Abstract
Protein ISGylation is unique among ubiquitin-like conjugation systems in that the expression and conjugation processes are induced by specific stimuli, mainly via the alpha/beta interferon signaling pathway. It has been suggested that protein ISGylation plays a special role in the immune response, because of its interferon-signal dependency and its appearance only in higher eukaryotic organisms. Here, we report the identification of an ISG15-conjugating enzyme, Ubc8. Like other components of the protein ISGylation system (ISG15, UBE1L, and UBP43), Ubc8 is an interferon-inducible protein. Ubc8 clearly mediates protein ISGylation in transfection assays. The reduction of Ubc8 expression by small interfering RNA causes a decrease in protein ISGylation in HeLa cells upon interferon treatment. Neither UbcH7/UbcM4, the closest homologue of Ubc8 among known ubiquitin E2s, nor the small ubiquitin-like modifier E2 Ubc9 supports protein ISGylation. These findings strongly suggest that Ubc8 is a major ISG15-conjugating enzyme responsible for protein ISGylation upon interferon stimulation. Furthermore, we established an assay system to detect ISGylated target proteins by cotransfection of ISG15, UBE1L, and Ubc8 together with a target protein to be analyzed. This method provides an easy and effective way to identify new targets for the ISGylation system and will facilitate related studies.
ISG15 is an interferon (IFN)-inducible ubiquitin-like protein (ubl), and its expression and conjugation to target proteins are highly induced upon viral or bacterial infection (20). Among several hundred proteins induced by alpha/beta interferons (IFN-α/β), ISG15 is one of the most predominant (8, 10). Upon IFN treatment, ISG15 is detected as a free protein and conjugated to target proteins in cells (28). In other cases, it is found as an extracellular free protein (21). The secreted form of ISG15 has been thought to function as a cytokine (6, 7, 44). So far, the known data regarding the ISG15 system indicate that protein ISG15 modification (ISGylation) occurs in a manner analogous to ubiquitylation or other ubl modifications (14, 20, 40). In human and rodent cells, ISG15 is expressed as a precursor protein with a C-terminal extension (45), which is processed prior to conjugation in order to expose Gly-Gly residues for conjugation (28). It has been observed that the expression of the isopeptidase for precursor processing seems to be independent of IFN signaling, though the exact nature of the protease has not been identified (41).
Recently, Yuan and Krug (55) identified UBE1L as an E1 enzyme for ISG15. UBE1L was originally cloned as a gene encoding a protein homologous to the ubiquitin-activating enzyme UBE1 (22). Interestingly, influenza B viral infection blocks protein ISGylation by inhibiting the activation step through the interaction of the NS1B viral protein with ISG15 (55). This finding suggests that ISGylation might be involved in setting up antiviral states in the cell. Additional evidence for UBE1L as an ISG15 E1 has been demonstrated by the reconstitution of UBE1L in K562 cells, which resulted in protein ISGylation upon IFN treatment (32). K562 cells not transfected with UBE1L express free ISG15 only in the presence of IFN (28). UBE1L is also hypothesized to be a protein involved in cancer development. The UBE1L gene is located on chromosome 3p21, whose deletion is associated with small cell lung cancer (22). UBE1L expression is not detectable in 14 human lung cancer cell lines (34), suggesting a possible role in lung cancer development.
Another enzyme identified for the ISG15 system is a de-ISGylating protease UBP43 (USP18) (31). Mouse UBP43 was initially cloned during the analysis of genes differentially expressed in both wild-type and leukemia fusion protein AML1-ETO knock-in mice (27). Porcine ubp43 was cloned in a differential screen from lung macrophages of virus-infected swine (57). Li et al. (26) and Kang et al. (18) cloned human ubp43 in screenings for RNase L substrates and for genes induced by IFN in melanoma cell lines, respectively. The primary amino acid sequence indicates that UBP43 is a member of the UBP family of ubiquitin-specific proteases (27, 47). UBP43 contains conserved Cys and His domains that are common in all UBP family members (3, 4) but shows enzymatic specificity for ISG15 (31). This enzyme specifically removes ISG15 from its conjugates but does not partake in the precursor processing, since UBP43-deficient cells can generate ISGylated proteins upon IFN treatment (32). The most intriguing phenotype of UBP43-deficient cells is their enhanced and prolonged activation of the Jak/Stat signaling pathway upon IFN-α/β treatment (32). The significant up-regulation of the UBP43 gene by viral infection (or double-stranded RNA), IFN, and lipopolysaccharide and the IFN hypersensitivity of UBP43 knockout mice all suggest that UBP43 might be involved in a number of processes, including the control of cell proliferation, inflammation, stress, and immune response.
Although ISG15 itself has been known for 25 years, the biological role of the modification has not been well studied. Limited numbers of ISGylation targets have been identified, including PLCγ1, ERK1, Jak1, Stat1, and serpin 2A, yet the biological consequence of the modification is still not well understood (12, 30). For the conjugation and deconjugation of ISG15, only two enzymes have been found so far. Here, we report that an IFN-inducible ubiquitin E2, Ubc8 (UbcH8 and UbcM8 for human and mouse, respectively) is responsible for protein ISGylation both in transfection experiments and in vivo. We were also able to set up a practical assay system to identify ISGylated proteins by the cotransfection of His-ISG15, UBE1L, and Ubc8 followed by the purification of conjugates by using Ni-nitrilotriacetic acid (NTA)-agarose and the detection of target proteins by Western blotting. This assay system provides a powerful and effective way to identify new targets for ISGylation and to confirm ISGylated proteins identified by other methods, thus facilitating the study of protein ISGylation.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
All mouse cDNA constructs except serpin 2A were generated by PCR from a cDNA library prepared from hepatitis B virus-infected hepatocytes (a kind gift of M. Robek and S. Uprichard, The Scripps Research Institute). The mouse ISG15 (mISG15) gene was subcloned into the pCAGGS vector without any tag (pCAGGS-mISG15), with five repeats of the hemagglutinin (HA) tag (pCAGGS-5HA-mISG15), or with a six-His tag (pCAGGS-6His-mISG15). UbcM8 and UbcM4 were also PCR amplified and subcloned into the pFlagCMV2 vector (Sigma, St. Louis, Mo.). UbcM4 is 100% identical to UbcH7 in amino acid sequences. Serpin 2A cDNA was amplified from lipopolysaccharide-treated macrophage RNA and subcloned into the pFlagCMV2 vector. UbcH8 cDNA was amplified from human erythrocyte leukemia RNA (from Eiki Kanbe, The Scripps Research Institute) and subcloned into the pFlagCMV2 vector. All of the constructs prepared by PCR were confirmed by sequencing. UBE1L cDNA was provided by Charles Buys (22) and subcloned into pCAGGS containing a 5′-end HA tag sequence generating pCAGGS-HA-UBE1L. PLCγ1 (from Demin Wang, Blood Research Institute, The Blood Center of Southeastern Wisconsin) was subcloned into the pCAGGS vector. Constructs of pFlagCMV2-UBE1 and pFlagCMV2-Ubc9 are from Chin Ha Chung (Seoul National University, Seoul, Korea), and pCEP-Erk1 was from Melanie Cobb (University of Texas Southwest Medical Center). Plasmid construct expressing Myc-Ub was obtained from Ron R. Kopito (52).
Mutagenesis for the replacement of the active-site cysteine to alanine of both UBE1L (UBE1LC598A) and UbcM8 (UbcM8C86A) was performed by using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. Mutated constructs were confirmed by sequencing.
Yeast two-hybrid screening.
The Matchmaker Library Construction & Screening kit (BD Biosciences Clontech, Palo Alto, Calif.) was used to generate and screen an IFN-β-induced library. IFN-β-induced RNA was generated by stimulating 129 SvEv bone marrow macrophages with 100 U of IFN-β/ml. RNA was harvested at 6, 12, 18, and 24 h posttreatment and pooled, and poly(A)+ RNA was isolated with a Poly A+ Spin mRNA isolation kit (New England BioLabs, Beverly, Mass.). First-strand cDNA was synthesized by using random priming of 1 μg of poly(A)+ RNA. To screen by mating, the IFN-β-induced library was transformed with pGADT7-Rec into AH109 and the transformants were pooled and aliquoted. The library was mated according to the manufacturer's protocols, with strain Y187 containing one of two forms of pGBKT7-ISG15 bait, full-length ISG15 (amino acids 1 to 161) or the first domain of ISG15 (amino acids 1 to 79). The mating mixture was plated on synthetic dropout (SD) plates lacking Ade, His, Leu, and Trp. Colonies larger than 2 mm were restreaked on SD X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside) plates lacking Ade, His, Leu, and Trp; clones that produced blue colonies and grew to 2 mm in diameter were selected for sequencing. After sequencing, isolated clones were retransformed into AH109 and mated with Y187 containing one of seven baits: pGBKT7 (empty vector), pGBKT7 expressing an irrelevant protein (p53 or lamin), or pGBKT7 containing one of the four forms of ISG15 (full-length, ISG15-GG, domain 1, or domain 2). Matings were initially plated on SD plates lacking Leu and Trp to select for mated yeast and were then restreaked onto SD X-Gal plates lacking Ade, His, Leu, and Trp.
Cell culture and transfections.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif.) with 10% fetal bovine serum (HyClone, Logan, Utah) and 2 mM l-glutamine (Invitrogen). Human IFN-α (Roferon-A) was from Roche Pharmaceuticals (Nutley, N.J.). Mouse IFN-β was from ICN Pharmaceuticals (Irvine, Calif.). For small-scale transfections, cells were grown on six-well plates and transfected by using PolyFect reagent (QIAGEN, Hilden, Germany). For large-scale transfections, cells were plated on 10-cm dishes and transfected by using calcium phosphate precipitation as described previously (56).
Generation of fibroblasts from mouse lung.
Primary lung fibroblasts were generated from wild-type and STAT1 knockout mice (9) according to previously published procedures (54). Briefly, lungs were aseptically excised from mice, washed with phosphate-buffered saline (PBS), cut into small pieces, and digested with trypsin (Invitrogen) for 30 min at 37°C. The resulting cell suspensions were collected and resuspended in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 2 mM l-glutamine. Cell suspensions were passed through a cell strainer and cultured in the same medium. Seven days later, cells were divided onto six-well plates. Twenty-four hours after replating, cells were treated with 500 U of IFN-β/ml for the indicated time periods.
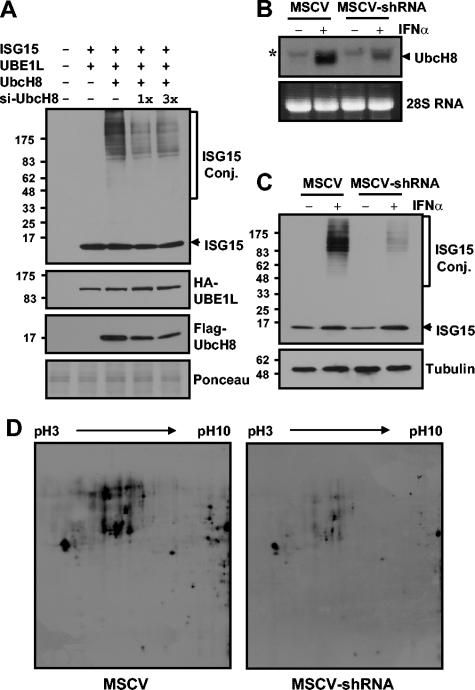
siRNA for UbcH8.
To construct MSCV-UbcH8 short hairpin RNA (shRNA), hairpin structured UbcH8 small interfering RNA (siRNA) was amplified with U6 promoter by PCR with primers 5′-TTTGGATCCCAAGGTCGGGCAGGAAGAGGGCCTATTTCC-3′ and 5′-TTTGAATTCAAAAAGCCTTGCACCAAGACTTGCTCTCTTGAAGCAAGTCTTGGTGCAAGGCTTGGTGTTTCGTCCTTTCCACAAGATATATAA-3′ (1). The PCR product was digested with BamHI and EcoRI and ligated to MSCV-PGKpuro (BD Biosciences Clontech) digested with BglII and EcoRI. UbcH8 knockdown experiments were performed by transiently expressing an shRNA construct for cells (29). To test the efficiency of UbcH8 shRNA in cotransfection experiments, 293T cells were transfected with MSCV-UbcH8 shRNA, pFlagCMV2-UbcH8, pCAGGS-mISG15, and pCAGGS-HA-hUBE1L. The expression of UbcH8 and protein ISGylation were checked 48 h posttransfection. To test the effect of UbcH8 shRNA in IFN-induced protein ISGylation in vivo, HeLa cells seeded in six-well plates were transfected with the control MSCV vector or MSCV-UbcH8 shRNA. Twenty-four hours posttransfection, cells were selected with 2 μg of puromycin/ml for 24 h to reduce the background ISGylation signal from untransfected cells. Dead cells were washed out with PBS, and attached cells were then treated with 5,000 U of IFN-α/ml for 24 h.
Immunoprecipitation and Ni-NTA-agarose pull-down assay.
Cell extract (500 μg) was subjected to either immunoprecipitation or Ni-NTA-agarose pull-down assays. For immunoprecipitation, cells were lysed in modified radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 0.25% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). Immunocomplexes were precipitated with a mixture of protein A-agarose and protein G-agarose (Amersham Biosciences, Piscataway, N.J.). Immunoprecipitates were washed three times with the same buffer and boiled in SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer. For Ni-NTA-agarose pull-down, cells were lysed in PBS containing 1% NP-40 and 20 mM imidazole. Precipitates were washed three times with the same buffer and subjected to SDS-PAGE followed by Western blotting.
Antibodies and Western blot analyses.
Antibodies against Flag (Sigma), HA (Covance, Denver, Pa.), Myc (Sigma), and PLCγ1 (BD Biosciences) were purchased from the respective manufacturers. Mouse anti-human ISG15 monoclonal antibody (clone 5.1) was kindly provided by Ernest C. Borden (The Cleveland Clinic Foundation, Cleveland, Ohio). Rabbit anti-mouse ISG15 polyclonal antibodies were generated by using full-length mouse ISG15 and were immunoaffinity purified by using immobilized antigen as described in detail previously (30). Rabbit anti-UbcM8 antibodies were generated by Covance by using glutathione S-transferase (GST)-UbcM8 and were affinity purified by using GST-UbcM8-agarose. Residual anti-GST antibodies were removed by passage through GST-agarose. Western blotting was performed as described previously (31).
Two-dimensional gel electrophoresis.
Protein samples were prepared in urea lysis buffer {9 M urea, 2.5 mM EDTA, 2.5 mM EGTA, 1% dithiothreitol, and 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)} and centrifuged at 50,000 rpm (Ti50 rotor; Beckman) for 90 min. Isoelectric focusing was performed by using 11-cm immobilized pH gradient gel strips (pH 3 to 10, nonlinear; Bio-Rad, Hercules, Calif.) with 200 μg of protein. Immobilized pH gradient gel strips were focused by ramping up the voltage in two steps, 0 to 250 V for 45 min and 250 to 8,000 V for 2.5 h, and by running at 8,000 V for a total of 20,000 Vh. After equilibration, the proteins on the gel strips were resolved on 8 to 16% polyacrylamide gradient SDS-PAGE. Proteins were transferred onto nitrocellulose membrane, and ISGylated proteins were detected by immunoblotting with anti-human ISG15 monoclonal antibody (clone 5.1).
RESULTS
Ubc8 expression is correlated with protein ISGylation through the Jak/Stat signaling pathway upon IFN-α/β treatment.
Based on the mechanism of protein ubiquitylation and other ubl conjugation (SUMOylation and Neddylation) (17, 53), protein ISGylation is predicted to involve the activity of E1, E2, and E3 enzymes. Currently, only the E1 enzyme, UBE1L, has been reported (55). To understand and regulate protein ISGylation, it is important to identify the E2 and E3 enzymes and to define their functions in a biological system. Each domain of ISG15 shares 30% homology with ubiquitin (20). Furthermore, UBE1L was originally identified as a member of the ubiquitin E1 family (22) and ISG15 protease UBP43 belongs to the ubiquitin protease family (27). Thus, it is highly possible that ISG15 E2 is a member of the ubiquitin E2 family. More importantly, both the expression of ISG15 and protein ISGylation are highly induced upon interferon stimulation. Therefore, we hypothesized that ISG15 E2 is not only a member of the ubiquitin-conjugating enzyme family but also an interferon-stimulated gene product. Based on published reports and the information available from websites, we identified one candidate protein, UbcM8, a mouse homologue of UbcH8, which fully matched the hypothesized criteria (37). We use Ubc8 to denote either the human or the mouse enzyme, unless otherwise specified. We also included 9-27 (Leu-13) and 1-8U proteins as candidates, because both of them are induced by IFN and show similarities to ubiquitin E2s (25, 42).
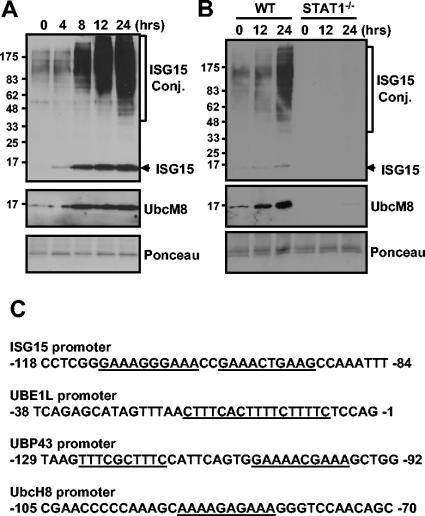
The binding of IFN-α/β to its receptors results in the activation of Jak tyrosine kinases, Jak1 and Tyk2, which are constitutively associated with the receptors (38, 48). The Jaks, in turn, phosphorylate specific tyrosine residues on the receptor, allowing Stats to bind via SH2-phosphotyrosine interaction. Jak kinases then phosphorylate Stat1 and Stat2. Phosphorylated Stat1 and Stat2, in addition to IRF9, form IFN-stimulated gene factor 3 complex, translocate to the nuclei, and activate gene expression via their interaction with the IFN-stimulated response element (ISRE) in the promoters of IFN-inducible genes (5). We tested whether the expression of UbcM8 correlates with protein ISGylation upon IFN-α/β treatment. As shown in Fig. 1A, UbcM8 expression and protein ISGylation are induced by IFN treatment and the two events are temporally well correlated with each other. In STAT-1-deficient fibroblasts, both protein ISGylation and UbcM8 expression were inhibited upon IFN treatment while the wild-type control showed UbcM8 expression and protein ISGylation (Fig. 1B). Interestingly, we could detect basal-level expression of UbcM8 and ISGylated proteins with wild-type cells but not with STAT1-deficient fibroblasts (Fig. 1), suggesting that the basal level of ISGylation and UbcM8 expression are dependent on the Jak/Stat signaling pathway. Furthermore, as shown in Fig. 1C, the promoter regions of genes related to ISG15 modification, including ISG15, UBE1L, UBP43, and UbcH8, contain the ISRE, which is responsible for the promoter activation by IFN-α/β (5, 11). Taken together, these results show that ISGylation and UbcM8 expression are dependent on the IFN signaling and are well coordinated, indicating a possible involvement of UbcM8 in protein ISGylation upon IFN signaling.
FIG. 1.
Correlation between Ubc8 expression and protein ISGylation. NIH 3T3 cells (A) and primary lung fibroblasts from wild-type (WT) and STAT1−/− mice (B) were treated with 500 U of mouse IFN-β/ml for the indicated times. Protein ISGylation and UbcM8 expression were detected by Western blotting. Molecular weight markers (in thousands) are indicated on the left side of each panel. Ponceau staining shows the relative amounts of protein in the samples. (C) Promoter regions of human genes for ISG15, UBE1L, UBP43, and UbcH8 contain ISRE (underlined). Conj., conjugate.
Yeast two-hybrid screening revealed the interaction between ISG15 and Ubc8.
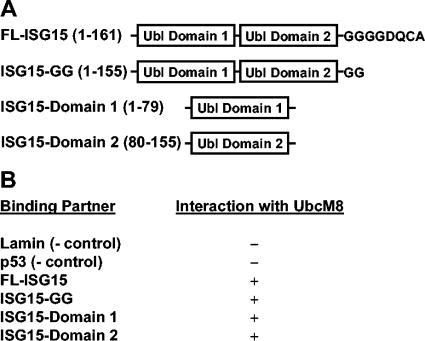
In order to identify ISG15-interacting proteins, we performed yeast two-hybrid screening with cDNA libraries generated from IFN-β-induced RNAs, which were constructed from 129 SvEv bone marrow macrophages stimulated with 100 U of IFN-β/ml. The library was screened by the yeast mating method with two ISG15 baits, full-length ISG15 (amino acids 1 to 161) or the first ubl domain of ISG15 (amino acids 1 to 79), as shown in Fig. 2A. For full-length ISG15 and the first domain of ISG15, 1.2 × 106 and 4.6 × 105 colonies were screened, respectively. From the initial screening, colonies larger than 2 mm were restreaked on minimal selection media containing X-Gal and clones that produced blue colonies and grew to at least 2 mm in diameter were selected for sequencing. A total of 107 clones were sequenced for the full-length ISG15 screening, and 78 clones were sequenced for the domain 1 screening. UbcM8 was identified once from the domain 1 ISG15 screening and once from the full-length ISG15 screening. The clone for UbcM8 from the domain 1 screening (amino acids 54 to 153) was used to test the interaction with lamin, p53, full-length ISG15, C-terminal LRGG-exposed ISG15 (ISG15-GG, amino acids 1 to 155), or domain 1 or domain 2 of ISG15. As indicated in Fig. 2B, UbcM8 interacted with all forms of ISG15 tested, while it did not interact with either lamin or p53. These results provided further support that UbcM8 might act as a conjugating enzyme in the protein ISGylation system.
FIG. 2.
Interaction between ISG15 and UbcM8 in yeast two-hybrid screening. (A) Full-length ISG15 (amino acids 1 to 161) or the first ubl domain of ISG15 (amino acids 1 to 79) was used as bait for yeast two-hybrid screening to identify ISG15-interacting proteins. (B) The clone from the domain 1 screening (UbcM8 amino acids 54 to 153) was retransformed into AH109 and mated withY187 expressing lamin, p53, full-length ISG15 (FL-ISG15), ISG15-GG, or domain 1 or domain 2 of ISG15. The − and + indicate negative and positive interactions of these proteins with UbcM8, respectively.
Ubc8 mediates protein ISGylation together with UBE1L.
In order to test whether UbcM8 can support protein ISGylation in cells, we performed transient transfection assays with UbcM8, 9-27, and 1-8U. For the experiments shown in Fig. 3A, mISG15, UBE1L, and two E2 candidates, UbcM8 and 9-27, were transfected into 293T cells and protein ISGylation was detected by Western blotting with an anti-mouse ISG15 antibody. UBE1L-transfected cells showed increased levels of ISGylated proteins compared to cells transfected with ISG15 only (Fig. 3A, lanes 2 and 3). Most importantly, cells expressing both UBE1L and UbcM8 showed a dramatic increase of protein ISGylation (Fig. 3A, lane 5), indicating that UbcM8 may function as an ISG15 E2. On the other hand, 9-27 expression did not support protein ISGylation (Fig. 3A, lane 4). We also tested 1-8U protein in the same assay but did not detect any increase in protein ISGylation (data not shown). Furthermore, to confirm that the ISGylation signal shown in Fig. 3A was specific for ISG15, we repeated the experiment with HA-tagged ISG15 and anti-HA antibody to detect ISGylated proteins. Again, the expression of UbcM8 together with UBE1L markedly enhanced protein ISGylation in cells (Fig. 3B). These results suggest that UbcM8 protein, but not 9-27 or 1-8U protein, functions as an E2 for protein ISGylation in our experimental system.
FIG. 3.
Ubc8 mediates protein ISGylation. Plasmid constructs pCAGGS-mISG15, pCAGGS-HA-hUBE1L, pcDNA3-HA-9-27, and pFlagCMV2-UbcM8 (A), and pCAGGS-5HA-mISG15, pCAGGS-HA-hUBE1L, and pFlagCMV2-UbcM8 (B) were transfected into 293T cells as indicated on the panels. Cells were harvested 48 h after transfection, and protein extracts were analyzed on an SDS-8 to 18% PAGE gel followed by Western blotting. Free or conjugated (Conj.) forms of ISG15 were detected with rabbit anti(α)-mISG15 polyclonal antibody (A) and with α-HA antibody (B). Other protein expressions were monitored with α-HA or α-Flag antibody. Plasmids are identified by the ending letters and/or numbers of each designation. The asterisk in panel B indicates nonspecific signal from α-HA antibody.
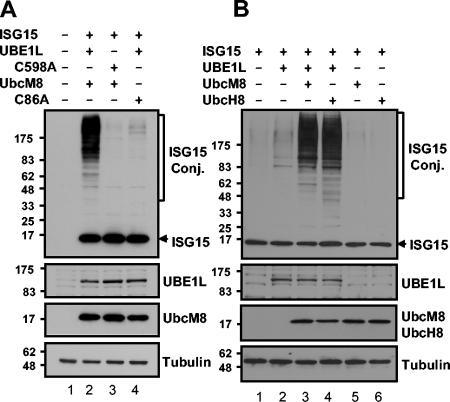
Enzymatic activities of both UBE1L and UbcM8 are required for the conjugation of ISG15.
To test if the protein ISGylation in the above transfection experiments was directly related to the enzymatic activity of UBE1L and UbcM8, we mutated the cysteine residue located at the theoretical active sites of the E1 (C598) and the E2 (C86), which are possibly involved in thioester formation with ISG15 (13, 50), to alanine. As shown in Fig. 4A, cells transfected with UBE1L-C598A or UbcM8-C86A did not show protein ISGylation, indicating that the cysteine is critical for the enzymatic activity of UBE1L and UbcM8. Furthermore, UbcM8 expression in the absence of UBE1L activity did not support protein ISGylation (Fig. 4A, lane 3; Fig. 4B, lane 5). This result also illustrated that protein ISGylation in the transfection assays results from the enzymatic function of both UBE1L and UbcM8.
FIG. 4.
Mutational analysis of active-site cysteine on both UBE1L and UbcM8. Plasmid constructs pCAGGS-mISG15, pCAGGS-HA-hUBE1L, pCAGGS-HA-hUBE1LC598A, pFlagCMV2-UbcM8, and pFlagCMV2-UbcM8C86A (A) and pCAGGS-mISG15, pCAGGS-HA-hUBE1L, pFlagCMV2-UbcM8, and pFlagCMV2-UbcH8 (B) were transfected into 293T cells in combinations as indicated. The expression of proteins and protein ISGylation were analyzed as described for Fig. 3. Plasmids are identified by the ending letters and numbers of each designation. Conj., conjugate.
The species homology for all of the components of the protein ISGylation system, including Ubc8, is relatively low compared to that of the members of other ubl systems. ISG15 itself is only 64% identical in amino acid sequences for human and mouse, while other ubls (ubiquitin, small ubiquitin-like modifier 1 [SUMO-1], and Nedd-8) are 100% identical. UbcH8 and UbcM8 share 75% identity, but conjugating enzymes for other ubl systems show 95 to 100% identity. Therefore, we questioned whether the proteins for the ISGylation system are interchangeable between human and mouse. In transfection assays, the expression of UbcH8 increased protein ISGylation to the same extent as that of UbcM8 (Fig. 4B). This finding shows that there is no species specificity for the ISGylation reaction, despite the relatively low homology between human and mouse E2 compared to other ubl systems.
Ubc8, but not its closest homologue UbcH7/UbcM4, can support protein ISGylation.
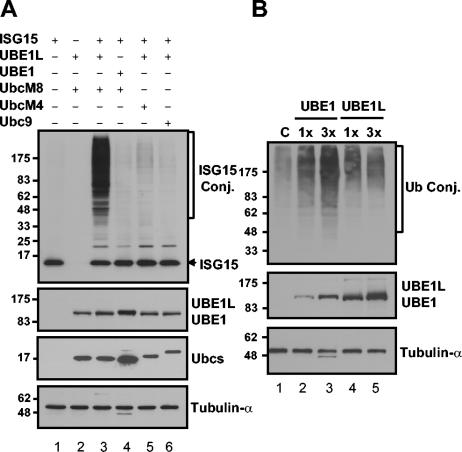
UbcH8 and its close homologue, UbcH7 (55% identical with UbcH8 in amino acid sequences) have been reported to interact with the same E3 ubiquitin ligases, such as E6AP and HHARI (24, 35). We therefore tested whether UbcH7/M4 or Ubc9, a well-characterized SUMO-specific E2, could support protein ISGylation in conjunction with UBE1L. We also checked whether UBE1 could replace UBE1L in a protein ISGylation reaction with UbcM8. The expression of UbcH7/M4 or Ubc9 with ISG15 and UBE1L caused very minor, if any, increases in protein ISGylation compared to the significant amounts of ISGylated proteins in UbcM8-expressing cells (Fig. 5A), indicating that only Ubc8 functions for a protein ISGylation system. Moreover, UBE1 could not replace UBE1L for protein ISGylation in this assay (Fig. 5A, lane 4). Similarly, we also tested whether the overexpression of UBE1L could promote protein ubiquitylation in the same transfection assays. 293T cells were transfected with Myc-tagged ubiquitin and with increasing amounts of either UBE1 or UBE1L. Expression of UBE1 increased ubiquitylated proteins in a dose-dependent manner, but UBE1L expression did not enhance protein ubiquitylation (Fig. 5B). Neither UbcM4 nor UbcM8 could increase the intensity of the ubiquitylation signal in the assays (data not shown), probably because the contribution of specific E2 to general ubiquitylation is too minor to be detected in these assays. Taken together, the homologies of UbcH7/M4 and Ubc8 and of UBE1 and UBE1L are insufficient for functional redundancy (at least for protein ISGylation), suggesting the existence of discrete specificities among the ubl systems. This finding supports the concept that interferon induction of the ISGylation system represents a specific mechanism for altering cell physiology via ISG15 functions independent of other ubls.
FIG. 5.
Ubc8 but not its closest homologue, UbcH7/UbcM4, can support protein ISGylation. (A) Plasmid constructs pCAGGS-mISG15, pCAGGS-HA-hUBE1L, pFlagCMV2-UBE1, pFlagCMV2-UbcM8, pFlagCMV2-UbcM4, and pFlagCMV2-Ubc9 were transfected into 293T cells. The expression of proteins and protein ISGylation were analyzed as described for Fig. 3. (B) Increasing amounts of Flag-UBE1 (pFlagCMV2-UBE1) or HA-UBE1L (pCAGGS-HA-hUBE1L) were transfected into 293T cells together with Myc-Ub-expressing plasmid. After 40 h of transfection, cells were treated with 10 μM MG132 for 8 h. Protein ubiquitylation was analyzed by Western blotting with anti-Myc antibody. Plasmids are identified by the ending letters and numbers of each designation. Conj., conjugate.
siRNA for UbcH8 blocks IFN-induced protein ISGylation.
After proving that Ubc8 could support protein ISGylation in transfection assays, we next tested the hypothesis that Ubc8 depletion in vivo would cause decreased levels of ISGylated proteins in IFN-treated cells. We designed an shRNA to specifically disrupt UbcH8 mRNA in an MSCV vector containing a puromycin selection marker. First, we tested whether this shRNA could decrease the expression of Flag-UbcH8 protein as well as protein ISGylation in 293T cells. Plasmids for ISG15, UBE1L, and UbcH8 were transfected into 293T cells with increasing amounts of the MSCV-UbcH8 shRNA construct. The expression of shRNA reduced the levels of Flag-UbcH8 protein by up to 70% and resulted in decreases in protein ISGylation, while the amounts of ISG15 and UBE1L remained unchanged (Fig. 6A). We then investigated the effect of UbcH8 shRNA on IFN-induced protein ISGylation in HeLa cells. As shown in Fig. 6B, shRNA expression reduced the induction of IFN-induced UbcH8 mRNA by 70 to 80% in comparison to the control. As a result, the amounts of ISGylated proteins decreased significantly while the amount of free ISG15 remained similar to that of the control (Fig. 6C). These results strongly indicate that Ubc8 is a bona fide ISG15 E2, responsible for increased ISGylation upon IFN treatment.
FIG. 6.
siRNA for UbcH8 blocks IFN-induced protein ISGylation. Increasing amounts of MSCV-UbcH8 shRNA construct were cotransfected with pFlagCMV2-UbcH8, pCAGGS-mISG15, and pCAGGS-HA-hUBE1L to 293T cells. Forty-eight hours after transfection, cell extracts were subjected to Western blot analysis to examine protein expression for Flag-UbcH8 and HA-UBE1L and also to examine protein ISGylation. Plasmids are identified by the ending letters and numbers of each designation (A). HeLa cells were transfected with control MSCV vector or MSCV-UbcH8 shRNA construct. Twenty-four hours posttransfection, cells were selected with 2 μg of puromycin/ml for 24 h. Dead cells were washed out with PBS, and the remaining HeLa cells were treated with 5,000 U of IFN-α/ml. RNA and protein samples were prepared after treating with IFN for 12 and 24 h, respectively. RNA samples were analyzed by Northern blotting with an UbcH8 probe (B), and proteins were subjected to Western blotting for ISGylation (C). The asterisk in panel B indicates nonspecific signal. For 2D gel electrophoresis, HeLa extracts were prepared as described for panel C, separated on an 11-cm 2D gel until the 25-kDa marker reached the bottom of the gel in order to get a better separation of high-molecular-weight species, and subsequently probed with anti-ISG15 antibody (D). Ponceau staining of the transferred membranes showed equal loading of proteins (data not shown). Conj., conjugate.
We also examined the effect of UbcH8 shRNA in more detail to analyze whether the decreased expression of UbcH8 affects ISGylation of all target proteins equally or only blocks ISGylation of specific substrates upon IFN treatment. Since we cannot clearly distinguish individual ISGylated proteins in one-dimensional gels, we used two-dimensional (2D) electrophoresis to separate and detect individual ISGylated proteins. HeLa cells transfected with MSCV vector or MSCV-UbcH8 shRNA were prepared as described for Fig. 6C. The protein samples were used for 2D gel electrophoresis, followed by immunoblotting with ISG15 antibodies. As shown in Fig. 6D, we detected individual ISGylated proteins. The decreased expression of UbcH8 caused a general reduction of ISGylated proteins without an obvious substrate preference. In summary, these results indicate that Ubc8 is a major ISG15 E2 in HeLa cells for general protein ISGylation upon IFN-α/β signaling.
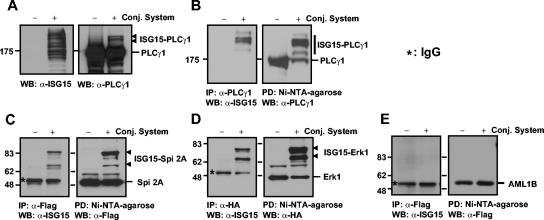
Detection of ISG15 conjugation to specific target proteins by using an artificial conjugation system.
One of the most critical objectives in ubl protein modification systems is the establishment of an easy assay system for detecting modification. We took advantage of the identification of an E2 enzyme for ISG15 to determine whether we could detect ISG15 conjugation of known target proteins (12, 30) by using a transfection-based, artificial conjugation system. His-ISG15, UBE1L, and UbcM8 were transfected into 293T cells together with either PLCγ1, Flag-serpin 2A, or HA-Erk1. As shown in Fig. 7A, we detected two major additional bands of PLCγ1 from the cell extract in which PLCγ1 was transfected with the ISG15 conjugation system only. We were also able to see ISG15-modified forms of serpin 2A or Erk1 from direct Western blots in the same set of experiments, but only with long exposures of blots (data not shown). Thus, we performed reciprocal pull-down and immunoblot studies to detect clearly the ISG15 conjugation of these proteins. Cell extracts were immunoprecipitated with anti-PLCγ1, anti-Flag (for serpin 2A), or anti-HA (for Erk1) antibodies and blotted with anti-mISG15 antibody. Likewise, the same extracts were pulled down with Ni-NTA-agarose for His-ISG15-conjugated proteins and blotted with anti-PLCγ1, anti-Flag, or anti-HA antibodies. As shown in Fig. 7B to D, we clearly detected ISG15-modified PLCγ1, serpin 2A, and Erk1 only from the extracts transfected with ISG15 conjugation system only. To check the specificity of the conjugation system, we tested AML1B in the same transfection experiments, followed by reciprocal pull-down and Western blot analyses. No ISG15-conjugated AML1B was detected (Fig. 7E), indicating that ISG15 conjugation to targets in the transfection system is a specific process.
FIG. 7.
Detection of ISG15 conjugation to specific target proteins. Plasmid constructs for ISG15 targets (pCAGGS-PLCγ1, pFlagCMV2-serpin 2A, pCEP-HA-Erk1, or pFlagCMV2-AML1B) were transfected to 293T cells in the presence or absence of a protein ISGylation system (6His-mISG15, HA-hUBE1L, and Flag-UbcM8). Cells were harvested 48 h after transfection and analyzed either by direct Western blotting (A) or by reciprocal pull-down assay followed by Western blotting (B, C, D, and E). Plasmids are identified by the ending letters and numbers of each designation. Conj., conjugate; IgG, immunoglobulin G; WB, Western blot; PD, pull-down assay; IP; immunoprecipitation.
DISCUSSION
IFNs are well-known cytokines that play pivotal roles in antiviral, antibacterial, cell proliferation and differentiation, and antitumor responses (46, 49). IFN-α/β induces several hundred genes through the Jak/Stat signaling pathway (8). IFN-α/β also triggers a dramatic increase of protein ISGylation in most types of cells (20). However, neither the biological effects of ISGylation nor enzymes involved in the conjugation process have been well studied. In this paper, we identified a new function of Ubc8, an IFN-inducible ubiquitin E2, as an ISG15-conjugating enzyme. Interestingly, all of the proteins so far identified for the ISGylation system, i.e., ISG15, UBE1L, UBP43, and Ubc8, are induced by IFN (8, 10, 26, 37, 55), suggesting their specialized functions in the innate immune response. Based on the characteristics of UBE1L and UBP43, we hypothesized that ISG15 E2 may be a member of the ubiquitin-conjugating enzyme family and also an IFN-stimulated protein. Ubc8 fits these criteria very well. Furthermore, we also identified UbcM8 as an ISG15-interacting protein in independent yeast two-hybrid screening (Fig. 2), which supported our original assumption. It is worth noting that domain 1 of ISG15, which is farther away from the ISG15 C-terminal conjugating site, is sufficient for the interaction with Ubc8 in this assay. In a cotransfection study, Ubc8 clearly mediated protein ISGylation in the presence of UBE1L (Fig. 3), but the inactive mutant form did not (Fig. 4B), indicating that Ubc8 is a bona fide ISG15 E2. The variation of ISGylation in ISG15- and UBE1L-transfected cells in each experiment most probably depends on transfection efficiencies. UBE1L is an enzyme required for the initiation of ISGylation reaction. In that context, we do not see a significant increase of ISGylation by transfecting Ubc8 only. However, we always detect highly increased amounts of ISGylated proteins in the presence of both UBE1L and Ubc8, suggesting that the ISGylation reaction requires both E1 and E2 enzymes. In our assay, we did not detect ISGylated proteins in the presence of 9-27 or 1-8U proteins (Fig. 3A and data not shown). These proteins have been suggested as putative ISG15 E2s based on the limited homology of the putative active-site cysteine region with Ubcs, but no experimental evidence was provided (42). Both 9-27 and 1-8U are IFN-inducible genes and are located on a genomic DNA fragment of less than 18 kb in humans (16, 25). The 9-27 protein has been reported to be localized to the surface of normal T cells (43), and the 1-8U protein also contains a putative transmembrane region (42). Therefore, we cannot rule out the possibility that these proteins still function as ISG15 E2s under other conditions.
UBE1 and UbcH7/M4 or Ubc9 could not act as E1 and E2 to support protein ISGylation, suggesting that there is specificity among ubl conjugation systems. UbcH7/M4 shares 55 and 50% amino acid sequence identity with UbcH8 and UbcM8, respectively. Furthermore, these E2s interact with the same E3 ligases, such as E6AP, HHARI, Parkin, and Dorfin (15, 24, 35, 36), to mediate protein ubiquitylation. However, UbcH7/M4 is not functional in an ISGylation system (Fig. 5). Thus, it is unlikely that general homology is closely related to functional specificity among ubl systems. It has been suggested that differences in surface charge distribution are critical for the discrimination between ubiquitin and SUMO conjugation systems (19). To date, we do not have any structural information for ISG15 and Ubc8. It will be interesting to see if a specific domain of ISG15 is responsible mainly for interacting with UBE1L and Ubc8 and to see which major structural factors are able to distinguish ISG15 from other ubls. In the ubiquitin system, the interaction between E2 enzymes and target proteins should be mediated by E3 enzymes (19). Thus, without E3 enzymes, ubiquitylation could not occur. On the other hand, Ubc9 can directly interact with target proteins and conjugate SUMO on the consensus sequence, which enables conjugation reactions without E3, although the efficiency of SUMOylation increases dramatically in the presence of E3. Ubc8 can mediate protein ISGylation without coexpression of E3 enzymes in our transfection assays. From this perspective, ISGylation seems to be closer to SUMOylation than ubiquitylation. However, it is still possible that E3 enzymes already exist in the cell and that they enhance protein ISGylation upon transfection with UBE1L and Ubc8.
UbcH8 was identified in a yeast two-hybrid screening through its interaction with E6AP and was suggested to be an ubiquitin E2 acting in conjunction with E6AP (24) and also with other E3 proteins, such as Parkin, Dorfin, Staring, Efp, and RLIM, based mainly on in vitro interaction between Ubc8 and E3s (2, 15, 23, 36, 51, 58). In our experiment, the reduction of UbcH8 expression by shRNA was directly linked to down-regulation of ISGylation upon IFN treatment in HeLa cells, indicating that Ubc8 is responsible for protein ISGylation in vivo (Fig. 6). Although we did not detect any change in protein ubiquitylation in the same experiment, this result does not prove that Ubc8 is not involved in ubiquitylation since it is difficult to see changes in general ubiquitylation by reducing the expression of a single E2. It is still possible that Ubc8 works for both the ubiquitin system and the ISGylation system, depending on the availability of other components. Ubiquitin and other ubl systems are closely related to each other in their functions. Nedd8 regulates the function of ubiquitin by modifying cullin family proteins that are molecular scaffolds responsible for assembling the ROC1/Rbx1 RING-based E3 ubiquitin ligases (39). SUMO also modulates the ubiquitylation of certain proteins, such as PCNA and IκBα, by competing for identical lysine residues (33). Thus, it will be interesting to analyze whether there are possible interplays between the ISGylation and ubiquitin pathways.
While we were preparing the present report, Zhao et al. came to a similar conclusion by identifying UbcH8 as a thioester forming E2 with ISG15; they showed, using an in vitro conjugation system, a dual functionality of UbcH8 towards both ubiquitin and ISG15 (59). They also found, using an siRNA approach, that UbcH8 is a major ISG15 E2 for IFN-induced protein ISGylation. Our report confirms this finding and provides additional characterizations to demonstrate that Ubc8 functions as an ISG15 E2. In the present paper, we showed (i) the direct interaction between ISG15 and Ubc8 with a yeast two-hybrid assay, (ii) the enzymatic function of Ubc8 as an ISG15 E2 together with UBE1L with a transfection-based in vivo assay, and (iii) the importance of Ubc8 in IFN-induced ISGylation with siRNA technology. In particular, using a combination of siRNA and 2D Western blotting, we were able to show that the decreased expression of UbcH8 protein caused a reduction of general protein ISGylation for all of the targets. Furthermore, it is important to emphasize that UbcM8 expression in vivo is dependent on the IFN-signaling pathway. Together with data showing that Ubc8 is capable of directing ISGylation, the spatial-temporal availability of this enzyme in vivo strongly argues that Ubc8 is a predominant E2 for IFN-induced ISG15 conjugation to target proteins. More importantly, we set up an in vivo assay system to detect ISG15 target proteins based on the transfection of ISG15, UBE1L, and Ubc8. In our system, we can detect protein ISGylation of specific proteins without knowing the matching E3 enzyme, which was indispensable information for use of the in vitro assay system (59). The novel protein ISGylation assay system presented in this report provides a useful and effective method for the identification and characterization of ISG15 target proteins, which are critical for understanding the biological function of the protein ISGylation system.
Acknowledgments
We thank Ernest Borden for antibodies; Chin Ha Chung, Melanie Cobb, Ian Kerr, Ron Kopito, and Demin Wang for plasmid constructs; Michael Robek, Susan Uprichard, and Eiki Kanbe for cDNAs; and members of the D.-E.Z. laboratory for valuable discussions and critical readings of the manuscript.
This work is supported by National Institutes of Health grants CA079849 (D.-E.Z.), GM066955 (D.-E.Z.), and AI057160 (H.W.V.). The Stein Endowment Fund has partially supported the departmental molecular biology service laboratory for DNA sequencing and oligonucleotide synthesis.
Footnotes
This is manuscript 16650-MEM from The Scripps Research Institute.
REFERENCES
- 1.Chen, C. D., D. S. Welsbie, C. Tran, S. H. Baek, R. Chen, R. Vessella, M. G. Rosenfeld, and C. L. Sawyers. 2004. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10:33-39. [DOI] [PubMed] [Google Scholar]
- 2.Chin, L. S., J. P. Vavalle, and L. Li. 2002. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J. Biol. Chem. 277:35071-35079. [DOI] [PubMed] [Google Scholar]
- 3.Chung, C. H., and S. H. Baek. 1999. Deubiquitinating enzymes: their diversity and emerging roles. Biochem. Biophys. Res. Commun. 266:633-640. [DOI] [PubMed] [Google Scholar]
- 4.D'Andrea, A., and D. Pellman. 1998. Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33:337-352. [DOI] [PubMed] [Google Scholar]
- 5.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 6.D'Cunha, J., E. Knight, Jr., A. L. Haas, R. L. Truitt, and E. C. Borden. 1996. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. USA 93:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Cunha, J., S. Ramanujam, R. J. Wagner, P. L. Witt, E. Knight, Jr., and E. C. Borden. 1996. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J. Immunol. 157:4100-4108. [PubMed] [Google Scholar]
- 8.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443-450. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, P. J., R. J. Broeze, and P. Lengyel. 1979. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 279:523-525. [DOI] [PubMed] [Google Scholar]
- 11.Fujii, Y., T. Shimizu, M. Kusumoto, Y. Kyogoku, T. Taniguchi, and T. Hakoshima. 1999. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 18:5028-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamerman, J. A., F. Hayashi, L. A. Schroeder, S. P. Gygi, A. L. Haas, L. Hampson, P. Coughlin, R. Aebersold, and A. Aderem. 2002. Serpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog. J. Immunol. 168:2415-2423. [DOI] [PubMed] [Google Scholar]
- 13.Hatfield, P. M., and R. D. Vierstra. 1992. Multiple forms of ubiquitin-activating enzyme E1 from wheat. Identification of an essential cysteine by in vitro mutagenesis. J. Biol. Chem. 267:14799-14803. [PubMed] [Google Scholar]
- 14.Hochstrasser, M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2:E153-E157. [DOI] [PubMed] [Google Scholar]
- 15.Imai, Y., M. Soda, and R. Takahashi. 2000. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275:35661-35664. [DOI] [PubMed] [Google Scholar]
- 16.Jaffe, E. A., D. Armellino, G. Lam, C. Cordon-Cardo, H. W. Murray, and R. L. Evans. 1989. IFN-gamma and IFN-alpha induce the expression and synthesis of Leu 13 antigen by cultured human endothelial cells. J. Immunol. 143:3961-3966. [PubMed] [Google Scholar]
- 17.Jentsch, S., and G. Pyrowolakis. 2000. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10:335-342. [DOI] [PubMed] [Google Scholar]
- 18.Kang, D., H. Jiang, Q. Wu, S. Pestka, and P. B. Fisher. 2001. Cloning and characterization of human ubiquitin-processing protease-43 from terminally differentiated human melanoma cells using a rapid subtraction hybridization protocol RaSH. Gene 267:233-242. [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. I., S. H. Baek, and C. H. Chung. 2002. Versatile protein tag, SUMO: its enzymology and biological function. J. Cell Physiol. 191:257-268. [DOI] [PubMed] [Google Scholar]
- 20.Kim, K. I., and D. E. Zhang. 2003. ISG15, not just another ubiquitin-like protein. Biochem. Biophys. Res. Commun. 307:431-434. [DOI] [PubMed] [Google Scholar]
- 21.Knight, E., Jr., and B. Cordova. 1991. IFN-induced 15-kDa protein is released from human lymphocytes and monocytes. J. Immunol. 146:2280-2284. [PubMed] [Google Scholar]
- 22.Kok, K., R. Hofstra, A. Pilz, A. van den Berg, P. Terpstra, C. H. C. M. Buys, and B. Carritt. 1993. A gene in the chromosomal region 3p21 with greatly reduced expression in lung cancer is similar to the gene for ubiquitin-activating enzyme. Proc. Natl. Acad. Sci. USA 90:6071-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer, O. H., P. Zhu, H. P. Ostendorff, M. Golebiewski, J. Tiefenbach, M. A. Peters, B. Brill, B. Groner, I. Bach, T. Heinzel, and M. Gottlicher. 2003. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 22:3411-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, S., W. H. Kao, and P. M. Howley. 1997. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J. Biol. Chem. 272:13548-13554. [DOI] [PubMed] [Google Scholar]
- 25.Lewin, A. R., L. E. Reid, M. McMahon, G. R. Stark, and I. M. Kerr. 1991. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199:417-423. [DOI] [PubMed] [Google Scholar]
- 26.Li, X. L., J. A. Blackford, C. S. Judge, M. Liu, W. Xiao, D. V. Kalvakolanu, and B. A. Hassel. 2000. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2-5A system in attenuation of the interferon response. J. Biol. Chem. 275:8880-8888. [DOI] [PubMed] [Google Scholar]
- 27.Liu, L. Q., R. Ilaria, Jr., P. D. Kingsley, A. Iwama, R. A. van Etten, J. Palis, and D. E. Zhang. 1999. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol. Cell. Biol. 19:3029-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeb, K. R., and A. L. Haas. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 267:7806-7813. [PubMed] [Google Scholar]
- 29.Lu, A., H. Zhang, X. Zhang, H. Wang, Q. Hu, L. Shen, B. S. Schaffhausen, W. Hou, and L. Li. 2004. Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA-dependent RNA polymerase. Virology 324:84-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malakhov, M. P., K. I. Kim, O. A. Malakhova, B. S. Jacobs, E. C. Borden, and D. E. Zhang. 2003. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 278:16608-16613. [DOI] [PubMed] [Google Scholar]
- 31.Malakhov, M. P., O. A. Malakhova, K. I. Kim, K. J. Ritchie, and D. E. Zhang. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277:9976-9981. [DOI] [PubMed] [Google Scholar]
- 32.Malakhova, O. A., M. Yan, M. P. Malakhov, Y. Yuan, K. J. Ritchie, K. I. Kim, L. F. Peterson, K. Shuai, and D. E. Zhang. 2003. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 17:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matunis, M. J. 2002. On the road to repair: PCNA encounters SUMO and ubiquitin modifications. Mol. Cell 10:441-442. [DOI] [PubMed] [Google Scholar]
- 34.McLaughlin, P. M., W. Helfrich, K. Kok, M. Mulder, S. W. Hu, M. G. Brinker, M. H. Ruiters, L. F. de Leij, and C. H. Buys. 2000. The ubiquitin-activating enzyme E1-like protein in lung cancer cell lines. Int. J. Cancer 85:871-876. [DOI] [PubMed] [Google Scholar]
- 35.Moynihan, T. P., H. C. Ardley, U. Nuber, S. A. Rose, P. F. Jones, A. F. Markham, M. Scheffner, and P. A. Robinson. 1999. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J. Biol. Chem. 274:30963-30968. [DOI] [PubMed] [Google Scholar]
- 36.Niwa, J., S. Ishigaki, M. Doyu, T. Suzuki, K. Tanaka, and G. Sobue. 2001. A novel centrosomal ring-finger protein, dorfin, mediates ubiquitin ligase activity. Biochem. Biophys. Res. Commun. 281:706-713. [DOI] [PubMed] [Google Scholar]
- 37.Nyman, T. A., S. Matikainen, T. Sareneva, I. Julkunen, and N. Kalkkinen. 2000. Proteome analysis reveals ubiquitin-conjugating enzymes to be a new family of interferon-alpha-regulated genes. Eur. J. Biochem. 267:4011-4019. [DOI] [PubMed] [Google Scholar]
- 38.O'Shea, J. J., M. Gadina, and R. D. Schreiber. 2002. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109(Suppl.):S121-S131. [DOI] [PubMed] [Google Scholar]
- 39.Pan, Z. Q., A. Kentsis, D. C. Dias, K. Yamoah, and K. Wu. 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23:1985-1997. [DOI] [PubMed] [Google Scholar]
- 40.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 41.Potter, J. L., J. Narasimhan, L. Mende-Mueller, and A. L. Haas. 1999. Precursor processing of pro-ISG15/UCRP, an interferon-β-induced ubiquitin-like protein. J. Biol. Chem. 274:25061-25068. [DOI] [PubMed] [Google Scholar]
- 42.Pru, J. K., K. J. Austin, A. L. Haas, and T. R. Hansen. 2001. Pregnancy and interferon-τ upregulate gene expression of members of the 1-8 family in the bovine uterus. Biol. Reprod. 65:1471-1480. [DOI] [PubMed] [Google Scholar]
- 43.Pumarola-Sune, T., F. Graus, Y. X. Chen, C. Cordon-Cardo, and R. L. Evans. 1986. A monoclonal antibody that induces T cell aggregation reacts with vascular endothelial cells and placental trophoblasts. J. Immunol. 137:826-829. [PubMed] [Google Scholar]
- 44.Recht, M., E. C. Borden, and E. Knight, Jr. 1991. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J. Immunol. 147:2617-2623. [PubMed] [Google Scholar]
- 45.Ritchie, K. J., and D. E. Zhang. 2004. ISG15: the immunological kin of ubiquitin. Semin. Cell Dev. Biol. 15:237-246. [DOI] [PubMed] [Google Scholar]
- 46.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwer, H., L. Q. Liu, L. Zhou, M. T. Little, Z. Pan, C. J. Hetherington, and D. E. Zhang. 2000. Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (Usp18). Genomics 65:44-52. [DOI] [PubMed] [Google Scholar]
- 48.Shuai, K., and B. Liu. 2003. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3:900-911. [DOI] [PubMed] [Google Scholar]
- 49.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, M. L., and R. D. Vierstra. 1991. Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J. Biol. Chem. 266:23878-23885. [PubMed] [Google Scholar]
- 51.Urano, T., T. Saito, T. Tsukui, M. Fujita, T. Hosoi, M. Muramatsu, Y. Ouchi, and S. Inoue. 2002. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature 417:871-875. [DOI] [PubMed] [Google Scholar]
- 52.Ward, C. L., S. Omura, and R. R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83:121-127. [DOI] [PubMed] [Google Scholar]
- 53.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640-643. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, D.-E., C. J. Hetherington, H.-M. Chen, and D. G. Tenen. 1994. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol. Cell. Biol. 14:373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, X., J. Shin, T. W. Molitor, L. B. Schook, and M. S. Rutherford. 1999. Molecular responses of macrophages to porcine reproductive and respiratory syndrome virus infection. Virology 262:152-162. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., J. Gao, K. K. Chung, H. Huang, V. L. Dawson, and T. M. Dawson. 2000. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. USA 97:13354-13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, C., S. L. Beaudenon, M. L. Kelley, M. B. Waddell, W. Yuan, B. A. Schulman, J. M. Huibregtse, and R. M. Krug. 2004. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-α/β-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 101:7578-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]