Abstract
Small DNA tumor viruses typically encode proteins that either inactivate or degrade p53. Human adenoviruses encode products, including E4orf6 and E1B55K, that do both. Each independently binds to p53 and inhibits its ability to activate gene expression; however, in combination they induce p53 degradation by the ubiquitin pathway. We have shown previously that p53 degradation relies on interactions of E4orf6 with the cellular proteins Cul5, Rbx1, and elongins B and C to form an E3 ligase similar to the SCF and VBC complexes. Here we show that, like other elongin BC-interacting proteins, including elongin A, von Hippel-Lindau protein, and Muf1, the interaction of E4orf6 is mediated by the BC-box motif; however, E4orf6 uniquely utilizes two BC-box motifs for degradation of p53 and another target, Mre11. In addition, our data suggest that the interaction of E1B55K with E4orf6 depends on the ability of E4orf6 to form the E3 ligase complex and that such complex formation may be required for all E4orf6-E1B55K functions.
The tumor suppressor p53 plays a major regulatory role in cells (for review, see reference 53). Its activation by stress, including DNA damage and expression of activated oncogenes, can induce either cell cycle arrest or apoptosis. In most human tumors, p53 (or components of the p53 pathway) is inactivated, making such cells resistant to growth arrest and cell death. Many small DNA tumor viruses, including simian virus 40 (SV40), human papillomavirus (HPV), and human adenoviruses, encode proteins that target p53 for inactivation, thus presumably enhancing viral replication (reviewed by Roulston et al. [41a]). SV40 large T antigen binds to p53 (43) and inhibits its ability to bind to DNA and activate transcription (2). The HPV E6 protein induces the ubiquitination and degradation of p53 (45). Human adenoviruses produce two early proteins, E1B55K and E4orf6, that are capable independently of interacting with p53 and inhibiting transactivational activity (7, 11, 24, 30, 33, 54). In addition, they cooperate to cause the ubiquitination and degradation of p53 (7, 18, 37, 38, 41, 47).
Inactivation of proteins by degradation commonly involves the ubiquitin-dependent proteasome pathway. Target proteins are first conjugated to a chain of ubiquitin molecules through the action of a series of enzymes termed E1 (ubiquitin activating), E2 (ubiquitin conjugating), and E3 (ubiquitin ligase). It is the role of the E3 ligase, often a multiprotein complex, to recognize the substrates and bridge them with E2 for ubiquitin conjugation. The HPV E6 protein acts as an adapter to bring p53 to a cellular E3 ligase, termed E6AP (3, 40). E6AP—which has been shown in noninfected cells to ubiquitinate HHR23A, a human Rad23 homologue (28), Mcm7 (27), and Blk, a Src family member (34)—is recruited by E6 to new substrates. In addition to p53, E6 redirects E6AP to ubiquitinate hDlg (15), hScrib (32), c-Myc (17), and Bak (51). Mutation of E6AP represents one cause of Angelman syndrome (26).
The adenovirus proteins E4orf6 and E1B55K also redirect a cellular E3 ligase to target p53 for degradation: in this case one that contains the cullin family member Cul5, elongins B and C, and the RING protein Rbx1, which were shown to interact with E4orf6 but not E1B55K (18, 37). Cullin-based E3 ligases are part of the RING-type group of E3 ligases, which unlike the HECT-type group do not form an E3-ubiquitin thioester intermediate (55). These ligases are composed of a cullin family member and the RING protein Rbx1 linked to a protein that acts as a substrate specificity factor. These complexes interact with an E2-conjugating enzyme, often either Cdc34 or Ubc5, to conjugate ubiquitin chains to its substrates. In the case of the SCF complex, for which a crystal structure has been determined (55), Cul1 is linked by Skp1 to one of many F-box proteins, which serve as the substrate recognition protein (for a review of SCF, see reference 20). In the VBC complex, the linker proteins elongins B and C link Cul2 to the von Hippel-Lindau (VHL) substrate specificity protein. Elongins B and C also serve as a linker for complexes of Cul5, which have been found to exist with presumed substrate specificity proteins Muf1, elongin A, WSB, and SOCS1 (21), although substrates for these complexes have not yet been identified. Interestingly, VHL has been found to form complexes with both Cul2 and Cul5. Thus far, VHL is the only protein found to interact with more than one cullin family member (21).
Like the HPV E6 protein, which was initially discovered to degrade p53 but later found to induce the degradation of other substrates, the E4orf6 complex probably induces the degradation of other cellular proteins in addition to p53. It has recently been suggested that Rad50 and Mre11, components of the double-stranded break repair complex, are substrates of the E4orf6 ligase complex (48).
In addition to their function in degradation of p53, E4orf6 and E1B55K form a complex that has been implicated in several late viral functions. Both are required for efficient transport of late viral mRNA transcripts and the block of nuclear export of cellular mRNAs (reviewed in references 12 and 14). Mutant viruses defective in either of these genes exhibit reduced replication and progeny production.
Elongin C-interacting proteins, including elongin A, VHL, and SOCS1, have been shown to contain a conserved sequence, termed the BC-box motif, which is the only region of significant homology between elongin A and VHL and which is required for interaction with elongin C (21, 23). In the present studies on the E4orf6 E3 ligase complex, we determined if E4orf6 contains such a motif and if it mediates interactions with elongins B and C in complex formation. We show that E4orf6 contains two functional near-consensus BC-box motifs and that both are needed for efficient E1B55K/E4orf6-induced degradation of p53 and Mre11. In addition, our results suggest that E1B55K does not interact directly with E4orf6 as previously believed, but rather that it requires E4orf6 to form a complex with Cul5 and elongins B and C to bind and to elicit p53 degradation and other functions involved in viral replication. These results suggest new functions for the E4orf6 E3 ligase activity related to control of mRNA transport and stability.
MATERIALS AND METHODS
Cells and cell lines.
Human small cell carcinoma H1299 cells (ATCC CRL-5803), carrying a homologous deletion of the p53 gene (31), were cultured in alpha minimal essential medium (Gibco) supplemented with 10% fetal calf serum (FCS; Gibco) containing 100-U/ml penicillin and streptomycin, 0.292-mg/ml l-glutamine (Gibco), and 10% FCS (Gibco). H1299-HACul5 stable cell lines were generated by transfection with plasmid DNAs pcDNA3-HACul5 and pcDNA3-puro at a ratio of 10:1, followed by selection in medium containing 2-mg/liter puromycin. Individual colonies were picked and expanded, and the level of expression of HA-Cul5 was verified by Western blotting using antihemagglutinin (anti-HA) antibodies. A high-expression clone was chosen for further experiments.
Adenovirus vectors and plasmids.
All adenovirus vectors used in this study have been described previously, including Adp53 expressing wild-type (wt) human p53 under the cytomegalovirus promoter (37), AdHH55K expressing HMK- and histidine-tagged Ad5 E1B55K (38), and AdE4orf6 expressing Ad5 E4orf6 (38). Plasmids pcDNA3-E1B55K wt and C184T were obtained from Thomas Dobner (unpublished data), and HA-Cul5 was described previously (37). Putative zinc domain mutants of E4orf6 were described previously (5). BC-box E4orf6 mutants were generated by PCR-based mutagenesis using the following oligonucleotide primers: L47G forward, CTGGAGGATCATCCCGGGCTGCCCGAATGTAAC, and reverse GTTACATTCGGGCAGCCCGGGATGATCCTCCAG, with wt pcDNA3-E4orf6 as template; L47G/C51 forward, CGGGCTGCCCGAAGTTAACACTTTGACAATG, and reverse, CATTGTCAAAGTGTTAACTTCGGGCAGCCCG, with L47G as template; L122S forward, CGAGTCCTGGGCTAGCCACTGTCATTGTTCC, and reverse, GGAACAATGACAGTGGCTAGCCCAGGACTCG, with wt pcDNA3-E4orf6 as template; and L122S/C126M forward, GCTAGCCACTGTCATATGTCCAGTCCCGGTTCC, and reverse, GGAACCGGGACTGGACATATGTCAGTGGCTAGC, with L122S as template. L47G/L122S was created by digesting single mutants L47G and L122S with EcoRV and combining the mutants. A similar approach was taken for the quadruple mutant L47G/C51V/L122S/C126M. p53-HA was generated by PCR with the following oligonucleotide primers: forward, CCCGGATCCACCATGGAGGAGCCGCAGTCA, and reverse, CCCGAATTCGTCTGAGTCAGGCCCTTC. The resulting PCR product was then digested with BamHI and EcoRI and cloned in frame into the pcDNA3HAtag vector.
Antisera.
E4orf6-specific rabbit polyclonal antibody 1807 was described in reference 4. Anti-p53 pAb1801 hybridoma supernatants were prepared as described previously (39). E1B55K was detected with the 2A6 monoclonal antibody (44), elongin C with mouse monoclonal antibody SIII p15 (Transduction Laboratories), Mre11 with rabbit polyclonal antiserum NB 100-142D3 (NOVUS Biologicals), and HA epitopes with anti-HA mouse monoclonal HA.11 (BabCO).
DNA transfection.
Cells were transfected in 60- or 100-mm-diameter dishes with the liposome reagent DMRIE-C (Gibco-BRL), as described by the manufacturer. For p53 degradation assays, a plasmid DNA ratio of 2:1 was used with 0.75 μg of pcDNA3-p53, 1.5 μg of pcDNA3-E4orf6 (wt or mutants), and 1.5 μg of pcDNA3-E1B55K in 60-mm-diameter dishes with 11.5 μl of reagent for 24 h. When required, the total amount of DNA was made up to 3.75 μg by addition of pcDNA3 empty vector DNA. For binding assays, 2.5 μg of each of the plasmid DNAs was transfected in 100-mm-diameter dishes using 15 μl of reagent.
Expression of recombinant proteins in Sf21 insect cells.
PCR was used to amplify wt elongin C, and elongin C deletion and point mutants containing an HPC4 tag were amplified from the corresponding M13mpET-elongin C constructs (50). The resulting fragments were subcloned into pBacPAK8. Baculoviruses encoding human elongin B and VHL and adenovirus type 5 E4orf6 were described previously (21, 37). Sf21 cells were cultured at 27°C in Sf-900 II SFM (Gibco) with 5% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). Plates containing 106 Sf21 cells were infected with the recombinant baculoviruses indicated in the figures. Sixty hours after infection, cells were collected and lysed in 1 ml of ice-cold buffer containing 40 mM HEPES-NaOH (pH 7.9), 150 mM NaCl, 1 mM dithiothreitol (DTT), 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 5-μg/ml leupeptin, 5-μg/ml antipain, 5-μg/ml pepstatin A, and 5-μg/ml aprotinin.
Immunoprecipitation and Western blotting.
Cells were lysed for 20 min on ice with lysis buffer (20 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 5% glycerol, 2 mM DTT, 4 mM NaF, 2 mM NaPP, 500 μM Na3VO4, 200-μg/ml PMSF, 2-μg/ml aprotinin, 5-μg/ml leupeptin). Aliquots of cell extract containing 30 μg of protein were loaded on sodium dodecyl sulfate (SDS)-polyacrylamide gels containing 10 or 15% polyacrylamide for Western blotting analysis, or between 800 and 1,300 μg (constant within each experiment), were used for immunoprecipitation using 5 μl of rabbit anti-E4orf6 polyclonal antibody 1807 or 7 μl of anti-E1B55K mouse monoclonal antibody 2A6, followed by incubation with protein G Sepharose (Amersham Bioscience). The beads were extensively washed in lysis buffer and examined by SDS-polyacrylamide gel electrophoresis (PAGE), as described previously (37, 39). For analysis of the interaction of elongin C mutants with elongin B and either VHL or E4orf6, approximately 100 μg of Sf21 cell lysates were incubated for 2 h at 4°C with 10 μl of protein G Sepharose and 2 μg of HPC4 monclonal antibody. Protein G Sepharose was washed three times in buffer containing 40 mM HEPES-NaOH (pH 7.9), 150 mM NaCl, 1 mM DTT, and 0.5% (vol/vol) Triton X-100. Immunoprecipitated proteins were analyzed by SDS-PAGE (13% polyacrylamide) and transferred to Hybond P membranes (Amersham-Pharmacia Biotech). Membranes were incubated with the indicated antibodies in TBS buffer (40 mM Tris-HCl, pH 7.6, 100 mM NaCl) containing 3% nonfat dry milk overnight at 4°C, followed by incubation of appropriate peroxidase-conjugated secondary antibodies (Sigma). Membranes were then visualized by Western blotting with Supersignal West Pico or Dura chemiluminescent reagent (Pierce) or were visualized and quantitated with the Storm Gel and Blot Imaging System following treatment by enhanced chemiluminescence with ECL plus Western blotting reagents (Amersham-Pharmacia Biotech).
Detection of in vivo ubiquitinated forms of p53.
H1299 cells were infected with adenovirus vectors at a multiplicity of infection of 50 PFU per cell for 1 h in 1 ml of serum-free medium (100-mm dishes) with occasional shaking before addition of normal medium. When indicated, proteasome inhibitor MG132 was added at 18 h postinfection (p.i.) at a final concentration of 20 μM. Cells were lysed at 24 h p.i., and extracts were immunoprecipitated with the 1807 (E4orf6) antibody.
RESULTS
Adenovirus E4orf6 and E1B55K proteins contain putative BC-boxes.
We have shown previously that the early human adenovirus protein E4orf6 associates with the cellular proteins Cul5, elongins B and C, and Rbx1 to form a complex similar to the SCF-type ligase in the presence or absence of the viral protein E1B55K (37). The complex has even stronger similarity to the VBC complex in which Cul2 is linked to the substrate specificity protein via interactions with elongins B and C (19, 22, 29). Interaction of elongin C with VHL, elongin A, SOCS1, and other interacting proteins is mediated through a motif termed the BC-box, containing a consensus sequence (A,P,S,T)LxxxCxxx(A,I,L,V) in which the leucine and cysteine residues (boldface) are almost invariable (23). To determine if the E4orf6 complex has a similar architecture to the VBC complex, the presence of a functional BC-box motif in E4orf6 was investigated. The E4orf6 protein sequence contains two near-consensus BC-box motifs, each containing both the highly conserved leucine and cysteine residues (Fig. 1B). A putative BC-box motif was also found in E1B55K (Fig. 1B). To determine if any of these motifs are functional in forming the E3 complex, site-specific mutations were generated in the leucine and cysteine residues of each of these sequences, as shown in Fig. 1C. With E4orf6, mutants were generated either individually in boxes 1 (mutants 1 and 2) and 2 (mutants 3 and 4) or in combination (mutants 5 and 6).
FIG. 1.
BC-box sequences and E4orf6 mutants. (A) Consensus BC-box sequence. (B) Putative BC-box sequences in E4orf6 and E1B55K. The sequences of two putative BC-boxes in E4orf6 and one in E1B55K have been presented with residue numbers within the proteins noted. (C) List of BC-box mutants created in E4orf6.
Both BC-box motifs of E4orf6 are functional and mediate interaction with elongin C.
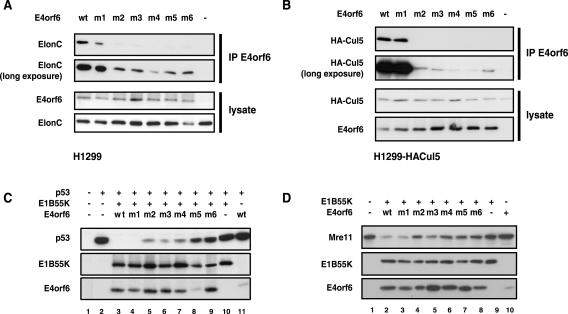
The effect of the E4orf6 BC-box mutations was first examined on stable interactions of E4orf6 with elongin C. p53-null H1299 cells were transfected with plasmid DNAs expressing wt or mutant forms of E4orf6 listed in Fig. 1C, and interactions of E4orf6 species with elongin C were determined by coimmunoprecipitation using anti-E4orf6 antibody followed by Western blotting using anti-elongin C antibodies (Fig. 2A). As seen previously, immunoprecipitation of wt E4orf6 resulted in the coimmunoprecipitation of elongin C. All mutant forms of E4orf6 were expressed at high levels; however, with the exception of mutant 1 (L47G) in E4orf6 BC-box 1, all failed to form stable interactions with elongin C. We also examined the ability of these E4orf6 proteins to associate with another member of the complex, Cul5. Because high-quality antibodies against Cul5 were not available, a stable H1299 cell line expressing HA-tagged Cul5 was generated and such cells were transfected with plasmid DNAs expressing wt or mutant forms of E4orf6. Cell extracts were immunoprecipitated with anti-E4orf6 antibody followed by Western blotting using anti-HA antibodies. Figure 2B shows that wt E4orf6 and E4orf6 mutant 1 (L47G) associated with high levels of HA-Cul5, whereas with the other mutants binding was greatly reduced or eliminated. Binding of E4orf6 to elongin C was also studied in this cell line, and the results were identical to those shown in Fig. 2A (data not shown). Thus, although the invariant Leu47 residue in BC-box 1 was not essential, it appeared that high levels of binding of E4orf6 to elongin C and the formation of the E3 ligase complex containing Cul5 require both E4orf6 BC-boxes 1 and 2.
FIG. 2.
Effects of BC-box mutations on the activity of E4orf6. H1299 cells (A, C, and D) or H1299 cells stably expressing HA-tagged Cul5 (B) were transfected with plasmid DNAs encoding wt or mutant E4orf6. (A and B) Whole-cell lysates were immunoprecipitated (IP) with anti-E4orf6 antibody 1807 and then subjected to Western blotting analysis with anti-elongin C (ElonC) (A) or anti-HA (B) antibodies. The use of wt or mutant E4orf6 (mutants identified in Fig. 1C) or no E4orf6 (−) has been indicated at the top of each lane. The top two panels represent Western blotting analyses for elongin C or Cul5 (A and B, respectively), using shorter and longer exposures of the gel. The bottom two panels represent Western blotting analyses of whole-cell extracts for E4orf6 and elongin C levels, as indicated. (C and D) Whole-cell lysates were analyzed for p53 or Mre11 expression levels by Western blotting using anti-p53 (C) or anti-Mre11 (D) antibodies. The plasmid DNAs used to transfect cells have been indicated at the top of each figure, and Western blotting analyses have been indicated for each panel.
To examine the effect of putative BC-box mutations in E4orf6 and E1B55K on the functionality of the E3 ligase complex in promoting degradation of p53, a degradation assay was used in which H1299 cells were transiently transfected with plasmid DNAs encoding human wt p53 and wt E4orf6 and E1B55K or the mutant E4orf6 forms listed in Fig. 1C. Degradation of p53 was assessed at 24 h posttransfection by Western blotting using anti-p53 antibodies. Complete degradation of p53 with wt viral proteins was observed at a DNA transfection ratio of 2:2:1 (or higher) for E4orf6-E1B55K-p53 (data not shown), and thus this ratio was used for further experiments. A representative result from many p53 degradation assays is shown in Fig. 2C. As with the binding assays, mutant 1, which contains a single L47G point mutation in box 1, was found to be functionally wt in its ability to degrade p53 (lane 4 versus 3); however, all other E4orf6 mutants were to various degrees defective in promoting p53 degradation. Interestingly, mutations in both BC-boxes were required for complete loss of p53 degradation (mutants 5 and 6). (Note that additional studies [not shown] in which higher levels of mutant 5 were expressed yielded a similar failure to support p53 degradation.) Mutation in only one of the boxes resulted in no effect (mutant 1) or a partial effect (mutants 2 to 4).
In addition to p53, two other substrates of the adenoviral E4orf6 protein have recently been identified, Rad50 and Mre11 (6, 48), which are subunits of a double-stranded DNA repair complex. We have been unable to demonstrate the E4orf6-E1B55K-dependent degradation of Rad50 in our system (data not shown); however, Fig. 2D shows results obtained with Mre11. Again, degradation was determined in extracts from cells expressing E1B55K and wt or mutant E4orf6 that were analyzed by Western blotting using anti-Mre11 antibodies. The results showed a pattern similar to that obtained with p53 in that E4orf6-dependent degradation of Mre11 occurred at high levels with wt E4orf6 and mutant 1 (L47G), but was severely reduced in mutants 5 and 6 in which the conserved leucines and cysteines in both BC-boxes 1 and 2 were altered. Taken together, these results confirmed the functional importance of both E4orf6 BC-boxes.
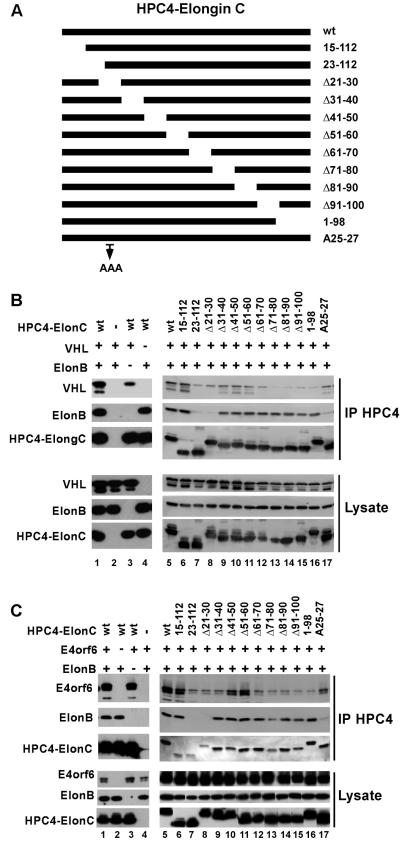
To study the nature of the E4orf6-elongin C interaction, the ability of a series of elongin C deletion mutants (see the list in Fig. 3A) to form complexes with BC-box proteins, including E4orf6, was tested. Sf21 insect cells were coinfected with baculoviruses expressing wt or mutant forms of HPC4-tagged elongin C, elongin B, and with BC-box protein VHL or E4orf6. Elongin C was immunoprecipitated from whole-cell extracts with anti-HPC4 antibodies, and the coprecipitation of the other proteins was analyzed by Western blotting using appropriate antibodies. Figure 3 shows the results of this study. As shown previously (49), a region of elongin C near the amino terminus, between residues 15 and 30, appears to be required for binding elongin B, as two deletion mutants, 23-112 and Δ21-30 both failed to interact significantly with elongin B, whereas the other amino-terminal mutants, including 15-112 and Δ31-40, retained the interaction. (Fig. 3B and C). In addition, elongin C mutants 23-112 and Δ21-30 interact with both VHL and E4orf6 less efficiently than the wt, consistent with previous observations that stable binding of elongin C to VHL, elongin A, and other BC-box proteins depends on elongin B (1, 13, 23, 25). The region of elongin C deleted from these mutants forms strands 1 and 2, which have been shown to participate in interactions with elongin B (46). While there are no direct contacts between strands 1 and 2 and VHL, loss of interaction with elongin B could destabilize the structure of the region that interacts with VHL. The results obtained with VHL (Fig. 3B) and E4orf6 (Fig. 3C) all indicated that the carboxy-terminal region of elongin C appears to be required for interaction with BC-box motifs, as deletion mutants starting at residues 71 to 80 resulted in the loss of stable interactions with both VHL and E4orf6. These results are consistent with published data for VHL (50) and with the VHL-elongin BC crystal structure (46), which showed that the carboxy-terminal region of elongin C forms helices H3 and H4 and loop L5, which are critical for the interaction with VHL. Taken together, our comparison of the abilities of the elongin C mutants to assemble into complexes with VHL and E4orf6 suggests that the interaction between E4orf6 and elongin C resembles that of VHL and that E4orf6 forms an E3 ligase complex in a manner similar to VHL in the VBC complex.
FIG. 3.
Analysis of complex formation using elongin C mutants. (A) Representation of elongin C deletion mutants. (B and C) Sf21 insect cells were coinfected with baculovirus vectors encoding HPC4-tagged elongin C or elongin C mutants, elongin B (ElonB), and either VHL (B) or E4orf6 (C). Sf21 cell lysates were immunoprecipitated (IP) with HPC4 monoclonal antibody, which recognizes the epitope tag on elongin C, and analyzed by Western blotting against the indicated proteins.
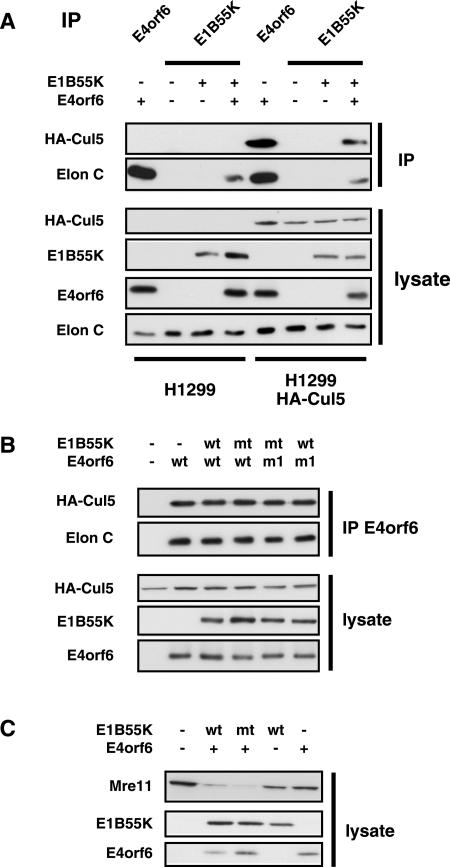
E1B55K does not interact with elongin C in the absence of E4orf6.
Because a putative BC-box motif was noted in the E1B55K, the ability of E1B55K to bind elongin C and Cul5 in the absence of E4orf6 was investigated in coimmunoprecipitation and Western blotting studies similar to those in Fig. 2 and 3. Figure 4A shows that, as shown before, E4orf6 interacts strongly with elongin C (lanes 1 and 5) and Cul5 (lane 5), whereas these species were only associated with E1B55K when E4orf6 was coexpressed (lane 3 versus 4 for elongin C and lane 7 versus 8 for Cul5). As in Fig. 2B, the Cul5 interaction was determined by using H1299 cells that express HA-Cul5 (lanes 5 to 9). Figure 4B shows that, as expected from the lack of interaction of wt E1B55K with elongin C, the presence of a mutation in the conserved Cys residue within the putative BC-box motif (C184T) of E1B55K had no effect on the interaction of E4orf6 or E4orf6 mutant L47G (m1) with either Cul5 or elongin C. In studies similar to those in Fig. 2D, E1B55K mutant C184T was also found to be fully functional in the induction of Mre11 degradation (Fig. 4C) and p53 (data not shown). These results suggested that only E4orf6 contributes to E3 ligase complex formation and that E1B55K is present in the complex through its interaction with E4orf6.
FIG. 4.
Requirement of E1B55K BC-box. (A) H1299 or H1299-HACul5 cells (lanes noted at bottom of the figure) were transfected with plasmid DNAs expressing E1B55K and/or E4orf6, as indicated at the top of the figure. Whole-cell lysates were either first immunoprecipitated (IP) (top two panels) using either 1807 anti-E4orf6 or anti-E1B55K antibodies as indicated on top of the figure or examined directly (bottom four panels) by Western blotting with antibodies against HA (for HA-Cul5), E1B55K, E4orf6, or elongin C (Elon C), as indicated in the figure. (B) H1299-HACul5 cells were transfected with combination of plasmid DNAs expressing wt or BC-box mutants of E1B55K (C184T) and E4orf6 (functional mutant 1, L47G), as indicated at the top of the figure. As in panel A, immunoprecipitates prepared using 1807 anti-E4orf6 antibodies or whole-cell extracts were examined by Western blotting using antibodies against HA (HA-Cul5), elongin C, E1B55K, or E4orf6, as indicated in the figure. (C) H1299 cells were transfected with the combination of plasmid DNAs expressing E4orf6 and the wt or BC-box mutant of E1B55K (C184T), as indicated at the top of the figure. Whole-cell lysates were analyzed for Mre11 expression levels by using anti-Mre11 antibodies.
Function of E1B55K in the ubiquitination of p53 and Mre11.
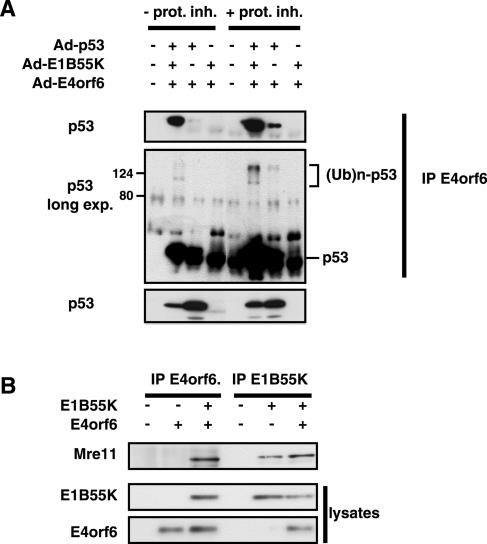
To confirm that the degradation of p53 induced by E4orf6 and E1B55K was due to ubiquitin-dependent proteasomal degradation, the existence of ubiquitin chains on p53 was examined in the presence of proteasome inhibitors. To detect only ubiquitin chains of p53 molecules produced due to the action of the E4orf6-E1B55K complex, extracts from cells that had been coinfected with adenoviral vectors expressing p53 and E4orf6 and/or E1B55K were immunoprecipitated with anti-E4orf6 antibodies and the precipitates were immunoblotted for p53 with anti-p53 antibodies. Figure 5A shows clearly that under conditions that result in significant degradation of p53 (bottom panel without proteasome inhibitor), more p53 was present in association with E4orf6 when E1B55K was also coexpressed (top panel). As the ratio of adenovirus vectors used for this experiment was a 1:1:1 ratio (compared to 1:2:2 in transfection experiments), complete degradation of p53 is not observed at this 24-h time point. When this immunoblot was exposed for a longer period, the presence of ubiquitin chains on p53 became evident through the appearance of heterogeneous material migrating more slowly than p53. Although this ubiquitinated material was more evident when E1B55K was also coexpressed, it was nevertheless also evident in extracts from cells expressing E4orf6 in the absence of E1B55K. Moreover, the ratio of slower-migrating ubiquitinated material between samples with and without E1B55K was approximately the same as the ratio of p53 bound to E4orf6 in the presence or absence of E1B55K. This observation may also provide an explanation to the question of what the role of E1B55K is in the degradation of p53, as E4orf6 can both bind to p53 and form the E3 ligase complex. Thus the function of E1B55K would be to enhance the introduction of the p53 substrate into the E4orf6 complex for ubiquitination and degradation. In the absence of E1B55K, p53 was less efficiently recruited to the E3 ligase, but nevertheless, approximately the same fraction of those p53 molecules that did bind E4orf6 could be ubiquitinated.
FIG. 5.
Role of E1B55K in the binding and ubiquitination of p53 and the binding of Mre11. (A) p53-H1299 cells were infected with combinations of adenoviral vectors expressing p53, E1B55K, or E4orf6, as indicated at the top of the figure. Half of the cultures were treated with proteasome inhibitor (prot. inh) MG132 at 50 nM from 18 to 24 h p.i. At 24 h p.i. whole-cell extracts were immunoprecipitated (IP) using 1807 anti-E4orf6 antibodies and the precipitates were analyzed by Western blotting using anti-p53 antibodies. The presence of slower-migrating ubiquitinated forms of p53 [(Ub)n-p53] has been noted. Shorter and longer exposures (exp.) of the gel have been included, as indicated. (B) H1299 cells were transfected with plasmid DNAs expressing E4orf6 and/or E1B55K, as indicated. Whole-cell extracts were immunoprecipitated with either anti-E4orf6 or anti-E1B55K antibodies followed by Western blotting analysis using anti-Mre11 antibodies.
This model for the function of E1B55K was then tested with the other known substrate of the E4orf6-E1B55K complex, Mre11. Figure 5B shows that immunoprecipitation of E1B55K resulted in the coimmunoprecipitation of Mre11. Figure 5B also shows that in the case of E4orf6, Mre11 was coimmunoprecipitated in the presence of anti-E4orf6 antibodies only when E1B55K was also expressed. Detection of Mre11 coprecipitating with E1B55K in the presence of both viral proteins is possible due to the incomplete degradation of endogenous Mre11 at this time point of 24 h. Near-complete (since not all cells are transfected) degradation of Mre11 (as in Fig. 2D) is only seen at the 48-h time point. Thus with Mre11, which does not interact directly with E4orf6, E1B55K functions to introduce the Mre11 substrate into the E4orf6 complex for ubiquitination and degradation.
Stable binding of E1B55K to E4orf6 requires intact BC-box motifs in E4orf6.
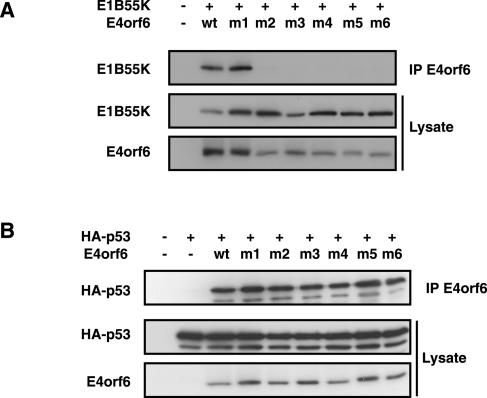
The BC-box mutants were also tested for their ability to interact stably with E1B55K and p53. H1299 cells were cotransfected with plasmid DNAs expressing mutant or wt E4orf6 and either E1B55K or human wt p53. Following immunoprecipitation with anti-E4orf6 antibodies, the presence of E1B55K and p53 was assessed by Western blotting using the appropriate antibodies. Figure 6A shows that, somewhat surprisingly, all BC-box mutants except BC-box 1 mutant 1 (L47G), which was shown to be wt for p53 degradation, lost the ability to interact with E1B55K. This loss of interaction could not simply be due to misfolding of these E4orf6 mutant proteins as Fig. 6B shows that all E4orf6 mutant proteins were still able to bind to p53.
FIG. 6.
E4orf6-E1B55K interactions correlate with elongin C-Cul5 complex formation. (A and B) Binding of E1B55K and p53 by E4orf6 mutants. H1299 cells were transfected with plasmid DNAs expressing wt or mutant (as identified in Fig. 1C) E4orf6 and E1B55K (A) or p53 (B). Whole-cell extracts were immunoprecipitated (IP) with 1807 anti-E4orf6 antibodies, and interactions with E1B55K (A) or p53 (B) were verified by Western blotting using appropriate antibodies.
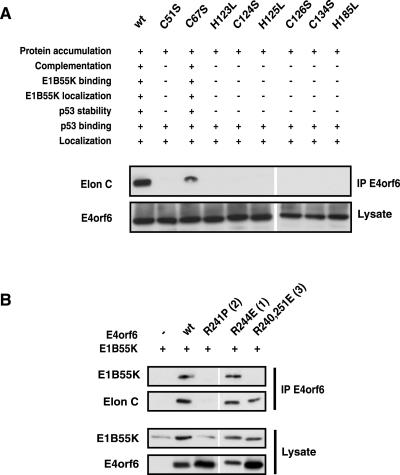
The characteristics demonstrated for the E4orf6 BC-box mutants (specifically the loss of 55K binding, loss of the ability to degrade p53, and retention of p53 binding) were highly reminiscent of a set of E4orf6 mutants generated to disrupt a putative zinc-binding domain of E4orf6 (5), as summarized in Fig. 7A. Thus it was possible that these mutants also had lost the ability to form stable complexes with elongin C and Cul5. This possibility was tested in coimmunoprecipitation experiments similar to those in Fig. 2. Figure 7A shows that all such mutants that had lost the ability to bind E1B55K and to degrade p53 had also lost the ability to form stable interactions with elongin C. These results further confirmed the importance of formation of the E3 ligase complex for the interaction of E4orf6 with E1B55K. As noted at the top of Fig. 7A, results from previous studies (5) indicated a correlation between the ability of E4orf6 to form the complex and to complement growth of a mutant adenovirus that fails to express all E4 products. Complex formation was also examined with an extensive series of E4orf6 point mutants in the amphipathic α-helical region, for which similar viral growth complementation data had been obtained (36). The results of this experiment and the complementation data are summarized in Table 1, with binding results presented with representative mutants shown in Fig. 7B. Table 1 shows that these mutants can be separated into three groups. Group 1 consists of mutants that complement an E4 deletion for normal virus production and have wt or nearly wt binding activities with both elongin C and E1B55K. Group 2 mutants do not complement nor bind either E1B55K or elongin C. Group 3 mutants both complement and bind to elongin C; however, interaction with E1B55K was undetectable. These results demonstrate that the formation of E3 ligase complex for functional activity of E4orf6 in the growth of the virus is paramount and perhaps even more important than its interaction with E1B55K.
FIG. 7.
Formation of the cullin complex correlates with E4orf6 functional activity in adenovirus-infected cells. (A) Binding to elongin C (Elon C) of E4orf6 mutants in the putative zinc-binding domain. The top panel shows a summary of results obtained by Boyer and Ketner (5) with putative zinc-binding domain E4orf6 mutants. The bottom two panels show results obtained with these mutants following Western blotting analysis using anti-elongin C antibodies of immunoprecipitates (IP) prepared using 1807 anti-E4orf6 antibodies (middle panel) or 1807 anti-E4orf6 antibodies using whole-cell extracts. (B) Binding to elongin C and E1B55K of representative members (with mutant group indicated in parentheses) of E4orf6 mutants in the amphipathic α-helical region. H1299 cells were transfected with plasmid DNAs expressing wt or the indicated E4orf6 mutants and E1B55K. Whole-cell extracts were immunoprecipitated with 1807 antibodies, and interactions with E1B55K and elongin C were verified by Western blotting with appropriate antibodies.
TABLE 1.
Analysis of the interaction of a series of E4orf6 mutants in the amphipathic α-helix domaina
| Mutation(s) | Complemen- tation | Binding
|
Group | |
|---|---|---|---|---|
| Elongin C | E1B55K | |||
| wt | Yes | +++ | +++ | 1 |
| L245P | No | − | − | 2 |
| R241P | No | − | − | 2 |
| AE255APE | Yes | +++ | +++ | 1 |
| R241A | Yes | ++ | + | 1 |
| R241E | Yes | ++ | − | 3 |
| R243A | No | − | − | 2 |
| R243E | Yes | + | − | 3 |
| R244E | Yes | ++ | ++ | 1 |
| R248E | Yes | ++ | ++ | 1 |
| R251A | Yes | +++ | +++ | 1 |
| R240, 251E | Yes | ++ | − | 3 |
| R248, 251A | Yes | +++ | +++ | 1 |
| R240, 244, 251A | Yes | ++ | + | 1 |
| R240, 244, 248, 251A | No | − | − | 2 |
| R268, 269, 270A | ? | ++ | ++ | 1 (complementation?) |
The data summarized in the table are from Orlando and Ornelles (36) and the present report.
DISCUSSION
The present results indicate that the adenovirus E4orf6 complex represents, both structurally and functionally, an E3 ubiquitin ligase complex that greatly resembles the VBC complex. As with the VHL tumor suppressor protein, the interaction of E4orf6 with elongin C also appears to be mediated through a BC-box motif; however, E4orf6 is unusual in that it appears to possess two functional BC-box motifs. While it remains formally possible that only one of these BC-box motifs is functional, mutation of each box individually was found to be sufficient to greatly reduce interactions with elongin C; some transient in vivo interactions must still exist with these single-motif mutants as full loss of function required mutation of both BC-boxes. Although data from the binding and activity assays of the different E4orf6 mutants (Fig. 2) were not fully quantitative, it was clearly apparent in these semiquantitative assays (Fig. 2C) that degradation activity is significantly reduced when both BC-box motifs are mutated (mutants 5 and 6) as compared to single-motif mutants (mutants 1 to 4). The reason for the requirement for two BC-box motifs in E4orf6 is not clear. According to the crystal structure from Stebbins et al. (46), elongin C can interact with only one BC-box motif, leaving a distinct possibility that only one of the E4orf6 motifs indeed interacts with elongin C, as in the case of VHL, while mutation in the other presumptive BC-box may disrupt other aspects of E4orf6 structure that are important for its interaction with elongin C. It is also possible that both sequences are bona fide BC-box motifs. While only one may be interacting with elongin C at any time, the presence of two motifs in adenovirus E4orf6 may have evolved to better compete with cellular BC-box proteins for binding to elongin C. In which case, when dissociation of the interaction occurs, elongin C would be more likely to reassociate with E4orf6, through use of either of the motifs. The same carboxy-terminal region of elongin C was shown to be involved in interactions with VHL and E4orf6, thus further demonstrating the similarity of these E3 ligase complexes.
It has been recently shown that adenovirus infection induces the degradation of Mre11 and Rad50, two components of the double-strand break repair complex Mre11-Rad50-NBS1. Studies with virus mutants implicated both E1B55K and E4orf6 in this degradation process and furthermore suggested a role for the viral E4orf3 protein in their relocalization from the viral inclusion bodies to PML-containing nuclear tracks (48). Using our degradation system involving transient coexpression of E4orf6 and E1B55K in the absence of other adenovirus proteins, we were unable to confirm the degradation of Rad50 by E4orf6 and E1B55K; however, degradation of Mre11 induced by these proteins was quite marked. It is possible that one or more additional viral proteins are required for efficient degradation of Rad50; however, it seems more likely that degradation of Rad50 is not a direct effect of the E4orf6/E1B55K E3 ligase complex, but rather results from a decrease in Rad50 due to the disruption of the Mre11-Rad50-NBS1 complex when Mre11 is degraded.
Unlike the BC-box motifs present in E4orf6, that present in E1B55K does not appear to be necessary for the function of the E3 ligase. Furthermore, it did not appear to be capable of interacting with elongin C or recruiting Cul5 (Fig. 4A) (18). It is possible that this BC-box motif may not be exposed and thus is not accessible to elongin C following formation of the final tertiary structure of E1B55K. In this regard, it is of interest to note that E1B55K proteins containing two other mutations in this motif (L180A/C184S and L180P/C184F) are highly unstable (data not shown).
In addition to forming the Cul5 complex, E4orf6 has been known for some time to bind to p53 in the absence of E1B55K. Thus what is the role of E1B55K in the degradation of p53 by the E4orf6 complex? Clearly E1B55K is required, as E4orf6 degrades p53 very poorly in its absence. Two explanations appear plausible. First, although p53 may bind to E4orf6 in the E3 ligase complex, E1B55K may be required to achieve an optimal orientation for the ubiquitination reaction. And second, although E4orf6 binds p53, much higher levels of p53 substrate may be introduced into the E4orf6-E3 ligase complex when E1B55K is present. The results presented in Fig. 5A suggest that the second explanation is the most likely. In the absence of E1B55K, only a small amount of p53 is associated with the E4orf6 complex. Thus the presence of E1B55K greatly increases the levels of p53 available for degradation; however, a similar fraction of the p53 associated with the E4orf6 complex was found to be ubiquitinated in the presence and absence of E1B55K. Thus, E1B55K probably functions to introduce high levels of p53 into the complex. Our favored model is that E4orf6 forms the E3 ligase complex involved in ubiquitination followed by proteasomal degradation and that E1B55K introduces substrates for degradation, including p53, Mre11, and most likely additional as yet unidentified cellular or viral proteins. As shown in coimmunoprecipitation experiments, only E1B55K (and not E4orf6) binds Mre11. The ability of E4orf6 to interact directly with p53 may be unique among targets of the E4orf6 degradation complex. Thus the ability of E4orf6 to block the transactivational activity of p53 may serve some function to the virus in addition to E3 ligase complex formation and ubiquitination.
An unexpected result was the failure of E1B55K to interact with E4orf6 BC-box mutants that were defective in E3 ligase complex formation. We believed it likely that the E4orf6 BC-box mutations would abolish complex formation with the cellular proteins elongin C and Cul5; however, the interaction between E4orf6 and E1B55K has been known for some time and believed to be direct. This lack of binding cannot simply be attributed to general misfolding of the E4orf6 BC-box mutants, as all formed stable interactions with p53. In explaining this effect, we propose that E1B55K does not interact directly with E4orf6, but rather interacts with the E3 ligase complex that E4orf6 has assembled. In other words, E1B55K only associates with E4orf6 once it has assembled the cellular E3 ligase complex, as shown in Fig. 8. Complex formation may alter the conformation of E4orf6 to expose the E1B55K binding site or stabilize the E4orf6-E1B55 interaction. It is also possible that E1B55K makes contact with both E4orf6 and one or more members of the cellular Cul5-containing complex. Whatever the case, these results may explain some previous observations about E4orf6. It had been observed that newly synthesized E4orf6 only associates with E1B55K 1 to 2 h following its synthesis (10). This effect might be explained by the time required for the newly synthesized E4orf6 to be assembled into the E3 ligase complex. In addition, other groups have reported difficulty in detecting interactions between E1B55K and E4orf6 in vitro (24, 42). Further, E4orf6 and E1B55K were not found to interact in the yeast two-hybrid system (42). The last two observations would make sense if cellular proteins were required for the interaction. In addition, little interaction between E4orf6 and E1B55K is evident in many rodent cell lines, suggesting that a primate-specific factor may be required (16). This component is probably not Cul5 or elongin B or C as (i) these proteins are nearly identical in rodents and humans and (ii) the genes encoding these proteins are not localized on human chromosome 21, to which the major primate-specific activity was mapped (8). These possibilities may explain some controversy about the location of the E1B55K binding site on E4orf6. Some evidence has suggested that the E1B55K binding site resides in the first 55 amino-terminal residues of E4orf6 (39, 42), whereas others have suggested that the amphipathic α-helical region towards the carboxy terminus of E4orf6 plays a role (7, 35, 52). Future studies will be required to define such interactions further.
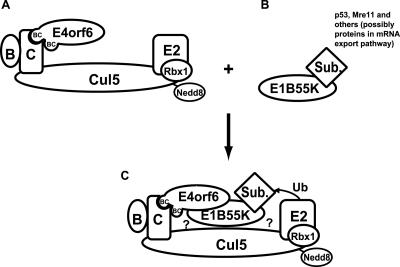
FIG. 8.
Schematic representation of E4orf6-E1B55K-elongin-Cul5 complex formation. (A) Interaction of E4orf6 with elongin C (C) is predicted to occur via either one of the E4orf6 BC-boxes to form a complex also containing elongin B (B), Cul5 modified by addition of Nedd8, Rbx1, and an E2 ligase. (B) E1B55K interacts with substrates to be ubiquitinated and degraded, including p53, Mre11, and others, possibly those involved in mRNA export. (C) Formation of the entire complex and the positioning of the substrate for ubiquitination occur primarily through the interaction between E4orf6 and E1B55K. E4orf6 may not interact directly with Mre11 and other substrates, apart from p53. E1B55K may interact with one or more members of the elongin-Cul5 complex (denoted by ?).
Other published observations also support the present model for E4orf6-E1B55K interaction. Harada et al. (18) showed using gel filtration of nuclear extracts that E1B55K and E4orf6 are part of a very large complex of 800 to 900 kDa that includes Cul5, elongin B, elongin C, Rbx1, and other proteins and that there was no E1B55K complex of the size expected for an E4orf6-E1B55K dimer or tetramer. Thus these data support the contention that E1B55K interacts with E4orf6 only after the formation of complexes with cellular proteins, including Cul5 and elongins B and C.
Studies on the three sets of E4orf6 point mutants presented herein have also led to additional insights about E4orf6-E1B55K function and complex structure. From the combined data with the three sets of E4orf6 mutants, it appears that in addition to the elongin C-BC-box interaction, other regions of E4orf6 may form interactions with other members of the complex. Most or all of these interactions would be required for a strong stable interaction detectable by coimmunoprecipitation, since mutations in any of these domains result in a loss of detectable binding. One of these additional interactions may be with Cul5 as E4orf6 forms a complex only with Cul5 and not with Cul2 (37). As Cul2 is also known to interact with VHL through elongins B and C, our results suggest that the specificity of E4orf6 for Cul5 may be regulated through direct interactions with Cul5. Further studies will be required to confirm this possibility. A series of point mutations in a putative zinc finger domain yielded E4orf6 mutant proteins with properties similar to those of the BC-box mutants. These mutants exhibited loss of E1B55K binding and relocalization to the nucleus, and they were defective for p53 degradation. Like the E4orf6 BC-box mutants, they were still able to bind p53. These similarities suggested that like the BC-box mutants, the putative zinc-binding domain mutants may be unable to form the E3 ligase complex, a prediction that was largely confirmed in the present studies. These mutants had also been shown previously to fail to complement an adenovirus deletion mutant lacking the complete E4 region for expression of high levels of late viral proteins. Also, a series of point mutations in the amphipathic α-helical region of E4orf6 yielded three mutant phenotypes (Table 1). Again a good correlation was seen between complex formation and binding to E1B55K as group 2 mutants that failed to bind elongin C also failed to bind E1B55K. Thus E3 ligase complex formation may be required for all functions requiring both E4orf6 and E1B55K, including export of late viral mRNA and inhibition of export of cellular mRNAs. This conclusion agrees well with the observations of Corbin-Lickfett and Bridge (9), who conclude that active proteasomes are required for E4orf6 to promote late gene expression. This possibility suggests that degradation of as yet unidentified cellular proteins by the E3 ligase complex may function in the control of mRNA metabolism. Lastly, it is worth noting that group 3 mutants in the amphipathic α-helical region (Table 1) complement the E4 adenovirus deletion mutant even though they do not appear to interact with E1B55K. While it remains possible that E1B55K interacts transiently or at low undetectable levels with these mutant species, it is also possible that the E4orf6 E3 ligase complex in the absence of E1B55K can provide some of the functions required to support adenovirus replication. In fact, with all the mutants tested in this study, a perfect correlation was evident between E3 ligase complex formation and complementation of the E4 defect, whereas such was not with the case with binding to E1B55K. Further studies are under way to examine these possibilities using appropriate E4orf6 and E1B55K mutant viruses.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (P.E.B.), the National Cancer Institute of Canada (P.E.B.), and the U.S. National Institute of General Medicine (R.C.C.). Paola Blanchette had a Canderel Fellowship from the McGill Cancer Centre.
REFERENCES
- 1.Aso, T., W. S. Lane, J. W. Conaway, and R. C. Conaway. 1995. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science 269:1439-1443. [DOI] [PubMed] [Google Scholar]
- 2.Bargonetti, J., I. Reynisdottir, P. N. Friedman, and C. Prives. 1992. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 6:1886-1898. [DOI] [PubMed] [Google Scholar]
- 3.Beer-Romero, P., S. Glass, and M. Rolfe. 1997. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene 14:595-602. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, D., M. R. Morison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, J. L., and G. Ketner. 2000. Genetic analysis of a potential zinc-binding domain of the adenovirus E4 34k protein. J. Biol. Chem. 275:14969-14978. [DOI] [PubMed] [Google Scholar]
- 6.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chastain-Moore, A. M., T. Roberts, D. A. Trott, R. F. Newbold, and D. A. Ornelles. 2003. An activity associated with human chromosome 21 permits nuclear colocalization of the adenovirus E1B-55K and E4orf6 proteins and promotes viral late gene expression. J. Virol. 77:8087-8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315:234-244. [DOI] [PubMed] [Google Scholar]
- 10.Cutt, J. R., T. Shenk, and P. Hearing. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61:543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 12.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25-54. [DOI] [PubMed] [Google Scholar]
- 13.Duan, D. R., A. Pause, W. H. Burgess, T. Aso, D. Y. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, and R. D. Klausner. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 14.Flint, S. J., and R. A. Gonzalez. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287-330. [DOI] [PubMed] [Google Scholar]
- 15.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 16.Goodrum, F. D., T. Shenk, and D. A. Ornelles. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 70:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross-Mesilaty, S., E. Reinstein, B. Bercovich, K. E. Tobias, A. L. Schwartz, C. Kahana, and A. Ciechanover. 1998. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA 95:8058-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 21.Kamura, T., D. Burian, Q. Yan, S. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276:29748-29753. [DOI] [PubMed] [Google Scholar]
- 22.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 23.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao, C. C., P. R. Yew, and A. J. Berk. 1990. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology 179:806-814. [DOI] [PubMed] [Google Scholar]
- 25.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269:1444-1446. [DOI] [PubMed] [Google Scholar]
- 26.Kishino, T., M. Lalande, and J. Wagstaff. 1997. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 15:70-73. [DOI] [PubMed] [Google Scholar]
- 27.Kuhne, C., and L. Banks. 1998. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem. 273:34302-34309. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., A. L. Talis, and P. M. Howley. 1999. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 274:18785-18792. [DOI] [PubMed] [Google Scholar]
- 29.Lisztwan, J., G. Imbert, C. Wirbelauer, M. Gstaiger, and W. Krek. 1999. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13:1822-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, M. E. D., and A. J. Berk. 1998. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 72:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7:171-180. [PubMed] [Google Scholar]
- 32.Nakagawa, S., and J. M. Huibregtse. 2000. Human Scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 1997. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc. Natl. Acad. Sci. USA 94:1206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oda, H., S. Kumar, and P. M. Howley. 1999. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 96:9557-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlando, J. S., and D. A. Ornelles. 1999. An arginine-faced amphipathic alpha helix is required for adenovirus type 5 E4orf6 protein function. J. Virol. 73:4600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando, J. S., and D. A. Ornelles. 2002. E4orf6 variants with separate abilities to augment adenovirus replication and direct nuclear localization of the E1B 55-kilodalton protein. J. Virol. 76:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Querido, E., M. R. Morison, H. Chu-Pham-Dang, S. W.-L. Thirlwell, D. Boivin, and P. E. Branton. 2001. Identification of three functions of the adenovirus E4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rolfe, M., P. Beer-Romero, S. Glass, J. Eckstein, I. Berdo, A. Theodoras, M. Pagano, and G. Draetta. 1995. Reconstitution of p53-ubiquitinylation reactions from purified components: the role of human ubiquitin-conjugating enzyme UBC4 and E6-associated protein (E6AP). Proc. Natl. Acad. Sci. USA 92:3264-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth, J., C. König, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 72:8510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoplosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 42.Rubenwolf, S., H. Schutt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruppert, J. M., and B. Stillman. 1993. Analysis of a protein-binding domain of p53. Mol. Cell. Biol. 13:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510-517. [DOI] [PubMed] [Google Scholar]
- 45.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 46.Stebbins, C. E., W. G. Kaelin, Jr., and N. P. Pavletich. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 47.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 48.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 49.Takagi, Y., R. C. Conaway, and J. W. Conaway. 1996. Characterization of elongin C functional domains required for interaction with elongin B and activation of elongin A. J. Biol. Chem. 271:25562-25568. [DOI] [PubMed] [Google Scholar]
- 50.Takagi, Y., A. Pause, R. C. Conaway, and J. W. Conaway. 1997. Identification of elongin C sequences required for interaction with the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 272:27444-27449. [DOI] [PubMed] [Google Scholar]
- 51.Thomas, M., and L. Banks. 1998. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene 17:2943-2954. [DOI] [PubMed] [Google Scholar]
- 52.Weigel, S., and M. Dobbelstein. 2000. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J. Virol. 74:764-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woods, D. B., and K. H. Vousden. 2001. Regulation of p53 function. Exp. Cell Res. 264:56-66. [DOI] [PubMed] [Google Scholar]
- 54.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]