Abstract
Transcriptional mechanisms remain poorly understood in trypanosomatid protozoa. In particular, there is no knowledge about the function of basal transcription factors, and there is an apparent rarity of promoters for protein-coding genes transcribed by RNA polymerase (Pol) II. Here we describe a Trypanosoma brucei factor related to the TATA-binding protein (TBP). Although this TBP-related factor (TBP-related factor 4 [TRF4]) has about 31% identity to the TBP core domain, several key residues involved in TATA box binding are not conserved. Depletion of the T. brucei TRF4 (TbTRF4) by RNA interference revealed an essential role in RNA Pol I, II, and III transcription. Using chromatin immunoprecipitation, we further showed that TRF4 is recruited to the Pol I-transcribed procyclic acidic repetitive genes, Pol II-transcribed spliced leader RNA genes, and Pol III-transcribed U-snRNA and 7SL RNA genes, thus supporting a role for TbTRF4 in transcription performed by all three nuclear RNA polymerases. Finally, a search for TRF4 binding sites in the T. brucei genome led to the identification of such sites in the 3′ portion of certain protein-coding genes, indicating a unique aspect of Pol II transcription in these organisms.
One of the most intriguing aspects in protozoa of the family Trypanosomatidae, which include African and South American trypanosomes and Leishmania, remains the regulation of gene expression (5). It appears that these organisms do not use transcription initiation as a major regulatory step to control the output of mRNA on a per gene basis; the majority of protein-coding genes are organized as polycistronic rather than monocistronic transcription units. Indeed, the 5′ ends of all mature mRNAs are formed by trans-splicing, an RNA processing reaction, rather than by transcription initiation as in most eukaryotic organisms.
Another curiosity in trypanosomatids is the apparent rarity of promoters for protein-coding genes transcribed by RNA polymerase (Pol) II. Although transcription is performed by three RNA polymerases with α-amanitin sensitivities similar to those of higher eukaryotic Pol I, II, and III, thus far it has been challenging to map Pol II transcription initiation sites for protein-coding genes (5). To date, there is a single report where transcriptional analysis of Leishmania major chromosome 1 suggests the presence of a bidirectional Pol II promoter (26). The only other characterized Pol II-dependent promoter drives expression of the spliced leader (SL) RNA (11). Genetic and biochemical studies delineated various promoter elements of the SL RNA gene and led to the isolation of the first transcription factor (promoter-binding protein 1 [PBP-1]) in trypanosomatids (8). PBP-1 interacts with the promoter element located between 60 and 80 bp upstream of the transcription start site. Interestingly, the 57-kDa subunit of PBP-1 is orthologous to the 50-kDa component of the small nuclear RNA-activating protein complex (SNAPc), involved in transcription of human small nuclear RNA (snRNA) genes (8). Studies on Pol I and Pol III transcription units in trypanosomatids have so far been limited to the identification and fine mapping of promoter elements (22, 24, 35), and no transcription factors have been identified either biochemically or by database mining.
The brief summary above underscores our rather limited knowledge of transcriptional mechanisms in trypanosomatids. In particular, nothing is known about the presence and/or function of basal transcription factors. TATA-binding protein (TBP) is the only known basal factor that is involved in transcription by all three eukaryotic nuclear RNA polymerases and functions on promoters with or without a TATA box (15, 31). Consistent with this general role, TBPs from a wide variety of organisms share both a high degree of amino acid conservation and very similar crystal structures. TBP has a variable N-terminal domain and a highly conserved C-terminal core domain (3). The latter domain has an approximate twofold intramolecular symmetry that through crystallographic studies revealed a saddle-shaped structure (21). In addition to TBP, all multicellular animals also express a TBP-like protein (TLP), also called TLF, TRF2, TRP, or TRF (2, 7). It was shown that TLP has a role in embryonic development and differentiation in Caenorhabditis elegans, Xenopus, and mice, but its function in transcriptional regulation remains to be determined (6, 18, 25, 37, 41). More recent data revealed that TLP regulates cell cycle progression and stress response (33). In Drosophila melanogaster, TRF1 functions in both Pol II (17) and Pol III (34) transcription, whereas TRF2, a third member of the TBP family, coordinates transcription of a subset of genes, including genes involved in DNA replication and cell proliferation (16).
Here, we describe the functional characterization of a Trypanosoma brucei factor related to TBP using RNA interference (RNAi) and chromatin immunoprecipitation (ChIP). We show that the T. brucei TBP-related factor is recruited to the Pol II-transcribed SL RNA gene, as well as to Pol I and Pol III transcription units, providing evidence that this TBP-related factor has a universal role in transcription in trypanosomes.
MATERIALS AND METHODS
Trypanosome cell lines.
Procyclic T. brucei cells were transfected as previously described (30). To generate the T. brucei TBP-related factor 4 (TbTRF4) RNAi cell line, a 597-bp fragment (nucleotides [nt] 29 to 626) of the T. brucei TRF4 translated region was assembled as two inverted repeats separated by a stuffer fragment and inserted downstream of a tetracycline (TET)-inducible promoter from the procyclic acidic repetitive protein (PARP) gene. The construct was linearized with NotI for integration at the rRNA gene nontranscribed spacer region of strain 29.13.6, expressing the TET repressor and T7 RNA polymerase (39). Transformed cells were selected in the presence of 2.5 μg of phleomycin per ml and cloned by limiting dilutions.
A PCR-based method (32) was used to establish a cell line in which one allele of TbTRF4 was replaced with the blasticidin (BSR) drug resistance gene. The second allele was tagged with an epitope at the N terminus; the epitope was BB2, corresponding to 10 amino acids from the immunologically well-characterized major structural protein of the Saccharomyces cerevisiae Ty1 virus-like particle. Mouse monoclonal antibodies against this epitope were generated in Keith Gull's laboratory (1). Similarly, one allele of the largest subunit of Pol II was tagged with the BB2 epitope in a background in which the other three alleles had been replaced by drug resistance genes, namely, for BSR, phleomycin, and hygromycin.
ChIP procedure.
The ChIP methodology was adopted from protocols provided by the laboratories of S. Buratowski (20) and P. J. Farnham (38). A procyclic T. brucei cell culture of 600 ml at a density of 6 × 106 to 8 × 106 cells/ml was mixed with 60 ml of 11% formaldehyde (final concentration, 1%) and incubated at room temperature between 15 and 40 min. Glycine was then added to a final concentration of 250 mM, and the culture was incubated for 5 min. The cells were washed twice with cold phosphate-buffered saline (8.1 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.5], 2.5 mM KCl, 140 mM NaCl) and then washed once with cold precell lysis wash buffer [1 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 7.4), 1 mM CaCl2, 5 mM MgCl2, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml]. The cells were resuspended in 15 ml of cold cell lysis buffer (1 mM PIPES [pH 7.4], 1 mM CaCl2, 5 mM MgCl2, 0.5 M hexylene glycol, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml), incubated on ice for 10 min, and broken twice using a French press set at 2,000 lb/in2. The lysate was centrifuged at 400 × g for 5 min at room temperature, the supernatant was removed, and the lysate was centrifuged again at 2,400 × g for 30 min at room temperature. The pellet was washed with 10 ml of nucleus wash buffer (0.25 M sucrose, 50 mM Tris-HCl, 5 mM MgCl2, 1% Triton X-100, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml) and centrifuged at 2,400 × g for 20 min at room temperature. The pellet was resuspended with 8 ml of cold formaldehyde (FA) lysis buffer (50 mM HEPES-KOH [pH 8], 150 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 1% Triton X-100, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml) plus 0.3% sodium dodecyl sulfate (SDS) and 0.5% Sarkosyl, Dounce homogenized about 30 times using a B-type pestle in a 7-ml glass Dounce homogenizer tube, and centrifuged at 100,000 rpm for 20 min at 4°C. The pellet was resuspended in 6 ml of cold FA lysis buffer containing 0.3% SDS. The sample was sonicated to an average size of about 400 bp and centrifuged at 100 × g for 25 min at 4°C, and the soluble sheared chromatin was adjusted to final concentrations of 275 mM NaCl and 0.1% SDS.
Before immunoprecipitation, the sheared chromatin was precleared with protein G beads in FA buffer containing 0.1% SDS. Immunoprecipitation was performed in FA buffer containing 0.1% SDS and 275 mM NaCl, and the precipitates were washed once with FA buffer containing 0.1% SDS, twice with FA buffer containing 0.1% SDS and 500 mM NaCl, once with a solution consisting of 10 mM Tris-HCl (pH 8.0), 0.25 M LiCl, 1 mM EDTA, 0.5% Nonidet P-40, and 0.5% sodium deoxycholate, and twice with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The beads were then incubated in a solution consisting of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 300 mM NaCl, and 1 μl of DNase-free RNase (10 μg/ml) at 65°C for 30 min. An equal volume of a solution consisting of 100 mM Tris-HCl (pH 8.0), 20 mM EDTA, and 2% SDS was then added to the solution, and the solution was incubated for 10 min. Proteinase K was added to a final concentration of 500 μg/ml, and the solution was incubated for 1 h at 42°C. The beads were then removed, and the supernatant was incubated at 52°C overnight. The DNA samples were subjected to agarose gel electrophoresis, and fragments in the size range of 100 to 500 bp were isolated using the Qiagen gel purification kit and analyzed by PCR.
PCR mixtures contained 3 μl of a 1:20 dilution of the immunoprecipitate or 3 μl of a 1:3,000 to 1:10,000 dilution of the input sample and 25 ng of each primer in a total volume of 10 μl. The PCR cycles were empirically adjusted for each primer pair to be in the linear range of the amplification. Further details about the ChIP methodology, PCR conditions, and oligonucleotide primers used are available upon request.
Identification of TRF4 targets.
Chromatin immunoprecipitates were labeled by random priming (Amersham Biosciences) and used to screen a T. brucei genomic library constructed in the Lambda ZAP II vector (Stratagene). Briefly, genomic DNA was partially digested with a combination of HaeIII, AluI, and RsaI, and DNA fragments of 4 to 5 kb were gel purified. EcoRI adapters were attached, and the resulting fragments were cloned into Lambda ZAP II (E. Ullu, unpublished data).
Analysis of newly synthesized RNA.
The TbTRF4 RNAi cell line was induced with tetracycline (10 μg/ml) for 1 or 2 days, and then the induced and uninduced cells were permeabilized with lysolecithin as described previously (36). After incubation at 28°C for 15 min, total RNA was extracted with TRIZOL reagent (Gibco-BRL) and processed for dot blot hybridization as described previously (10). The following DNA fragments of TRF4 and Pol II chromatin immunoprecipitate (TPChIP) clones 5 and 8 were subcloned into pBluescript and used for dot blotting: 1, nt 55 to 377 of the pteridine reductase 1 (Ptr1) translated region; 2, nt 789 to 1108 of the Ptr1 translated region; 3, nt 16 to 328 of the Ptr1 3′ untranslated region (3′ UTR); 4, nt 329 to 702 of the Ptr1 3′ UTR; 5, nt 703 of the Ptr1 3′ UTR to the ATG initiation codon of ORF2; 6, nt 17 to 890 of ORF2; a, nt −173 to 152 with respect to the tryparedoxin (TR) ATG initiation codon; b, nt 153 of the TR open reading frame (ORF) to 136 of the TR 3′ UTR; c, nt 137 to 468 of the TR 3′ UTR; d, nt 469 of the TR 3′ UTR to 156 nt upstream of the DNA helicase (DH) ATG initiation codon; e, nt 581 to 1021 of the DH translated region.
RESULTS
Sequence analysis of a TBP-related factor in trypanosomatids.
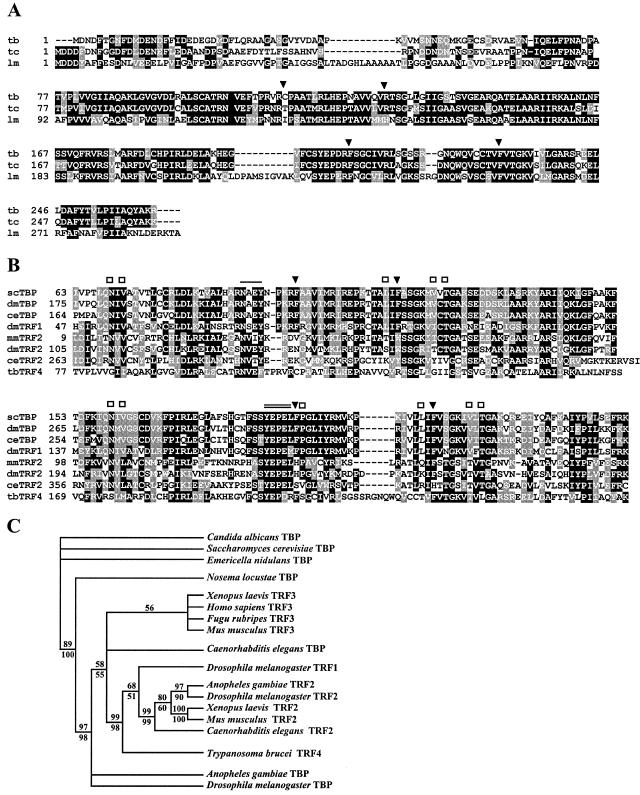
The current databases of the T. brucei, Trypanosoma cruzi, and L. major genome projects contain a single protein related to the TBP family (Fig. 1A). We will use the term TRF4 (TBP-related factor 4) to avoid confusion with the trypanosome lytic factor or TLF and to distinguish this protein from the existing family of TBP-type factors (Fig. 1C). As in all other members of this family, the trypanosomatid TRF4s have the typical TBP signature domain at the C terminus with two imperfect direct repeats, each encompassing about 85 amino acids. Overall, the three polypeptides are approximately 60% identical, with most of the conserved amino acids residing in the TBP signature domain; fewer identical amino acids are seen in the N-terminal domain of about 80 amino acids.
FIG. 1.
(A) ClustalW alignment of T. brucei TRF4 (tb; GeneDB accession no. Tb10.61.0330), T. cruzi TRF4 (tc; GeneDB accession no. Tc00.1047053503809.150) (note that the TRF4 ORF is encoded in the −1 reading frame), and L. major TRF4 (lm; GeneDB accession no. LmjF19.1390). Conserved amino acids (white letters on black background) and similar amino acids (white letters on shaded background) are shown. Gaps introduced to maximize alignment are indicated by dashes. The locations of the two phenylalanine pairs conserved in TBPs (solid arrowheads) are indicated. (B) Alignment of the C-terminal core domain of a selected number of TBPs with the corresponding domains of TBP-related factors. scTBP, S. cerevisiae TBP (accession no. P13393); dmTBP, D. melanogaster TBP (accession no. AAA68629); ceTBP, C. elegansTBP (accession no. AAA03582); dmTRF1, D. melanogaster TRF1 (accession no. Q27896); mmTRF2, Mus musculus TRF2 (accession no. AB017697); dmTRF2, D. melanogaster TRF2 (accession no. AF136569); ceTRF2, C. elegans TRF2 (accession no. AJ233443); tbTRF4, T. brucei TRF4 (GeneDB accession no. Tb10.61.0330). The locations of the two phenylalanine pairs conserved in TBPs (solid arrowheads), amino acids involved in DNA recognition (open squares), and TBP residues interacting with TFIIA (single line) and TFIIB (double line) are indicated. (C) Strict consensus of 278 most parsimonious trees from the analysis of a partial alignment of 127 parsimony informative characters. The number above the line represents the bootstrap resampling results using a heuristic parsimony analysis (percentage of 1,000 replicates). The number below the line is based on quartet puzzling steps (percentage of 10,000 replicates). Only values above 50% are shown. The analysis was done using PAUP 4.0b10.
To determine the relationship between the trypanosomatid proteins and other TBP family members, the T. brucei protein was aligned with a selected number of TBPs and TBP-like proteins using the ClustalW multiple-sequence alignment algorithm (Fig. 1B), and a phylogenetic tree was constructed (Fig. 1C). As expected, the N-terminal domain of the T. brucei protein did not reveal significant similarities with other members of this family. The C-terminal core domain, on the other hand, was 31% identical (53% similar) to D. melanogaster TBP and 30% identical (54% similarity) to D. melanogaster TRF1. This contrasts with 21% identity (41% similarity) of the T. brucei core domain to the D. melanogaster TRF2. Thus, it appears that the T. brucei polypeptide is somewhat more closely related to TBPs and TRF1 than to TRF2s.
However, the trypanosomatid proteins have characteristics that clearly distinguish them from TBPs and TRF1. TBPs and TRF1, but not TRF2s, have two pairs of highly conserved phenylalanine residues that play a key role in the interaction with the TATA box. Intriguingly, in T. brucei, T. cruzi, and L. major, the phenylalanine pair in the first repeat is replaced with a cysteine-arginine, threonine-arginine, and isoleucine-histidine pair, respectively (Fig. 1A). Moreover, whereas several other amino acids involved in DNA recognition are conserved in TBPs and TRFs, very few of these residues are present in the trypanosomatid proteins (Fig. 1B). Taken together, these findings suggest that the trypanosomatid TBP-related factors will recognize different DNA sequences than the TBPs or TRF1. However, what is conserved in the trypanosomatid proteins are the motifs in TBP known to interact with TFIIA and TFIIB (Fig. 1B).
T. brucei TRF4 is encoded by an essential gene.
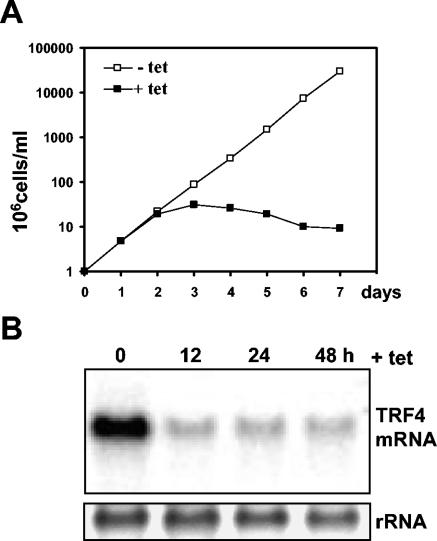
To begin to address the roles of the trypanosomatid TRF4s in transcription, T. brucei mRNA was downregulated by RNAi. We generated a construct containing the TET-inducible promoter from the PARP genes driving expression of hairpin double-stranded RNA (dsRNA). After integration at the rRNA gene nontranscribed spacer region, stable clonal cell lines were established. The production of dsRNA was monitored 12 h after TET was added, and the cell line generating the largest amount of dsRNA was chosen for further analysis (data not shown). Next, TET was maintained in the culture medium for up to 7 days, and the fate of TRF4 mRNA was monitored by Northern blotting (Fig. 2B). This showed a drastic decrease of TbTRF4 mRNA within 12 h of dsRNA induction with very little change thereafter. During the first 48 h of TRF4 silencing, there was no noticeable effect on cell growth (Fig. 2A). However, after 72 h, the cells gradually stopped growing and eventually died, indicating that T. brucei TRF4 is essential for viability.
FIG. 2.
RNAi of the T. brucei TRF4. (A) Growth curve of the TRF4 RNAi cell line in the absence (−) or presence (+) of tetracycline (tet). (B) The TRF4 RNAi cell line was induced with tetracycline for the indicated number of hours, and poly (A+) RNA was probed for TRF4 mRNA. Ethidium bromide staining of rRNA served as a loading control.
Silencing of TRF4 affects transcription of most genes.
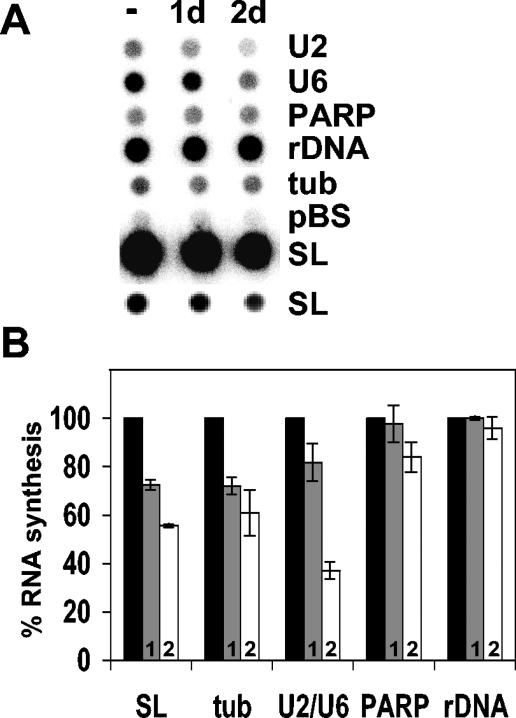
The requirement for cell viability and the conservation of both motifs and the primary amino acid sequence suggest an essential, housekeeping role for TRF4 in the cell. To assay what effect TRF4 depletion had on transcription, lysolecithin-permeabilized cells were prepared after TET induction, and newly synthesized transcripts were labeled with [α-32P]UTP. Since cell death became apparent after 72 h of dsRNA induction, we restricted our analysis to 1 and 2 days after induction. At these time points, we were able to reproducibly measure the transcriptional activity of a variety of genes by dot blot hybridization. Newly synthesized [32P]RNA was hybridized to gene probes immobilized on nitrocellulose filters representing genes transcribed by Pol I (rRNA genes and PARP), Pol II (SL RNA and tubulin), and Pol III (U6 and U2 snRNA). Quantitation of the dot blot results shown in Fig. 3A revealed that under these conditions, TRF4 depletion did not significantly change Pol I-mediated transcription of the rRNA genes but had a slight and reproducible effect on PARP synthesis (Fig. 3B). On the other hand, Pol II transcription of the SL RNA and tubulin genes and Pol III transcription of the U6 and U2 snRNA genes was affected, with SL and U2 or U6 RNA synthesis reduced by 45 and 65%, respectively, after 2 days of induction. These depletion experiments using RNAi supported the conclusion that TbTRF4 participates in transcription by Pol I, Pol II, and Pol III but that the synthesis of the large rRNAs did not appear to require TRF4.
FIG. 3.
RNA synthesis in the TbTRF4 RNAi cell line. (A) Dot blot hybridization with RNA synthesized in uninduced cells (−) or after 1 or 2 days (1d and 2d) of TET induction. Two different exposure times are shown for the SL RNA hybridization. rDNA, rRNA genes; tub, tubulin; pBS, plasmid vector control. (B) Quantitation of three independent experiments as shown in panel A. Percent RNA synthesis compared to no induction (100%) (black bars) is shown after 1 day (bars 1, or grey bars) or 2 days of induction (bars 2, or white bars). Since the values for the U2 and U6 snRNA genes were almost identical, they are shown together. tub, tubulin gene probe (see reference 10 for a description of the plasmids); rDNA, rRNA genes.
Recruitment of TRF4 to the Pol II-transcribed SL RNA genes.
Although the results of the RNAi experiments back the notion that TbTRF4 plays an essential role in transcription in vivo, they fail to provide a direct mechanistic link between TRF4 and cellular promoters. Therefore, we monitored the association of the T. brucei TRF4 polypeptide with transcribed genes in living cells using ChIP. To this end, we first generated a procyclic T. brucei cell line expressing solely an epitope-tagged version of TRF4. We deleted one allele by homologous recombination with a PCR-generated cassette encoding the blastocydin (BSR) resistance gene and then introduced an N-terminal BB2 epitope in the second allele at its original chromosomal locus. This approach ensured that the epitope-tagged version of the protein was functional and expressed at a level comparable to that of the endogenous protein. In addition, a second cell line in which the largest subunit of RNA Pol II was tagged with an epitope at the N terminus was generated. Since the gene coding for this subunit is present in two copies per haploid genome, three of the alleles were replaced with drug resistance markers, and a BB2 epitope tag was placed at the N terminus of the fourth allele, thus generating a cell line exclusively expressing a BB2-tagged Pol II. Next, living cells were exposed to 1% formaldehyde, and fixed chromatin served as a substrate for immunoprecipitation with anti-BB2 antibodies. Precipitated DNA fragments with an average length of 100 to 500 bp were analyzed by PCR using gene-specific oligonucleotide primers (see Materials and Methods).
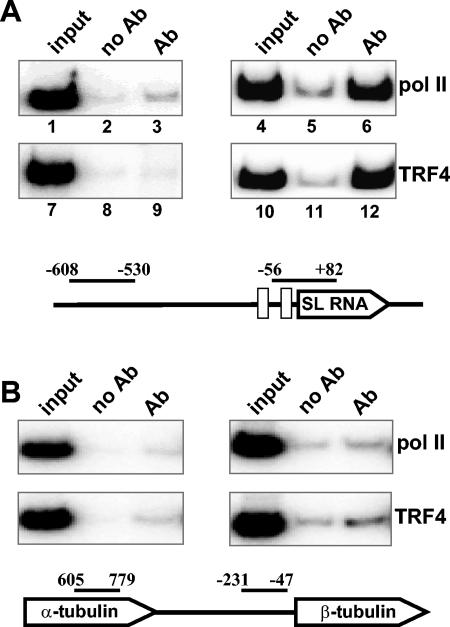
To validate the ChIP methodology, we first examined the Pol II-transcribed SL RNA genes (11), which are tandemly repeated about 200 times in T. brucei. Each gene has its own Pol II promoter, and the transcription unit encompasses 140 nt of coding region and 100 nt of upstream sequences containing promoter elements. After immunoprecipitation and reversal of the cross-links, samples were analyzed by PCR using primers amplifying the region from nt −56 to +82 of the SL RNA transcription unit, as well as a region upstream of the gene (nt −530 to −608), which was not expected to recruit factors required for transcription (Fig. 4A). For a negative control, we included a reaction mixture lacking the primary antibody. For each primer pair, the PCR conditions were defined experimentally in such a way that the final products were in the linear amplification range (data not shown).
FIG. 4.
(A) Characterization of proteins bound in vivo to the SL RNA gene. ChIP was performed with antibodies (Ab) directed against the BB2 epitope tags present on the largest subunit of Pol II (lanes 3 and 6) or on TRF4 (lanes 9 and 12) or with no antibody (no Ab) as a control (lanes 2, 5, 8, and 11). PCR amplification of the immunoprecipitates was done with primers upstream of the promoter (nt −608 to −530) or within the promoter (nt −56 to +82). Nucleotides are numbered with respect to the SL RNA transcription start site. An aliquot of the total input is also shown (lanes 1, 4, 7, and 10). (B) ChIP assay of the tubulin gene cluster. Immunoprecipitates for Pol II or TRF4 were PCR amplified with tubulin-specific primers as indicated. Nucleotides are numbered with respect to the translation start site. The drawings are not to scale.
As predicted, Pol II was found associated with the SL RNA transcription unit, but not with the upstream region (Fig. 4A). Immunoprecipitates of the epitope-tagged largest subunit of Pol II were enriched 10-fold for the fragment from nt −56 to +82 (Fig. 4A, lane 6), compared to the no-antibody control (lane 5). In contrast, no enrichment was seen for the upstream negative-control fragment in the immunoprecipitates (compare lanes 2 and 3). Similarly, PCR analysis of TbTRF4 immunoprecipitates resulted in the amplification of the SL RNA gene-specific fragment (lane 12), whereas the amplification of the upstream region was similar to the no-antibody control (lanes 8 and 9). These ChIP results strongly supported our permeable cell experiments and established a specific role for TbTRF4 in the transcription of the SL RNA genes. Although downregulation of TRF4 by RNAi appeared to indicate a role for this polypeptide in tubulin transcription (Fig. 3), so far we have not been able to specifically detect TRF4 at these genes by ChIP (Fig. 4B and data not shown). However, it should be pointed out that we have not yet done an extensive search for TRF4 binding sites in the tubulin gene locus, which consists of about 15 repeated gene copies.
Pol I- and Pol III-transcribed genes.
We next extended the ChIP experiments to Pol I- and Pol III-transcribed genes. For Pol I transcription, we surveyed the promoters of the large rRNA genes and the developmentally regulated procyclin or PARP genes. In agreement with the results of RNAi experiments, TRF4 was not recruited to the promoter regions of the rRNA genes (Fig. 5). However, using PARP promoter-specific primers, a positive PCR signal was detected in the immunoprecipitated sample, whereas only a background signal was seen for the upstream negative-control fragment (Fig. 5).
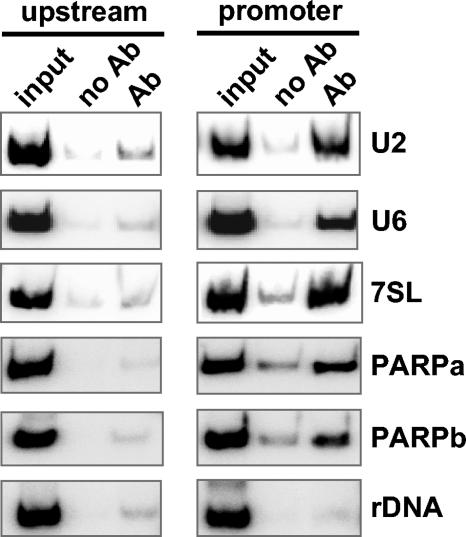
FIG. 5.
TRF4 binding in vivo. ChIP was performed with antibodies (Ab) directed against the BB2 epitope tag present on TRF4, and precipitates were analyzed by PCR with primers specific for the promoter or an upstream negative-control region. A control with no antibody (no Ab) and an aliquot of the total input (input) is also shown. rDNA, rRNA genes.
In contrast to most eukaryotes, where Pol II is responsible for the synthesis of the U2 snRNA, in T. brucei the single-copy U2 snRNA gene is transcribed by Pol III (35). As shown in Fig. 5 and in support of the results of RNAi experiments, TRF4 was found associated with the U2 transcription unit, but not with an upstream control region. The involvement of TRF4 in Pol III transcription was also observed at the U6 snRNA and 7SL RNA gene locus (Fig. 5).
Identification of genomic sequences interacting with TRF4 and Pol II.
The above experiments established a role for TbTRF4 in transcription by all three RNA polymerases. However, they did not provide a link between TRF4 and Pol II-mediated transcription of protein-coding genes (Fig. 4B), i.e., addressing the question of whether conventional Pol II promoters exist in trypanosomes.
To begin to tackle this issue, we combined the ChIP assay with a hybridization analysis to search for genomic regions occupied by both TRF4 and Pol II. After reversal of the cross-links, DNA purified independently from TRF4 and Pol II immunoprecipitates was labeled with 32P and hybridized to duplicate filters of a phage library of T. brucei genomic sequences with an average insert size of 4 kb. In a pilot screen of approximately 2,000 clones (representing about 8 Mb or 30% of the trypanosome genome), we identified 27 clones that hybridized to both TRF4 and Pol II chromatin immunoprecipitates.
To validate our approach, we hybridized a third filter to a SL RNA gene probe, since according to our ChIP experiments (Fig. 4A), these genes should be represented among the positive signals. Indeed, all the 17 phages identified as SL positive were among the 27 clones that hybridized to both the TRF4 and Pol II chromatin immunoprecipitates. We felt it critical to confirm that the remaining 10 clones were indeed bound by TRF4 and Pol II in vivo and did not represent DNAs that gave false- positive results due to repeated elements and/or nonspecific precipitation. Therefore, we randomly chose five clones for a more detailed analysis. The identities of these clones were determined by end sequencing followed by BLAST analysis of the T. brucei GeneDB database at the Sanger Institute (http://www.genedb.org/).
Subsequently, the sequences hybridizing to both the TRF4 and Pol II chromatin were narrowed down to about 1,000 bp using a combination of restriction enzyme digests and Southern blots (see Fig. 6). Next, the recruitment of Pol II and TRF4 to the genomic regions identified by hybridization was confirmed with standard ChIP experiments for all five isolates (see Fig. 7B; also data not shown), and the mapped regions were shown to be present in single copies in the genome by BLAST searches and Southern blot hybridizations.
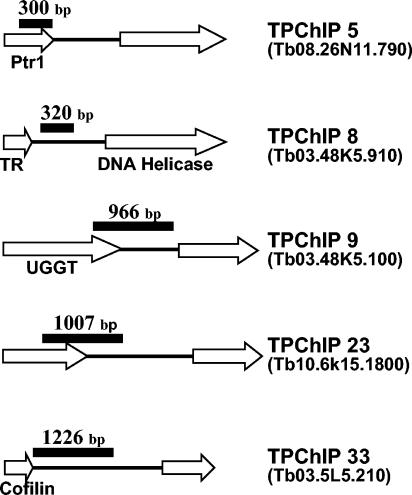
FIG. 6.
Genomic organization of TPChIP clones. Genomic clones hybridizing to both TRF4 and Pol II chromatin immunoprecipitates (TPChIP) were end sequenced and analyzed by GeneDB searches (www.genedb.org/) (accession numbers are shown in parentheses to the right of the clones). ORFs are shown as open arrows with the known genes indicated: Ptr1, pteridine reductase 1; TR, tryparedoxin; UGGT, UDP-glucose glycoprotein:glucosyltransferase. The region hybridizing to both TRF4 and PolII chromatin immunoprecipitates is shown as a black bar. Drawings are not to scale.
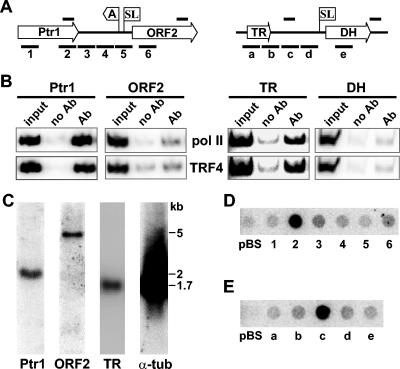
FIG. 7.
Characterization of two TPChIP clones. (A) Graphic representations of TPChIP clones 5 (left) and 8 (right). ORFs are shown as open arrows (Ptr1, pteridine reductase 1; TR, tryparedoxin; and DH, DNA helicase) and the poly(A) and SL addition sites are indicated. For Ptr1, the poly(A) and SL addition sites were determined by sequencing reverse transcription-PCR products, whereas the SL addition site for DH was predicted from the sequence. The solid bars above the drawing indicate the regions amplified in the ChIP assay shown in panel B, and the solid bars below the drawing indicate the DNA fragments used in the dot blot hybridization shown in panels D and E. Drawings are not to scale. (B) ChIP assay for Pol II and TRF4. Ab, antibody. (C) Northern blot analysis with probes specific for the Ptr1 gene, ORF2, TR, and α-tubulin (α-tub). The positions of molecular size standards (in kilobases) are indicated to the right of the blots. (D) Dot blot hybridization of newly synthesized RNA to DNA fragments of about 300 bp, cloned into pBluescript, and spotted onto a nitrocellulose membrane. The numbers refer to the regions indicated below the left drawing in panel A. pBS, plasmid control. (E) Dot blot hybridizations to DNA fragments covering the regions as depicted below the right drawing in panel A. pBS, plasmid control.
Inspection of the five regions binding Pol II and TRF4 revealed that they were all within transcription units of protein-coding genes. The surprising result was that they were either located at the end of a translated region or within 3′ UTRs (Fig. 6). This somewhat unorthodox result prompted us to have a closer look at two such regions, namely, Ptr1 and TR. In the case of Ptr1, the Pol II/TRF4 region was mapped to the last 300 bp of the translated region. Ptr1 is part of large directional cluster of protein-coding genes on chromosome 8 and is followed downstream by a hypothetical 4.5-kb ORF (ORF2 [Fig. 7A]). Northern blot analysis of steady-state RNA revealed that Ptr1 and ORF2 mRNAs of the predicted size accumulated to approximately similar levels, and there was no evidence for additional RNAs from this region (Fig. 7C).
As an alternative experimental approach to assay Pol II density, we examined newly synthesized RNA originating from this genomic region. As shown in Fig. 7D, DNA fragments of equal size (300 bp) covering the beginning and end of the Ptr1 translated region, the 3′ UTR of Ptr1, the intergenic region, the 5′ UTR of ORF2, and the translated region of ORF2 hybridized weakly and with approximately similar intensities to 32P-labeled RNA made in permeable cells. In contrast, the 300-bp region identified as a binding site for Pol II and TRF4 (fragment 2) had a 10-fold increase in the hybridization signal. As predicted, this hybridization signal was decreased in TRF4-depleted cells (data not shown). It is important to note that fragment 2 is present as a single copy in the genome, as judged by BLAST searches and Southern blot hybridization; therefore, the signal was not due to a repetitive sequence.
Using strand-specific probes, we further determined that transcription occurred in the same direction as that of the Ptr1 gene (data not shown). Attempts to resolve the size of the transcripts, by either hybrid selection, Northern blotting, or using DNA oligonucleotides for dot blots, were not successful. Furthermore, we found no evidence for the presence of RNAs smaller than the Ptr1 mRNA in steady-state RNA by Northern blot or RNase protection experiments (Fig. 7C and data not shown). Nevertheless, a similar analysis of the Pol II/TRF4 binding site in the TR gene locus (Fig. 7E) underscored the emerging picture that the isolated regions have striking local increases in transcriptional activity.
DISCUSSION
The experiments described in this report highlight a fundamental role for a T. brucei TBP-related factor (TRF4) in transcription mediated by Pol I, Pol II, and Pol III. Although we cannot discount the possibility that another TBP-related gene exists in the trypanosome genome, we think this is highly unlikely for the following reasons. First, the coverage of the trypanosomatid genome is quite extensive and includes not only three finished trypanosomatid protozoan genomes, namely, T. brucei, L. major, and T. cruzi, but also considerable coverage of the genomes of two other African trypanosomes, which are closely related to T. brucei, namely, Trypanosoma congolense and Trypanosoma vivax. Thus, we think it is significant that we could identify only TRF4 in the combined genomic information of the family Trypanosomatidae. Second, by performing low-stringency Southern hybridization and PCR amplification with degenerate oligonucleotides, we did not obtain evidence for the presence of TBP or an additional TBP-related factor (unpublished data).
For Pol I, the ChIP experiments placed TRF4 at the promoter region of the developmentally regulated PARP genes, but interestingly, we could not detect TRF4 at the promoters of the rRNA genes. The latter result was already apparent in RNAi experiments, where downregulation of TRF4 did not have a noticeable effect on rRNA gene transcription (Fig. 3). One concept emerging from these observations is that the composition of the transcriptional machineries assembling on these two promoters is distinct. Surveying the current T. brucei database did not produce candidate factors that could be tested by ChIP for their involvement in Pol I transcription (C. Tschudi, unpublished data). Thus, resolution of this intriguing possibility will have to come from a biochemical analysis of the available in vitro transcription system (23).
To assess whether TRF4 is involved in Pol II-dependent transcription in T. brucei, we chose the SL RNA gene, which remains the only Pol II unit in trypanosomatids characterized in some detail (8, 11). Initial experiments in a variety of systems identified two regulatory elements by mutational analysis: one close to the transcription start site and the other located between 60 and 80 bp upstream (35). Elegant biochemical experiments in V. Bellofatto's laboratory have extended these studies to isolate a transcription factor, referred to as PBP-1, which specifically binds to the upstream element (8). This factor is composed of three polypeptides, and one of these proteins, with a molecular mass of 57 kDa, is an orthologue of the human 50-kDa subunit of SNAPc, also known as PBP or PTF. SNAPc binds to the proximal sequence element of Pol II- and Pol III-transcribed snRNA genes. Thus, the transcriptional machinery assembling on the SL RNA promoter appears to resemble snRNA transcription in higher eukaryotes. It is well established that TBP is required for both Pol II- and Pol III-transcribed snRNA genes (14). The results of our experiments presented here showed that TRF4 is recruited to the SL RNA genes and thus is involved in the transcription of these genes. This result raises the intriguing possibility that TRF4 has taken the place of TBP in the assembly of a transcription initiation complex on the SL RNA genes. At this time, we do not know whether TRF4 recognizes SL RNA promoter elements directly or whether it is recruited by interaction with other component(s) of the transcriptional machinery, such as PBP-1. Nevertheless, since several key residues, including two of the four highly conserved phenylalanines, known to interact with the TATA box are replaced in the T. brucei TRF4 (Fig. 1), it is likely that TRF4 does not bind to a canonical TATA box. Indeed, studies of a dinoflagellate TBP-like protein, in which all four phenylalanines have been replaced, revealed that a TTTT box was a better binding substrate than a TATA box (12). So far, we have not succeeded in producing T. brucei TRF4 in a soluble recombinant form to test its binding to DNA.
We have also shown that TRF4 is recruited to the Pol III-transcribed U-snRNA and 7SL RNA genes. The regulatory elements of these genes are unusual in that a gene internal element acts in concert with upstream A and B boxes located in a divergently oriented tRNA gene (10, 29). At this time, TRF4 is the only known transcription factor involved in Pol III-mediated transcription in trypanosomatids. Similar to the SL RNA genes, we do not know whether TRF4 binds directly to DNA in the U-snRNA or 7SL RNA promoters or whether the observed recruitment is mediated by auxiliary factors.
Having established that T. brucei TRF4 binds to promoters mediating transcription by all three nuclear RNA polymerases, we went on to ask whether TRF4 is also involved in Pol II transcription of protein-coding genes. Genome sequencing projects have underscored that protein-coding genes in trypanosomatids are organized in large directional clusters that most likely give rise to polycistronic pre-mRNAs (9, 13, 28, 40). Although the genome data also exposed putative Pol II promoters at points of diverging clusters, experimental evidence for the existence of promoters in such regions is restricted to a single report (26). In addition, at this time Pol II is the only known component of the machinery transcribing protein-coding genes in trypanosomes.
Due to the apparent absence of a bona fide TBP in the kinetoplastid databases, we hypothesized that TRF4 might lead us to putative transcription initiation sites by Pol II. Thus, we took a limited global approach and searched for regions in the genome that recruit both Pol II and TRF4. Using ChIP DNA to screen a genomic phage library, we indeed uncovered 27 regions in the T. brucei genome that bind Pol II and TRF4. As predicted, a subset of the positive results corresponded to the SL RNA gene. The regions in the remaining 10 isolates were all mapped to clusters of protein-coding regions.
The unexpected result came when we realized that Pol II and TRF4 were binding to sequences near the 3′ ends of genes. Although perplexing at first, very recent studies in S. cerevisiae and humans have uncovered scenarios, where transcription can initiate within coding regions. In one study, a mutation in Spt6, believed to be involved in transcription elongation, mRNA processing, and interaction with nucleosomes resulted in aberrant transcription initiation within coding regions (19). In a second report, conditional inactivation of the Spt16 subunit of the FACT complex (named FACT for facilitates chromatin transcription) increased Pol II density, transcription, and TBP recruitment in the 3′ portion of certain genes (27). Taken together, these observations were interpreted to mean that these factors contribute to the fidelity of Pol II transcription by repressing initiation at cryptic promoters. In both studies, transcription initiation at these cryptic sites generated a stable poly(A)-containing transcript.
In humans, the mapping of the binding sites for transcription factors Sp1, c-Myc, and p53 on chromosomes 21 and 22 uncovered a surprising number of sites (36%) lying within or near the 3′ ends of genes (4). Furthermore, these regions produce stable noncoding RNAs, some of them overlapping with protein-coding transcripts. This is quite different from our results, since we have no evidence for the production of stable RNAs initiating at the 3′ ends of the Ptr1 and TR genes. One explanation for this difference could be that Pol II elongation is somehow arrested at these two loci, as evidenced by the synthesis of short nascent transcripts. It is worth pointing out that the Pol II, as well as TRF4, density at the 3′ ends of the Ptr1 and TR genes is extraordinarily high, compared to the surrounding coding regions, which remained at background levels and thus could not be measured (Fig. 7B and data not shown). The significance of this pileup of Pol II and TRF4 is not clear at this time, but it would be interesting to test whether the recruitment of Pol II and TRF4 to these 3′-end regions can be manipulated, for instance by downregulating subunits of the T. brucei FACT complex. Nevertheless, our results highlight once more that Pol II transcription of protein-coding genes in these organisms is highly unusual and far from understood.
Acknowledgments
This work was supported in part by Public Health Service grants AI43594 to C.T. and AI28798 to E.U. from NIAID. G.K.A. was supported through a training grant from NIH (T32 AIO7404).
We are grateful to Torsten Ochsenreiter for help with the phylogenetic analysis and to Sarah Renzi for excellent technical assistance.
REFERENCES
- 1.Bastin, P., Z. Bagherzadeh, K. R. Matthews, and K. Gull. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77:235-239. [DOI] [PubMed] [Google Scholar]
- 2.Berk, A. J. 2000. TBP-like factors come into focus. Cell 103:5-8. [DOI] [PubMed] [Google Scholar]
- 3.Burley, S. K., and R. G. Roeder. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65:769-799. [DOI] [PubMed] [Google Scholar]
- 4.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499-509. [DOI] [PubMed] [Google Scholar]
- 5.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dantonel, J. C., S. Quintin, L. Lakatos, M. Labouesse, and L. Tora. 2000. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol. Cell 6:715-722. [DOI] [PubMed] [Google Scholar]
- 7.Dantonel, J. C., J. M. Wurtz, O. Poch, D. Moras, and L. Tora. 1999. The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem. Sci. 24:335-339. [DOI] [PubMed] [Google Scholar]
- 8.Das, A., and V. Bellofatto. 2003. RNA polymerase II-dependent transcription in trypanosomes is associated with a SNAP complex-like transcription factor. Proc. Natl. Acad. Sci. USA 100:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sayed, N. M., E. Ghedin, J. Song, A. MacLeod, F. Bringaud, C. Larkin, D. Wanless, J. Peterson, L. Hou, S. Taylor, A. Tweedie, N. Biteau, H. G. Khalak, X. Lin, T. Mason, L. Hannick, E. Caler, G. Blandin, D. Bartholomeu, A. J. Simpson, S. Kaul, H. Zhao, G. Pai, S. Van Aken, T. Utterback, B. Haas, H. L. Koo, L. Umayam, B. Suh, C. Gerrard, V. Leech, R. Qi, S. Zhou, D. Schwartz, T. Feldblyum, S. Salzberg, A. Tait, C. M. Turner, E. Ullu, O. White, S. Melville, M. D. Adams, C. M. Fraser, and J. E. Donelson. 2003. The sequence and analysis of Trypanosoma brucei chromosome II. Nucleic Acids Res. 31:4856-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fantoni, A., A. O. Dare, and C. Tschudi. 1994. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol. Cell. Biol. 14:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilinger, G., and V. Bellofatto. 2001. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 29:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillebault, D., S. Sasorith, E. Derelle, J. M. Wurtz, J. C. Lozano, S. Bingham, L. Tora, and H. Moreau. 2002. A new class of transcription initiation factors, intermediate between TATA box-binding proteins (TBPs) and TBP-like factors (TLFs), is present in the marine unicellular organism, the dinoflagellate Crypthecodinium cohnii. J. Biol. Chem. 277:40881-40886. [DOI] [PubMed] [Google Scholar]
- 13.Hall, N., M. Berriman, N. J. Lennard, B. R. Harris, C. Hertz-Fowler, E. N. Bart-Delabesse, C. S. Gerrard, R. J. Atkin, A. J. Barron, S. Bowman, S. P. Bray-Allen, F. Bringaud, L. N. Clark, C. H. Corton, A. Cronin, R. Davies, J. Doggett, A. Fraser, E. Gruter, S. Hall, A. D. Harper, M. P. Kay, V. Leech, R. Mayes, C. Price, M. A. Quail, E. Rabbinowitsch, C. Reitter, K. Rutherford, J. Sasse, S. Sharp, R. Shownkeen, A. MacLeod, S. Taylor, A. Tweedie, C. M. Turner, A. Tait, K. Gull, B. Barrell, and S. E. Melville. 2003. The DNA sequence of chromosome I of an African trypanosome: gene content, chromosome organisation, recombination and polymorphism. Nucleic Acids Res. 31:4864-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez, N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276:26733-26736. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 16.Hochheimer, A., S. Zhou, S. Zheng, M. C. Holmes, and R. Tjian. 2002. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420:439-445. [DOI] [PubMed] [Google Scholar]
- 17.Holmes, M. C., and R. Tjian. 2000. Promoter-selective properties of the TBP-related factor TRF1. Science 288:867-870. [DOI] [PubMed] [Google Scholar]
- 18.Kaltenbach, L., M. A. Horner, J. H. Rothman, and S. E. Mango. 2000. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol. Cell 6:705-713. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 20.Keogh, M. C., and S. Buratowski. 2004. Using chromatin immunoprecipitation to map cotranscriptional mRNA processing in Saccharomyces cerevisiae. Methods Mol. Biol. 257:1-16. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. L., D. B. Nikolov, and S. K. Burley. 1993. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365:520-527. [DOI] [PubMed] [Google Scholar]
- 22.Laufer, G., and A. Günzl. 2001. In-vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei. Mol. Biochem. Parasitol. 113:55-65. [DOI] [PubMed] [Google Scholar]
- 23.Laufer, G., G. Schaaf, S. Bollgonn, and A. Günzl. 1999. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol. 19:5466-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. G., and L. H. Van der Ploeg. 1997. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu. Rev. Microbiol. 51:463-489. [DOI] [PubMed] [Google Scholar]
- 25.Martianov, I., G. M. Fimia, A. Dierich, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2001. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7:509-515. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Calvillo, S., S. Yan, D. Nguyen, M. Fox, K. Stuart, and P. J. Myler. 2003. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell 11:1291-1299. [DOI] [PubMed] [Google Scholar]
- 27.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myler, P. J., L. Audleman, T. deVos, G. Hixson, P. Kiser, C. Lemley, C. Magness, E. Rickel, E. Sisk, S. Sunkin, S. Swartzell, T. Westlake, P. Bastien, G. Fu, A. Ivens, and K. Stuart. 1999. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA 96:2902-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakaar, V., A. O. Dare, D. Hong, E. Ullu, and C. Tschudi. 1994. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 14:6736-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz, M. C., R. H. Reeder, and S. Hahn. 1992. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell 69:697-702. [DOI] [PubMed] [Google Scholar]
- 32.Shen, S., G. K. Arhin, E. Ullu, and C. Tschudi. 2001. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113:171-173. [DOI] [PubMed] [Google Scholar]
- 33.Shimada, M., T. Nakadai, and T. A. Tamura. 2003. TATA-binding protein-like protein (TLP/TRF2/TLF) negatively regulates cell cycle progression and is required for the stress-mediated G2 checkpoint. Mol. Cell. Biol. 23:4107-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takada, S., J. T. Lis, S. Zhou, and R. Tjian. 2000. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell 101:459-469. [DOI] [PubMed] [Google Scholar]
- 35.Tschudi, C., and E. Ullu. 2002. Unconventional rules of small nuclear RNA transcription and cap modification in trypanosomatids. Gene Expr. 10:3-16. [PMC free article] [PubMed] [Google Scholar]
- 36.Ullu, E., and C. Tschudi. 1990. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 18:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veenstra, G. J., D. L. Weeks, and A. P. Wolffe. 2000. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 290:2312-2315. [DOI] [PubMed] [Google Scholar]
- 38.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 40.Worthey, E. A., S. Martinez-Calvillo, A. Schnaufer, G. Aggarwal, J. Cawthra, G. Fazelinia, C. Fong, G. Fu, M. Hassebrock, G. Hixson, A. C. Ivens, P. Kiser, F. Marsolini, E. Rickell, R. Salavati, E. Sisk, S. M. Sunkin, K. D. Stuart, and P. J. Myler. 2003. Leishmania major chromosome 3 contains two long convergent polycistronic gene clusters separated by a tRNA gene. Nucleic Acids Res. 31:4201-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, D., T. L. Penttila, P. L. Morris, M. Teichmann, and R. G. Roeder. 2001. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292:1153-1155. [DOI] [PubMed] [Google Scholar]