Abstract
Notch proteins are transmembrane receptors that participate in a highly conserved signaling pathway that regulates morphogenesis in metazoans. Newly synthesized Notch receptors are proteolytically cleaved during transit to the cell surface, creating heterodimeric mature receptors comprising noncovalently associated extracellular (NEC) and transmembrane (NTM) subunits. Ligand binding activates Notch by inducing two successive proteolytic cleavages, catalyzed by metalloproteases and gamma-secretase, respectively, that permit the intracellular portion of NTM to translocate to the nucleus and activate transcription of target genes. Prior work has shown that the presence of NEC prevents ligand-independent activation of NTM, but the mechanisms involved are poorly understood. Here, we define the roles of two regions at the C-terminal end of NEC that participate in maintaining the integrity of resting Notch receptors through distinct mechanisms. The first region, a hydrophobic, previously uncharacterized portion of NEC, is sufficient to form stable complexes with the extracellular portion of NTM. The second region, consisting of the three Lin12/Notch repeats, is not needed for heterodimerization but acts to protect NTM from ligand-independent cleavage by metalloproteases. Together, these two contiguous regions of NEC impose crucial restraints that prevent premature Notch receptor activation.
Notch proteins define a unique class of highly conserved transmembrane receptors regulating cell growth, differentiation, and death in different tissues of multicellular organisms (reviewed in reference 1). Aberrations in Notch signaling have been linked to several diseases in mammals. The first human homologue of Notch, NOTCH1, was identified through its involvement in chromosomal translocations found in human T-cell acute lymphoblastic leukemias/lymphomas (T-ALL) (12). In nonhuman mammals, retroviral activation of each of the other three mammalian Notch homologues (NOTCH2, NOTCH3, and NOTCH4) has also been associated with neoplastic phenotypes (5, 9, 14). Additionally, mutations in NOTCH3 and the NOTCH ligand JAGGED1 cause two different inherited development disorders of humans, CADASIL syndrome (19) and Allagile syndrome (29), respectively.
Human NOTCH1 (hN1) has a crucial influence upon several stages of lymphocyte development (reviewed in reference 31). Expression of constitutively active hN1 in hematopoietic stem cells induces the development of CD4+ CD8+ double-positive (DP) immature T cells in the bone marrow, inhibits normal marrow B-cell development, and eventually leads to the appearance of fatal T-cell leukemias (32). Conversely, inducible notch1 knockout mice fail to develop mature T cells due to a requirement for notch1 during early stages of intrathymic T-cell development (33). Thus, although Notch receptors are essential for the development of multicellular organisms, their activation must be precisely regulated.
Notch receptors are first synthesized as 300- to 350-kDa type I single-pass transmembrane glycoproteins. During maturation, Notch precursor polypeptides are proteolytically processed by a furin-like convertase at a site called S1 (Fig. 1A), yielding two noncovalently associated subunits (26). The resulting two associated subunits, here termed extracellular Notch (NEC) and transmembrane Notch (NTM), constitute the mature heterodimeric form of the protein present at the cell surface (2, 7).
FIG. 1.
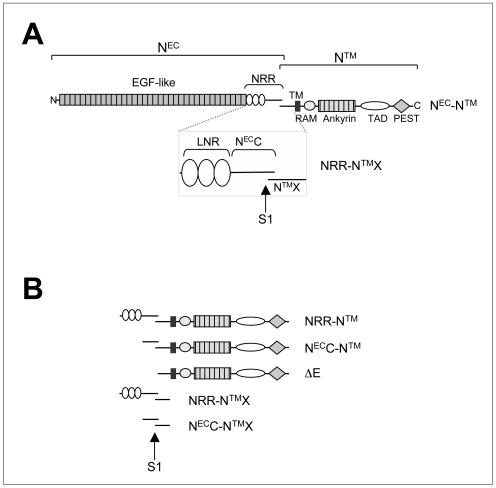
(A) Schematic representation of hN1. NEC, Notch extracellular subunit; NTM, Notch transmembrane subunit; NRR, negative regulatory region; LNR, domain comprising the three LIN12/Notch repeats; NECC, C-terminal segment of NEC; NTMX, NTM extracellular region; TM, transmembrane segment; ICN, intracellular Notch; TAD, transactivation domain; S1, furin cleavage site. (B) Truncated hN1 polypeptides used in these studies.
The NEC subunit of hN1 (Fig. 1A) contains 36 N-terminal epidermal growth factor (EGF)-like repeats that include the region responsible for ligand binding (23, 36). The EGF-like repeats are followed by a negative regulatory region (NRR), which participates in restraining premature activation of Notch receptors (15, 34). In hN1, the NRR includes the three LNR modules and an additional C-terminal tail of 103 residues (NECC). The NTM subunit (Fig. 1A) includes a short extracellular region (NTMX) containing a pair of conserved cysteines (25, 27), followed by a transmembrane segment, and an intracellular region (ICN). ICN consists of a RAM domain, seven ankyrin (ANK) repeats flanked by two nuclear localization signals, a transactivation domain (TAD), and a PEST region thought to regulate protein degradation (37).
Notch ligands are transmembrane proteins that are members of the DSL (Delta, Serrate, and LAG-2) family. These proteins contain a cysteine-rich DSL domain essential for binding to the EGF-like domain in the NEC subunit of Notch receptors (16). In the current model for Notch activation, ligand binding to repeats within the EGF-like domain induces two sequential proteolytic events termed S2 and S3 cleavages. S2 cleavage is catalyzed by disintegrin-metalloproteases like tumor necrosis factor alpha (TNF-α)-converting enzyme (TACE) at a site lying 13 residues external to the transmembrane segment of the NTM subunit (8, 27). Following cleavage at S2, γ-secretase cleavage within the transmembrane domain at site S3 releases active ICN (39, 46). Once released from the plasma membrane, ICN translocates to the nucleus, where it interacts with members of the CSL (for CBF-1/RBPJκ, Suppressor of Hairless, and Lag-1) family of transcription factors, resulting in transcriptional activation of target genes (4, 11, 18, 20, 24, 39, 40, 43, 47, 48). The RAM and ANK domains of ICN participate in CSL binding, while the ANK domain is also essential for recruitment of coactivators and transactivation (21, 28, 38).
Enforced expression of forms of Notch lacking the NEC subunit results in phenotypes characteristic of constitutively active Notch signaling in a variety of model organisms (10, 13, 25, 30, 35, 42). Additionally, disruption of the mature heterodimeric form of the receptor leads to nuclear localization of a polypeptide of the size of ICN and increased transcriptional activation of a CSL-specific reporter gene (34). In contrast, forms of Notch lacking the EGF-like repeats, but retaining the NRR, are restrained in an inactive state and are unable to respond to ligand (20, 25, 35, 42). Furthermore, certain deletions and point mutations within the NRR lead to gain-of-function phenotypes in Drosophila (25, 42) and Caenorhabditis elegans (6, 15). Taken together, these studies suggest that a major role of the NRR region of the NEC subunit is to restrain activation of Notch receptors in the absence of ligands, yet permit activation in response to ligand binding.
Although it is clear that the NRR participates in the regulation of Notch activity, there is limited information about the mechanism by which this region restrains signaling. Here, we establish that the NECC region within the NRR is sufficient to form a heterodimer with the extracellular region of NTM (NTMX) and show that the LNR domain is necessary for preventing metalloprotease cleavage at the S2 site in the absence of ligand, even though it is not required for heterodimerization. In addition, Notch polypeptides lacking both the ligand-binding and the LNR domains activate CSL reporter genes in cultured cells and drive the appearance of CD4+ CD8+ DP T cells in the peripheral blood when expressed in hematopoetic stem cells of mice, confirming that the removal of the LNR domain results in a ligand-independent gain-of-function phenotype. Together, these findings identify two distinguishable activities residing within the NRR: heterodimerization, mediated through the NECC region; and prevention of ligand-independent proteolysis, which also requires the LNR domain.
MATERIALS AND METHODS
Expression plasmids.
The various forms of mammalian Notch used in this study are summarized in Fig. 1. Constructs for expression of N1, N2, and N3 polypeptides were created by PCR amplification with human NOTCH1 (12), human NOTCH2 (41), and mouse NOTCH3 (22) cDNAs as templates. Eukaryotic hN1, hN2, and mouse N3 (mN3) constructs used for the subunit association studies were engineered in the plasmid vector pcDNA3 (Invitrogen). cDNAs encoding hN1 NRR-NTMX (residues Glu-1447 to Gln-1734), hN1 NECC-NTMX (residues His-1565 to Gln-1734), hN2 NECC-NTMX (residues Asp-1537 to Gln-1677), and mN3 NECC-NTMX (residues Glu-1504 to Ser-1641) were inserted in frame into an engineered BamHI site at the 3′ end of the sequence encoding the hN1 signal peptide (bp −15 to +69). FLAG and hemagglutinin (HA) epitope tags were then introduced at the N and C termini, respectively. For transcriptional activation studies, cDNAs encoding hN1 NEC-NTM (residues Met-1 to Lys-2556), hN1 NRR-NTM (residues Glu-1447 to Lys-2556), NECC-NTM (residues Ala-1563 to Lys-2556), and ΔE (residues Gly-1674 to Lys-2556) were engineered in frame at the 3′ end of the hN1 signal peptide in the pcDNA3 plasmid vector. For metalloprotease sensitivity experiments, three myc epitope tags were added in series at the C-terminal end of the NTM subunit of each construct. The bacterial hN2 NECC-NTMX-H6 expression construct (residues Asp-1537 to Gln-1677) was engineered in the plasmid vector pET-21a(+) (Novagen). The construct was designed such that only a single additional methionine was added at the N terminus of the encoded hN2 polypeptides and a hexahistidine tag was added to the C terminus. To permit stable expression in bone marrow cells, cDNAs encoding hN1 NRR-NTM and NECC-NTM were cloned into the backbone of the retroviral shuttle vector MigRI (32).
Expression of Notch proteins in cultured cells.
U2OS and 293T cells (American Type Culture Collection) were transiently transfected with Lipofectamine Plus reagent (Invitrogen). U2OS and 293T cells were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Sigma), 2 mM l-glutamine (Gibco), 100 U of penicillin/ml (Gibco), and 100 μg of streptomycin/ml (Gibco). Cells were grown at 37°C under 5% CO2.
NRR-NTMX association studies.
293T cells transfected with control vector or with vectors encoding different soluble forms of Notch heterodimers were grown on six-well plates for 72 h. Three days after transfection, the cells were washed twice with ice-cold Hanks balanced salt solution and collected by centrifugation at 500 × g for 5 min. Cell pellets were lysed by heating (100°C) for 10 min in sodium dodecyl sulfate (SDS) loading buffer in the presence of reducing agent and analyzed by Western blot after SDS-polyacrylamide gel electrophoresis (PAGE), using an antibody directed against the HA epitope tag (HA.11; Covance). Immunoprecipitates from conditioned media were prepared by mixing 1 ml of conditioned medium with anti-HA or anti-FLAG (Sigma) beads at 4°C for 2 h. Following incubation, the beads were washed three times with 1 ml of ice-cold buffer A (50 mM Tris, pH 7.5, 100 mM NaCl, 1% NP-40) and heated (100°C) for 10 min in SDS loading buffer in the presence of reducing agent. The recovered proteins were resolved by SDS-PAGE and analyzed by Western blotting with antibodies against the epitope tags.
Recombinant protein expression and purification.
The hN2 NECC-NTMX-H6 polypeptide (residues Asp-1537 to Gln-1677) was subcloned into the plasmid vector pET-21a(+) (Novagen) with a hexahistidine tag at the C-terminal end for affinity purification. The hN2 expression plasmid was then transformed into Escherichia coli strain BL21(DE3)pLysS. Protein expression was induced by addition of 0.4 mM isopropyl-1-thio-β-d-galactopyranoside to log-phase cell cultures grown in Luria-Bertani broth. After induction for 4 h, cells were recovered by centrifugation and resuspended in ice-cold 50 mM Tris buffer, pH 8.0, containing 300 mM NaCl, 20% (wt/vol) sucrose, 10 mM β-mercaptoethanol, 0.5 mM EDTA, 10 mM imidazole, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 2-μg/ml pepstatin, 5-μg/ml aprotinin, and 2-μg/ml leupeptin. Cells were lysed by freezing and thawing, followed by three cycles of sonication for 30 s on ice. Insoluble material was removed from the lysate by centrifugation, and the supernatant was adsorbed to nickel-nitrilotriacetic acid (NTA)-agarose resin (QIAGEN) at 4°C for 2 h. After extensive washing with buffer B (50 mM Tris buffer [pH 8.0], 300 mM NaCl, 10 mM imidazole), bound NECC-NTMX-H6 was eluted from the Ni-NTA agarose resin with buffer C (buffer B containing 250 mM imidazole). After dialysis at 4°C for 12 to 16 h against buffer D (50 mM Tris buffer [pH 8.0], 300 mM NaCl), NECC-NTMX-H6 was further purified by size exclusion chromatography on a Superdex 75 column (Amersham Biosciences) in buffer D.
In vitro furin processing of recombinant hN2 NECC-NTMX.
Purified NECC-NTMX-H6 was incubated in buffer E (50 mM Tris [pH 7.5], 1 mM CaCl2) with soluble furin (New England BioLabs) at 30°C for 4 h, using 1 U of furin per 50 μg of protein. After cleavage, the furin was removed by adsorbing the NECC-NTMX-H6 heterodimer to Ni-NTA-agarose at 4°C for 2 h after cleavage and then eluting it in buffer C. The processed NECC-NTMX-H6 heterodimer was purified and analyzed by size exclusion chromatography on a Superdex 75 column in buffer D. For analysis and identification of the cleaved products by reversed-phase high-performance liquid chromatography (HPLC), aliquots from the peak were acidified by addition of 5% (vol/vol) acetic acid, applied to an analytical-scale C18 column, and resolved with a water-acetonitrile gradient. The two eluted peptides (monitored by A229) were collected, lyophilized, resuspended in water, and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry.
Transcriptional activation assays.
Plasmids were introduced into U2OS cells by transfection with Lipofectamine (Invitrogen). To assay activation of CSL-dependent transcription, cells were cotransfected in triplicate with various Notch1 expression constructs in pcDNA3, a CSL firefly luciferase reporter plasmid (17), and an internal control sea pansy Renilla luciferase plasmid (Promega). Normalized firefly luciferase activities were measured in whole-cell extracts prepared 44 to 48 h after transfection with the Promega dual luciferase kit.
Metalloprotease sensitivity assay.
U2OS cells transiently transfected with myc-tagged forms of hN1 (NRR-NTM-myc and NECC-NTM-myc) were treated with the presenilin inhibitor compound E (CalBiochem) at a concentration of 1 μM or with carrier alone (0.01% dimethyl sulfoxide) as described previously (45). Cells were washed once with ice-cold Hanks buffered saline and lysed in buffer F (50 mM Tris [pH 7.5], 100 mM NaCl, 10 mM phenylmethylsulfonyl fluoride, 10-μg/ml aprotonin, 10-μg/ml leupeptin, 1% NP-40) for 15 min on ice. Lysates were cleared by centrifugation at 16,000 × g for 15 min at 4°C. hN1 polypeptides were immunoprecipitated from whole-cell lysates (2 × 106 cells) by incubation for 12 to 16 h at 4°C with anti-myc antibody (9E10) cross-linked to protein G-Sepharose beads (Sigma). After incubation, beads were washed twice with buffer F and once with a mixture of 50 mM Tris, pH 7.5, and 100 mM NaCl. Anti-myc immunoprecipitates were treated with lambda protein phosphatase (New England BioLabs) as recommended by the manufacturer prior to Western blot analysis.
Murine bone marrow transplantation assays.
All experiments were performed as described previously (4) and were conducted in accordance with National Institutes of Health guidelines for the care and use of animals and with an approved animal protocol from the University of Pennsylvania Animal Care and Use Committee. Briefly, cDNAs cloned into the MigRI vector were packaged into retroviruses by transient transfection of Bosc23 cells. After titering of viruses on NIH 3T3 cells, green fluorescent protein (GFP)-normalized retroviral supernatants were used to “spinoculate” bone marrow cells from female BALB/c mice (Taconic Farms). The retrovirally transduced bone marrow cells were then injected into lethally irradiated (900 rads) 4- to 8-week-old female syngeneic recipients. Three weeks posttransplantation, peripheral blood sampled from retro-orbital sinuses was assessed for the presence of GFP+ immature T cells by flow cytometric analysis (Becton Dickinson; FACSCalibur). Cells were incubated with biotinylated CD8α (53-6.7) and phycoerythrin-conjugated CD4 (RM4-5) (Pharmingen). Biotinylated antibodies were revealed with streptavidin-CyChrome. Dead cells, identified by forward scatter (FSC) and side scatter (SSC), were excluded from the analysis. Flow cytometry results were analyzed with CellQuest software.
RESULTS
The NRR-NTMX precursor from hN1 is properly processed into noncovalently associated subunits.
Several lines of evidence suggested that association between the two subunits of hN1 after S1 cleavage results from contacts between the NRR of the NEC subunit and the extracellular stub of the NTM subunit (NTMX). Therefore, we first designed a construct for expression of the NRR-NTMX precursor, predicting that this polypeptide would be processed at the S1 site during maturation and secreted into the culture medium when expressed in mammalian cells.
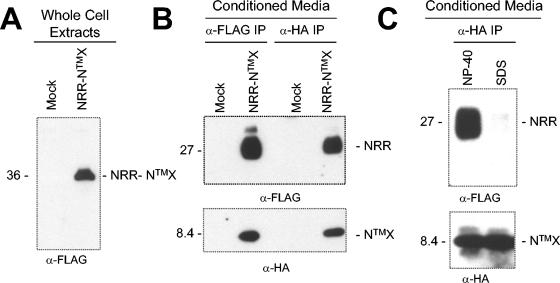
To determine whether the precursor is processed at the S1 site during maturation and whether the NRR and NTMX fragments remain associated with each other after cleavage at the S1 site during maturation, we expressed the NRR-NTMX precursor polypeptide of hN1 with FLAG and HA epitopes at the N and C termini, respectively, to facilitate detection of the individual subunits of the complex in immunoprecipitation assays (Fig. 1B). Following transient transfection into 293T cells, the single-chain precursor (NRR-NTMX) was detected in whole-cell extracts (Fig. 2A), whereas immunoprecipitates prepared from conditioned media using either FLAG or HA-coupled beads contained both epitope-tagged subunits, NRR and NTMX (Fig. 2B), but little or no uncleaved precursor. Thus, the two subunits remain stably associated as an NRR-NTMX heterodimer after furin cleavage and secretion into the conditioned media. The interaction between the NRR and NTMX subunits was disrupted by 0.1% SDS (Fig. 2C), demonstrating that association between the subunits is noncovalent, as seen with full-length hN1 (34).
FIG. 2.
Recovery of hN1 heterodimeric complexes secreted into conditioned media. Notch1-derived polypeptides with FLAG and HA epitope tags were resolved by SDS-PAGE under reducing conditions and detected on Western blots. Antibodies used to detect NRR and NTMX were anti-FLAG and anti-HA, respectively. (A) The NRR-NTMX single-chain precursor (36 kDa), detected in whole-cell extracts by Western blot with anti-FLAG. (B) Noncovalently associated NRR (27 kDa) and NTMX (8.4 kDa) subunits, coimmunoprecipitated (IP) from conditioned media with anti-FLAG or anti-HA as indicated, and detected by Western blot using anti-FLAG (top panel), or anti-HA (lower panel). (C) Dissociation of the heterodimer upon treatment with SDS. Coimmunoprecipitated NRR and NTMX subunits were treated with 1% NP-40, or 0.025% SDS prior to Western blotting with anti-FLAG (top panel) or anti-HA (lower panel).
LNR modules are not necessary for stable NRR-NTMX association.
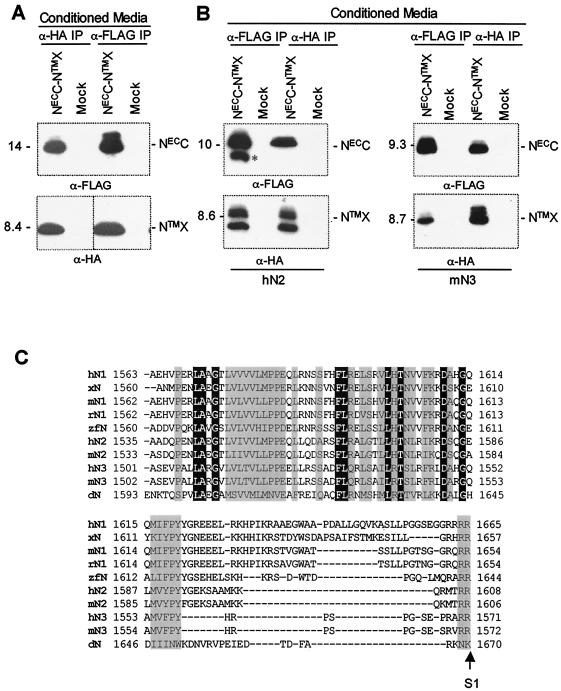
In order to determine whether the LNR modules are necessary for the association of the two subunits, we designed a shorter construct lacking the LNR modules. In this construct, the encoded polypeptide, termed NECC-NTMX, includes only the 103 residues C terminal to the LNR domain from the NECC subunit and the NTMX sequence, with epitope tags at both termini (Fig. 1B). When expressed transiently in 293T cells, the NECC-NTMX construct is properly processed and secreted into conditioned media as a stable heterodimer, demonstrating that the LNR domain is not required for stable association of the two subunits (Fig. 3A). In contrast, a construct lacking the NECC region (but retaining the LNR domain) exhibits poor expression and processivity by furin (data not shown), suggesting that the NECC region is essential for proper folding of the precursor, as well as for maintaining the integrity of the heterodimer. Analogous NECC-NTMX polypeptides from hN2 and mN3 are also sufficient for formation of stable heterodimers after cleavage (Fig. 3B), indicating that this is a common feature of mammalian Notch receptors (Fig. 3C).
FIG. 3.
The NECC region is sufficient for formation of stable heterodimers with NTMX. (A, B) NECC-NTMX heterodimers were recovered from conditioned media by immunoprecipitation (IP) with either anti-FLAG or anti-HA and were subjected to SDS-PAGE under reducing conditions. Each subunit was then detected by Western blotting for its respective epitope tag. (Top panel) Detection of NECC with anti-FLAG; (bottom panel) detection of NTMX with anti-HA. (A) hN1 heterodimers. (B) hN2 and mN3 heterodimers. The asterisk denotes a polypeptide of slightly smaller molecular weight than hN2 NECC that is not competent for association with NTMX. (C) Sequence alignment of the NECC region of various Notch proteins up to the putative furin site (S1). Identical and conserved residues among the sequences are enclosed in black and gray boxes, respectively. H, human; x, Xenopus; m, mouse; r, rat; and zf, zebra fish.
Recombinant furin-cleaved NECC-NTMX forms a stable heterodimer.
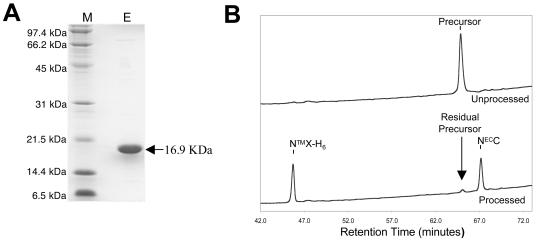
To prove unequivocally that other factors are not required for the formation of stable NECC-NTMX heterodimers, we purified a histidine-tagged, recombinant form of the NECC-NTMX precursor from hN2 (Fig. 4A) and tested whether the two subunits remained associated after furin cleavage. Before furin cleavage, a sample of the purified NECC-NTMX-H6 precursor elutes as a single peak when analyzed by denaturing HPLC on a C18 column (Fig. 4B, top). After furin cleavage, however, two peaks are present in the HPLC chromatogram (Fig. 4B, bottom), even though the two subunits copurify as a single peak on gel filtration with the same volume of elution as the precursor polypeptide. The first peak (retention time, 46 min) results from NTMX-H6 and the second from NECC (retention time, 68 min), based on masses determined by MALDI-TOF mass spectrometry (NECC expected molecular mass = 8,419 kDa, actual = 8,420 kDa; NTMX-H6 expected molecular mass = 8,494 kDa, actual = 8,489 kDa). These results demonstrate that the NECC region of hN2 is sufficient to form a stable heterodimer with NTMX.
FIG. 4.
(A) Purification of the NECC-NTMX-H6 precursor from hN2 after expression in bacteria. M, molecular mass standards; E, the NECC-NTMX-H6 uncleaved precursor after elution from nickel-NTA beads. (B) Denaturing HPLC chromatograms of the hN2 precursor (top) and NECC-NTMX heterodimer (bottom). Samples of both the precursor and intact heterodimer (after cleavage with recombinant furin) were recovered in a single peak after gel filtration. An aliquot of the peak from each sample was further analyzed by denaturing reversed-phase HPLC chromatography using a C18 column. The peaks corresponding to the precursor and each of the subunits are indicated.
LNR modules prevent ligand-independent Notch activation in cells.
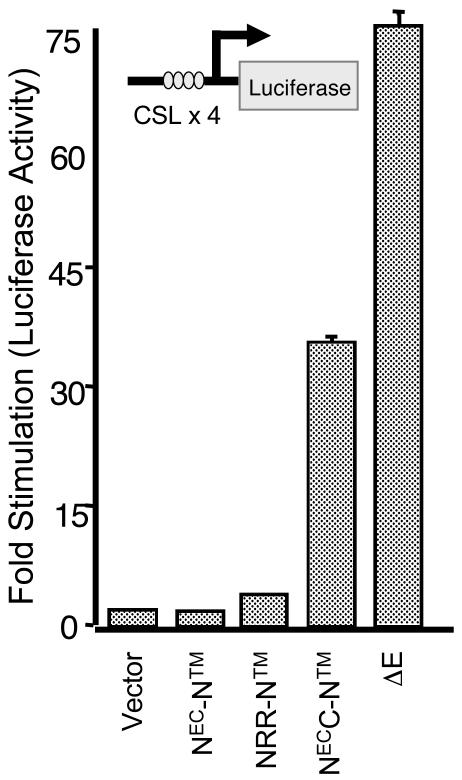
Prior studies suggesting that the LNR domain is needed to restrain Notch signaling were conducted with mutations or deletions involving the LNR domain within receptors that retained the EGF repeat ligand-binding region. Thus, increases in Notch function stemming from these mutations could have resulted from either ligand-independent signaling or hyperresponsiveness to ligand. To distinguish between these possibilities, we compared the activities of two hN1 constructs: NRR-NTM, which lacks the ligand-binding domain, and NECC-NTM, which lacks both the ligand binding and LNR domains. We first compared the abilities of these two constructs to activate transcription from a CSL-dependent luciferase reporter gene in cultured cells (Fig. 5). As anticipated, NRR-NTM had no intrinsic signaling activity in this assay. In contrast, NECC-NTM demonstrated substantial ability to activate CSL, confirming that the LNR domain is needed to prevent ligand-independent receptor activation. The extent of receptor activation at equivalent doses of input was more modest than that observed with a construct that lacks the entire NEC subunit (Fig. 5; designated ΔE). In Western blots, we consistently observe that NECC-NTM expresses at lower levels than does ΔE (as in Fig. 6B) (data not shown), possibly explaining the observed difference in activity.
FIG. 5.
Signaling activity of native hN1 and a series of hN1 NEC deletion variants in CSL reporter gene assays. Luciferase assays were performed on U2OS cell lysates prepared from cells transfected in triplicate with 50 ng of empty pcDNA3 plasmid or plasmids encoding the indicated forms of Notch1, along with a luciferase reporter plasmid containing iterated CSL-binding site and an internal control plasmid expressing Renilla luciferase from a thymidine kinase promoter. Firefly luciferase activity, normalized for variation in Renilla luciferase activity, is expressed relative to the activity in extracts prepared from cells transfected with empty pcDNA3, which is arbitrarily set to a value of 1.
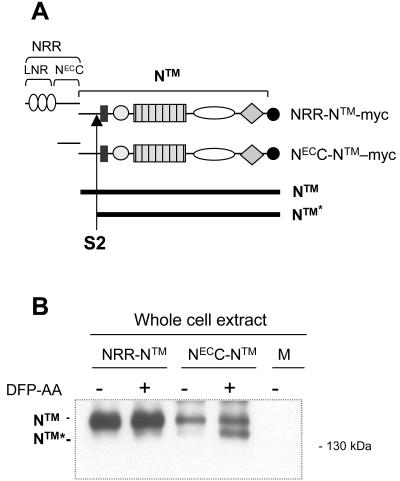
FIG. 6.
The LNR domain of hN1 confers resistance to proteolysis at the S2 site. (A) Schematic representation of the hN1 polypeptides used in the proteolysis experiments, noting the position of the myc epitope tag, the site of S2 cleavage, and the difference in polypeptide length between native NTM and the product after S2 cleavage, denoted NTM*. (B) The S2 proteolysis product NTM* is generated when the LNR domain is deleted from hN1. U2OS cells transfected with the indicated hN1 variants were incubated in the presence of a presenilin inhibitor (DFP-AA). NTM and NTM* polypeptides were then recovered by immunoprecipitation with an anti-myc antibody, resolved by SDS-PAGE under reducing conditions, and detected by Western blotting using anti-myc.
The LNR domain prevents ligand-independent proteolysis of Notch receptors.
Since the LNR domain is not necessary to maintain stable association of the two Notch subunits, we hypothesized that it protects NTMX from ligand-independent proteolysis. To test this idea, we compared the extents to which NRR-NTM and NECC-NTM forms of hN1 are cleaved at the S2 site (Fig. 6A).
Normally the product of S2 cleavage (NTM*, Fig. 6A) is rapidly cleaved at the S3 site by gamma-secretase to release ICN1 (i.e., the active form of Notch), a species that has a very short half-life. However, in the presence of gamma-secretase inhibitors, NTM* is stabilized, allowing it to accumulate and be detected (45). Cells transiently expressing NECC-NTM accumulate readily detectable amounts of NTM* in the presence of a gamma-secretase inhibitor, whereas cells expressing NRR-NTM, a molecule that still has the LNR modules but is quiescent in the reporter assay, do not (Fig. 6B). Together, these results show that although the LNR domain is not essential for association between the two Notch subunits, it does play a critical role in the regulation of receptor activity by preventing premature proteolytic activation in the absence of ligand.
Loss of the LNR modules causes Notch gain of function in a mouse model.
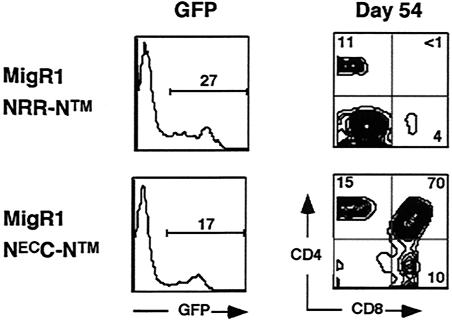
To assess the biological activity of NECC-NTM in vivo, we scored its activity in a murine bone marrow transplantation assay. When used to reconstitute irradiated mice, bone marrow stem cells transduced with gain-of-function forms of Notch1 rapidly give rise to an abnormal CD4+ CD8+ DP T-cell population in the bone marrow that spills into the peripheral blood by 3 weeks posttransplantation (4, 31). In these experiments, we used the retroviral vector MigRI, which also drives the expression of GFP from a bicistronic mRNA with an internal ribosomal entry sequence, in order to gate on transduced cells. The GFP+ cells in the peripheral blood of a mouse reconstituted with hematopoetic stem cells transduced with NRR-NTM virus on day 54 are mainly CD4− CD8− (Fig. 7); most of these cells are B220+ B cells and Gr1+ granulocytes (not shown), a result similar to that seen with empty MigRI virus (4, 32). In contrast, 70% of the GFP+ cells in the peripheral blood of a mouse reconstituted with NECC-NTM-transduced marrow cells are CD4+ CD8+ immature DP T cells (Fig. 7). Similar results were obtained with nine mice in each group (not shown). Thus, the LNR region is necessary to prevent ligand-independent receptor activation in vivo.
FIG. 7.
A form of Notch1 lacking both ligand-binding EGF repeats and the LNR domain, NEC-NTM, causes abnormal T-cell development. Peripheral blood was obtained from mice (n = 9 for each group) on day 54 after reconstitution with hematopoetic stem cells transduced with MigRI retroviruses encoding either NRR-NTM, which lacks only the ligand-binding domain, or NEC-NTM. Representative flow cytometric results are shown.
DISCUSSION
Because Notch signaling controls so many developmental events, its activation must be precisely controlled. Here, we define the roles of two regions at the C-terminal end of the NEC subunit that participate in maintaining the integrity of resting Notch receptors through distinct mechanisms. The domain mapping experiments define a previously uncharacterized region, termed NECC, that associates directly with NTMX to create the stable Notch heterodimer (Fig. 2 to 4). The LNR modules, which are not required for heterodimerization, are needed to prevent premature proteolytic cleavage at the S2 site of NTMX (Fig. 5 and 6).
Prior studies had shown that mutations involving the LNR domain result in inappropriate increases in Notch signals, implying an important regulatory role for these domains. We noted previously that the folding and structural integrity of individual LNR modules require calcium (3, 44); that Notch heterodimers immunoprecipitated from whole-cell lysates dissociate in the presence of EDTA (a metal ion chelator) (34); and that EDTA treatment of intact cells results in release of NTM, nuclear translocation of ICN, and activation of CSL reporter genes (34). While the simplest explanation for these observations was for the LNR domain to participate directly in binding to the NTM subunit, observations described here argue against such a role, as the region lying C terminal of the LNR domain, NECC, is sufficient to form stable heterodimers with NX in secreted and recombinant Notch “minireceptors.” This NECC region ranges in size from 70 to 103 residues among the mammalian Notch receptors and includes a number of highly conserved hydrophobic residues that might participate in the association with NTMX (Fig. 3C). Most striking among the conserved sequences is the cluster of aliphatic amino acids that lies near the NECC N-terminal end (Fig. 3C).
NECC is not as well conserved in Drosophila Notch, consistent with the possibility that fly Notch may not require S1 cleavage for activation. However, significant sequence similarity does exist in this region, both within the hydrophobic stretch near the N-terminal end and at a series of invariant residues scattered throughout the region. Efforts to determine a high-resolution structure of a mammalian Notch heterodimer, which will reveal the nature of the interface in detail, are now under way.
Mutations in NECC and in its heterodimerization partner, the extracellular stub of NTM (NTMX), have been previously associated with gain-of-function phenotypes in both flies and worms. In worms, the S642N mutation in NECC of the glp-1 notch homolog causes a gain-of-function phenotype (6). In flies, simultaneous replacement of the two conserved cysteines in NTMX with serine residues leads to gain of function, as assessed by the development of an antineurogenic phenotype (25), whereas point mutations elsewhere within NTMX are associated with gain of function in worms (15). The biochemical studies reported here provide a rationalization for the observed gain of function in the genetic studies; various mutations in either NECC or NTMX can destabilize the heterodimer, causing premature subunit dissociation after S1 cleavage and subsequent Notch activation by the normal proteolytic cascade of S2 and S3 cleavages.
LNR domains are a unique feature of Notch receptors, existing in all members of the family as three tandemly repeated modules. Although the NECC region is sufficient to form a stable heterodimer with NTMX, it is not sufficient to prevent proteolytic activation in the absence of the adjacent LNR modules (Fig. 5 and 6). The implication of these studies is that LNR mutations cause gain-of-function phenotypes because they allow proteolysis at the S2 site even in the absence of ligand, although without promoting premature dissociation of the subunits. The effects we observed previously showing that EDTA dissociates and activates Notch heterodimers may be explained by loss of this “anti-protease” activity, as these experiments were performed (whole-cell lysates or intact cells at 25 to 37°C) under conditions in which proteases may have been active.
More generally, our studies show that stable heterodimer association and prevention of ligand-independent activating cleavages are separable functions that can be attributed to two different regions of the extracellular domain of Notch and raise further mechanistic questions. Most notably, does the LNR domain prevent ligand-independent proteolysis merely by sterically preventing access of metalloproteases to the S2 site, or do more complex mechanisms come into play? It will also be important to resolve the puzzle of how physiologic ligand binding overcomes the intrinsic restraint on signaling imposed by the LNR domain.
ADDENDUM IN PROOF
A report coauthored by several of us now shows that activating mutations of human Notch1 are frequently found in human T-cell acute lymphocytic leukemia. The majority of these gain-of-function mutations are located within the heterodimerization domain defined by these studies (Science, in press).
Acknowledgments
We thank Vytas Patriub and Sai Hong for excellent technical support.
This research was supported by NIH grants CA92433 (to S.C.B.), CA82308 (to J.C.A.), AI47833 (to W.S.P.), and 1 F31 CA97547-01 (to C.S.I.). S.C.B. is a Pew Scholar in the Biomedical Sciences and an Established Investigator of the American Heart Association.
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 2.Aster, J., W. Pear, R. Hasserjian, H. Erba, F. Davi, B. Luo, M. Scott, D. Baltimore, and J. Sklar. 1994. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp. Quant. Biol. 59:125-136. [DOI] [PubMed] [Google Scholar]
- 3.Aster, J. C., W. B. Simms, Z. Zavala-Ruiz, V. Patriub, C. L. North, and S. C. Blacklow. 1999. The folding and structural integrity of the first LIN-12 module of human Notch1 are calcium-dependent. Biochemistry 38:4736-4742. [DOI] [PubMed] [Google Scholar]
- 4.Aster, J. C., L. Xu, F. G. Karnell, V. Patriub, J. C. Pui, and W. S. Pear. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by Notch1. Mol. Cell. Biol. 20:7505-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellavia, D., A. F. Campese, E. Alesse, A. Vacca, M. P. Felli, A. Balestri, A. Stoppacciaro, C. Tiveron, L. Tatangelo, M. Giovarelli, C. Gaetano, L. Ruco, E. S. Hoffman, A. C. Hayday, U. Lendahl, L. Frati, A. Gulino, and I. Screpanti. 2000. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 19:3337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry, L. W., B. Westlund, and T. Schedl. 1997. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124:925-936. [DOI] [PubMed] [Google Scholar]
- 7.Blaumueller, C. M., H. Qi, P. Zagouras, and S. Artavanis-Tsakonas. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90:281-291. [DOI] [PubMed] [Google Scholar]
- 8.Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5:207-216. [DOI] [PubMed] [Google Scholar]
- 9.Capobianco, A. J., P. Zagouras, C. M. Blaumueller, S. Artavanis-Tsakonas, and J. M. Bishop. 1997. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffman, C. R., P. Skoglund, W. A. Harris, and C. R. Kintner. 1993. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell 73:659-671. [DOI] [PubMed] [Google Scholar]
- 11.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518-522. [DOI] [PubMed] [Google Scholar]
- 12.Ellisen, L. W., J. Bird, D. C. West, A. L. Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649-661. [DOI] [PubMed] [Google Scholar]
- 13.Fortini, M. E., and S. Artavanis-Tsakonas. 1994. The suppressor of hairless protein participates in notch receptor signaling. Cell 79:273-282. [DOI] [PubMed] [Google Scholar]
- 14.Gallahan, D., and R. Callahan. 1997. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene 14:1883-1890. [DOI] [PubMed] [Google Scholar]
- 15.Greenwald, I., and G. Seydoux. 1990. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346:197-199. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, S. T., D. Gao, S. Christensen, and J. Kimble. 1997. Functional domains of LAG-2, a putative signaling ligand for LIN-12 and GLP-1 receptors in Caenorhabditis elegans. Mol. Biol. Cell 8:1751-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh, J. J. D., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 19.Joutel, A., C. Corpechot, A. Ducros, K. Vahedi, H. Chabriat, P. Mouton, S. Alamowitch, V. Domenga, M. Cecillion, E. Marechal, J. Maciazek, C. Vayssiere, C. Cruaud, E. A. Cabanis, M. M. Ruchoux, J. Weissenbach, J. F. Bach, M. G. Bousser, and E. Tournier-Lasserve. 1996. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383:707-710. [DOI] [PubMed] [Google Scholar]
- 20.Kopan, R., E. H. Schroeter, H. Weintraub, and J. S. Nye. 1996. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurooka, H., K. Kuroda, and T. Honjo. 1998. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 26:5448-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lardelli, M., J. Dahlstrand, and U. Lendahl. 1994. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech. Dev. 46:123-136. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, N., T. Klein, K. Brennan, and A. Martinez Arias. 2000. Structural requirements for notch signalling with delta and serrate during the development and patterning of the wing disc of Drosophila. Development 127:3185-3195. [DOI] [PubMed] [Google Scholar]
- 24.Lecourtois, M., and F. Schweisguth. 1998. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr. Biol. 8:771-774. [DOI] [PubMed] [Google Scholar]
- 25.Lieber, T., S. Kidd, E. Alcamo, V. Corbin, and M. W. Young. 1993. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 7:1949-1965. [DOI] [PubMed] [Google Scholar]
- 26.Logeat, F., C. Bessia, C. Brou, O. LeBail, S. Jarriault, N. G. Seidah, and A. Israel. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95:8108-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197-206. [DOI] [PubMed] [Google Scholar]
- 28.Nam, Y., A. P. Weng, J. C. Aster, and S. C. Blacklow. 2003. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 278:21232-21239. [DOI] [PubMed] [Google Scholar]
- 29.Oda, T., A. G. Elkahloun, B. L. Pike, K. Okajima, I. D. Krantz, A. Genin, D. A. Piccoli, P. S. Meltzer, N. B. Spinner, F. S. Collins, and S. C. Chandrasekharappa. 1997. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 16:235-242. [DOI] [PubMed] [Google Scholar]
- 30.Pear, W. S., J. C. Aster, M. L. Scott, R. P. Hasserjian, B. Soffer, J. Sklar, and D. Baltimore. 1996. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183:2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pear, W. S., and F. Radtke. 2003. Notch signaling in lymphopoiesis. Semin. Immunol. 15:69-79. [DOI] [PubMed] [Google Scholar]
- 32.Pui, J. C., D. Allman, L. Xu, S. DeRocco, F. G. Karnell, S. Bakkour, J. Y. Lee, T. Kadesch, R. R. Hardy, J. C. Aster, and W. S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299-308. [DOI] [PubMed] [Google Scholar]
- 33.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H. R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10:547-558. [DOI] [PubMed] [Google Scholar]
- 34.Rand, M. D., L. M. Grimm, S. Artavanis-Tsakonas, V. Patriub, S. C. Blacklow, J. Sklar, and J. C. Aster. 2000. Calcium depletion dissociates and activates heterodimeric Notch receptors. Mol. Cell. Biol. 20:1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rebay, I., R. G. Fehon, and S. Artavanis-Tsakonas. 1993. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74:319-329. [DOI] [PubMed] [Google Scholar]
- 36.Rebay, I., R. J. Fleming, R. G. Fehon, L. Cherbas, P. Cherbas, and S. Artavanis-Tsakonas. 1991. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67:687-699. [DOI] [PubMed] [Google Scholar]
- 37.Rechsteiner, M. 1988. Regulation of enzyme levels by proteolysis: the role of pest regions. Adv. Enzyme Regul. 27:135-151. [DOI] [PubMed] [Google Scholar]
- 38.Roehl, H., M. Bosenberg, R. Blelloch, and J. Kimble. 1996. Roles of the RAM and ANK domains in signaling by the C. elegans GLP-1 receptor. EMBO J. 15:7002-7012. [PMC free article] [PubMed] [Google Scholar]
- 39.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393:382-386. [DOI] [PubMed] [Google Scholar]
- 40.Schweisguth, F. 1995. Suppressor of Hairless is required for signal reception during lateral inhibition in the Drosophila pupal notum. Development 121:1875-1884. [DOI] [PubMed] [Google Scholar]
- 41.Stifani, S., C. M. Blaumueller, N. J. Redhead, R. E. Hill, and S. Artavanis-Tsakonas. 1992. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat. Genet. 2:119-127. [DOI] [PubMed] [Google Scholar]
- 42.Struhl, G., and A. Adachi. 1998. Nuclear access and action of notch in vivo. Cell 93:649-660. [DOI] [PubMed] [Google Scholar]
- 43.Struhl, G., and I. Greenwald. 1999. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398:522-525. [DOI] [PubMed] [Google Scholar]
- 44.Vardar, D., C. L. North, C. Sanchez-Irizarry, J. C. Aster, and S. C. Blacklow. 2003. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry 42:7061-7067. [DOI] [PubMed] [Google Scholar]
- 45.Weng, A. P., Y. Nam, M. S. Wolfe, W. S. Pear, J. D. Griffin, S. C. Blacklow, and J. C. Aster. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of Notch signaling. Mol. Cell. Biol. 23:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe, M. S. 2001. Presenilin and gamma-secretase: structure meets function. J. Neurochem. 76:1615-1620. [DOI] [PubMed] [Google Scholar]
- 47.Wolfe, M. S., W. Xia, B. L. Ostaszewski, T. S. Diehl, W. T. Kimberly, and D. J. Selkoe. 1999. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 398:513-517. [DOI] [PubMed] [Google Scholar]
- 48.Ye, Y., N. Lukinova, and M. E. Fortini. 1999. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature 398:525-529. [DOI] [PubMed] [Google Scholar]