF-κB-dependent gene expression requires the two independent events: nuclear relocalization of NF-κB and nuclear relocalization of SIMPL.
MATERIALS AND METHODS
Plasmid constructs and antibodies.
The IL-8-LUC and (NF-κB)3-LUC reporter constructs as well as the wild-type SIMPL expression construct have been described previously (27, 28). The SIMPL mutant (SIMPLΔNLS) was generated by PCR with the following primer set: 5′ primer (TCGGGCCGCGTCCGATGTCGCTG) and 3′ primer (AGTATCGATACCTACGAAGCTGCATGG). The PCR-amplified cDNA sequence was subcloned into the NotI/ClaI sites of the pFLAG-CMV-2 expression vector (Eastman Kodak Company, New Haven, Conn.), which introduced an amino-terminal Flag epitope. SIMPLΔNLS contains residues 1 to 241 of SIMPL. The nucleotide sequence of the SIMPLΔNLS construct was confirmed by sequencing (Biochemistry Biotechnology Facility; Indiana University School of Medicine). Wild-type and catalytically inactive versions of IKKα and IKKβ were kindly provided by Michael Karin (University of California, San Diego); the p65 construct was kindly provided by Warner Greene (University of California, San Francisco); the IκBαSR, c-Jun, and Gal4-estrogen receptor transactivation domain constructs were kindly provided by Harikrishna Nakshatri (Indiana University School of Medicine), and the C/EBP plasmid was kindly provided by Steven McKnight (University of Texas Southwestern [UTSW]). The p65-Gal4 and Gal4-LUC constructs were kindly provided by Richard Gaynor (UTSW). The mouse c-myc monoclonal antibody, 9E10, was purchased from Roche Biochemicals (Indianapolis, Ind.). Chromatographically purified mouse immunoglobulin G was purchased from Zymed Laboratories (South San Francisco, Calif.). IKKα, IKKβ, and p65 antisera were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Cell culture and transfections.
The human embryonic kidney epithelial cells (HEK 293 cell line) were maintained and transfected using Fugene as described previously (27). To monitor transfection efficiencies, DNA precipitates also included a construct containing Renilla luciferase cDNA. Cultures were harvested 24 or 48 h following transfection, and luciferase activities were determined using the Dual-Luciferase Reporter assay system (Promega, Madison, Wis.) according to the manufacturer's specifications. Individual assays were normalized for Renilla luciferase activity, and data are presented as n-fold increases in activity relative to empty vector control. Data are from two to three independent experiments performed in duplicate or triplicate with similar qualitative results and standard errors indicated.
Indirect immunofluorescence assays.
HEK 293 cells (3 × 104) were cultured directly on glass coverslips in 24-well plates. Twenty-four hours later, cells were transfected with the indicated constructs. After an additional 24 h, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) (for 10 min at room temperature), permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked with a solution of PBS, 15% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, Pa.), and 0.2% Tween 20. Monoclonal antibodies to Flag were applied for 1 h, followed by a 1-h incubation with Texas Red-conjugated donkey anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories). In some experiments, DNA staining (0.5 μg of Hoechst no. 33258/ml; SIGMA) was used to identify cell nuclei. For double immunofluorescence staining of SIMPL and IRAK/mPLK, primary mouse antibody that recognizes the Flag epitope was used and detected with mouse anti-Myc antibodies linked to fluorescein isothiocyanate (Zymed, San Francisco, Calif.). Coverslips were mounted in Fluoromount-G (Southern Biotechnology Associates, Inc, Birmingham, Ala.).
Confocal microscopy.
Samples were scanned with a Zeiss LSM 510 laser scanning confocal device attached to an Axiovert 100 microscope using a C-Apochromat ×40/1.2 W Corr objective or a Plan-Apochromat ×100/Oil DIC objective (Carl Zeiss). Hoechst 33258, fluorescein, or Texas Red was excited with laser light at wavelengths of 351, 488, or 543 nm, respectively. Fluorescence acquisitions were performed with the 351-, 488-, or 543-nm laser lines to excite UV, fluorescein isothiocyanate, or Texas Red, respectively. To avoid bleed-through effects in double-staining experiments, each dye was scanned independently using the multitracking function of the LSM 510 unit. Images were electronically merged using the LSM 510 software and stored as TIFF files. Figures were assembled from TIFF files by using Adobe Photoshop or Microsoft PowerPoint software.
Immunoprecipitations and Western blotting.
Immunoprecipitations, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western analysis were performed as described previously (27).
RNase protection assays.
Total cellular RNA was isolated from HEK 293 cells transfected with mammalian expression vectors encoding SIMPL, c-Jun, and/or p65 using a MicroRNA isolation kit according to manufacturer's specifications (Strategene). For detection and quantitation of mRNA species, RNase protection assays were performed, using the RiboQuant Multi-Probe RNase protection system (Pharmagin, San Diego, Calif.) according to the manufacturers' specifications. [32P]TTP-labeled RNA probes were prepared by in vitro transcription, using the Multi-probe template set hCK-5 (Pharmagin) as a template. Five to ten micrograms of total cellular RNA samples were hybridized overnight with the purified radiolabeled RNA probe, after which free probe and other single-stranded RNA were digested with an RNase A and T1 mix. The remaining RNase-protected probes were purified and resolved on 5% denaturating polyacrylamide gels, and autoradiograms were prepared. Developed autoradiograms were quantitated using a ChemiImager4000 (Alpha Innotech Corporation).
RESULTS
SIMPL contains a nuclear localization signal.
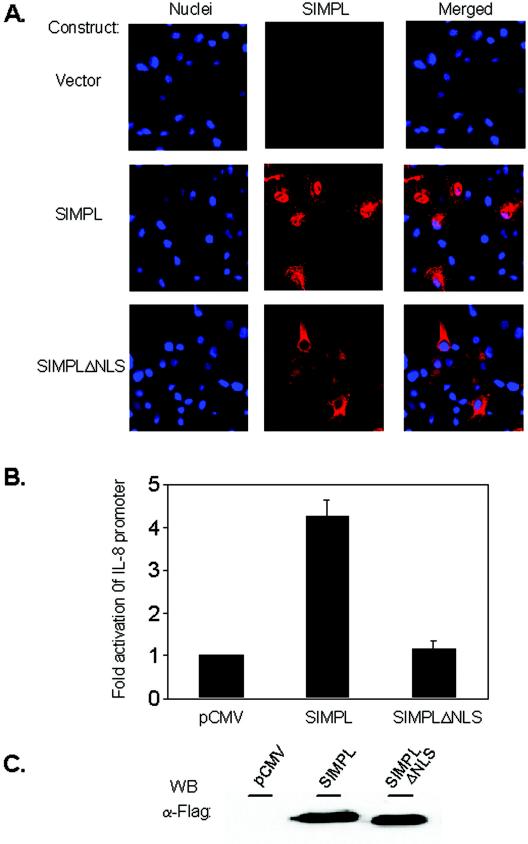
Sequence analysis of the SIMPL protein did not initially identify any known conserved functional domains. However, further inspection revealed a subcellular targeting sequence, specifically, a lysine-rich region, a motif common to nuclear proteins, in the SIMPL carboxyl terminus (4, 5, 10). Comparison of the lysine-rich sequence in SIMPL to sequence of other known nuclear proteins suggests that SIMPL contains either a mono- or bipartite nuclear localization signal (Table 1). To determine if the lysine-rich domain in SIMPL was necessary for nuclear localization, a SIMPL mutant was engineered in which the seven most carboxy-terminal lysine residues are removed (SIMPLΔNLS; contains amino acids 1 to 241). In HEK 293 cells expressing Flag-tagged wild-type SIMPL, the SIMPL protein is found in the cytoplasm and the nucleus (Fig. 1A, SIMPL); in contrast, Flag-tagged SIMPLΔNLS is detected only in the cytoplasm (Fig. 1A, SIMPLΔNLS). We also examined whether the lysine-rich domain derived from SIMPL was sufficient to direct the nuclear localization of an unrelated protein. A construct encoding a chimeric protein in which the SIMPL nuclear localization signal (SIMPL-NLS) (amino acids 239 to 259) is added to the carboxyl terminus of green fluorescent protein (GFP) to generate GFP-SIMPL-NLS (Clontech) was generated. Wild-type GFP is detected predominantly as a cytoplasmic protein, whereas the GFP-SIMPL-NLS chimera accumulates in the nucleus (data not presented). These data demonstrate that the carboxy-terminal domain of SIMPL contains a nuclear localization signal capable of working in trans.
TABLE 1.
SIMPL contains a nuclear localization signal
| Protein | NLS sequencea |
|---|---|
| SIMPL | KVFITFEVKGKEKKKKHL259 |
| Nucleoplasmin | RPAATKKAGQAKKKKLDK174 |
| N1N2 | KRKTEEESPLKDKAKKSK554 |
| Human RB | KRSAEGSNPPKPLKKLR877 |
| Mouse RB | KRSAEGPNPPKPLKNVR870 |
| Human CBP80 | RRRHSDENDGGQPHKRRK20 |
| Bipartite NLS consensus | K/R K/R (10-12 residues), 3 K/R in 5 residues |
| SIMPL | KVFITFEVKGKEKKKKHL259 |
| SV40 T-agb | PKKKRKV132 |
| NF-κB p50 | QRKRQK372 |
| NF-κB p65 | HRIEEKRKRTYE340 |
| Myc | PAAKRVKLD328 |
| Monopartite NLS consensus | K R/K×R/K |
Single-letter amino acid code is used. Bold letters indicate the basic residues of the monopartite and bipartite NLSs.
Simian virus 40 T antigen.
FIG.1.
SIMPL contains a nuclear localization signal that is required for activation of NF-κB. (A) HEK 293 cells were transfected with vector alone or a vector encoding Flag-tagged SIMPL or Flag-tagged SIMPLΔNLS. Twenty-four hours later, cultures were fixed, processed for indirect immunofluorescence with Flag-specific antisera (column labeled SIMPL), or stained with Hoechst no. 33258 to visualize the nuclei (column labeled Nuclei). (B and C) HEK 293 cells were transfected with an IL-8 promoter-firefly luciferase construct, a promoterless sea pansy luciferase construct, and 0.5 μg of the indicated plasmid constructs. Forty-eight hours later, cultures were harvested and either (B) subjected to dual-luciferase assays as described under Materials and Methods or (C) normalized for protein content and subjected to Western analysis using the indicated antisera.
Nuclear relocalization is required for SIMPL- and TNF RI-dependent activation of NF-κB.
We previously demonstrated that SIMPL is a novel component of a TNF-α and not an IL-1 signaling pathway leading to activation of NF-κB (28). Ectopic expression of SIMPL leads to activation of a reporter construct regulated by the NF-κB-dependent IL-8 promoter (28) but does not activate a reporter construct containing the activator protein 1-dependent IL-11 promoter (data not presented). We next examined whether nuclear localization of SIMPL is required for SIMPL-dependent activation of an NF-κB-dependent reporter construct. Ectopic expression of Flag-tagged wild-type SIMPL in HEK 293 cells results in a four- to fivefold increase in IL-8 promoter activity, whereas induction of IL-8 promoter activity by SIMPLΔNLS is equivalent to that detected in cultures transfected with vector alone (Fig. 1B). The lack of activity detected with the SIMPLΔNLS protein is not due to a diminished level of protein, since wild-type SIMPL and SIMPLΔNLS are expressed at comparable levels (Fig. 1C). To determine if SIMPLΔNLS functions as a dominant-negative mutant, SIMPLΔNLS and wild-type SIMPL were coexpressed, and activation of an NF-κB-dependent reporter was measured. SIMPLΔNLS, in a dose-dependent manner, decreased SIMPL-dependent activation of IL-8 promoter activity (data not presented). Thus, nuclear relocalization of SIMPL is required for SIMPL-dependent activation of NF-κB, and SIMPLΔNLS functions as a dominant-negative mutant.
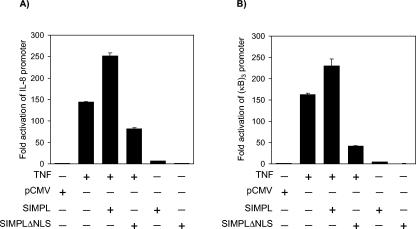
Since SIMPL activity is required for TNF RI-dependent activation of NF-κB-dependent reporter constructs (28), it was of interest to determine if nuclear localization of SIMPL was required. TNF-α treatment of cultures overexpressing wild-type SIMPL results in a greater increase in IL-8 promoter activity than is detected in cultures treated with TNF-α alone or in cultures ectopically expressing SIMPL (Fig. 2A). If cultures overexpress SIMPLΔNLS instead of wild-type SIMPL, the TNF-α-induced increase in IL-8 promoter activity decreases to a level below that detected when cultures are treated with TNF-α alone (Fig. 2A). The IL-8 promoter is a complex promoter composed of binding sites for several transcription factors in addition to NF-κB, including activator protein 1 and CCAAT/enhancer binding protein beta (C/EBPβ). Previous studies linked the SIMPL binding partner IRAK-1 activity to NF-κB-dependent changes in gene expression (28). Thus, it was of interest to determine if SIMPLΔNLS affected TNF-α-dependent activation of a reporter construct composed of NF-κB response elements (6). In comparison to the activity detected for the IL-8 promoter, ectopic expression of SIMPLΔNLS blocked a greater fraction of the TNF-α-induced activation of the (NF-κB)3-LUC reporter construct (compare Fig. 2A and B). These data support a role for SIMPL in the TNF RI signaling pathway coupled to the regulation of NF-κB activity.
FIG. 2.
Nuclear localization of SIMPL is required for TNF RI-dependent NF-κB activation. (A and B) HEK 293 cells were transfected with 0.5 μg of the indicated constructs, 20 ng of the promoterless sea pansy luciferase construct, and 0.2 μg of either (A) a firefly luciferase construct under the control of the IL-8 gene promoter or (B) an artificial promoter composed of three tandem repeats of the NF-κB response element from the human immunodeficiency virus long terminal repeat. Twenty-four hours later, cultures were harvested and processed for luciferase activity as described in Materials and Methods.
SIMPL modulates p65-dependent and not c-Jun- or C/EBP-dependent transcription.
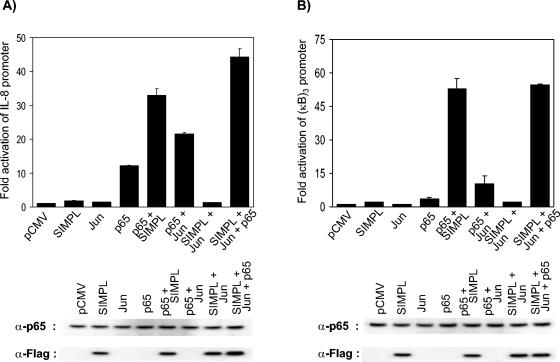
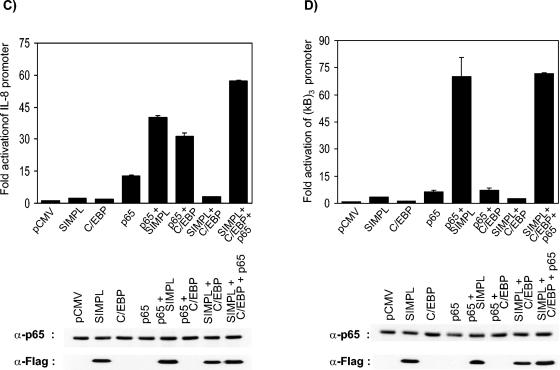
Ectopic expression of SIMPLΔNLS blocked TNF RI-dependent activation of a reporter construct under the control of trimerized NF-κB sites but only partially inhibited TNF RI-dependent activation of a reporter construct under the control of the IL-8 gene promoter (Fig. 3), suggesting that SIMPL specifically modulates p65 activity. To test this hypothesis, the ability of SIMPL to modulate p65-, c-Jun-, and/or C/EBPβ-dependent gene expression was measured. Ectopic expression of SIMPL, c-Jun, or C/EBPβ alone in HEK293 cells leads to a two- to threefold increase in IL-8 promoter activity, whereas ectopic expression of p65 results in a more dramatic increase in IL-8 promoter activity (13-fold) (3A and C). Cotransfection with either c-Jun or SIMPL enhances p65-dependent IL-8 promoter activity, whereas no enhancement in IL-8 promoter activity is seen if SIMPL and c-Jun are coexpressed (Fig. 3A). Similar results are obtained in IL-8 promoter reporter assays if C/EBPβ is used instead of c-Jun (Fig. 3C). These data suggest that SIMPL may enhance p65-dependent transcription by replacing the activity supplied by either c-Jun or C/EBPβ, since both have been shown to bind to NF-κB in the absence of DNA (22, 23). To test this hypothesis we performed an identical experiment, substituting (NF-κB)3-LUC for the IL-8-LUC reporter. Consistent with the knowledge that c-Jun and C/EBPβ are sequence-specific transcription factors, ectopic expression of c-Jun or C/EBPβ had no effect upon the (NF-κB)3-LUC activity (Fig. 3B and D). In contrast to the case with c-Jun or C/EBPβ, cotransfection of SIMPL with p65 resulted in a dramatic increase in (NF-κB)3-LUC activity. Taken together, the data presented thus far support a model in which SIMPL is a TNF-α signaling pathway component specifically involved in modulating p65 activity.
FIG. 3.
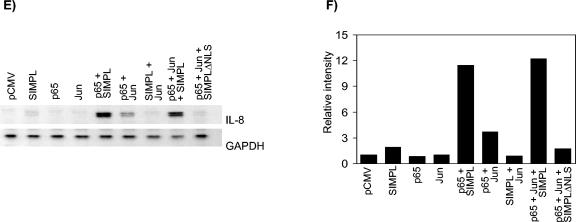
SIMPL modulates p65 and not c-Jun- or C/EBP-dependent transcription. (A to D) HEK 293 cells were transfected with 0.5 μg of the indicated constructs, 20 ng of the promoterless sea pansy luciferase construct, and 0.2 μg of either (A and C) a firefly luciferase construct under the control of the IL-8 gene promoter or (B and D) an artificial promoter composed of three tandem repeats of the NF-κB response element from the human immunodeficiency virus long terminal repeat. Twenty-four hours later, cultures were harvested and processed for luciferase activity as described in Materials and Methods. In all experiments, cell lysates normalized for protein content were subjected to Western analysis to confirm expression of the Flag-tagged SIMPL construct and the p65 construct. (E) HEK cells were transfected with 0.5 μg of the indicated constructs. Twenty-four hours later, cultures were harvested, RNA was isolated, and RNase protection assays were performed as described in Materials and Methods. (F) The autoradiogram used to generate the figure in panel E was quantitated by densitometry, and the ratio of the values obtained (IL-8/GAPDH) was replotted as a histogram.
SIMPL induces endogenous p65-dependent gene expression.
SIMPL appears to specifically modulate p65-dependent transcription. Therefore, we were interested in extending the results obtained with the reporter constructs to the regulation of endogenous gene expression. To determine if SIMPL is capable of modulating IL-8 gene expression, RNase protection assays were performed. In HEK 293 cells transfected with constructs encoding p65, SIMPL, or c-Jun, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene transcripts are detected but little IL-8 message is detected (Fig. 3E). In parallel to the results obtained in the IL-8-LUC and (NF-κB)3-LUC reporter assays, cotransfection of cells with p65 and either c-Jun or SIMPL enhances p65-dependent IL-8 gene transcription. Coexpression of p65 and SIMPL leads to a dramatic increase in endogenous IL-8 gene transcription that is enhanced slightly if c-Jun is also present (Fig. 3E). Consistent with the ability of SIMPLΔNLS to block TNF RI dependent activation of either the IL-8-LUC or the (NF-κB)3-LUC reporter constructs, SIMPLΔNLS decreases endogenous IL-8 gene transcription in cultures coexpressing p65 and c-Jun. Thus, SIMPL is required for endogenous p65-dependent gene expression.
SIMPL-p65 complexes form in response to TNF-α.
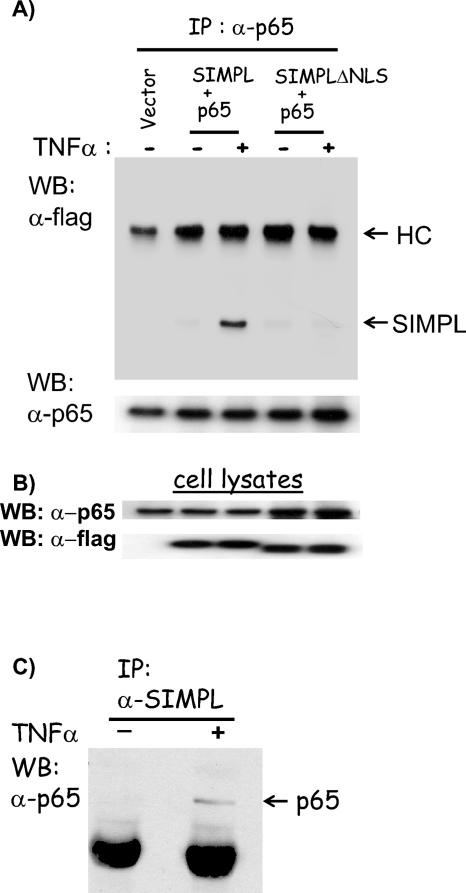
The ability of SIMPL to modulate p65-dependent gene expression suggested a physical interaction between p65 and SIMPL. Immunocomplexing assays were used to determine if p65-SIMPL complexes could be isolated. HEK 293 cells transfected with wild-type p65 and either wild-type SIMPL or SIMPLΔNLS were treated with TNF-α for 15 min, and immunocomplexes were generated with p65 antisera (Fig. 4A). In control cultures, a trace amount of wild-type SIMPL or SIMPLΔNLS could be found in the p65-containing complexes. However, in response to ΤNF-α, there was a significant increase in the amount of SIMPL but not SIMPLΔNLS present in immunocomplexes generated with p65 antisera. We next confirmed that the interaction between endogenous p65 and endogenous SIMPL occurs in cells stimulated with TNF-α (Fig. 4C). These data reveal that TNF-α treatment enhances SIMPL-p65 complex formation and suggest that SIMPL may directly modulate p65 transcriptional activity.
FIG. 4.
SIMPL-p65 complexes form in response to TNF-α. (A and B) HEK 293 cells were transfected with an expression vector encoding p65 and an expression vector encoding either wild-type SIMPL or SIMPLΔNLS. Twenty-four hours later, the indicated cultures were treated with recombinant human TNF-α and were harvested 15 min later. Cell lysates were prepared, and p65 antisera were used to generate immunocomplexes. (A) Immunocomplexed materials or (B) cell lysates used to generate the immunocomplexes were analyzed by Western analysis for SIMPL or p65. (C) Duplicate sets of HEK 293 cell cultures were plated; 24 h later, one set was not treated and the second set was treated with rhuTNFα for 15 min; all cultures were then harvested. Cell lysates were prepared, and immunocomplexes were generated with SIMPL antisera. Immunocomplexes (α-SIMPL) were subjected to SDS-PAGE, and Western blots were prepared and probed with p65 antisera.
Loss of SIMPL does not modulate p65 nuclear localization.
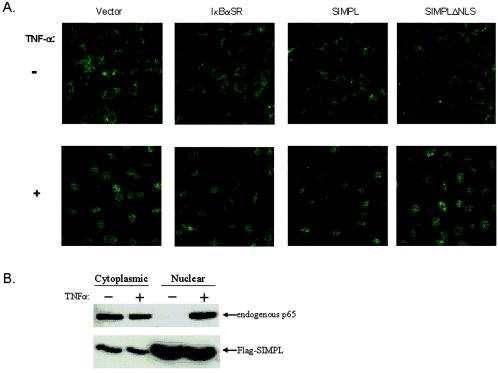
The requirement of SIMPL nuclear localization for ΤNF-α-induced NF-κB activity and the formation of SIMPL-p65 complexes in response to TNF-α led us to explore whether SIMPL could modulate TNF-α-dependent nuclear localization of p65. Under steady-state conditions, p65 is primarily cytoplasmic, and the addition of TNF-α leads to a significant increase in the nuclear p65 pool (Fig. 5, Vector) that is prevented if cells express a mutant IκBα protein that cannot be phosphorylated on serines 32 and 36 (Fig. 5, IκBαSR). Ectopic expression of wild-type SIMPL does not lead to an increase in the nuclear p65 pool in the absence of TNF-α (Fig. 5, SIMPL), and expression of SIMPLΔNLS does not block the TNF-α-dependent increase in nuclear p65 (Fig. 5, SIMPLΔNLS). To confirm that ectopic expression of SIMPL did not lead to signal-independent p65 nuclear relocalization, mouse embryo fibroblasts (MEFs) were transfected with Flag-SIMPL. Western analysis of cytoplasmic and nuclear fractions revealed that nuclear p65 was detected only if cultures were treated with TNF-α (Fig. 5B). Taken together, these data suggest that the increased NF-κB activity in cells ectopically expressing SIMPL is not due to SIMPL-mediated nuclear relocalization of p65. Furthermore, the ability of SIMPLΔNLS to block TNF-α-dependent activation of NF-κB is not the result of SIMPLΔNLS blocking TNF-α-dependent nuclear accumulation of p65. Taken together, these data suggest that SIMPL modulates nuclear p65 activity.
FIG. 5.
Loss of SIMPL does not prevent nuclear localization of p65. (A) Duplicate sets of C3H10T1/2 mouse embryo fibroblasts were transfected with 0.5 μg of the indicated constructs. Twenty-four hours later, half of the cultures were treated with TNF-α (+), and all cultures were fixed and washed with PBS 15 min later. Cultures were processed for indirect immunofluorescence by using antiserum that recognizes p65. (B) C3H10T1/Z mouse embryo fibroblasts were transfected with a construct encoding Flag-SIMPL. Forty-eight hours later, cultures were treated with recombinant human TNF-α (10 ng/ml) for 15 min. Cytoplasmic and nuclear fractions were prepared, and equal amounts of cellular protein were subjected to SDS-PAGE. A Western blot was prepared and probed with p65 or Flag-specific antiserum.
SIMPL activates the transactivation domain of p65.
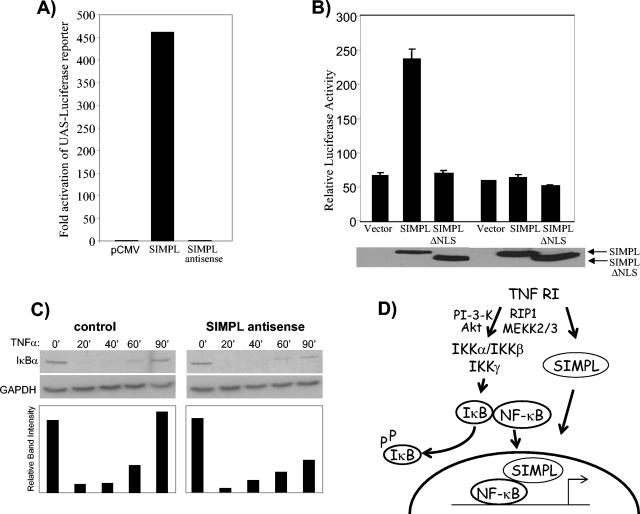
To explore SIMPL's ability to modulate nuclear p65 activity, we hypothesized that SIMPL either could be enhancing the ability of p65 to bind DNA or could be directly affecting the transactivation domain of p65. Synthetic biotinylated oligonucleotides containing NF-κB binding sites were used to explore the ability of p65 to bind DNA in the presence and absence of SIMPL. SIMPL did not appear to affect p65's ability to bind DNA (data not shown). To determine if SIMPL affected p65 transactivation activity, HEK 293 cells were transfected with constructs encoding a chimeric protein composed of the yeast Gal4 DNA binding domain fused in-frame to the p65 transactivation domain, a reporter construct containing a luciferase cDNA under the control of the Gal4-dependent upstream activator sequence, and either vector, wild-type SIMPL, or antisense SIMPL. Ectopic expression of wild-type SIMPL led to a dramatic increase in p65 transactivation activity (Fig. 6A). In parallel studies, ectopic expression of SIMPL did not enhance the activity of a Gal4 fusion protein containing the estrogen receptor transactivation domain (data not presented). The data presented in Fig. 3A to E and in Fig. 6 demonstrate that SIMPL specifically enhances p65 transactivation activity. To determine if SIMPL activity was dependent upon p65, relA−/− fibroblasts were transfected with SIMPL and IL-8 promoter activity was measured. Ectopic expression of SIMPL induced IL-8 promoter activity in wild-type but not relA−/− MEFs (Fig. 6B). We next examined whether SIMPL was required for TNF-α activation of the IκBα gene, which is known to be controlled by NF-κB (9, 13). Ectopic expression of a SIMPL antisense construct had no effect upon TNF-α-dependent IκBα degradation (Fig. 6C, compare control and antisense SIMPL panels). However, the reappearance of the IκBα protein was significantly diminished in cultures expressing the SIMPL antisense construct. Taken together, the data presented herein demonstrate that SIMPL interacts with nuclear p65 in a TNF-α-dependent manner to enhance NF-κB transactivation activity.
FIG. 6.
SIMPL functions as a p65 coactivator. (A) HEK 293 cells were transfected with 1.0 μg of the indicated constructs, 5 ng of a p65-Gal4 construct, and 200 ng of a Gal4-luciferase construct. Cultures were allowed to incubate for 24 to 36 h, at which time cells were harvested and lysates were made and assayed for luciferase activity. (B) Wild-type (first three bars) or RelA−/− (last three bars) MEFs were transfected with 250 ng of the indicated vectors plus 200 ng of an IL-8-luciferase construct and 20 ng of the promoterless sea pansy luciferase construct. Twenty-four hours later, cultures were harvested and luciferase activities were measured. (C) HEK 293 cells were transfected with empty vector (control) or a construct expressing antisense SIMPL. Twenty-four hours later, cultures were stimulated with recombinant human-TNF-α (10 ng/ml), and cultures were harvested at the indicated times. Cell lysates, normalized by protein concentration, were subjected to SDS-PAGE, Western blots were prepared and probed with IκBα and GAPDH antisera. Films were scanned and quantitated by using TotalLab (Nonlinear Dynamics). (D) Model of TNF control of NF-κB-dependent gene expression.
DISCUSSION
SIMPL is a TNF RI signaling pathway component originally identified in a screen for IRAK-1 binding partners. The interaction between the endogenous proteins was confirmed in immunocomplexing assays and independently by mass spectrometry (28). Initial studies suggested that SIMPL functioned as an adapter coupling IRAK-1 to the IKK-containing complex. However, further analysis revealed that a decrease in the steady-state level of SIMPL protein lead to a coordinate loss of IKKβ protein, and IRAK-1 complexes could be detected in cultures lacking SIMPL if IKKβ was ectopically expressed from a heterologous promoter (data not presented). These results suggested that SIMPL does not function as an adaptor molecule. Thus, we began searching for an alternative role for SIMPL in the regulation of NF-κB-dependent transcription.
Comparison of a lysine-rich region in the carboxyl terminus of SIMPL to other known nuclear proteins revealed a nuclear localization signal. SIMPL nuclear localization is necessary for SIMPL and for TNF RI activation of NF-κB-dependent gene expression. The promoters of genes whose expression is controlled by TNF-α are complex, with binding sites for multiple transcription factors. SIMPL specifically modulates p65-dependent transcription and has no effect upon the ability of either c-Jun or C/EBPβ to modulate gene transcription. SIMPL interacts with p65 in a TNF-dependent manner but is not necessary for the nuclear localization of p65 or p65 DNA binding activity. The ability of SIMPL to enhance p65-dependent endogenous IL-8 gene expression is most likely the result of SIMPL functioning as a p65-specific coactivator. The lack of SIMPL activity in the relA−/− MEFs further suggests that SIMPL does not function as a coactivator for either c-Rel or Rel-B, since both are present in RelA−/− MEFs (7).
The data present herein, combined with the knowledge that the IKK complex is required for TNF-dependent NF-κB-dependent gene expression, reveal that two distinct nuclear relocalization events are necessary for the activation of NF-κB-dependent promoters. Under steady-state conditions, NF-κB-IκBα complexes shuttle between the cytoplasm and the nucleus (26), and there is a modest level of NF-κB-dependent gene expression which appears to be IKKα dependent (15). In response to TNF-α treatment, there is a decrease in cellular IκBα protein levels and an accumulation of nuclear NF-κB. In cells derived from animals deficient in either IKKα or IKKβ, TNF-α treatment induces nuclear relocalization of p65, a response absent in cells deficient in both IKKα and IKKβ (15, 16, 24). Interestingly, nuclear accumulation of NF-κB is not lost in cells that have lost SIMPL function in spite of the fact that IKKβ protein levels are greatly diminished (28; unpublished observation), yet there is a significant decrease in the activation of NF-κB-dependent gene expression. These data argue that TNF RI-dependent changes in NF-κB gene expression require more than the targeted destruction of IκBα and the nuclear accumulation of p65. We propose a model in which two interrelated pathways are required for effective TNF-α induction of NF-κB-dependent gene expression (Fig. 6D). Binding of TNF-α to TNF RI results in the activation of the IKK complex, targeting IκBα for degradation and allowing NF-κB to be retained in the nucleus. Coordinate activation of SIMPL by TNF-α allows SIMPL to translocate into the nucleus. Thus, optimal TNF-α-dependent NF-κB activation requires nuclear p65 and SIMPL.
Many signals cause activation of p65-containing complexes, including TNF-α, IL-1, UV light, bacterial products, and viruses, yet the genes expressed in response to these stimuli differ (for a review, see reference 12). Since mammalian gene promoters contain binding sites for multiple transcription factors, the coordinate activation of additional signaling pathways culminating in activation of different combinations of transcription factors provides one mechanism to explain differences in the pattern of gene expression. The discovery of SIMPL as a coactivator specific for TNF-α-dependent NF-κB activation provides another mechanism to explain how TNF-α can share in common a signaling complex (e.g., the IKK complex) and generate a different pattern of gene expression. Since SIMPL activity is required for TNF-α-dependent and not IL-1-dependent NF-κB activation (28), this model provides a basis for examining the more complex issue of how cytokine-specific gene expression occurs in signaling pathways that require IKK activity.
Acknowledgments
This work was supported by Public Health Service grant AI/GM 42798 from the National Institutes of Health (M.A.H.) and National Science Foundation grant MCB 9728069 (M.G.G.).
REFERENCES
- 1.Baldwin, A. S. 2001. The transcription factor NF-κB and human diseases. J. Clin. Investig. 107:3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Z. J., L. Parent, and T. Maniatis. 1996. Site-specific phosphorylation of IkBa by a novel ubiquitination-dependent protein kinase activity. Cell 84:853-862. [DOI] [PubMed] [Google Scholar]
- 3.DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature 388:548-554. [DOI] [PubMed] [Google Scholar]
- 4.Efthymiadis, A., H. Shao, S. Hubner, and D. A. Jans. 1997. Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. J. Biol. Chem. 272:22134-22139. [DOI] [PubMed] [Google Scholar]
- 5.Fontes, M. R. M., T. The, and B. Kobe. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297:1183-1194. [DOI] [PubMed] [Google Scholar]
- 6.Hedin, K. E., M. P. Bell, K. R. Kalli, C. J. Huntoon, B. M. Sharp, and D. J. McKean. 1997. Delta-opioid receptors expressed by Jurkat T cells enhance IL-2 secretion by increasing AP-1 complexes and activity of the NF-AT/AP-1-binding promoter element. J. Immunol. 159:5431-5440. [PubMed] [Google Scholar]
- 7.Hoffmann, A., T. H. Leung, and D. Baltimore. 2003. Genetic analysis of NF-kB/Rel transcription factors defines functional specificities. EMBO J. 22:5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, Y., V. Baud, M. Delhase, P. Zhang, T. Deerinck, M. Ellisman, R. Johnson, and M. Karin. 1999. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of IκB kinase. Science 284:316-320. [DOI] [PubMed] [Google Scholar]
- 9.Ito, C. Y., A. G. Kazantsev, and A. S. Baldwin. 1994. Three NF-κB sites in the IκB-α promoter are required for induction of gene expression by TNFα. Nucleic Acids Res. 22:3787-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jans, D. A., C.-Y. Xiao, and M. H. C. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? BioEssays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 11.Kalb, A., H. Bluethmann, M. W. Moore, and W. Lesslauer. 1996. Tumor necrosis factor receptors (Tnfr) in mouse fibroblasts deficient in Tnfr1 or Tnfr2 are signaling competent and activate the mitogen-activated protein kinase pathway with differential kinetics. J. Biol. Chem. 271:28097-28104. [DOI] [PubMed] [Google Scholar]
- 12.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 13.Le Bail, O., R. Schmidt-Ullrich, and A. Israel. 1993. Promoter analysis of the gene encoding the I kappa B-alpha/MAD3 inhibitor of NF-kappa B: positive regulation by members of the rel/NF-kappa B family. EMBO J. 12:5043-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Q., and I. Verma. 2002. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2:725-734. [DOI] [PubMed] [Google Scholar]
- 15.Li, Q., G. Estepa, S. Memet, A. Israel, and I. M. Verma. 2000. Complete lack of NF-kappaB activity in IKK1 and IKK2 double-deficient mice: additional defect in neurulation. Genes Dev. 14:1729-1733. [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Q., Q. Lu, J. Y. Hwang, D. Buscher, K. F. Lee, J. C. Izpisua-Belmonte, and I. M. Verma. 1999. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 13:1322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Q., D. Van Antwerp, F. Mercurio, K. F. Lee, and I. M. Verma. 1999. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science 284:321-325. [DOI] [PubMed] [Google Scholar]
- 18.Medzhitov, R., and C. Janeway, Jr. 2000. Innate immunity. N. Engl. J. Med. 343:338-344. [DOI] [PubMed] [Google Scholar]
- 19.Mercurio, F., H. Zhu, B. W. Murray, A. Shevchenko, B. L. Bennett, J. W. Li, D. B. Young, M. Barbosa, M. Mann, A. Manning, and A. Rao. 1997. IKK-1 and IKK-2: cytokine-activated IkB kinases essential for NF-kB activation. Science 278:860-866. [DOI] [PubMed] [Google Scholar]
- 20.Regnier, C. H., H. Y. Song, X. Gao, D. V. Goeddel, Z. Cao, and M. Rothe, M. 1997. Identification and characterization of an IkB kinase. Cell 90:373-383. [DOI] [PubMed] [Google Scholar]
- 21.Schnare, M., G. M. Barton, A. C. Holt, K. Takeda, S. Akira, and R. Medzhitov. 2001. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2:947-950. [DOI] [PubMed] [Google Scholar]
- 22.Stein, B., A. S. Baldwin, Jr., B. W. Ballard, W. C. Greene, P. Angel, and P. Herrlich. 1993. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potential biological function. EMBO J. 12:3879-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein, B., P. C. Cogswell, Jr., and A. S. Baldwin. 1993. Functional and physical associations between NF-κB and C/EBP family members: Rel domain-b-ZIP interaction. Mol. Cell. Biol. 13:3964-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda, K., O. Takeuchi, T Tsujimura, S. Itami, O. Adachi, T. Kawai, H. Sanjo, K. Yoshikawa, N. Terada, and S. Akira. 1999. Limb and skin abnormalities in mice lacking IKKα. Science 284:313-316. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka, M., M. E. Fuentes, K. Yamaguchi, M. H. Durnin, S. A. Dalrymple, K. L. Hardy, and D. V. Goeddel. 1999. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity 10:421-429. [DOI] [PubMed] [Google Scholar]
- 26.Turpin, P., B. Ossareh-Nazari, and C. Dargemont. 1999. Nuclear transport and transcriptional regulation. FEBS Lett. 452:82-86. [DOI] [PubMed] [Google Scholar]
- 27.Vig, E., M. Green, Y. Liu, D. B. Donner, N. Mukaida, M. G. Goebl, and M. A. Harrington. 1999. Modulation of tumor necrosis factor and interleukin-1 dependent NF-kB activity by mPLK/IRAK. J. Biol. Chem. 274:13077-13084. [DOI] [PubMed] [Google Scholar]
- 28.Vig, E., M. Green, Y. Liu, K. Kang-Yeol, H.-J. Kwon, J. Tian, M. Goebl, and M. A. Harrington. 2001. SIMPL is a TNF RI-mPLK/IRAK dependent NF-κB activator. J. Biol. Chem. 276:7859-7866. [DOI] [PubMed] [Google Scholar]
- 29.Woronicz, J. B., Z. Gao, Z. Cao, M. Rothe, and D. V. Goeddel. 1997. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science 278:866-869. [DOI] [PubMed] [Google Scholar]
- 30.Zandi, E., D. M. Rothwarf, M. Delhase, M. Hayakawa, and M. Karin. 1997. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell 91:243-252. [DOI] [PubMed] [Google Scholar]