Abstract
Multiple enhancers govern developmental and tissue-specific expression of the H19-Igf2 locus, but factors that bind these elements have not been identified. Using chromatin immunoprecipitation, we have found two FoxA binding sites in the H19 E1 enhancer. Mutating these sites diminishes E1 activity in hepatoma cells. Additional chromatin immunoprecipitations show that FoxA binds to E1 in fetal liver, where H19 is abundantly expressed, but that binding decreases in adult liver, where H19 is no longer transcribed, even though FoxA proteins are present at both times. FoxA proteins are induced when F9 embryonal carcinoma cells differentiate into visceral endoderm (VE) and parietal endoderm (PE). We show that FoxA binds E1 in VE cells, where H19 is expressed, but not in PE cells, where H19 is silent. This correlation between FoxA binding and H19 expression indicates a role for FoxA in regulating H19, including developmental activation in the yolk sac and liver and postnatal repression in the liver. This is the first demonstration of a tissue-specific factor involved in developmental control of H19 expression. These data also indicate that the presence of FoxA proteins is not sufficient for binding but that additional mechanisms must govern the accessibility of FoxA proteins to their cognate binding sites within the H19 E1 enhancer.
The mouse H19 and insulin-like growth factor 2 (Igf2) genes are tightly linked on distal chromosome 7. These genes are in the same transcriptional orientation, with H19 downstream of Igf2, and are separated by a 70-kb region (57). H19 encodes a 2.4-kb untranslated mRNA of unknown function, whereas Igf2 encodes a fetal growth factor. These two genes are coordinately regulated and exhibit nearly identical spatial and temporal patterns of expression in both endodermal (including liver, gut, and yolk sac) and mesodermal (including skeletal and cardiac muscle) tissues; shortly after birth, their expression is significantly downregulated in most tissues (30, 36, 39). Moreover, expression of H19 and Igf2 is reciprocally imprinted, with H19 being transcribed exclusively from the maternal allele (4) and Igf2 being transcribed only from the paternal allele (13). Indeed, these genes have served as an important paradigm for genomic imprinting studies (reviewed in references 3 and 51).
The tight linkage, similar pattern of expression, and reciprocal imprinting suggest that the H19 and Igf2 genes share a common set of transcriptional regulatory elements. In support of this notion, multiple enhancers that regulate endodermal and mesodermal Igf2 and H19 expression have been identified. Two enhancers, called E1 and E2, are located 5 and 6.5 kb downstream, respectively, of the H19 gene polyadenylation site and were identified by transient transfections in Hep3B human hepatoma cells (55). When these enhancers were deleted by homologous recombination in embryonic stem cells, a dramatic reduction of H19 and Igf2 expression was seen in endodermal tissues, although mesodermal expression remained normal (32). Additional transgenic studies, genomic comparisons, and characterization of the radiation-induced minute (Mnt) mutation identified mesodermal enhancers located further downstream of E1 and E2 and in the Igf2-H19 intergenic region (1, 12, 14, 21, 26, 55).
The regulatory sequences of numerous genes expressed in the liver contain binding sites for endoderm-enriched transcription factors, including FoxA, C/EBP, hepatocyte nuclear factor 1 (HNF1), HNF6, and orphan nuclear receptors (reviewed in reference 10). FoxA proteins (previously called HNF3) were originally identified as liver-enriched factors that regulated the rat transthyretin (TTR) gene (9). Three FoxA isoforms (FoxA1, FoxA2, and FoxA3) are encoded by distinct genes (28, 29) and are defined by a novel “winged-helix” DNA binding motif (6) which also is found in related Fox proteins (27). FoxA proteins are among the earliest factors to be expressed during hepatogenesis (24, 35) and are known to play a role in regulating the transcription of many hepatic genes, including those for albumin and alpha-fetoprotein (AFP) (7). C/EBPs comprise a family of transcription factors that regulate the differentiation of hematopoietic cells, adipocytes, and hepatocytes as well as additional target genes in these cell lineages (reviewed in reference 49).
Despite numerous studies that have helped elucidate aspects of imprinting in the Igf2-H19 locus, little is known about developmental and tissue-specific regulation of these genes. Since H19 enhancers E1 and E2 govern endoderm (liver)-specific expression of H19-Igf2, it is likely that liver-enriched transactivating factors interact with these enhancers. As a rapid in vivo screen to test the binding of liver-enriched factors to H19 control regions, we performed chromatin immunoprecipitation (ChIP) (53) assays with antibodies to FoxA and C/EBP proteins. This analysis revealed no interactions between C/EBP and E1 or E2 but showed that FoxA proteins associate with E1. Electrophoretic mobility shift assays (EMSAs) identified two FoxA binding sites within E1. The importance of these FoxA sites for full E1 activity was confirmed using transient transfections. Finally, ChIP assays in fetal and adult liver, as well as F9 embryonal carcinoma cells, showed an absolute correlation among FoxA binding, H19 expression, and acetylation of histone H4 proteins in the H19 promoter. These findings indicate that FoxA proteins help govern H19 expression in endodermal cell lineages.
MATERIALS AND METHODS
Tissues and cell culture.
Three-month-old female mice with 14.5-day pregnancies were sacrificed. Liver tissues from adult female mice and liver and yolk sac from embryonic day 14.5 (e14.5) fetuses were collected. HepG2, Hep3B, HeLa, and F9 cells were maintained as described elsewhere (44, 46). Insulin, retinoic acid (RA), and dibutyryl cyclic AMP (cAMP) were obtained from Sigma Chemical Corporation (St. Louis, Mo.); all other tissue culture reagents were from Life Sciences (Gaithersburg, Md.). To differentiate into visceral endoderm (VE), F9 cells were grown in suspension as aggregates in petri dishes in the presence of 10−7 M RA. To differentiate into parietal endoderm (PE), F9 cells were grown as monolayers in media containing 10−7 M RA and 10−3 M cAMP. Cells were refed every other day and harvested at the designated time points.
ChIP assays.
ChIP assays were performed according to the protocol provided by Upstate Biotechnology with several modifications. Briefly, tissues (liver and yolk sac) were diced into small pieces using a scalpel blade and resuspended in phosphate-buffered saline (PBS) whereas undifferentiated and differentiated F9 cells were collected by centrifugation and resuspended in PBS. Cells (from tissues or cell culture) were fixed for 15 min by adding formaldehyde to a final concentration of 1%, followed by the addition of glycine to a final concentration of 0.125 M. Cells were then washed three times with ice-cold PBS containing protease inhibitors (1 μg of aprotinin/ml, 1 μg of pepstatin A/ml, 1 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and then lysed in cell lysis buffer (5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 8.0], 85 mM KCl, 0.5% NP-40, and proteinase inhibitors). After 10 min on ice, the cells were homogenized in a glass Dounce homogenizer and centrifuged at 2,500 × g rpm. The nuclear pellet was resuspended in sodium dodecyl sulfate lysis buffer containing proteinase inhibitors for 20 min on ice and was sonicated on ice to fragment the genomic DNA. The mixture was then centrifuged to remove debris, and aliquots (one 100-μl aliquot equaled 107 cells or 0.03 g of tissue) were stored at −80°C. For ChIP, aliquots were diluted in ChIP dilution buffer containing proteinase inhibitors. After preclearance with salmon sperm DNA-protein A agarose, samples were incubated overnight at 4°C with rabbit polyclonal antibodies to FoxA1 and FoxA2 (gifts from Robert Costa, University of Illinois at Chicago), C/EBPα, C/EBPδ proteins (obtained from Santa Cruz Biotechnology), or acetylated histone H4 (Upstate Biotechnology). Immune complexes were precipitated by the addition of salmon sperm DNA-protein A agarose slurry and low-speed spinning. Precipitates were sedimented at 100 × g for 2 min and were washed twice (5 min each at 4°C) with low-salt buffer, once (5 min at 4°C) with high-salt buffer, once (5 min at 4°C) with LiCl buffer, and three times (5 min each at 4°C) with Tris-EDTA. The DNA-protein-antibody complexes were eluted from the beads by vortexing in 250 μl of 0.1 M NaHCO3-1% sodium dodecyl sulfate and were pooled with a second extraction. NaCl was added to the eluates to a final concentration of 0.3 M, and the samples were incubated at 65°C for 4 h to reverse formaldehyde-induced cross-linking. After deproteinization, the DNA was extracted with phenol-chloroform followed by ethanol precipitation. The DNA pellet was resuspended in 50 μl of sterile H2O, and 4 μl of DNA solution was used as a template for PCR with an annealing temperature of 65°C. The linear range for each primer pair was determined empirically to be between 31 and 37 cycles. The following primer combinations were used for amplification of precipitated DNA: H19 enhancer E1, 5′-TCTAGAGTCCATGCATCTGAGG and 5′-AAGCTGCAGACATGGTGG; H19 enhancer E2, 5′-TGTTAACCTGGCTCTGCCCTGC and 5′-CTGCAGGTACAAACACGTAGG; H19 promoter, 5′-CTGTGGATCCGTAGGCTGTTCATACTCC and 5′-ATTAGATCTCCCACACCCGGTGCTTCG; TTR promoter, 5′-AGCGAGTGTTCCGATACTC and 5′-ACCCCCTCCTTCCAACCCA; albumin promoter, 5′-CTCCAGATGGCAAACATACG and 5′-TCTGTGTGCAGAAAGACTCG. The PCR products were separated on a 1.5% agarose gel, visualized by Syber Green staining, and quantified using ImageQuant.
Plasmids and oligonucleotides.
The fragment corresponding to the mouse Igf2-H19 enhancer E1 was generated by PCR amplification, with the primers 5′-TCTAGAGTCCATGCATCTGAGG-3′ and 5′-AAGCTGCAGACATGGTGG-3′ and cloned into pGEM-T (Promega, Madison, Wis.). This fragment was confirmed by restriction digest and DNA sequencing and then excised from pGEM-T and inserted into the BamHI site of the Δ44-lacZ reporter vector (45). PCR-directed mutagenesis was performed as described previously (20) to change site 1 and site 2, either alone or together, in the context of the entire E1. For site 1, the sequence was changed from GTGTACACAAAGCCCTC to AGACTCGAGCCAATAGA; for site 2, the sequence was changed from CACTCCTGTGTCAAC to AGTGAAGCTTCAGCT. Mutated constructs were inserted into the BamHI site of the Δ44-lacZ reporter vector. The pGL2-promoter luciferase plasmid was obtained from Promega. The cytomegalovirus-based rat FoxA1 and FoxA2 expression vectors (provided by Robert Costa) and empty vector (EV) were described previously (20, 40, 41). Oligonucleotides used for PCR amplification of DNA fragments for EMSAs were as follows: H19 promoter, 5′-CTGTGGATCCGTAGGCTGTTCATACTCC and 5′-TATTAGATCTCCCACACCCGGTGCTTCG; H19E1, 5′-TCTAGAGTCCATGCATCTGAGG and 5′-AAGCTGCAGACATGGTGG; H19E2, 5′-TGTTAACCTGGCTCTGCCCTGC and 5′-CTGCAGGTACAAACACGTAGG; H19E1A, 5′-TCTAGAGTCCATGCATCTGAGG and 5′-AGCACTACCCTGAGAGG; H19E1B, 5′-AGGGTAGTGCTGTGAGG and 5′-AACCAGTCTTTCCCAGC; H19E1C, 5′-GACTGGTTTATTCAGG and 5′-TTTGTGTACACCTCCC; H19E1D, 5′-TGTACACAAAGCCCTC and 5′-AAGCTGCAGACATGGTGG. Oligonucleotides that were annealed to generate transcription factor binding sites for use in EMSAs were as follows: FoxA-TTR, 5′-GATCGTTGACTAAGTCAATAATCAGAATCA and 5′-GATCTGATTCTGATTATTGACTTAGTCAAC; C/EBP-Alb, 5′-GATCCTATGATTTTGTAATGGGGC and 5′-GATCGCCCCATTACAAAATCATAG; E1D-a (FoxA site 1), 5′-GGGAGGTGTACACAAAGCCCTC and 5′-GAGGGCTTTGTGTACACCTCCC; E1D-a (FoxA site1mut), 5′-GGAGAGACTCGAGCCAATAGAAATA and 5′-TATTTCTATTGGCTCGAGTCTCTCC; E1D-b, 5′-CACAAAGCCCTCAATAAGTT and 5′-AACTTATTGAGGGCTTTGTG; E1D-c, 5′-CCTCAATAAGTTCCCCATTG and 5′-CAATGGGGAACTTATTGAGG; E1D-d, 5′-TCCCCATTGGCTACTGGTGA and 5′-TCACCAGTAGCCAATGGGGA; E1D-e, 5′-GCTACTGGTGACTTCACTCCTG and 5′-CAGGAGTGAAGTCACCAGTAGC; E1D-f (FoxA site 2), 5′-TTCACTCCTGTGTCAACACC and 5′-GGTGTTGACACAGGAGTGAA; E1D-f (FoxA site2mut), 5′-TTAGTGAAGCTTCAGCTACC and 5′-GGTAGCTGAAGCTTCACTAA; E1D-g, 5′-TGTCAACACCACCATGTCTGCA and 5′-TGCAGACATGGTGGTGTTGACA.
Cell transfection.
Transient transfections were carried out in 6-cm-diameter plates by the calcium phosphate method with a total of 7.5 μg of DNA consisting of 5 μg of the Δ44-lacZ reporter construct and 2.5 μg of the pGL2 luciferase expression vector. For HeLa-synthesized FoxA proteins, cells in 10-cm-diameter plates were transfected with 15 μg of FoxA vectors by using Lipofectamine. Forty-eight hours after the addition of DNA, cells were washed three times in PBS, scraped from plates into 1.5 ml of PBS, and transferred to 1.5-ml microcentrifuge tubes. Cells were pelleted by centrifugation and used for β-galactosidase (β-Gal)-luciferase assays or the preparation of nuclear extracts as described below.
β-Gal-luciferase assays.
Cell pellets from transient transfections were resuspended in 400 μl of reporter lysis buffer (Promega). Cells were lysed by one cycle of freeze-thawing with the use of liquid nitrogen, followed by centrifugation at 14,000 × g for 1 min. The β-Gal activity was measured using the colorimetric substrate chlorophenol red-β-d-galactopyranoside (CPRG; Boehringer Mannheim, Indianapolis, Ind.) as described previously (45). Luciferase assays were performed in triplicate according to the Promega protocol with 20 μl of cell extract and 100 μl of the luciferase assay reagent. The β-Gal activity was normalized to luciferase to control for variations in transfection efficiency.
Preparation of nuclear extracts-EMSAs.
Nuclear extracts were prepared from Hep3B or HeLa cells. Briefly, monolayers of untransfected or transfected cells were harvested by scraping as described above and resuspended in buffer containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, and 0.2 mM PMSF. After a 10-min incubation on ice, nuclei were collected by centrifugation at 14,000 × g for 2 min. The nuclear pellet was resuspended in buffer containing 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% (vol/vol) glycerol, 0.5 mM dithiothreitol, and 0.2 mM PMSF. After a 20-min incubation on ice, samples were centrifuged at 14,000 × g for 2 min. The supernatant was removed, and nuclear extracts were stored in aliquots at −80°C until use. Protein concentrations were determined using the BCA assay kit (Pierce Biochemicals, Rockford, Ill.).
EMSAs were carried out as described previously (20). Briefly, 1 μg of extract and 0.25 μg of poly(dI:dC) were incubated with indicated probes (labeled with [γ-32P]ATP by using T4 polynucleotide kinase) with or without cold competitors. Reaction mixtures were resolved on nondenaturing 8% polyacrylamide gels in 1× TBE (2.2 mM Tris, 2.2 mM boric acid, 0.5 mM EDTA) running buffer. Gels were dried and subjected to autoradiography by using phosphorimaging screens. Oligonucleotides used as probes or cold competitors in EMSAs were either chemically synthesized and annealed oligonucleotides (IDT, Coralville, Iowa) or PCR-amplified products. Supershift antibodies used in EMSAs were obtained from Santa Cruz Biotechnology (anti-FoxA2, p-19; anti-C/EBPδ, c-22).
Reverse transcription-PCR (RT-PCR) assays.
Total RNA was isolated from undifferentiated or differentiated F9 cells (from tissues-cell culture) with Trizol according to the instructions of the manufacturer (Invitrogen); total RNA from liver tissues was prepared using the LiCl-urea method (36). One micromolar oligo(dT) primers were used to synthesize first-strand cDNA from 1 to 2 μg of total RNA with the Ominiscript RT kit (Qiagen). PCRs were carried out using 2.5 μl of the RT reaction mixture and the following primers designated to amplify transcripts of H19, TTR, Igf2, AFP, and β-actin: H19, 5′-CGTTCTGAATCAAGAAGATGCTGC-3′ and 5′-TTTGAGTCTCTCAAGCAAGGAAGG-3′; TTR, 5′-ACAAGCTCCTGACAGGATGG-3′ and 5′-CAGCATCCAGGACTTTGACC-3′; AFP, 5′-AAAACTCGTGATGCTTTGGGCG-3′ and 5′-GGACATCTTCACCATGTGG-3′; Igf2, 5′-TCAGTTTGTCTGTTCGGACC-3′ and 5′-ATTGGAAGAACTTGCCCACG; β-actin, 5′-GTGGGCCGCTCTAGGCACCA-3′ and 5′-CGGTTGGCCTTAGGGTTCAGGGGGG-3′.
RESULTS
Identification of FoxA binding to E1 by ChIP analysis.
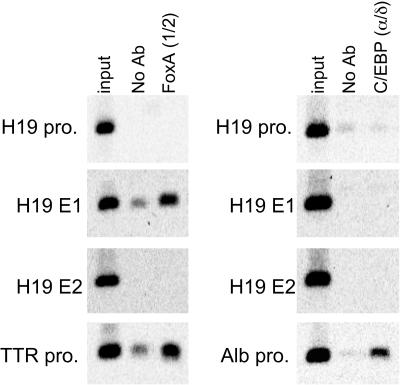
The H19 and Igf2 genes are highly expressed in fetal endoderm tissues, including liver, gut, and yolk sac, and are repressed shortly after birth. Although two enhancers (E1 and E2) direct endoderm-specific expression of both H19 and Igf2 (55), transcription factors that control tissue specificity have not been identified. To screen for factors that bind these enhancers, ChIP assays were performed with e14.5 liver cells by using antibodies to the well-known liver-enriched transcription factors FoxA and C/EBP. Precipitated DNA samples were PCR amplified with primers specific for H19 enhancers E1 and E2 and promoters of the H19, TTR, and albumin genes (Fig. 1). Antibodies to FoxA1 and FoxA2 precipitated DNA containing the TTR promoter, as expected (8). These antibodies also precipitated DNA from E1 but not from E2 or the H19 promoter. Although antibodies to C/EBP (α and δ) precipitated DNA from the positive-control albumin promoter (33, 34), anti-C/EBP antibodies did not bind any of the H19 control regions tested. These data demonstrated an in vivo association between FoxA proteins and H19 E1.
FIG. 1.
FoxA proteins bind to the H19-Igf2 enhancer E1 in vivo. Sheared formaldehyde-fixed chromatin was prepared from fetal (e14.5) liver tissues and precipitated with no antibody (No Ab) or a mixture of antibodies to FoxA1 and FoxA2 [FoxA (1/2)] or C/EBPα and C/EBPδ [C/EBP (α/δ)] followed by PCR amplification with primers that spanned the H19 promoter, E1, or E2; the TTR or albumin (Alb) promoter was amplified as a positive control for FoxA or C/EBP antibodies, respectively. Primers are described in Materials and Methods.
Localization of FoxA binding to E1 by EMSAs.
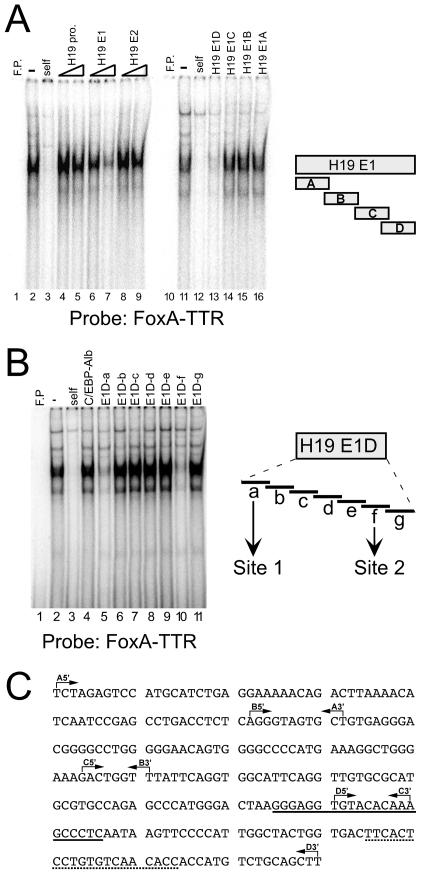
To confirm FoxA binding to E1, EMSAs were performed using the well-characterized FoxA binding site from the rat TTR promoter (FoxA-TTR) as a radiolabeled probe (Fig. 2). Nuclear extracts from human hepatoma Hep3B cells, which express H19, contained proteins that bind the FoxA-TTR site (Fig. 2A, lane 2); binding of these proteins was effectively competed by nonradioactive self fragment (Fig. 2A, lane 3). Consistent with the ChIP data, the E1 fragment, but not E2 or the H19 promoter, could compete for binding to FoxA-TTR in a dose-dependent manner (Fig. 2A, lanes 4 to 9). EMSAs were then used to localize the regions within E1 (shown in Fig. 2C) that could compete for binding to FoxA-TTR. Of the four H19 E1 subfragments (shown on the right side of Fig. 2A), the “D” fragment (E1D) was the only one that could effectively compete for FoxA binding to the FoxA-TTR probe (Fig. 2A, lanes 10 to 16). This indicates that FoxA proteins bind to the 3′ end of E1. EMSAs with the H19 E1D fragment used as a radiolabeled probe confirmed binding of FoxA proteins to this region (data not shown).
FIG. 2.
Two FoxA binding sites are present in the 3′ end of the E1 enhancer. EMSAs were performed with Hep3B nuclear extracts and a radiolabeled probe containing the well-characterized FoxA binding site from the rat TTR promoter (FoxA-TTR). (A) EMSAs were performed with no extract (free probe [F.P.], lanes 1 and 10); extract alone (“-,” lanes 2 and 11); or extract with a 100-fold excess of self as a cold competitor (lanes 3 and 12) or a 50- or 100-fold excess of unlabeled DNA, respectively, from the H19 promoter (lanes 4 and 5), E1 (lanes 6 and 7), or E2 (lanes 8 and 9), or a 100-fold excess of unlabeled DNA from E1 subfragments as shown at the right side of the figure. Oligonucleotides used to amplify E1A, E1B, E1C, and E1D are described in Materials and Methods. (B) EMSAs were performed with no extract (free probe [F.P.], lane 1), probe alone (lane 2), or probe with a 100-fold excess of the following cold competitors: FoxA-TTR (self, lane 3), C/EBP site from the albumin promoter (lane 4), or oligonucleotides E1D-a to E1D-g, which spanned the E1D fragment as shown at the right side of the figure. Oligonucleotides corresponding to these regions were annealed to generate double-stranded fragments and are listed in Materials and Methods. (C) Sequence of H19 enhancer E1 as defined by Yoo-Warren et al. (55). The 5′ and 3′ endpoints of the A, B, C, and D subfragments are designated by the rightward and leftward arrows, respectively. The E1D-a and E1D-f regions are designated by a solid line and a dotted line, respectively.
Computer analysis failed to identify obvious FoxA sites within the E1D fragment. Therefore, to localize further FoxA binding in E1D, seven double-stranded oligonucleotides spanning this 80-bp region were used as cold competitors in EMSAs with the FoxA-TTR site as a radiolabeled probe (Fig. 2B). The control albumin promoter C/EBP oligonucleotide and fragments b, c, d, e, and g could not compete for binding (Fig. 2B). However, oligonucleotides E1D-a and E1D-f (Fig. 2B, lanes 5 and 10; designated by solid underline and dotted underline, respectively, in Fig. 2C) were effective in competing for binding, indicating the presence of two FoxA sites in this enhancer (the sites in E1D-a and E1D-f will be referred to as site 1 and site 2, respectively). To directly test whether these two putative FoxA sites have the capacity to bind FoxA proteins, oligonucleotides corresponding to these regions were used as radiolabeled probes (Fig. 3). The E1D-a fragment bound several complexes (Fig. 3A, lane 2). Of these, the fastest-migrating complex was effectively competed by self and by the FoxA-TTR site but not by the E1D-g and albumin promoter C/EBP site (Fig. 3A, lanes 3 to 6). The presence of FoxA in this complex was confirmed further by the addition of anti-FoxA2 antibodies, which abolished this complex, whereas the addition of anti-C/EBP antibodies had no effect (Fig. 3A, lanes 7 and 8). EMSAs with extracts from HeLa cells transfected with an empty vector or vectors expressing FoxA1 or FoxA2 proteins provided additional evidence that the E1D-a fragment can bind FoxA1 and FoxA2 proteins, although the faintness of the bands suggests that FoxA binding to site 1 is not very strong (Fig. 3A, lanes 9 to 11). Similar experiments were performed with the E1D-f fragment as a radiolabeled probe (Fig. 3B). The cold competitions with self, FoxA-TTR, and albumin C/EBP sites along with experiments with anti-FoxA and anti-C/EBP antibodies demonstrate that FoxA proteins can bind to site 2 (Fig. 3B, lanes 2 to 7). EMSAs with HeLa extracts confirm that FoxA proteins bind site 2 and indicate that FoxA2 binding is stronger than FoxA1 binding (Fig. 3B, lanes 8 to 10). Experiments using the FoxA-TTR site as a radiolabeled probe confirm that equal amounts of FoxA1 and FoxA2 proteins are present in the extracts prepared from transfected HeLa cells (Fig. 3C).
FIG. 3.
FoxA proteins bind directly to site 1 and site 2 in the E1D fragment in vitro. (A) EMSAs were performed using the E1D-a fragment (site 1) as a radiolabeled probe with no extract (free probe [F.P.], lane 1), Hep3B nuclear extracts (lanes 2 to 8), or nuclear extracts from HeLa cells that were transfected with an EV (lane 9) or expression vectors for FoxA1 (lane 10) or FoxA2 (lane 11). Cold competitions were performed with the Hep3B extracts by using a 100-fold molar excess of either no competitor (“-, ” lane 2), E1D-a fragment (self, lane 4), the E1D-g fragment (lane 5), or the albumin promoter C/EBP site (lane 6). Supershift experiments were performed with antibodies to FoxA2 (lane 7) or C/EBPδ (lane 8). (B) EMSAs performed using the E1D-f fragment (site 2) as the radiolabeled probe. Cold competitions and supershift experiments were performed as described for panel A. (C) EMSA with the TTR-FoxA site (TTR-FoxA) as a radiolabeled probe and nuclear extracts from HeLa cells transfected with a control EV (lane 2), FoxA1 expression vector (lane 3), or FoxA2 expression vector (lane 4) demonstrating equal amounts of FoxA1 and FoxA2 proteins in the HeLa nuclear extracts. Lane 1, free probe (F.P.).
Functional analysis of FoxA binding sites in E1.
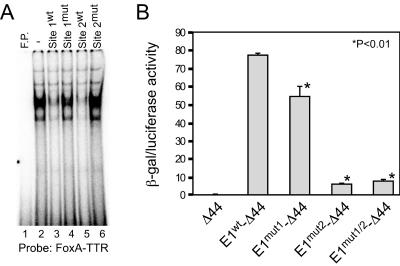
The functional activity of the E1 FoxA binding sites was evaluated further by mutational analysis. To confirm that the mutations eliminated FoxA binding, cold competitions were performed using the FoxA-TTR probe with Hep3B extracts (Fig. 4A). The FoxA-TTR and wild-type fragments containing site 1 or site 2 could effectively compete for binding, whereas the fragments containing mutations in either of these sites eliminated this competition. Reporter constructs were generated by linking wild-type or mutated E1 (each mutation alone or both mutations combined) to the Δ44-lacZ expression vector that we have used previously to monitor the transcriptional activity of enhancer fragments (45). Constructs were transiently transfected into HepG2 human hepatoma cells along with a luciferase vector to normalize for variations in transfection efficiency. The wild-type E1 dramatically activated the Δ44 promoter (Fig. 4B). The site 1 mutation reduced enhancer activity about 30%, indicating that this FoxA site contributes to full E1 activity. A much more dramatic effect was seen with the site 2 mutation, which reduced enhancer activity to roughly 10% of wild-type levels. The double mutant had the same activity as the site 2 mutant. These data confirm the importance of these sites for full E1 activity and indicate that site 2 is more important than site 1 in HepG2 cells. Similar results were seen when transfections were performed in Hep3B cells (data not shown).
FIG. 4.
H19 enhancer E1 site 1 and site 2 are functionally significant as determined by EMSAs and transient transfections. (A) EMSAs were performed with Hep3B nuclear extracts and the TTR-FoxA site as a radiolabeled probe. Cold competitions were performed with no competitor (“-,” lane 2) or with a 50-fold molar excess of oligonucleotides corresponding to the wild-type and mutated putative FoxA site 1 and site 2 (as shown on the left). Lane 1, free probe (F.P.). (B) Reporter constructs carrying wild-type enhancer E1 (E1wt) or E1 with mutations in FoxA site 1 (E1mut1), FoxA site 2 (E1mut2), or both sites (E1mut1/2) linked to the Δ44-lacZ expression vector were transfected into human hepatoma HepG2 cells. The enhancerless Δ44-lacZ vector (Δ44) was included as a control. Cells were also transfected with pGL2 luciferase to control for variations in transfection efficiency. Forty-eight hours after transfection, cell extracts were collected and β-Gal and luciferase activities were determined. Data shown (mean values ± standard deviations) are from two experiments that were each performed in quadruplicate. Statistical significance was determined by the Student t test; the P value was used to indicate a significant difference from E1wt-Δ44.
Developmental changes in FoxA binding to E1.
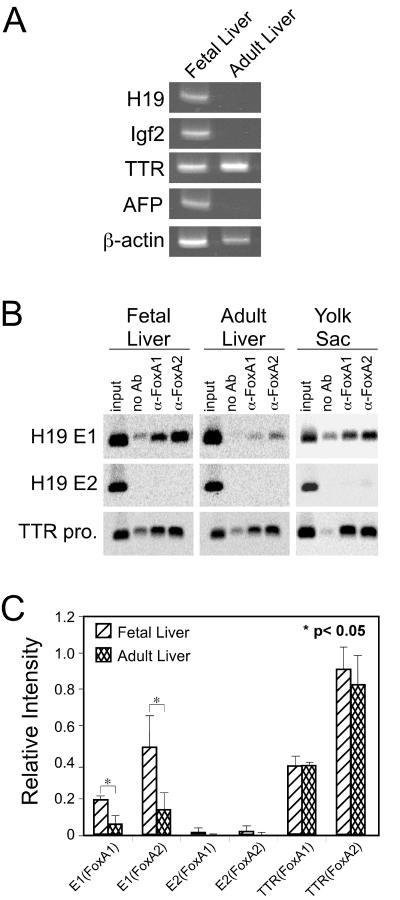
H19 and Igf2 are highly expressed in fetal liver and yolk sac VE, with transcription in the liver dramatically repressed soon after birth; this pattern of expression is similar to that of the mouse AFP gene (Fig. 5A) (31, 32, 36). FoxA proteins are present in fetal and adult liver and yolk sac VE (35). To monitor FoxA binding to E1 in these tissues in vivo, single-cell suspensions were prepared from e14.5 fetal liver and yolk sac and adult liver for use in ChIP assays with antibodies to FoxA1 and FoxA2. The TTR promoter, which contains a strong FoxA binding site, and H19 E2, which does not bind FoxA proteins, were used as positive and negative controls, respectively. The TTR gene is expressed at high levels in yolk sac and fetal and adult liver (Fig. 5A) (11). Consistent with this expression, FoxA1 and FoxA2 proteins were bound to the TTR promoter in all three cell populations (Fig. 5B). Binding to E2 was not detected in any of these cells, consistent with our previous results that this enhancer lacks FoxA binding sites. Products amplified with E1 primers were present in all three cell populations but were more abundant in the fetal liver tissue than in the adult liver tissue (Fig. 5B). Quantitative analysis of the amplified products was used to more accurately compare FoxA binding between fetal and adult liver (Fig. 5C). The amount of TTR promoter product amplified after immunoprecipitation with anti-FoxA1 or anti-FoxA2 was essentially unchanged between fetal and adult liver. In contrast, more FoxA1 and FoxA2 proteins bound to E1 in the fetal liver than in the adult liver. Overall, these results indicate that the amount of FoxA proteins bound to E1 correlates with the level of H19-Igf2 expression during liver development, even though FoxA proteins are equally abundant in fetal and adult liver.
FIG. 5.
The amount of E1-bound FoxA proteins correlates with the H19-Igf2 expression levels in fetal liver, adult liver, and yolk sac. (A) RT-PCR was used to monitor expression of H19, Igf2, TTR, AFP, and β-actin in fetal and adult liver. (B) ChIP assays were performed with chromatin immunoprecipitated from e14.5 fetal liver, adult liver, and e14.5 yolk sac and with no antibody (no Ab) or antibodies to FoxA1 or FoxA2. Oligonucleotides from the TTR promoter or H19 enhancer E1 or E2 were used to amplify the immunoprecipitated DNA. (C) The intensity of PCR-amplified bands in fetal and adult liver with antibodies to FoxA1 and FoxA2 was quantitated using ImageQuant 5.0 software. The intensity of input bands for a DNA fragment in both tissues was first normalized to the input DNA; the relative level of DNA-bound protein was then calculated by subtracting the band intensity in the no-antibody sample from that in the sample precipitated with a specific antibody. The data shown are from three independent experiments, the results from one of which are shown in panel B; significance was determined using the Student t test.
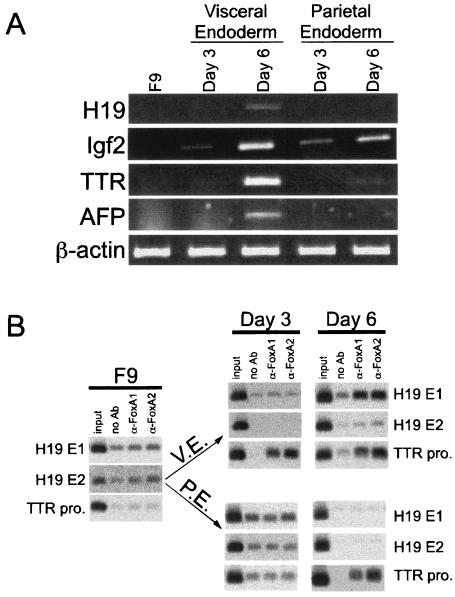
Embryonal carcinoma cells, including F9 cells, have served as tissue culture models to study yolk sac gene expression (18). F9 cells resemble primitive yolk sac endoderm cells under normal conditions (48). When grown in suspension in the presence of RA, F9 cells differentiate into structures known as embryoid bodies (19). Most of the cells on the outer surface of these aggregates closely resemble yolk sac VE in cellular phenotype and gene expression (17). When maintained as adherent cells in the presence of RA and cAMP, F9 cells differentiate into PE-like cells (48). Since H19 is activated in an enhancer-dependent manner in F9 cells that are differentiated into VE cells (36, 55), we used these cells to monitor FoxA binding to E1 during H19 induction. To ensure that the F9 cells were differentiating along the VE and PE pathways, RNA was isolated from F9 cells that were untreated or treated with RA alone or RA and cAMP, for 3 or 6 days, and H19 expression was determined by RT-PCR (Fig. 6A). H19 mRNA was present in day 6 VE cells but not in other cells examined, consistent with previous studies showing that H19 is induced only in VE cells and not in undifferentiated F9 or PE cells. The expression of AFP, a marker for VE, was also evident only in day 6 VE cells. TTR expression could be detected in VE cells after 3 days of RA treatment, and it increased after 6 days. RT-PCR analysis also could detect TTR mRNA in day 6 PE cells. Previous Northern studies failed to detect TTR in the PE lineage (43); our results may be due to the increased sensitivity of RT-PCR compared to Northern analysis. Igf2 expression has not previously been analyzed in differentiated F9 cells. We expected Igf2 to be induced similarly to H19 but found Igf2 to be activated equally well in both the VE and PE pathways (Fig. 6A). Primers specific for the β-actin gene were used to control for RNA loading.
FIG. 6.
FoxA binding to H19 E1 correlates with H19 activation during F9 cell differentiation into VE-like cells. (A) RT-PCR analysis was performed to detect H19, Igf2, and TTR transcripts in undifferentiated F9 cells (F9) or F9 cells treated for 3 and 6 days with RA (VE cells) or RA and cAMP (PE cells). AFP transcripts served as a marker for VE cells; β-actin was used as a control for RNA loading. Oligonucleotides used for PCR amplification were from different exons to eliminate the possibility of amplified products from DNA contamination. (B) ChIP assays were performed with chromatin immunoprecipitated from undifferentiated F9 cells or cells differentiated into VE or PE for 3 or 6 days, with no antibody (no Ab) or antibodies to FoxA1 or FoxA2. Oligonucleotides from the TTR promoter or H19 enhancer E1 or E2 were used to amplify the immunoprecipitated DNA.
FoxA proteins are absent in undifferentiated F9 cells but are induced when F9 cells differentiate along both the VE and PE pathways (22). We performed ChIP assays to monitor FoxA binding to E1 in untreated and differentiating F9 cells (Fig. 6B). FoxA binding to E1, E2, or the TTR promoter was not detected in undifferentiated F9 cells, consistent with previous EMSA data and Western analysis (22). As expected, FoxA proteins did not bind to E2 in VE or PE cells at either time point. Binding of FoxA1 and FoxA2 to TTR promoter DNA could be readily detected in day 3 VE cells and remained high at day 6. Interestingly, we found FoxA proteins bound to the TTR promoter in day 6 PE cells, which correlated with the onset of low TTR expression in PE cells at this time point (Fig. 6B). However, despite the fact that FoxA proteins are present in both VE and PE cells, FoxA1 and FoxA2 binding to E1 was evident only in day 6 VE cells, which is also the only situation where H19 expression was detected. Taken together, the binding of FoxA proteins to H19 E1 and TTR promoter in vivo occurs simultaneously with the expression of these target genes during F9 cell differentiation into either VE or PE-like cells.
Changes in H19 promoter acetylation during liver development.
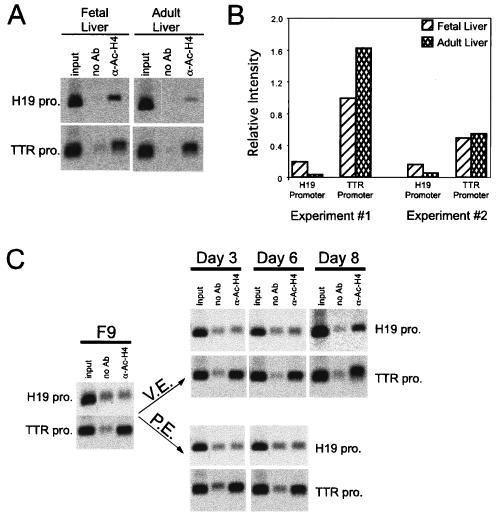
Histone proteins are normally hyperacetylated in the promoters of actively transcribed genes (reviewed in reference 47). Since H19 transcription changes during liver development and F9 cell differentiation, we examined whether H4 acetylation within the H19 promoter correlates with changes in H19 transcription. Chromatin was isolated from fetal and adult liver and F9, VE, and PE cells and immunoprecipitated with antibodies to acetylated histone H4 (Fig. 7). The extent of histone H4 acetylation at the H19 promoter is higher in fetal liver than in adult liver (Fig. 7A and B). In contrast, H4 acetylation of the TTR promoter is slightly higher in the adult liver than the fetal liver. When similar assays were performed using F9 cells, increased H4 acetylation of the H19 promoter was seen only in cells differentiated into VE, where H19 is activated; this change was evident at day 6 and more pronounced by day 8 (Fig. 7C). In contrast, H4 histones on the TTR promoter were acetylated in all cells examined, including undifferentiated F9 cells. H4 hyperacetylation in the TTR promoter in undifferentiated F9 cells might explain why this gene is activated earlier than H19 in the VE pathway and in the PE pathway.
FIG. 7.
Histone acetylation of the H19 promoter correlates with the H19 expression in the liver development and during F9 cell differentiation. (A) ChIP assays were performed with chromatin immunoprecipitated from e14.5 fetal and adult liver with no antibody (no Ab) or antibodies to acetylated histone H4 proteins (α-Ac-H4). Oligonucleotides from the TTR promoter or H19 promoter were used to amplify the immunoprecipitated DNA. (B) The intensity of PCR-amplified bands was quantitated using ImageQuant 5.0 software. The intensity of input bands for a DNA fragment in both tissues was first normalized to the input DNA; the relative level of DNA-bound protein was then calculated by subtracting the band intensity in the no-antibody sample from that in the sample precipitated with anti-Ac-H4 antiserum. Data from two independent experiments are shown; the data shown in panel A correspond to experiment 1 in panel B. (C) ChIP assays were performed with chromatin immunoprecipitated from undifferentiated F9 cells or cells differentiated into VE or PE for 3, 6, or 8 days, with no antibody (no Ab) or antibodies to acetylated histone H4 proteins. Oligonucleotides from the TTR promoter or H19 promoter were used to amplify the immunoprecipitated DNA.
DISCUSSION
The mouse H19 and Igf2 genes are expressed in multiple mesodermal and endodermal tissues and are transcribed at higher levels in fetal tissues than in adult tissues. Standard expression studies in cultured cells and transgenic mice, along with genomic comparisons, have identified multiple enhancers that are upstream and downstream of the mouse H19 gene. Several of these enhancers function in endodermal tissues, including the liver, whereas others are active in mesoderm. The two enhancers first identified, E1 and E2, are essential for developmental activation of H19 and Igf2 genes; the absence of these two enhancers resulted in a dramatic reduction of H19 and Igf2 expression in endodermal tissues, although expression in nonendodermal tissues was essentially unchanged (32). Reactivation of these genes in tumors, which often occurs, was not seen when these enhancers were deleted (50). While these studies demonstrate a critical role for these enhancers during developmental gene regulation and reactivation during carcinogenesis, trans-acting factors that regulate E1 and E2 have not been identified.
Using ChIP assays as a rapid in vivo screen for factors bound to the H19 regulatory regions, we have shown here that FoxA proteins bind to H19 enhancer E1. The data also indicate that FoxA proteins do not bind to the H19 promoter or E2 and that C/EBP proteins bind none of these three regulatory regions. This is the first identification of a tissue-specific factor involved in H19 regulation. Interestingly, our computer analysis of H19 regulatory regions failed to reveal FoxA sites within E1. Thus, while computer-based screening offers one strategy to identify potential transcription factor binding sites (37, 38), ChIP assays provides a valuable alternative to in silico approaches for identifying factor binding sites. Furthermore, ChIP assays provide a direct in vivo measure of transcription factor occupancy in a control region, whereas factor binding sites based on computer-based predictions must be confirmed using in vivo or in vitro strategies.
Our studies identified two FoxA sites in E1. FoxA proteins can bind both sites as judged by EMSA analysis, although these factors bound site 2 with a higher avidity than they bound site 1 (Fig. 3). Functional analysis of these two sites by transient transfection revealed that site 2 contributed more to enhancer activity than did site 1 (Fig. 4). Thus, by two criteria, site 2 is more important than site 1 for FoxA binding and enhancer activity. Multiple FoxA binding sites are found in other regulatory regions, including the TTR promoter, which also has both a strong and a weak FoxA binding site (8). In addition, site 2 binds FoxA2 more strongly than it binds FoxA1 as judged by EMSA (Fig. 3B) and ChIP analysis of fetal liver (Fig. 5). Taken together, these data suggest that FoxA2 may be more important than FoxA1 in regulating E1. FoxA1 and FoxA2 proteins, though structurally similar, have distinct activities. These two proteins have different patterns of expression in various tissues during fetal life, although their patterns of expression in the developing liver are the same (35). FoxA2-knockout mice die early in development and lack an organized node and notochord (2, 52), whereas FoxA1-knockout mice die shortly after birth and have impaired glucose homeostasis (25). Furthermore, nuclear localization of FoxA2, but not FoxA1, is regulated by phosphorylation via the Akt signaling pathway (54).
FoxA proteins possess a forkhead or winged-helix DNA binding domain, which structurally resembles the globular domain of linker histones H1 and H5 (5). FoxA proteins are able to open and reposition chromatin by displacing linker histones in an ATP-independent manner (42). Elegant studies investigating albumin activation during hepatogenesis indicate that FoxA proteins bind to the albumin enhancer prior to albumin activation (15). This has led to the concept of genetic potentiation, whereby FoxA binding facilitates the binding of additional factors that subsequently activate albumin transcription at a later developmental stage (56). Using F9 cells as a tissue culture model of differentiation, we tested whether FoxA binding to E1 occurred prior to H19 activation. In contrast to what Zaret and colleagues saw with albumin, our data with differentiated F9 cells suggest that FoxA binding to H19 E1 occurs concomitantly with H19 activation (Fig. 6). This would indicate that FoxA binding does not act to potentiate H19 transcription but instead is more directly involved in gene activation. This raises the interesting possibility that other factors exist to potentiate the H19 gene prior to transcriptional activation. In contrast to H19, we found that the Igf2 gene was activated in both the VE and PE pathways. This suggests that additional mechanisms can facilitate Igf2 activation in the PE pathway that cannot act upon the H19 promoter.
Histone modification plays an important role in gene regulation by remodeling chromatin structure (23, 47). Previous studies showed that parental origin-specific acetylation exists at the promoters of the Igf2-H19 genes and correlates with the parental origin-specific expression of the Igf2-H19 genes (16). This suggested a role for histone acetylation in regulating monoallelic H19 and Igf2 expression. Our studies indicate a decrease in histone H4 acetylation and H19 gene expression during the transition from fetal to adult liver (Fig. 7A and B) and increased histone H4 acetylation and H19 gene expression during F9 cell differentiation (Fig. 7C). Whether proteins that bind the H19 enhancers, including FoxA, influence promoter acetylation, or whether histone modifications in the H19 promoter region influence factor binding to E1 or E2, will require additional studies.
Although FoxA proteins are equally abundant in the fetal and adult livers, our data show that FoxA proteins are bound to H19 enhancer E1 at higher levels in the fetal liver than in the adult liver. This reduction in binding correlates with postnatal H19 repression in the liver. This is specific for the H19 gene; FoxA binding to the TTR promoter continues to be high in the adult liver, which correlates with continued TTR transcription after birth. The decrease in FoxA binding to E1 in the adult liver raises the possibility that loss of enhancer activity may contribute to postnatal H19-Igf2 repression. Furthermore, our studies with F9 cells also show that FoxA binding occurs only in VE cells, in which H19 is active, but not in PE cells, which do not express H19, even though FoxA proteins are activated in both cell lineages (this study with TTR and reference 22). These data in both liver and F9 cells provide in vivo evidence that the presence of FoxA proteins, by itself, is not sufficient for FoxA binding to H19 E1. Additional mechanisms must restrict the accessibility of FoxA proteins to this enhancer during gene activation (F9 → VE but not F9 → PE) and repression (fetal to adult liver). The presence of negative regulators, or inaccessibility due to chromatin modification, could block FoxA binding to E1 in adult liver and PE cells. Further studies will be needed to investigate these possibilities.
Acknowledgments
We thank Martha Peterson, Jeff Davidson, and members of our laboratory for helpful discussions and critical reading of the manuscript; Marisa Bartolomei for providing H19 plasmids; Rob Costa for providing expression vectors and antiserum; and Michelle Glenn for technical assistance.
This research was supported by Public Health Service grant DK-51600 from the National Institute of Diabetes and Digestive and Kidney Diseases.
REFERENCES
- 1.Ainscough, J. F., L. Dandolo, and M. A. Surani. 2000. Appropriate expression of the mouse H19 gene utilises three or more distinct enhancer regions spread over more than 130 kb. Mech. Dev. 91:365-368. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S.-L., A. Wierda, D. Wong, K. A. Stevens, S. Cascio, J. Rossant, and K. Zaret. 1993. The formation and maintenance of the definitive endoderm lineage of the mouse: involvement of HNF3-forkhead proteins. Development 119:1301-1315. [DOI] [PubMed] [Google Scholar]
- 3.Arney, K. L. 2003. H19 and Igf2—enhancing the confusion? Trends Genet. 19:17-23. [DOI] [PubMed] [Google Scholar]
- 4.Bartolomei, M., S. Zemel, and S. M. Tilghman. 1991. Parental imprinting of the mouse H19 gene. Nature 351:153-155. [DOI] [PubMed] [Google Scholar]
- 5.Burley, S. K., X. Xie, K. L. Clark, and F. Shu. 1997. Histone-like transcription factors in eukaryotes. Curr. Opin. Struct. Biol. 7:94-102. [DOI] [PubMed] [Google Scholar]
- 6.Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Cocrystal structure of the HNF3/fork head DNA-recognition motif resembles histone H5. Nature 364:412-418. [DOI] [PubMed] [Google Scholar]
- 7.Costa, R. H. 1994. Hepatocyte nuclear factor 3/forkhead protein family: mammalian transcription factors that possess divergent cellular expression patterns and binding specificities, p. 183-205. In R. Tronche and M. Yaniv (ed.), Liver gene expression. R. G. Landes Company, New York, N.Y.
- 8.Costa, R. H., and D. R. Grayson. 1991. Site-directed mutagenesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactions with its enhancer region. Nucleic Acids Res. 19:4139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa, R. H., D. R. Grayson, and J. E. Darnell, Jr. 1989. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and α1-antitrypsin genes. Mol. Cell. Biol. 9:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa, R. H., V. V. Kalinichenko, A. X. Holterman, and X. Wang. 2003. Transcription factors in liver development, differentiation, and regeneration. Hepatology 38:1331-1347. [DOI] [PubMed] [Google Scholar]
- 11.Costa, R. H., T. A. Van Dyke, C. Yan, F. Kuo, and J. E. Darnell, Jr. 1990. Similarities in transthyretin gene expression and differences in transcription factors: liver and yolk sac compared to choroid plexus. Proc. Natl. Acad. Sci. USA 87:6589-6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, K., L. Bowden, P. Smith, W. Dean, D. Hill, H. Furuumi, H. Sasaki, B. Cattanach, and W. Reik. 2002. Disruption of mesodermal enhancers for Igf2 in the minute mutant. Development 129:1657-1668. [DOI] [PubMed] [Google Scholar]
- 13.DeChiara, T., E. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849-859. [DOI] [PubMed] [Google Scholar]
- 14.Drewell, R. A., K. L. Arney, T. Arima, S. C. Barton, J. D. Brenton, and M. A. Surani. 2002. Novel conserved elements upstream of the H19 gene are transcribed and act as mesodermal enhancers. Development 129:1205-1213. [DOI] [PubMed] [Google Scholar]
- 15.Gauldi, R., P. Bossard, M. Zheng, Y. Hamada, J. R. Coleman, and K. S. Zaret. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10:1670-1682. [DOI] [PubMed] [Google Scholar]
- 16.Grandjean, V., L. O'Neill, T. Sado, B. Turner, and A. Ferguson-Smith. 2001. Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted Igf2-H19 domain. FEBS Lett. 488:165-169. [DOI] [PubMed] [Google Scholar]
- 17.Grover, A., R. Oshima, and E. Adamson. 1983. Epithelial layer formation in differentiating aggregates of F9 embryonal carcinoma cells. J. Cell Biol. 96:1690-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogan, B. L. M., D. P. Barlow, and R. Tilly. 1983. F9 teratocarcinoma cells as a model for the differentiation of parietal and visceral endoderm in the mouse embryo. Cancer Surv. 2:115-140. [Google Scholar]
- 19.Hogan, B. L. M., A. Taylor, and E. Adamson. 1981. Cell interactions modulate embryonal carcinoma cell differentiation into parietal or visceral endoderm. Nature 291:235-237. [DOI] [PubMed] [Google Scholar]
- 20.Huang, M.-C., K. K. Li, and B. T. Spear. 2002. The mouse alpha-fetoprotein promoter is repressed in HepG2 hepatoma cells by hepatocyte nuclear factor-3 (FOXA). DNA Cell Biol. 21:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishihara, K., N. Hatano, H. Furuumi, R. Kato, T. Iwaki, K. Miura, Y. Jinno, and H. Sasaki. 2000. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in Igf2/H19 imprinting. Genome Res. 10:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob, A., S. Budhiraja, X. Qian, D. Clevidence, R. Costa, and R. R. Reichel. 1994. Retinoic acid-mediated activation of HNF3α during EC stem cell differentiation. Nucleic Acids Res. 22:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 24.Kaestner, K. H., H. Hiemisch, B. Luckow, and G. Schutz. 1994. The HNF-3 gene family of transcription factors in mice: gene structure, cDNA sequence, and mRNA distribution. Genomics 20:377-385. [DOI] [PubMed] [Google Scholar]
- 25.Kaestner, K. H., J. Katz, Y. Liu, D. J. Drucker, and G. Schutz. 1999. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 13:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaffer, C. R., A. Grinberg, and K. Pfeifer. 2001. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell. Biol. 21:8189-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, E., K. L. Clark, S. K. Burley, and J. E. Darnell, Jr. 1993. Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proc. Natl. Acad. Sci. USA 90:10421-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, E., V. R. Prezioso, E. Smith, O. Litvin, R. H. Costa, and J. E. J. Darnell. 1990. HNF-3A, a hepatocyte enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 4:1427-1436. [DOI] [PubMed] [Google Scholar]
- 29.Lai, E., V. R. Prezioso, W. Tao, W. S. Chen, and J. E. Darnell. 1991. Hepatocyte nuclear factor 3α belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 5:416-427. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J. E., J. Pintar, and A. Efstratiadis. 1990. Pattern of the insulin-like growth factor II gene expression during early mouse embryogenesis. Development 110:151-159. [DOI] [PubMed] [Google Scholar]
- 31.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34-39. [DOI] [PubMed] [Google Scholar]
- 32.Leighton, P. A., J. R. Saam, R. S. Ingram, C. L. Stewart, and S. M. Tilghman. 1995. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9:2079-2089. [DOI] [PubMed] [Google Scholar]
- 33.Lichtsteiner, S., J. Waurin, and U. Schibler. 1987. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell 51:963-973. [DOI] [PubMed] [Google Scholar]
- 34.Milos, P. M., and K. S. Zaret. 1992. A ubiquitous factor is required for C/EBP-related proteins to form stable transcription complexes on an albumin promoter segment in vitro. Genes Dev. 6:991-1004. [DOI] [PubMed] [Google Scholar]
- 35.Monaghan, A. P., K. H. Kaestner, E. Grau, and G. Schutz. 1993. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 α, β and γ genes in determination of the definitive endoderm, chordamesoderm, and neuroectoderm. Development 119:567-578. [DOI] [PubMed] [Google Scholar]
- 36.Pachnis, V., A. Belayew, and S. M. Tilghman. 1984. Locus unlinked to α-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. USA 81:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pennacchio, L. A., and E. M. Rubin. 2003. Comparative genomic tools and databases: providing insights into the human genome. J. Clin. Investig. 111:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pennacchio, L. A., and E. M. Rubin. 2001. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2:100-106. [DOI] [PubMed] [Google Scholar]
- 39.Poirier, F., C. T. Chan, P. M. Timmons, E. J. Robertson, M. J. Evans, and P. W. Rigby. 1991. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development 113:1105-1114. [DOI] [PubMed] [Google Scholar]
- 40.Qian, X., and R. H. Costa. 1995. Analysis of hepatocyte nuclear factor-3b protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res. 23:1184-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian, X., U. Samadani, A. Porcella, and R. H. Costa. 1995. Decreased expression of hepatocyte nuclear factor 3α during the acute phase response influences transthyretin gene transcription. Mol. Cell. Biol. 15:1364-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shim, E. Y., C. Woodcock, and K. S. Zaret. 1998. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 12:5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soprano, D., K. Soprano, M. Wyatt, and D. Goodman. 1988. Induction of the expression of retinol-binding protein and transthyretin in F9 embryonal carcinoma cells differentiated into embryoid bodies. J. Biol. Chem. 263:17897-17900. [PubMed] [Google Scholar]
- 44.Spear, B. T., and A. W. Ellis. 1995. Endogenous and transfected mouse alpha-fetoprotein genes in undifferentiated F9 cells are activated in transient heterokaryons. Somat. Cell Mol. Genet. 21:19-31. [DOI] [PubMed] [Google Scholar]
- 45.Spear, B. T., T. Longley, S. Moulder, S. L. Wang, and M. L. Peterson. 1995. A sensitive lacZ-based expression vector for analyzing transcriptional control elements in eukaryotic cells. DNA Cell Biol. 14:635-642. [DOI] [PubMed] [Google Scholar]
- 46.Spear, B. T., and S. M. Tilghman. 1990. Role of α-fetoprotein regulatory elements in transcriptional activation in transient heterokaryons. Mol. Cell. Biol. 10:5047-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer, V. A., and J. R. Davie. 1999. Role of covalent modifications of histones in regulating gene expression. Gene 240:1-12. [DOI] [PubMed] [Google Scholar]
- 48.Strickland, S., and V. Mahdavi. 1978. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell 15:393-403. [DOI] [PubMed] [Google Scholar]
- 49.Takiguchi, M. 1998. The C/EBP family of transcription factors in the liver and other organs. Int. J. Exp. Pathol. 79:369-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vernucci, M., F. Cerrato, N. Besnard, S. Casola, P. V. Pedone, C. B. Bruni, and A. Riccio. 2000. The H19 endodermal enhancer is required for Igf2 activation and tumor formation in experimental liver carcinogenesis. Oncogene 19:6374-6385. [DOI] [PubMed] [Google Scholar]
- 51.Verona, R. I., M. R. Mann, and M. S. Bartolomei. 2003. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 19:237-259. [DOI] [PubMed] [Google Scholar]
- 52.Weinstein, D., A. Altaba, W. Chen, P. Hoodless, V. Prezioso, T. Jessell, and J. E. Darnell, Jr. 1994. The winged-helix transcription factor HNF-3 is required for notochord development in the mouse embryo. Cell 78:575-588. [DOI] [PubMed] [Google Scholar]
- 53.Wells, J., and P. J. Farnham. 2002. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 26:48-56. [DOI] [PubMed] [Google Scholar]
- 54.Wolfrum, C., D. Besser, E. Luca, and M. Stoffel. 2003. Insulin regulates the activity of forkhead transcription factor Hnf-3beta/Foxa-2 by Akt-mediated phosphorylation and nuclear/cytosolic localization. Proc. Natl. Acad. Sci. USA 100:11624-11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoo-Warren, H., V. Pachnis, R. S. Ingram, and S. M. Tilghman. 1988. Two regulatory domains flank the mouse H19 gene. Mol. Cell. Biol. 8:4707-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaret, K. 1999. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev. Biol. 209:1-10. [DOI] [PubMed] [Google Scholar]
- 57.Zemel, S., M. S. Bartolomei, and S. M. Tilghman. 1992. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat. Genet. 2:61-65. [DOI] [PubMed] [Google Scholar]