Abstract
The ARF tumor suppressor is widely regarded as an upstream activator of p53-dependent growth arrest and apoptosis. However, recent findings indicate that ARF can also regulate the cell cycle in the absence of p53. In search of p53-independent ARF targets, we isolated nucleophosmin (NPM/B23), a protein we show is required for proliferation, as a novel ARF binding protein. In response to hyperproliferative signals, ARF is upregulated, resulting in the nucleolar retention of NPM and concomitant cell cycle arrest. The Mdm2 oncogene outcompetes NPM/B23 for ARF binding, and introduction of Mdm2 reverses ARF's p53-independent properties: in vitro, NPM is released from ARF-containing protein complexes, and in vivo S phase progression ensues. ARF induction by oncogenes or replicative senescence does not alter NPM/B23 protein levels but rather prevents its nucleocytoplasmic shuttling without inhibiting rRNA processing. By actively sequestering NPM in the nucleolus, ARF utilizes an additional mechanism of tumor suppression, one that is readily antagonized by Mdm2.
The murine INK4a/ARF locus, encoding both the p16INK4a and p19ARF (p14ARF in humans) tumor suppressors, exhibits an unparalleled efficiency of organization within a mammalian genome. Specifically, p16INK4a and p19ARF contain distinct promoters and first exons yet splice into a shared second exon that is translated in alternative reading frames (ARF) (33). While both proteins clearly contribute to tumor surveillance in mice and humans, they appear to play coordinate, yet independent, roles within the cell cycle. p16INK4a imposes a G1/S phase block via direct inhibition of the cdk4 and cdk6 cyclin-dependent kinases, maintaining the active, hypophosphorylated state of the retinoblastoma (Rb) tumor suppressor (36). ARF, in response to hyperproliferative signals relayed by the expression of oncoproteins, such as Myc, E2F, E1A, and Ras, binds and sequesters Mdm2 in the nucleolus, thereby promoting p53-dependent pathways of growth arrest or apoptosis through stabilization of the nucleoplasmic pool of p53 (3, 9, 48). Additionally, ARF directly inhibits the ubiquitination of p53 by Mdm2, suggesting that nucleolar sequestration might not be a requisite step in ARF's activation of p53 (13, 46).
Mounting evidence suggests that the ARF-p53-Mdm2 pathway is not strictly linear. Mice engineered to overexpress a Myc transgene under the control of the immunoglobulin heavy chain enhancer (Eμ) develop B-cell lymphomas that exhibit biallelic ARF deletion, mutation of p53, or Mdm2 overexpression (11). Additional molecular analysis revealed that several tumors which lacked functional p53 also displayed Mdm2 overexpression, arguing against a simple epistatic relationship among ARF, p53, and Mdm2 (11). Additionally, Carnero and colleagues showed that diminished ARF expression resulted in bypassing of replicative senescence, whereas induction of ARF restored ARF's tumor-suppressive properties, even in the presence of a dominant-negative p53 mutant (7). In support of these initial findings, two other groups have demonstrated the physiological significance of the p53-independent ARF pathway through examination of relevant mouse model systems. Mice lacking ARF primarily develop lymphomas and fibrosarcomas (18, 19); in contrast, p53-null animals consistently display lymphomas and osteosarcomas (16). Surprisingly, ARF/p53 double-null mice exhibit multiple tumors with distinct origins, namely, the simultaneous presentation of carcinomas with lymphomas (45). In a corroborative study, wild-type and p53-null mice displayed normal development of the hyaloid vascular system in the eye, yet ARF-null and ARF/p53 double-null animals failed to show proper regression of this structure (27). Collectively, these data reinforce the fact that ARF and p53 play shared, as well as unique roles, as sensors of inappropriate cell growth.
Clearly, genetic studies have confirmed the existence of an alternative, p53-independent ARF tumor surveillance pathway. In an effort to uncover novel players in this pathway, we isolated ARF-containing protein complexes from cells devoid of p53 and Mdm2. Here, we report the identification of nucleophosmin (NPM), a nucleolar phosphoprotein and clinical marker of highly proliferative cells, as a bona fide ARF-interacting protein. Recently, two studies have also described the formation of ARF-NPM complexes in mouse fibroblasts but drew two differing conclusions: ARF targets NPM/B23 for degradation (15) and/or ARF prevents rRNA processing (4). Our data offer a contrasting view. Specifically, ARF binds and retains NPM in the cell nucleolus, effectively impeding the nucleocytoplasmic shuttling of NPM, resulting in subsequent growth arrest. Importantly, Mdm2 antagonizes these effects, thereby preventing ARF-induced withdrawal from the cell cycle. Two intriguing implications of our findings are that (i) ARF, through its interaction with NPM, may directly access and inhibit additional cytoplasmic growth-promoting events and (ii) Mdm2, a well-established mediator of oncogenesis, dictates ARF's tumor-suppressive capacity in the absence of functional p53.
MATERIALS AND METHODS
Cell culture.
Primary mouse embryonic fibroblasts (MEFs) (initially obtained from Gerard Zambetti, St. Jude Children's Research Hospital) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM glutamine, 0.1 mM nonessential amino acids, and 100 U of penicillin and streptomycin (GIBCO/BRL, Gaithersburg, Md.). Virus production and infection of primary MEFs were carried out according to methods described previously, using retroviral helper and vector plasmids (34) provided by Charles Sawyers (University of California—Los Angeles).
Plasmid constructs.
Vectors encoding p19ARF, p19ARF Δ1-14, and Mdm2(210-304) were used as previously described (44) or were subcloned into the EcoRI sites of pEGFP-C1 plasmids (Clontech). A retroviral plasmid encoding full-length Mdm2 was a generous gift from Martine Roussel (St. Jude's Children's Research Hospital). Full-length NPM was cloned from wild-type MEF reverse-transcribed RNAs using the following PCR primers: 5′-GCGCATATGGAAGACTCGATGGATATGGAC-3′ (sense) and 5′-GCGGGATCCTTAAAGAGATTTCCTCCACTGCCAGAG-3′ (antisense). The NPM PCR product was digested with the NdeI and BamHI restriction enzymes and subcloned into the NdeI and BamHI sites of the pET28a vector (Novagen, Madison, Wis.). The pET28a six-histidine-tagged NPM insert was PCR subcloned using the following primers: 5′-GCGGAATTCATGGGCAGCAGCCATCATCAT-3′ (sense) and 5′-GCGGAATTCTTAAAGAGATTTCCTCCACTG-3′ (antisense). The resultant PCR product was subcloned into the EcoRI site of the pSRα-MSV-tkneo retroviral vector. A retroviral vector encoding DMP1 was a generous gift from Charles J. Sherr (St. Jude's Children's Research Hospital).
Nucleolar isolation and MALDI-TOF.
TKO MEFs labeled with [35S]methionine and infected for 96 h with retroviruses encoding hemagglutinin-tagged ARF (HA-ARF) were harvested in ice-cold phosphate-buffered saline (PBS) and homogenized with a glass douncer. Nucleoli were isolated as previously described (2). Nucleoli were verified by staining them in 0.1% azure C dye (Sigma, St. Louis, Mo.). The purified nucleoli were lysed in 25 mM Tris-HCl, pH 7.4-150 mM NaCl-1 mM MgCl2-1 mM EDTA-1% Tween-20 and sonicated with three 10-s bursts at 20% output. HA-ARF complexes were immunoprecipitated with antibodies against the HA epitope conjugated to protein A-Sepharose (Amersham, Piscataway, N.J.). Proteins were eluted with 100 mM glycine (pH 3.0), neutralized in 1 M Tris-HCl (pH 7.4), and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to polyvinylidine difluoride (PVDF) membranes (Millipore, Bedford, Mass.) and exposed to film. Identified bands were excised and digested with 0.1 μg of trypsin per ml (Promega, Rockford, Ill.) overnight at 30°C. Tryptic peptide fragments were extracted with 60% acetonitrile and 0.1% trifluoroacetic acid using an automated Multiprobe II system (Packard Biosciences, Meriden, Conn.). The extracted peptides were dried under vacuum, purified with Zip Plates (Millipore), resuspended in matrix, and subjected to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis using a Voyager DE Pro spectrometer (Applied Biosystems, Foster City, Calif.).
Immunoprecipitation and Western blot analysis.
Wild-type, DKO (p53/Mdm2 null), and TKO (ARF/p53/Mdm2 null) MEFs were infected with retroviruses encoding MycER (a gift from J. Bishop), DMP1, ARF, or ARF Δ1-14 and lysed in binding buffer (25 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, and 0.4 U of aprotinin) either 48 (wild type and DKO) or 96 (TKO) h after infection. For MycER infections, cells were lysed 48 h after the addition of 4-hydroxytamoxifen (4-HT) (1 μM). Antibody to the ARF C terminus (a gift from Charles Sherr), antibody to NPM (Zymed, San Francisco, Calif.), or nonimmune rabbit serum (NRS) or nonimmune mouse serum was added to the binding reaction mixtures for 1 h at 4°C. Immune complexes were precipitated with protein A-Sepharose and washed with binding buffer. The precipitated proteins, as well as direct protein lysates, were separated by SDS-PAGE and transferred to PVDF membranes preactivated in methanol. Mdm2, ARF, NPM, and γ-tubulin proteins were visualized by direct immunoblotting with 2A10, ab80 (abcam, Cambridge, United Kingdom), NPM (Zymed), and γ-tubulin (Santa Cruz, Santa Cruz, Calif.) antibodies, respectively.
Immunofluorescence and confocal microscopy.
Wild-type or TKO MEFs (3 × 104) were seeded onto glass coverslips and infected with retroviruses encoding MycER, tkNeo (vector), ARF, or ARF in combination with Mdm2 or NPM. The cells were fixed 96 h after infection (TKO MEFs) or 48 h after the addition of 4-hydroxytamoxifen (MycER-infected wild-type MEFs) with ice-cold methanol-acetone (1:1 [vol/vol]) and stained for 1 h with either a rabbit ARF C-terminal antibody (4 μg per ml), human fibrillarin (Sigma), or mouse NPM antibody (4 μg per ml), followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse or anti-rabbit immunoglobulin (Pierce) or tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit or anti-human immunoglobulin (Pierce). For measurement of DNA replication, 5-bromodeoxyuridine (BrdU) (Sigma) was added to the culture medium 72 h after infection at a final concentration of 10 μM. Twenty-four hours after the addition of BrdU, the cells were fixed in ice-cold methanol-acetone as described above, treated for 10 min with 1.5 N HCl, and stained for 1 h with a mouse monoclonal anti-BrdU antibody (Amersham), followed by FITC-conjugated anti-mouse immunoglobulin (Pierce). Nuclei were visualized with DAPI (4′,6-diamidino-2-phenylindole) (Sigma). At least 100 cells were counted on each of three coverslips for all experimental conditions. Fluorescence signals were detected using a Nikon epifluorescent compound microscope (100×) fitted with a Nikon FDX-35 charge-coupled device camera.
FPLC.
ARF synthetic peptides were coupled to cyanogen bromide-activated Sepharose (Amersham) and equilibrated as previously described (44). TKO lysates (200 μg) were injected at a flow rate of 1.0 ml per min, washed with 20 ml of 25 mM Tris-HCl (pH 7.4) at 1.0 ml per min, and eluted with a 25 ml of NaCl gradient (0 to 1.5 M) at 1.0 ml per min, followed by 20 ml of 100 mM glycine (pH 3.0) at 1.0 ml per min, using BioLogic fluid phase liquid affinity chromatography (FPLC) and BioLogic HR software (Bio-Rad, Hercules, Calif.). The 1.0-ml collected fractions were precipitated with trichloroacetic acid, resuspended in 1 M Tris-HCl (pH 8.0), and electrophoretically separated on denaturing polyacrylamide gels containing SDS. The separated proteins were transferred to PVDF membranes, and individual proteins were detected using antibodies to cyclin A (Santa Cruz), fibrillarin (Sigma), NPM (Zymed), and Mdm2 (2A10).
Heterokaryon assay.
HeLa cells (2 × 105) were seeded onto glass coverslips in six-well plates and transfected with His-NPM alone or in combination with green fluorescent protein (GFP)-ARF, GFP-ARF N62, or GFP-ARF Δ1-14. As a positive shuttling control, Myc-tagged NPC-M9 (a gift from J. Alan Diehl, University of Pennsylvania) was transfected in combination with GFP-ARF N62. NIH 3T3 cells (6 × 105) were seeded onto the HeLa cells 24 h posttransfection and cocultured for an additional 16 h at 37°C. The cocultures were then incubated for 30 min with DMEM containing cycloheximide (100 μg per ml). For fusion of mouse and human plasma membranes, the medium was removed and the cells were incubated with 50% polyethylene glycol in PBS (prewarmed to 37°C) for 105 s, followed by three PBS washes (prewarmed to 37°C). The cocultures were incubated with DMEM containing cycloheximide (100 μg per ml) for an additional 4 h to permit protein shuttling. Heterokaryons were fixed with formalin-methanol (10% [vol/vol] in H2O) for 15 min and permeabilized with 1% NP-40 in PBS for 5 min at room temperature, followed by three PBS washes. The cells were then blocked for 1 h with 5% fetal bovine serum in PBS and stained for 1 h with a rabbit anti-His antibody (1:25; Santa Cruz) or mouse anti-myc antibody (1:10; Santa Cruz), followed by either FITC-conjugated anti-rabbit or -mouse immunoglobulin (Pierce) or rhodamine X-conjugated anti-rabbit or -mouse immunoglobulin (Pierce) for 30 min. Nuclei were stained with DAPI. Fluorescent signals were detected using a Nikon epifluorescent compound microscope (100×) fitted with a Nikon FDX-35 charge-coupled device camera.
rRNA processing.
HeLa cells (106) were seeded onto 60-mm-diameter dishes and transfected with plasmids encoding GFP or GFP-ARF. The cells were pulse-labeled for 45 min with [5,6-3H]uridine (Amersham) and chased with complete medium for 1 h as previously described (39). Total RNA was harvested from cells using Trizol (GIBCO), and labeled rRNAs (2,000 cpm/lane) were separated on 1% formaldehyde-agarose gels. The gels were denatured for 20 min (0.05 N NaOH, 0.15 M NaCl) and neutralized for 30 min (0.1 M Tris, pH 7.5, 0.15 M NaCl) before being transferred to Hybond N+ membranes. Labeled rRNAs were visualized on EN3HANCE (Perkin-Elmer)-sprayed membranes using standard autoradiography.
RNA interference.
The following 19-nucleotide duplex, corresponding to nucleotides 133 to 151 downstream of the murine NPM transcriptional start site, was synthesized and cloned into the pSUPER.retro plasmid vector (OligoEngine, Seattle, Wash.) according to the manufacturer's instructions: 5′-AGAACGGTCAGTTTAGGAG-3′ (sense) and 5′-CTCCTAAACTGACCGTTCT-3′ (antisense). Retroviruses encoding NPM-targeting small interfering RNAs (siRNAs) were generated via cotransfection of pSUPER.retro-NPM RNA interference (RNAi) and helper plasmids into packaging cells as described above. TKO MEFs seeded onto 100-mm-diameter dishes were infected with pSUPER-NPM RNAi retroviruses and subjected to Western blotting and indirect immunofluorescent analyses 96 h after retroviral transduction, using antibodies described above. For indirect immunofluorescent analysis of NPM expression and BrdU incorporation, cells infected on 100-mm-diameter dishes were seeded onto glass coverslips (105 per coverslip) 24 h after infection, followed by addition of BrdU (10 μM), fixation, and staining for NPM and BrdU at 96 h posttransduction, as described above. Nuclei were demarcated with DAPI, and fluorescence was visualized as described above. As a control, TKO MEFs were infected with empty siRNA vector and scrambled siRNAs (gifts from Helen Piwnica-Worms) (5′-AGGGATGTGTCCCCTTGTG-3′ [sense] and 5′-CTCTTGGGGTCTCTTCCC-3′ [antisense]) and were assayed in parallel with those infected with pSUPER-NPM RNAi retroviruses.
RESULTS
MALDI-TOF analysis reveals that NPM is a component of the ARF complex in vivo.
ARF antagonizes the numerous functions of Mdm2, including the ubiquitination (13, 28, 46) and nuclear export of p53 (41), via direct binding of two conserved domains within ARF to two central regions within Mdm2 (6, 10, 26, 44). Through this interaction, ARF mobilizes Mdm2 into the cell nucleolus, permitting nucleoplasmic accumulation of active p53 molecules (25, 26, 41, 43). However, in limited settings, nucleolar sequestration of Mdm2 appears not to be required for ARF's activation of p53 (21, 24).
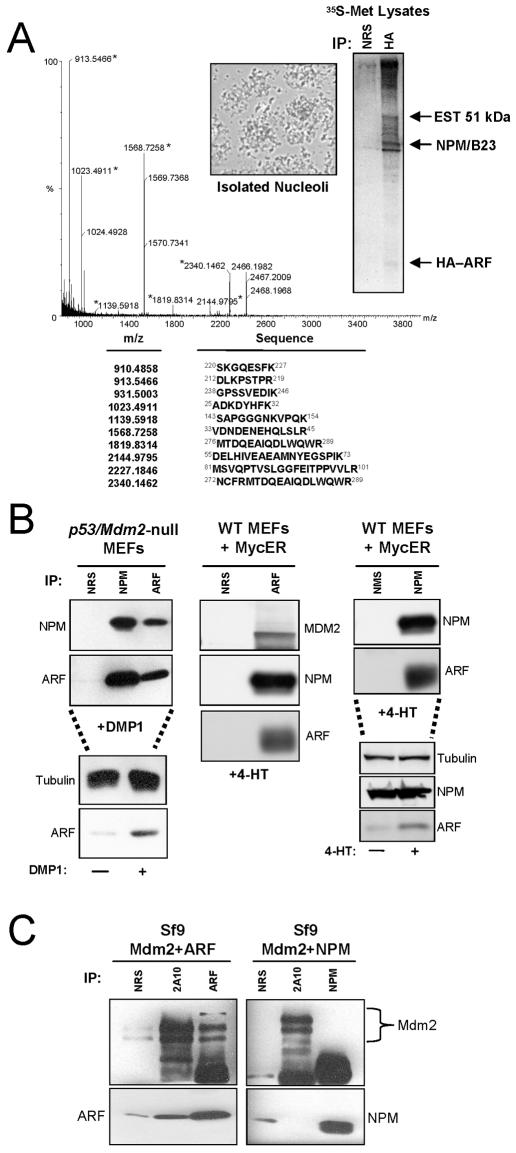
To establish the biochemical composition of an Mdm2-free ARF complex, we infected MEFs derived from mice lacking ARF, p53, and Mdm2 (TKO) with retroviruses encoding HA-ARF and labeled cellular proteins with [35S]methionine. To reduce nonspecific binding of abundant cytoplasmic and nuclear proteins during cell lysis, HA-ARF complexes were isolated from purified nucleoli. Intact nucleoli (Fig. 1A, inset) were lysed, and ARF-containing protein complexes were precipitated using antibodies against the HA moiety (Fig. 1A). HA-ARF is difficult to detect at ∼20 kDa due to its extreme lack of methionine and cysteine residues. Proteins within ARF complexes were separated via SDS-PAGE, digested with trypsin to obtain individual peptide fragments, and combined with matrices. Dried matrix-peptide mixtures were subjected to MALDI-TOF analysis. Peptide masses obtained from a protein band at ∼38 kDa matched the peptide fingerprint for murine NPM (Fig. 1A). An abundant nucleolar phosphoprotein, NPM is upregulated in response to mitogenic signals (12, 20) and is necessary for entry into S phase of the cell cycle in established fibroblast cell lines (17). NPM's reported role as a marker of cell proliferation prompted us to further investigate its association with the ARF tumor suppressor.
FIG. 1.
NPM interacts with ARF in response to hyperproliferative signals. (A) TKO (p53/ARF/Mdm2 null) MEFs infected with retroviruses encoding HA-tagged ARF in the presence of [35S]methionine were harvested in cold PBS. Nucleoli were isolated from TKO extracts (inset), and purified nucleolar proteins were immunoprecipitated (IP) with nonimmune rabbit serum or antibodies recognizing the HA epitope. The indicated radiolabeled protein band at ∼38 kDa was excised, digested with trypsin, and resuspended in matrices. The sample was subjected to MALDI-TOF analysis, and resultant tryptic peptide molecular masses were entered into the Prospector Database search engine for protein identification. (B) DKO (p53/Mdm2-null) and wild-type (WT) MEFs infected with retroviruses encoding DMP1 or tamoxifen-inducible MycER were harvested and lysed 48 h after infection (DMP1) or induction (MycER) with 4-HT. NPM and ARF proteins were immunoprecipitated with NRS, normal mouse serum (NMS), monoclonal antibody to NPM, or a rabbit polyclonal antibody directed to the ARF C terminus (ARF). Precipitated proteins were electrophoretically separated on denaturing gels, transferred to PVDF membranes, and immunoblotted with the indicated antibodies. Protein induction and loading controls are shown below each IP for each condition. (C) Sf9 insect cells infected for 48 h with baculoviruses encoding Mdm2 and ARF or Mdm2 and NPM were harvested and lysed. Mdm2, ARF, and NPM proteins were immunoprecipitated with NRS, monoclonal antibody to Mdm2 (2A10), monoclonal antibody to NPM, or a polyclonal antibody directed to the ARF C terminus (ARF). Precipitated proteins were electrophoretically separated on denaturing gels, transferred to PVDF membranes, and immunoblotted with the same antibodies.
We next examined the ARF-NPM interaction in a physiologically relevant in vivo setting, in which hyperproliferative signals were used to induce endogenous ARF expression in MEFs possessing an intact ARF locus. Specifically, MEFs derived from p53/Mdm2-null mice (DKO) were infected with retroviruses encoding the DMP1 transcription factor, a direct activator of the ARF promoter in response to hyperproliferative signals such as myc (14). DMP1 transduction resulted in the upregulation of ARF protein levels, and subsequent coimmunoprecipitation experiments, using antibodies raised against ARF or NPM, demonstrated the in vivo formation of the ARF-NPM protein complex (Fig. 1B, left). Similarly, wild-type MEFs infected with retroviruses encoding a tamoxifen-inducible form of the myc oncoprotein (MycER) displayed increased ARF protein levels upon treatment with 4-HT, and coimmunoprecipitation experiments again verified the ARF-NPM interaction (Fig. 1B, middle and right). As expected, induction of ARF in wild-type MEFs also resulted in the coimmunoprecipitation of Mdm2 with ARF (Fig. 1B, middle). To ensure that Mdm2 was not mediating the ARF-NPM interaction in wild-type MEFs via potential formation of ternary complexes, we investigated whether NPM could bind directly to Mdm2. Whereas ARF readily complexed with Mdm2, NPM failed to bind to Mdm2 (Fig. 1C), demonstrating that the ARF-NPM and ARF-Mdm2 interactions represent distinct and independent protein complexes.
ARF colocalizes with NPM in the granular region of the nucleolus.
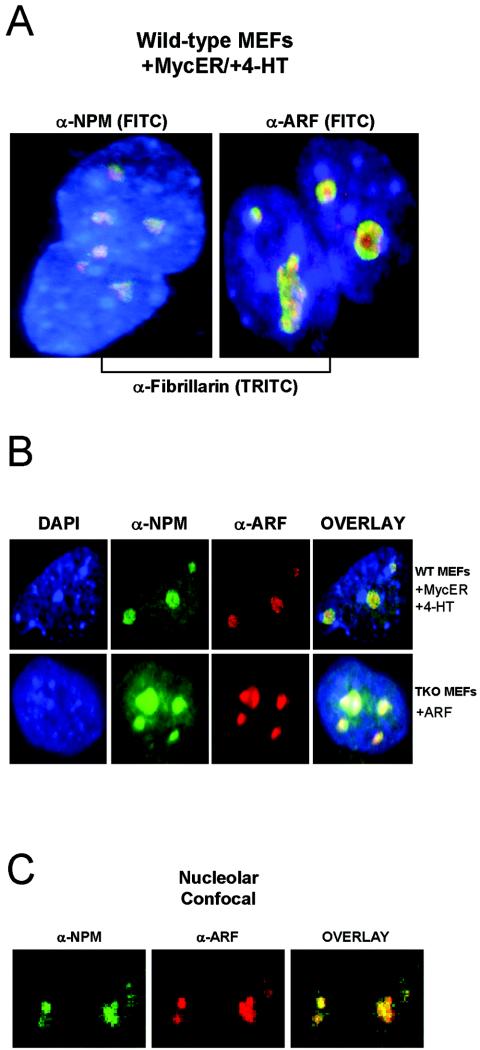
In mouse and primary diploid human cells, ARF is expressed at low basal levels and localizes to the granular region of the nucleolus (21, 25, 32, 43), a subnuclear organelle that is readily detected upon immunostaining for fibrillarin. NPM, a highly abundant nucleolar protein, localizes primarily to fibrillarin-positive nucleoli, but additional staining throughout the nucleus is observed (Fig. 2A and C). To corroborate our finding that ARF and NPM interact and reside within the same protein complex, we next wanted to verify that NPM colocalizes with ARF in the granular region of the nucleolus under conditions of ARF induction in vivo. As expected, myc-mediated upregulation of endogenous ARF in wild-type MEFs resulted in the accumulation of ARF in nucleoli, as indicated by the colocalization of ARF with fibrillarin (Fig. 2A). Similarly, retroviral transduction of exogenous ARF into TKO MEFs produced a robust nucleolar pattern of ARF expression (Fig. 2B). Coimmunostaining for NPM and ARF in myc-transduced wild-type MEFs and ARF-transduced TKO MEFs revealed that NPM and ARF colocalize to the granular region of nucleoli, as confirmed by indirect immunofluorescent (Fig. 2B, overlay) and confocal (Fig. 2C, overlay) microscopy.
FIG. 2.
ARF colocalizes with NPM in the granular region of the nucleolus. (A) Wild-type (WT) MEFs were fixed and immunostained for NPM, fibrillarin, and ARF to mark nucleolar structures. WT MEFs infected with MycER were treated with 4-HT for 48 h to induce ARF expression. (B) WT MEFS infected with MycER retroviruses and treated with 4-HT were fixed and stained with antibodies against ARF (red) and NPM (green) 48 h after 4-HT addition. TKO MEFs infected with retroviruses encoding ARF were fixed and stained using antibodies against NPM (green) and the ARF C terminus (red) 96 h after infection. Nuclei were demarcated with DAPI staining. All immunofluorescent microscopy images in (A) and (B) were captured at ×100 magnification. (C) Confocal microscopy was performed on the same ARF-infected TKO MEFs shown in (B). Overlap of staining was verified using Adobe Photoshop (yellow). The staining pattern shown was exhibited by >95% of cells viewed in four independent experiments.
ARF utilizes its extreme amino terminus in binding NPM.
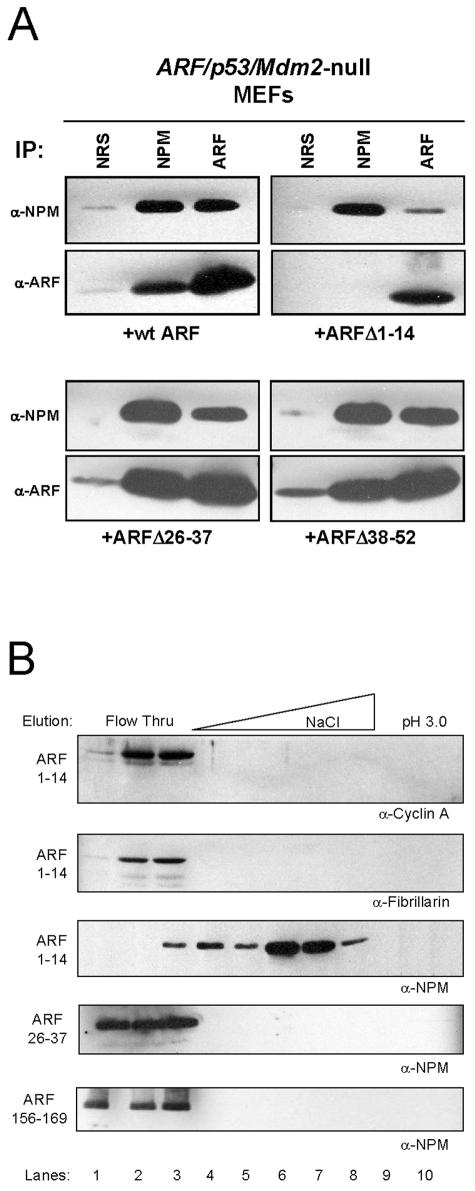
The first 14 amino-terminal residues of murine ARF (ARF 1 to 14) represent the most highly conserved domain among all ARF orthologs and are indispensable for ARF's p53-dependent and -independent properties (44, 45). To test our hypothesis that NPM interacts with these functionally conserved residues within ARF, we investigated whether an amino-terminal deletion mutant lacking ARF's first 14 residues (ARF Δ1-14) could interact with NPM in vivo. TKO MEFs were infected with retroviruses encoding full-length ARF or ARF Δ1-14, followed by coimmunoprecipitation against ARF and NPM to assess formation of the ARF-NPM complex. Whereas full-length murine ARF readily bound NPM (Fig. 3A, upper left) ARF Δ1-14 failed to exhibit a significant in vivo interaction with NPM (Fig. 3A, upper right), consistent with its inability to induce complete cell cycle arrest in TKO MEFs (45). However, we did observe some residual binding of this mutant (Δ1-14) with NPM, leaving open the possibility that other regions within ARF may be important in mediating ARF-NPM interactions in vivo. Contiguous deletion of ARF residues 26 to 37 (which define the low-affinity Mdm2 binding site) or 38 to 52 did not alter ARF-NPM complex formation (Fig. 3A, bottom), and unlike ARF Δ1-14, these mutants are fully capable of inducing cell cycle arrest (43).
FIG. 3.
NPM binds to the amino terminus of ARF. (A) TKO MEFs infected with ARF, ARF Δ1-14, ARF Δ26-37, or ARF Δ38-52 were harvested and lysed 96 h after infection. NPM and ARF proteins were immunoprecipitated (IP) with NRS, monoclonal antibody to NPM, or a polyclonal antibody directed to the ARF C terminus (ARF). Precipitated proteins were electrophoretically separated on denaturing gels, transferred to PVDF membranes, and immunoblotted with the same antibodies. (B) TKO lysates were injected onto the indicated ARF peptide FPLC columns, washed (lanes 1 to 3), and eluted with an NaCl gradient (lanes 4 to 8), followed by acid (lanes 9 and 10) as shown. Trichloroacetic acid-precipitated proteins were separated on denaturing polyacrylamide gels and immunoblotted with antibodies against cyclin A, fibrillarin, and NPM.
Interestingly, ARF's amino terminus, which contains several conserved arginine residues, exhibits a local charge (pI = 10.8) comparable to that of the full-length ARF protein (pI = 11.2), suggesting that ARF, in the absence of its amino terminus, could utilize its other positively charged regions to mediate critical protein-protein interactions. However, previous studies of the ARF-Mdm2 complex have demonstrated that ARF's selection of physiologically relevant targets is based on more than simple electrostatic affinity (6, 10). To define the specificity of NPM's interaction with the ARF amino terminus, we injected whole-cell lysates from TKO MEFs onto affinity columns comprised of similarly charged ARF peptides and eluted bound proteins. Cyclins (A, B, D, and E) and their respective catalytic cdk partners (2, 1, 4/6, and 2, respectively) do not specifically bind to ARF (J. D. Weber, unpublished observation), and elution profiles from ARF peptide affinity columns confirmed this (Fig. 3B, lanes 1 to 3). NPM bound to the first 14 residues of ARF and eluted in the salt gradient (Fig. 3B, lanes 4 to 8), whereas other abundant and equally acidic nucleolar proteins, such as fibrillarin, did not (Fig. 3B, lanes 1 to 3). Importantly, NPM did not interact with ARF residues 26 to 37, the low-affinity site for Mdm2 association, and also failed to bind amino acids 156 to 169 of ARF, a region with a basic charge content equal to that of residues 1 to 14 (Fig. 3B).
Mdm2 releases NPM from ARF-containing protein complexes.
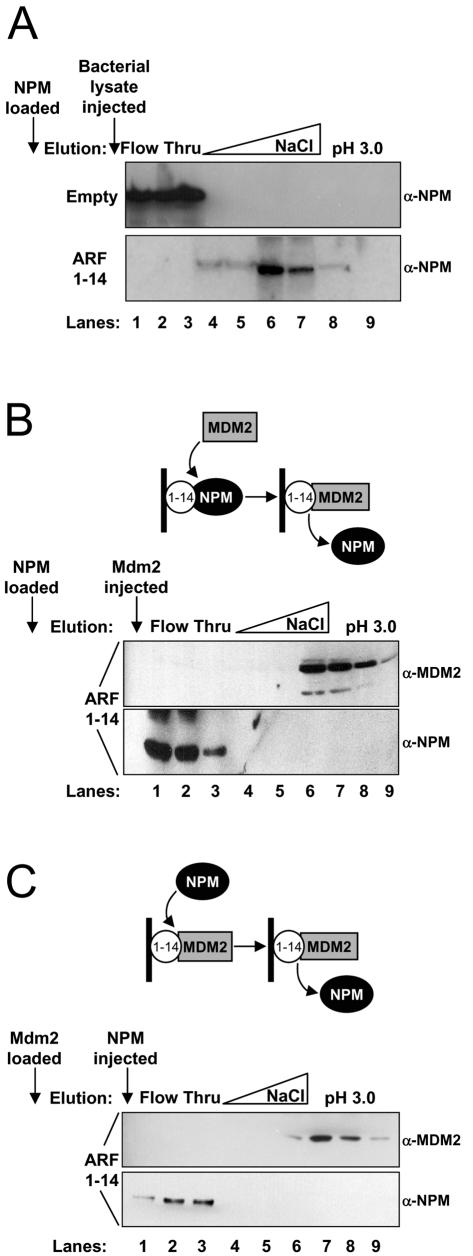
Previous work established ARF's reasonably strong affinity for a central region of Mdm2 (residues 210 to 304) (6, 10), and interestingly, both Mdm2 and NPM interact with the first 14 amino acids of ARF (Fig. 3B) (44). To compare the relative binding strength of the ARF-NPM association with that of ARF-Mdm2, we injected purified full-length NPM onto a peptide column composed of ARF residues 1 to 14. NPM did not interact with the Sepharose used to make the column (Fig. 4A, top, lanes 1 to 3) and was not nonspecifically outcompeted once it was bound to the ARF 1 to 14 peptide column (Fig. 4A, bottom, lanes 4 to 8). Once NPM had bound to the ARF column (Fig. 4B, diagram), the column was washed with buffer containing an excess molar amount of purified Mdm2 residues 210 to 304, which corresponds to the ARF binding domain (6, 44). Bound proteins were eluted with a salt gradient followed by an acid wash, and the resultant fractions were subjected to Western blot analysis for detection of NPM and Mdm2. Initially, NPM was retained on the ARF column yet was exchanged for Mdm2 in subsequent wash steps and eluted in the flowthrough (Fig. 4B, lanes 1 to 3). Newly formed ARF-Mdm2 complexes were stable but eventually eluted under high-salt or acidic conditions (Fig. 4B, lanes 6 to 9), demonstrating ARF's preference for Mdm2 over NPM as a binding partner. In agreement with this finding, Mdm2 remained bound to the ARF peptide column when the column was washed with buffer containing an excess molar amount of purified full-length NPM (Fig. 4C, lanes 6 to 9).
FIG. 4.
NPM and Mdm2 compete for the conserved ARF amino terminus. Affinity-purified (A and B) NPM (1 μM) or (C) Mdm2 (1 μM) containing residues 210 to 304 was initially loaded onto either an empty or ARF 1 to 14 peptide affinity column and washed. Subsequently, (A) nonspecific bacterial lysate, purified recombinant (B) Mdm2 (1 μM), or (C) NPM (1 μM) was injected onto the ARF 1 to 14 column (arrows), washed (lanes 1 to 3), and eluted with an NaCl gradient (0.5 to 1.5 M) (lanes 4 to 6), followed by acid (pH 3.0) (lanes 7 to 9). Fractions from the flowthrough and elution steps were collected, separated on denaturing polyacrylamide gels, and immunoblotted with antibodies against NPM and Mdm2 (H221).
Mdm2 antagonizes ARF's p53-dependent and -independent functions.
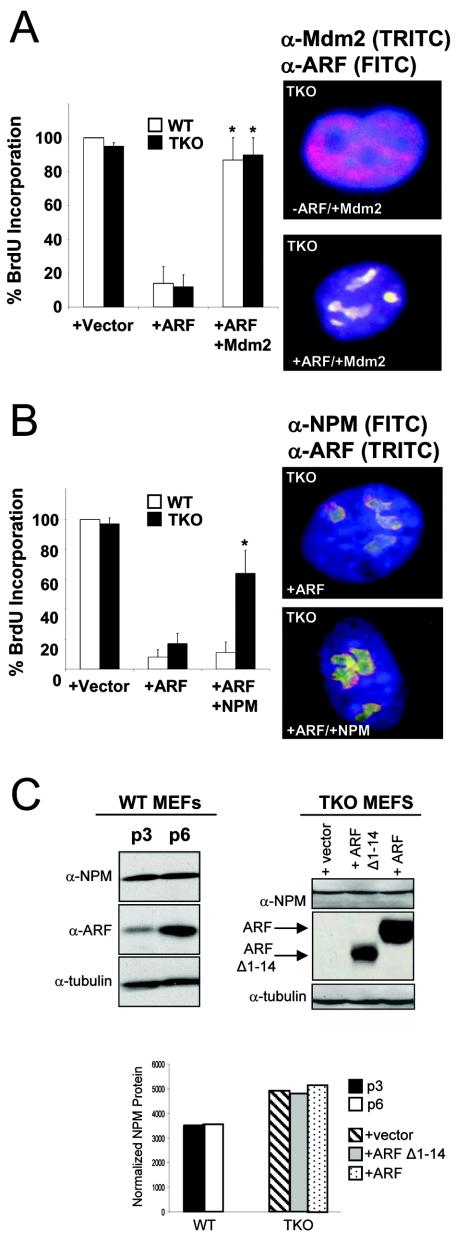
ARF induction evokes a G1 phase cell cycle arrest in wild-type and TKO MEFs via p53-dependent and -independent mechanisms, respectively (45). Importantly, both lines of MEFs failed to arrest when Mdm2 was transduced in concert with ARF (Fig. 5A), presumably due to Mdm2's capacity to (i) terminate the p53 response in wild-type cells and (ii) release NPM from ARF complexes in TKO MEFs via preferential binding of Mdm2 to nucleolar ARF (Fig. 4B and C). Consistent with these findings, introduction of exogenous NPM into ARF-transduced TKO MEFs significantly rescued the cells' capacity to proceed through G1 and into S phase (Fig. 5B). However, overexpression of NPM in ARF-transduced wild-type MEFs was not sufficient to override ARF's p53-dependent cell cycle inhibition (Fig. 5B). Notably, this could be attributed to ARF's ability to induce the p53 effector, p21CIP1, in wild-type MEFs but not in TKOs (45, 48). These findings suggest that restoration of Mdm2 expression in ARF-transduced TKO MEFs triggers a switch in ARF's binding partners, prompting NPM's release from ARF-containing protein complexes.
FIG. 5.
Overexpression of Mdm2 or NPM antagonizes ARF-induced cell cycle arrest. (A) Primary wild-type (WT) and TKO MEFs were infected with retroviruses encoding tk Neo (vector), ARF, or ARF in combination with Mdm2. The cells were labeled with BrdU 24 h prior to fixation (48 h for WT; 96 h for TKO [postinfection]). The cells were stained with antibodies recognizing incorporated BrdU. The error bars indicate standard deviations of at least 100 cells in three independent experiments (*, P > 0.005). TKO MEFs infected with ARF in combination with Mdm2 were fixed 96 h postinfection and stained with antibodies recognizing Mdm2 (red) and ARF (green); regions of yellow indicate overlap of Mdm2 and ARF. Nuclei were stained with DAPI (blue). (B) Primary wild-type (WT) and TKO MEFs were infected with retroviruses encoding tkNeo (vector), ARF, or ARF in combination with NPM. The cells were labeled with BrdU 24 h prior to fixation (48 h for WT; 96 h for TKO [postinfection]). The cells were stained with antibodies recognizing incorporated BrdU. The error bars indicate standard deviations of 100 cells counted in three independent experiments (*, P > 0.005). TKO MEFs infected with ARF in combination with NPM were fixed 96 h postinfection and stained with antibodies recognizing NPM (green) and ARF (red); regions of yellow indicate overlap of NPM and ARF. Nuclei were stained with DAPI (blue). (C) Wild-type MEFs were passaged on a 3T3 protocol and collected at passages 3 and 6. TKO MEFs infected with retroviruses encoding tkNeo (vector), ARF Δ1-14, or ARF were harvested and lysed 96 h postinfection. Whole-cell lysates were separated on denaturing polyacrylamide gels, transferred to PVDF membranes, and immunoblotted with antibodies recognizing NPM, ARF, and γ-tubulin (loading control). NPM protein was normalized to γ-tubulin using ImageJ software and graphed as arbitrary normalized units.
Given that overexpression of NPM was sufficient to bypass ARF-induced growth arrest in TKO MEFs but not wild-type MEFs (Fig. 5B), we wanted to verify that ARF's cell cycle arrest was not mediated through simple downregulation of NPM protein expression. In contrast to a recent report (15), NPM protein levels were not reduced in response to ARF transduction in TKO MEFs (Fig. 5C, right). A hallmark of mouse cell culture-induced cellular senescence is the gradual induction of ARF (48). ARF accumulation leads to cell cycle arrest, and appropriately, mouse fibroblasts lacking ARF are immortal and can be passaged indefinitely (18). To test whether ARF accumulation lowered NPM protein levels in this physiological setting, we passaged wild-type MEFs on a 3T3 protocol (18) and assayed for ARF and NPM protein expression. As cells accumulated ARF protein, protein levels of NPM failed to decline and remained steady throughout fibroblast passaging, demonstrating that ARF does not reduce NPM protein levels to arrest cell growth (Fig. 5C, left). This finding, in combination with others' observations that NPM shuttles rapidly into the nucleus (5, 47), led us to hypothesize that nucleolar ARF might inhibit NPM's transit throughout the cell, effectively negating NPM's growth-promoting potential via topological restraint rather than protein degradation or turnover.
ARF prevents NPM's nucleocytoplasmic shuttling.
As an upstream activator of p53-dependent pathways of growth arrest and apoptosis, ARF stabilizes p53 protein levels through binding and sequestration of Mdm2 in the nucleolus, away from nucleoplasmic pools of p53, suggesting that regulation of protein topology may be an underlying feature of ARF-mediated tumor suppression (25, 41, 43). The notion that subcellular compartmentalization can either enable or disable a protein's function is further supported by our present understanding of NPM dynamics within the cell. Previous studies have established NPM's participation in nucleocytoplasmic shuttling (5, 47), while more recent work indicates that a relatively small fraction of NPM appears to transit from the nucleolus to the cytosol in a cell cycle-dependent manner (29, 42). Given that nucleolar ARF limits Mdm2's oncogenic potential by preventing its access to, and export from, the nucleus (41, 43), we hypothesized that ARF may utilize a similar strategy to inhibit NPM function, thereby achieving p53-independent command of the cell cycle.
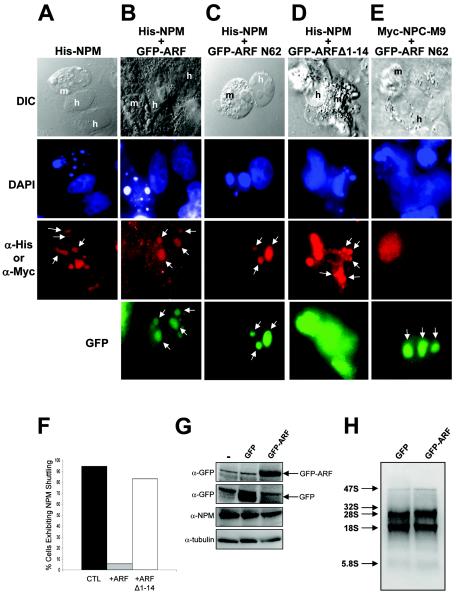
To address ARF's potential role in regulating NPM trafficking throughout the cell, we conducted in vivo heterokaryon shuttling assays (41). An expression construct encoding six-histidine-tagged full-length murine NPM (His-NPM) was transiently transfected alone or in combination with green fluorescent protein-tagged constructs encoding either full-length murine ARF (GFP-ARF) or ARF deletion mutants that either retain (GFP-ARF N62) or lack (GFP-ARF Δ1-14) the conserved amino terminus into asynchronously growing HeLa cells. Mouse NIH 3T3 fibroblasts were seeded onto transfected HeLa cells, yielding a heterogeneous human-mouse cell population, followed by fusion of plasma membranes and subsequent immunofluorescent detection of protein-shuttling events. As evidenced by staining of DNA with DAPI, the nuclei of NIH 3T3 and HeLa cells are easily distinguished by the greater heterochromatin focus content of mouse cells (speckled pattern) (Fig. 6, DAPI). In the absence of GFP-ARF, NPM readily migrated from human nucleoli to mouse nucleoli, as visualized in interspecies heterokaryons (Fig. 6A). However, in the presence of nucleolar GFP-ARF or GFP-ARF N62, NPM failed to shuttle and was restricted to human nucleoli within heterokaryons (Fig. 6B and C). Conversely, GFP-ARF Δ1-14, devoid of the NPM binding domain, was unable to restrict NPM shuttling between human and mouse nucleoli (Fig. 6D). As GFP-ARF did not hinder the nucleocytoplasmic trafficking of Myc-NPC-M9, an hnRNP protein that readily shuttles (Fig. 6E) (31), our combined body of data indicates that ARF specifically binds and sequesters NPM into nucleolar ARF complexes to completely impede NPM nucleocytoplasmic shuttling (Fig. 6F). Inhibition of NPM shuttling was not due to reduction of NPM protein, as GFP-ARF had no effect on NPM protein expression in HeLa cells (Fig. 6G). Additionally, GFP-ARF did not inhibit rRNA processing of 28S, 18S, and 5.8S subunits in HeLa cells (Fig. 6H), indicating that NPM's roles in rRNA processing and nuclear export are distinct and separate events.
FIG. 6.
ARF prevents the nucleocytoplasmic shuttling of NPM. NIH 3T3 cells (ARF null) were seeded onto HeLa cells that had been transfected with (A) His-NPM in combination with (B) GFP-ARF, (C) GFP-ARF N62, or (D) GFP ARF Δ1-14. (E) Myc-tagged NPC-M9 was transfected in combination with GFP-ARF N62 as a shuttling control. NIH 3T3 and HeLa cell cocultures were incubated with cycloheximide for 30 min prior to membrane fusion with polyethylene glycol. Fused cells (heterokaryons) were incubated in medium containing cycloheximide for an additional 4 h before fixation. Heterokaryon formation was verified under phase-contrast microscopy using a conventional differential interference contrast (DIC) filter, while His-NPM and ARF proteins were visualized with antibodies against His (red [A to D]) or Myc (red [E]) and naturally emitting GFP spectra (green) using tetramethyl rhodamine isothiocyanate and FITC filters, respectively. The arrows indicate the staining pattern of nucleoli. The data are representative of at least five independent heterokaryons formed for each transfection condition in three independent experiments with the aggregate NPM shuttling events of all experiments plotted (F). HeLa cells transfected with GFP or GFP-ARF were harvested and lysed 48 h after transfection. (G) Proteins were separated by SDS-PAGE and immunoblotted with antibodies recognizing γ-tubulin, NPM, and GFP. (H) For analysis of rRNA processing, transfected cells were labeled with [5,6-3H]uridine for 45 min, followed by a 1-h chase. RNA was harvested and separated on agarose-formaldehyde gels, transferred to membranes, and visualized by autoradiography. The arrows indicate sizes of rRNA components.
NPM expression is required for cell cycle progression.
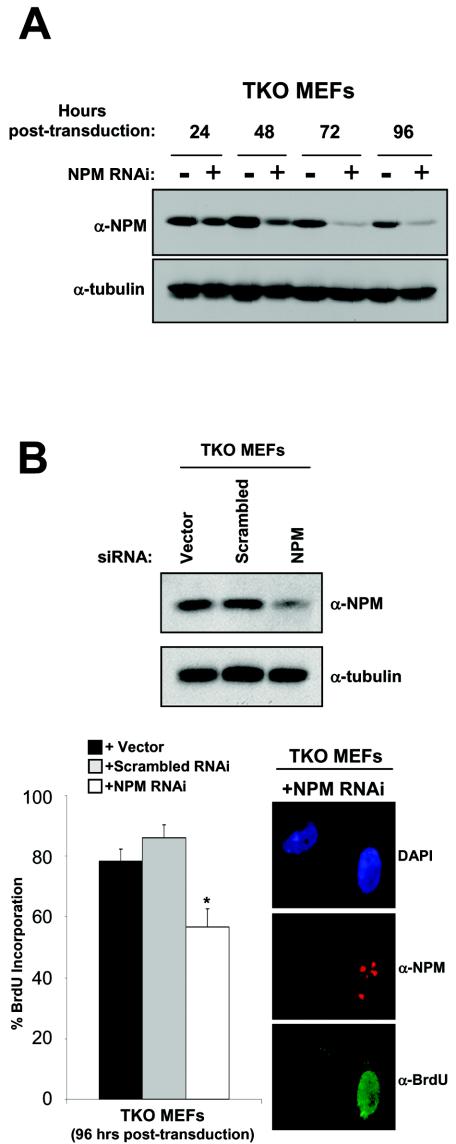
Previous investigations into NPM function have correlated NPM mRNA and protein levels with the cell's growth state (12, 17), and our data further indicate that NPM may be essential for S phase entry. Based on our finding that NPM shuttling is required for cell cycle progression, we utilized RNA interference methodologies to more directly assess the contribution of NPM to cell proliferation. TKO MEFs transduced with retroviruses encoding NPM-targeting siRNA duplexes (pSUPER-NPM RNAi) displayed a significant knockdown in NPM protein expression by 72 h postinfection (Fig. 7A) with no observed increase in apoptosis (data not shown). In contrast, the levels of γ-tubulin (Fig. 7A), as well as other nucleolar proteins (data not shown), remained unchanged, demonstrating the specificity of our siRNA targeting sequence. Additionally, a scrambled siRNA did not reduce NPM levels (Fig. 7A), underscoring the fact that NPM protein expression is not influenced by introduction of nonspecific RNAi molecules. Importantly, pSUPER-NPM RNAi-infected TKO MEFs that displayed reduced NPM protein expression were significantly impaired in their ability to enter S phase compared to cells infected with control viruses that exhibited normal levels of NPM (Fig. 7B). Given that the reduction of NPM protein expression and the inhibition of NPM's nucleocytoplasmic shuttling each trigger growth arrest in MEFs, we conclude that NPM is an essential player in the process of cell cycle progression.
FIG. 7.
Loss of NPM hinders cell cycle progression. (A) TKO MEFs infected with retroviruses encoding NPM-targeting siRNA duplexes (NPM RNAi) were harvested and lysed at 24-, 48-, 72-, and 96-h time points postinfection; as a control, uninfected cells were harvested and lysed in parallel. Whole-cell lysates were separated on denaturing polyacrylamide gels, transferred to PVDF membranes, and immunoblotted with antibodies recognizing NPM and γ-tubulin (loading control). (B) TKO MEFs transduced with either empty vector (dark bar), scrambled (shaded bar), or NPM (open bar) RNAi-encoding retrovi-ruses were seeded onto 100-mm-diameter dishes and glass coverslips at 24 h postinfection, followed by addition of BrdU (10 μM) at 72 h postinfection (for coverslips only). At 96 h postinfection, the cells were either harvested for Western blot analysis (NPM and γ-tubulin) or fixed, and subjected to indirect immunofluorescent detection of NPM expression (red) and BrdU incorporation (green) using the antibodies described above. Nuclei were demarcated with DAPI. The error bars indicate standard deviations of 100 cells counted three times for each slide (*, P > 0.005).
DISCUSSION
The nucleolus was originally described as the cell's command center for ribosomal biosynthesis and assembly, with a host of proteins being implicated in these processes. Nucleoli are not constrained by a membrane but rather actively recruit and retain proteins via arginine-lysine-rich nucleolar localization signal domains, such as those encoded within ARF, Mdm2, and NPM (22, 25, 43). In recent years, we have come to appreciate that proteins can actively shuttle from the nucleolus to various subcellular compartments in a regulated manner, providing evidence that the nucleolus is not merely a static site of ribosome biogenesis. While the nuclear and nucleolar functions of ARF have been heavily debated, several groups have shown that ARF can utilize nucleocytoplasmic transport to negate Mdm2-mediated degradation of p53 in the cytoplasm; specifically, ARF prevents the nuclear export of Mdm2 by actively sequestering it in the nucleolus (6, 10, 25, 26, 41, 43). It is reasonable to speculate that ARF, in its capacity as a tumor suppressor, may employ a similar tactic to regulate other growth-promoting proteins.
The key to ARF's proficiency in arresting cell proliferation, irrespective of p53 status, resides within its extreme amino terminus, namely, the first 14 residues of the ARF amino acid sequence. In search of additional interacting partners of this domain, we have verified the formation of both endogenous and exogenous ARF-NPM nucleolar complexes in primary mouse fibroblasts. Notably, ARF utilizes these exact residues to establish its high-affinity association with Mdm2, raising the possibility that ARF may affiliate with two distinct nucleolar complexes and/or represent a source of competition between Mdm2 and NPM. Upon its induction by hyperproliferative signals, ARF readily draws both Mdm2 and NPM into seemingly independent, distinct nucleolar complexes, evidenced by the absence of a ternary complex. However, overexpression of Mdm2 results in the release of NPM from ARF-containing protein complexes, suggesting that ARF's ability to dictate growth arrest in the presence or absence of p53 is largely determined by the stoichiometry of its binding partners. In light of this finding, it is appealing to think of Mdm2 as a target of ARF suppression, as well as a dampener of persistent ARF function. Specifically, Mdm2 may negatively regulate the p53 response through a dual mechanism: degradation of its direct transcriptional activator, p53 (38, 40), and inhibition of ARF function via preferential binding over NPM. We show here that Mdm2 can antagonize both the p53-dependent and -independent functions of ARF. By uncovering a pathway through which Mdm2 can promote cell growth independently of its classic upstream activator, p53, our study has exposed an unforeseen capacity of the Mdm2 oncoprotein.
While NPM is an abundant nucleolar phosphoprotein, data from our laboratory and others indicate that it is distributed throughout the cell in discrete pools, and factors such as NPM's state of posttranslational modification, bound protein partners, and subcellular localization may determine the composition and activity of any given NPM pool (5, 8, 22, 23, 30, 35, 47). As revealed by our data and the results of others, we hypothesize that ARF sequesters a specific pool of NPM in the nucleolus, preventing its transit and intended function(s) elsewhere in the cell. The intrinsic growth-promoting potential of the NPM pool immobilized within the ARF complex remains unclear, but recent studies support several possibilities. ARF transduction into MEFs lacking ARF and p53 (ARF/p53-null) was shown to significantly impair the processing of rRNAs, and this effect was strictly dependent upon the highly conserved first 14 amino acids within ARF's extreme amino terminus (39). Importantly, transduction of p53 failed to reproduce this result (39), indicating that ARF's ability to down-regulate rRNA processing is distinct from ARF's established roles within the classical ARF/p53/Mdm2 epistatic pathway. Additionally, two studies recently reported that NPM was a nucleolar ARF binding partner (4, 15). Both studies indirectly demonstrate that ARF can prevent proper rRNA processing, but how this might affect proper protein translation and how this event signals proliferative arrest remain unanswered. Nonetheless, these findings, in combination with our data, suggest that ARF's interaction with NPM may facilitate contact between ARF and the nucleolar ribosomal processing machinery, given that NPM appears to function as an integral component of ribosome maturation and export (30). We have further shown that NPM shuttling is required for cell proliferation, suggesting that ARF, via nucleolar sequestration of NPM, may not only target rRNA processing but might also prevent the nucleolar or nuclear export of processed rRNAs. This would be analogous to an earlier hypothesis in which protein targets of the ARF tumor suppressor may “ride the ribosome” out of the nucleolus and into the cytoplasm to execute their growth-promoting functions (37).
In an unrelated study, NPM was shown to be phosphorylated by cyclin E/cdk2 at the centrosome, resulting in the initiation of centrosome duplication (29). Hence, it is possible that nucleolar ARF, in response to hyperproliferative signals, binds and immobilizes the NPM pool that is designated for transit to the centrosome, consistent with our findings. However, a more recent report failed to detect NPM in isolated centrosomes (1), raising the possibility that other functional targets of NPM reside within or are transported to the cytoplasm, one of which might be the maturing ribosome itself. The models described above warrant further investigation, but we cannot overlook the possibility that NPM may, in addition to its ascribed roles in ribosome biogenesis and centrosome duplication, transmit additional growth-promoting commands and that Mdm2-mediated release of NPM from ARF complexes potentiates these functions. Nonetheless, ARF, in its capacity as a tumor suppressor, employs redundant mechanisms of protein binding and topological sequestration to inhibit both p53-dependent and -independent targets, thereby achieving control over cell proliferation.
Acknowledgments
We thank J. Alan Diehl, Martine Roussel, Charles Sherr, and Gerard Zambetti for plasmid constructs, antibodies, and primary DKO and TKO MEFs, as well as Sarah Mullen, Loryn Rikimaru, and Joanne Doherty for their excellent assistance. We also thank Martine Roussel and Charles Sherr for their support while J.D.W. was an associate of the Howard Hughes Medical Institute. We are extremely grateful to J. Alan Diehl, Helen Piwnica-Worms, John Cleveland, and Gerard Zambetti for insightful discussions; Charles Sherr and Dawn Quelle for sharing of unpublished data; and Joseph Baldassare and members of his laboratory for helpful comments.
S.N.B. is supported through the Siteman Center Cancer Biology Pathway. Y.Y. is the recipient of a grant-in-aid from the Department of Defense Breast Cancer Research Program (BC030793). L.B.M. is supported by institutional training grant T32-HL07873 from the National Institutes of Health. J.D.W. is a Pew Scholar and the recipient of grants-in-aid from the Edward Mallinckrodt, Jr., Foundation, American Cancer Society (IRG-58-010-46), and National Institutes of Health (GM066032).
REFERENCES
- 1.Andersen, J. S., C. J. Wilkinson, T. Mayor, P. Mortensen, E. A. Nigg, and M. Mann. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426:570-574. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 3.Bates, S., A. C. Phillips, P. A. Clark, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. P14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 4.Bertwistle, D., M. Sugimoto, and C. J. Sherr. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borer, R. A., C. F. Lehner, H. M. Eppenberger, and E. A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379-390. [DOI] [PubMed] [Google Scholar]
- 6.Bothner, B., W. S. Lewis, E. L. DiGiammarino, J. D. Weber, S. J. Bothner, and R. W. Kriwacki. Defining the molecular basis of Arf and Hdm2 interactions. J. Mol. Biol. 314:263-277. [DOI] [PubMed]
- 7.Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. P16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2:148-155. [DOI] [PubMed] [Google Scholar]
- 8.Chou, Y.-H., and Y. M. Yung. 1995. Cell cycle phase-dependent changes of localization and oligomerization states of nucleophosmin/B23. Biochem. Biophys. Res. Commun. 217:313-325. [DOI] [PubMed] [Google Scholar]
- 9.De Stanchina, E., M. E. McCurrach, F. Zindy, S. Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiGiammarino, E. L., I. Filippov, J. D. Weber, B. Bothner, and R. W. Kriwacki. 2001. Solution structure of the p53 regulatory domain of the p19ARF tumor suppressor protein. Biochemistry 40:2379-2386. [DOI] [PubMed] [Google Scholar]
- 11.Eischen, C. M., J. D. Weber, M. F. Roussel, C. J. Sherr, and J. L. Cleveland. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13:2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feuerstein, N., P. K. Chan, and J. J. Mond. 1988. Identification of numatrin, the nuclear matrix protein associated with induction of mitogenesis, as the nucleolar protein B23. Implication for the role of the nucleolus in early transduction of mitogenic signals. J. Biol. Chem. 263:10608-10612. [PubMed] [Google Scholar]
- 13.Honda, R., and H. Yasuda. 1999. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue, K., M. F. Roussel, and C. J. Sherr. 1999. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc. Natl. Acad. Sci. USA 96:3993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itahana, K., K. P. Bhat, A. Jin, Y. Itahana, D. Hawke, R. Kobayashi, and Y. Zhang. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12:1151-1164. [DOI] [PubMed] [Google Scholar]
- 16.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, P. S., and B. Y. Yung. 1999. Down-regulation of nucleophosmin/B23 mRNA delays the entry of cells into mitosis. Biochem. Biophys. Res. Commun. 257:865-870. [DOI] [PubMed] [Google Scholar]
- 18.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 19.Kamijo, T., S. Bodner, E. van de Kamp, D. H. Randle, and C. J. Sherr. 1999. Tumor spectrum in ARF-deficient mice. Cancer Res. 59:2217-2222. [PubMed] [Google Scholar]
- 20.Kondo, T., N. Minamino, T. Nagumura-Inoue, M. Matsumoto, T. Taniguchi, and N. Tanaka. 1997. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene 15:1275-1281. [DOI] [PubMed] [Google Scholar]
- 21.Korgaonkar, C., L. Zhao, M. Modestou, and D. E. Quelle. 2002. ARF function does not require p53 stabilization or Mdm2 relocalization. Mol. Cell. Biol. 22:196-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindstrom, M. S., U. Klangby, R. Inoue, P. Pisa, K. G. Wiman, and C. E. Asker. 2000. Immunolocalization of human p14(ARF) to the granular component of the interphase nucleolus. Exp. Cell Res. 256:400-410. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Q.-R., and P. K. Chan. 1991. Formation of nucleophosmin/B23 oligomers requires both the amino- and carboxyl-terminal domains of the protein. Eur. J. Biochem. 200:715-721. [DOI] [PubMed] [Google Scholar]
- 24.Llanos, S., P. A. Clark, J. Rowe, and G. Peters. 2001. Stabilization of p53 by p14ARF without relocalization of MDM2 to the nucleolus. Nat. Cell. Biol. 3:445-452. [DOI] [PubMed] [Google Scholar]
- 25.Lohrum, M. A., M. Ashcroft, M. H. Kubbutat, and K. H. Vousden. 2000. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2:179-181. [DOI] [PubMed] [Google Scholar]
- 26.Lohrum, M. A., M. Ashcroft, M. H. Kubbutat, and K. H. Vousden. 2000. Contribution of two independent MDM2-binding domains in p14ARF to p53 stabilization. Curr. Biol. 10:539-542. [DOI] [PubMed] [Google Scholar]
- 27.McKeller, R. N., J. L. Fowler, J. J. Cunningham, N. Warner, R. J. Smeyne, F. Zindy, and S. X. Skapek. The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc. Natl. Acad. Sci. USA 99:3848-3853. [DOI] [PMC free article] [PubMed]
- 28.Midgley, C. A., J. M. Desterro, M. L. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 29.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127-140. [DOI] [PubMed] [Google Scholar]
- 30.Okuwaki, M., M. Tsujimoto, and K. Nagata. 2002. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell 13:2016-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 32.Pomerantz, J., N. Schreiber-Agus, N. J. Liegois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H. W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19ARF, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 33.Quelle, D. E., F. Zindy, R. A. Ashmun, and C. J. Sherr. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993-1000. [DOI] [PubMed] [Google Scholar]
- 34.Roussel, M. F., A. M. Theodoras, M. Pagano, and C. J. Sherr. 1995. Rescue of defective mitogenic signaling by D-type cyclins. Proc. Natl. Acad. Sci. USA 92:6837-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt-Zachmann, M. S., and W. W. Franke. 1998. DNA cloning and amino acid sequence determination of a major constituent protein of mammalian nucleoli. Correspondence of the nucleoplasmic-related protein NO38 to mammalian protein B23. Chromosoma 96:417-426. [DOI] [PubMed] [Google Scholar]
- 36.Serrano, M., G. J. Hannon, and D. Beach. 1993. A new regulatory motif in cell cycle control causing specific inhibition of cyclin D/CDK4. Nature 366:704-707. [DOI] [PubMed] [Google Scholar]
- 37.Sherr, C. J., and J. D. Weber. 2000. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 38.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugimoto, M., M. L. Kuo, M. F. Roussel, and C. J. Sherr. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11:415-424. [DOI] [PubMed] [Google Scholar]
- 40.Tao, W., and A. J. Levine. 1999. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2 mediated degradation of p53. Proc. Natl. Acad. Sci. USA 96:3077-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao, W., and A. J. Levine. 1999. P19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokuyama, Y., H. F. Horn, K. Kawamura, P. Tarapore, and K. Fukasawa. 2001. Specific phosphorylation of nucleophosmin on Thr199 by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 276:21529-21537. [DOI] [PubMed] [Google Scholar]
- 43.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 44.Weber, J. D., M. L. Kuo, B. Bothner, E. L. DiGiammarino, R. W. Kriwacki, M. F. Roussel, and C. J. Sherr. 2000. Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 20:2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber, J. D., J. R. Jeffers, J. E. Rehg, D. H. Randle, G. Lozano, M. F. Roussel, C. J. Sherr, and G. P. Zambetti. p53-independent functions of the p19ARF tumor suppressor. Genes Dev. 14:2358-2365. [DOI] [PMC free article] [PubMed]
- 46.Xirodimas, D., M. K. Saville, C. Edling, D. P. Lane, and S. Lain. 2001. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 20:4972-4983. [DOI] [PubMed] [Google Scholar]
- 47.Zatsepina, O. V., A. Rousselet, P. K. Chan, M. O. Olson, E. G. Jordan, and M. Bornens. 1999. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J. Cell Sci. 112:455-466. [DOI] [PubMed] [Google Scholar]
- 48.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed]