Abstract
Calcineurin, which binds to the Z-disc in cardiomyocytes via α-actinin, promotes cardiac hypertrophy in response to numerous pathologic stimuli. However, the endogenous mechanisms regulating calcineurin activity in cardiac muscle are not well understood. We demonstrate that a muscle-specific F-box protein called atrogin-1, or muscle atrophy F-box, directly interacts with calcineurin A and α-actinin-2 at the Z-disc of cardiomyocytes. Atrogin-1 associates with Skp1, Cul1, and Roc1 to assemble an SCFatrogin-1 complex with ubiquitin ligase activity. Expression of atrogin-1 decreases levels of calcineurin A and promotes its ubiquitination. Moreover, atrogin-1 attenuates agonist-induced calcineurin activity and represses calcineurin-dependent transactivation and NFATc4 translocation. Conversely, downregulation of atrogin-1 using adenoviral small interfering RNA (siRNA) expression enhances agonist-induced calcineurin activity and cardiomyocyte hypertrophy. Consistent with these cellular observations, overexpression of atrogin-1 in hearts of transgenic mice reduces calcineurin protein levels and blunts cardiac hypertrophy after banding of the thoracic aorta. These studies indicate that the SCFatrogin-1 ubiquitin ligase complex interacts with and represses calcineurin by targeting calcineurin for ubiquitin-mediated proteolysis, leading to inhibition of cardiac hypertrophy in response to pathologic stimuli.
Introduction
Cardiac hypertrophy is an adaptive change in heart size due to an increase in cardiomyocyte cell volume (1). Hypertrophy is a major contributor to diastolic dysfunction of the heart and symptomatic congestive heart failure in humans and is a potent prognostic indicator of all-cause and cardiac mortality independent of symptomatology (2). Sustained cardiac hypertrophy develops in response to a variety of pathologic stimuli, including pressure overload, hypertension, and contractile protein abnormalities. In order for any form of hypertrophic remodeling to occur, stress stimuli must activate specific signaling pathways that lead to an increase in size of cardiac myocytes, protein synthesis, sarcomeric assembly and organization, and activation of a fetal cardiac gene expression program (3).
In recent years, substantial progress has been made in defining intracellular signaling pathways that control transduction of hypertrophic stimuli. Among these pathways, a role for the calcineurin/nuclear factor of activated T cells–dependent (calcineurin/NFAT–dependent) signaling pathway as a regulator of cardiac hypertrophy has been clearly established (4). Calcineurin A is a calcium-activated serine/threonine phosphatase that dephosphorylates and activates NFAT family members. First described as a regulator of immune function in T cells, calcineurin A lies downstream of G protein–coupled receptor activation in a signaling cascade that leads to cardiac hypertrophy. Calcineurin A is tethered to α-actinin-2 at the Z-disc in myocytes, which may allow coupling of contractile status and calcineurin activation (5). In cultured cardiomyocytes, the calcineurin inhibitors cyclosporin A and FK506 prevent hypertrophy of myocytes in the presence of hypertrophic agonists (1). In transgenic mice, cardiac-specific overexpression of activated calcineurin A promotes a hypertrophic response that progresses to heart failure and death (4). Conversely, calcineurin A–deficient mice show a dramatic reduction in calcineurin activity and hypertrophic responses, induced by pressure overload or agonist infusion (6). Moreover, proteins that inhibit calcineurin activation — Cabin/Cain, A kinase–anchoring protein 79 (AKAP79), calcineurin B homology protein, and modulatory calcineurin–interacting protein 1 (MCIP1) — can abrogate cardiac hypertrophy in response to hypertrophic stimuli both in vitro and in vivo (7–10). However, the mechanisms through which calcineurin activity is regulated in cardiomyocytes remain unclear at the present time.
Atrogin-1, also known as muscle atrophy F-box, is a skeletal muscle– and cardiac muscle–specific F-box protein that binds to Skp1, Cul1, and Roc1, components of the SCF family of ubiquitin ligases (11, 12). The F-box proteins function as adaptors that bind specific substrates through protein-protein interaction domains to target them for ubiquitin-dependent degradation (13, 14). Atrogin-1 lacks common domains found in other F-box proteins that are known to interact with substrates (e.g., WD-40 repeats, leucine-rich repeats), so that identification of putative atrogin-1 targets has proven elusive. However, atrogin-1 contains conserved domains such as a PDZ-binding motif and a cytochrome c family heme-binding site signature at its carboxyl terminus that may be involved in protein-protein interactions (12). Overexpression of atrogin-1 in skeletal myotubes leads to atrophy, and mice deficient in atrogin-1 are resistant to denervation atrophy of skeletal muscle (11, 12). These observations suggest that atrogin-1 may regulate muscle cell size via its participation in a muscle-specific ubiquitin ligase complex, although such an activity is yet to be proven and signaling pathways regulated by atrogin-1 have not been determined. In addition, neither the function of atrogin-1 nor its downstream targets in the heart are known. It is logical to speculate that atrogin-1 should regulate pathways that are shared by skeletal and cardiac muscle.
In order to resolve these issues, we have searched for potential partner proteins of atrogin-1 that might indicate a functional role for this F-box protein in the heart. We report that atrogin-1 binds α-actinin-2 and calcineurin A and localizes at the Z-disc of cultured cardiomyocytes. Atrogin-1 attenuates agonist-induced calcineurin activity, hypertrophy, and fetal gene expression and also represses calcineurin-dependent transactivation and NFATc4 translocation in vitro. The F-box domain of atrogin-1 binds other components of the SCF ubiquitin ligase complex and has intrinsic ubiquitin ligase activity. Consistent with its role as a ubiquitin ligase component, atrogin-1 downregulates levels of endogenous calcineurin in vivo and promotes its ubiquitination in vitro. Finally, overexpression of atrogin-1 in the heart blunts induction of calcineurin activity and cardiac hypertrophy after aortic banding in transgenic mice, indicating that atrogin-1 acts as a calcineurin signaling repressor both in vitro and in vivo. Cardiac-specific regulation of calcineurin activity via ubiquitin-dependent proteolysis by atrogin-1 provides a mechanism for rapid regulation of calcineurin A specifically and without concomitant effects on immune function.
Results
Atrogin-1 associates with α-actinin-2 and calcineurin A.
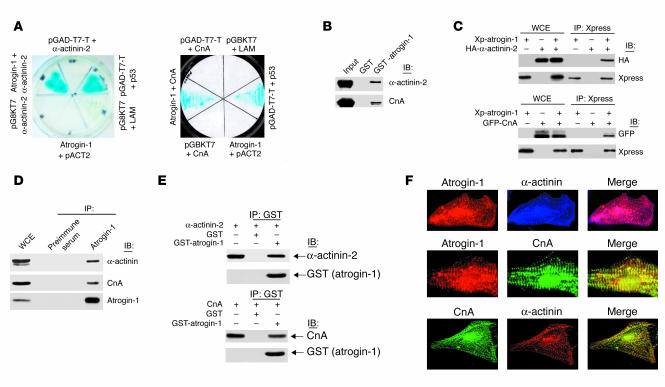
As a first step to investigate the functions of atrogin-1, we searched for partner proteins that interact directly with atrogin-1 via a yeast 2-hybrid assay. A systematic search for functional domains within atrogin-1 indicated the presence of potential protein-protein interaction domains, including sequences adjacent to the F-box that might be required for substrate recognition, in the C-terminus of atrogin-1 (12). We therefore expressed the C-terminus of atrogin-1 as a fusion protein with the GAL4 DNA-binding domain in pGBKT7 vector as bait to screen an adult human heart cDNA library, which was cloned in pACT2. Among the potential binding partners found in this screen, the Z-disc–localizing actin-binding protein α-actinin-2 exhibited strong and specific interaction (Figure 1A). The interaction between atrogin-1 and α-actinin-2 was confirmed by in vitro glutathione-S-transferase (GST) pull-down assays (Figure 1B). The interaction of atrogin-1 with α-actinin-2 was further tested by coimmunoprecipitation of epitope-tagged proteins in COS-7 cells. Xpress-tagged atrogin-1 efficiently coprecipitated hemagglutinin-tagged (HA-tagged) α-actinin-2 (Figure 1C). Atrogin-1 overexpression did not affect levels of α-actinin-2 protein in cardiomyocytes, which suggested to us that α-actinin-2 is not a ubiquitination target of atrogin-1 and might instead participate in multiprotein complexes with atrogin-1 and other proteins that are themselves targets for the activity of atrogin-1.
Figure 1.
Molecular interaction of atrogin-1 with α-actinin-2 and calcineurin A (CnA). (A) Yeast 2-hybrid analysis of atrogin-1 interaction with α-actinin-2 (left) and calcineurin A (right). pGBKT7 and pACT2 are empty plasmids. pGAD-T7 vector expresses a T antigen–GAL4 activation domain fusion that interacts with p53 (positive control reaction). (B) In vitro interactions of atrogin-1 with α-actinin-2 and calcineurin A in GST pull-down assays. The ability of α-actinin-2 (top) or calcineurin A (bottom) expressed in COS-7 cells to be retained by GST or a GST–atrogin-1 fusion protein was analyzed by immunoblotting after binding reactions. (C) COS-7 cells were transfected with Xpress-tagged (Xp) atrogin-1 and GFP-tagged calcineurin A or HA-tagged α-actinin-2 expression plasmids as indicated. Equal amounts of cell extract were immunoprecipitated with the Xpress antibody and analyzed by immunoblotting with antibodies directed against Xpress (to detect atrogin-1) and HA (to detect α-actinin-2, top) or against GFP (to detect calcineurin A, bottom). WCE, whole cell extract. (D) Endogenous protein interactions were examined in cardiomyocyte cell lysates that were immunoprecipitated with preimmune serum or anti–atrogin-1 antibody and analyzed by immunoblotting with antibodies to detect α-actinin (top), calcineurin A (middle), and atrogin-1 (bottom). (E) Direct protein interactions were detected by incubation of GST–atrogin-1 or GST (1 μg) with 1 μg of recombinant calcineurin A or α-actinin-2 proteins. Mixtures were precipitated with anti-GST antibody and analyzed by immunoblotting with antibodies against α-actinin, calcineurin A, and GST. (F) Coimmunostaining analysis of endogenous atrogin-1, calcineurin A, and α-actinin-2 in neonatal cardiomyocytes. The overlay shows that atrogin-1 colocalizes with both α-actinin (top) and calcineurin A (middle).
To explore the relevance of this association, we tested whether endogenous atrogin-1 can interact with α-actinin-2 under physiologic conditions. We performed immunoprecipitation and immunoblotting experiments using extracts prepared from rat neonatal cardiomyocytes and found that α-actinin-2 efficiently coimmunoprecipitated with endogenous atrogin-1 (Figure 1D). Immunostaining indicated that endogenous atrogin-1 expression is concentrated at the Z-disc in a pattern that overlaps with that of α-actinin-2 (Figure 1E). Among known interaction partners of α-actinin-2 in striated muscle, calcineurin A stands out because of its role in coordinating myocyte gene expression programs that determine cell size (5). We therefore compared the immunolocalization of atrogin-1 and calcineurin A and found that these proteins were also similarly distributed and concentrated at the Z-disc. This expression pattern suggested that atrogin-1 and calcineurin A may also directly interact. The association of atrogin-1 and calcineurin A was confirmed in yeast 2-hybrid and GST-binding assays (Figure 1, A and B) and coimmunoprecipitation experiments (Figure 1, C and D). To test whether these interactions are direct, we performed in vitro binding reactions using recombinant proteins. Both α-actinin-2 and calcineurin A bound efficiently to a GST–atrogin-1 fusion protein in these assays (Figure 1F). Taken together, these experiments indicate that atrogin-1 is a bona fide interaction partner of both α-actinin-2 and calcineurin A within the cardiac myofibril, most likely participating in a ternary multiprotein complex.
Interaction domains of atrogin-1, calcineurin A, and α-actinin-2.
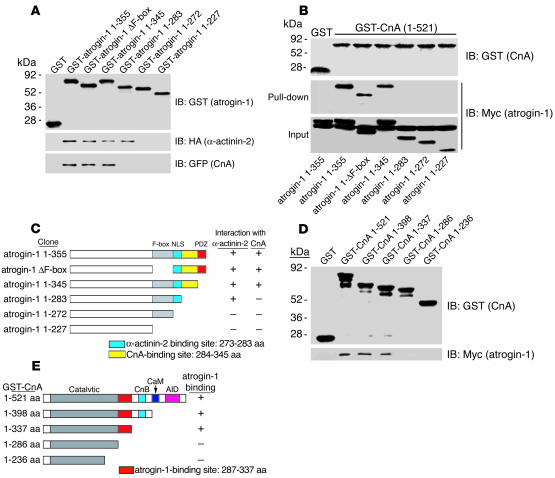
To define the regions in atrogin-1 that are responsible for binding with calcineurin A, we tested serially deleted atrogin-1 mutants expressed in bacteria as GST fusion proteins for their ability to bind calcineurin A (Figure 2A, top). GST pull-down assays revealed that the F-box domain of atrogin-1 is dispensable for protein-protein interactions; amino acids 272–283 of atrogin-1 are necessary for the interaction of atrogin-1 with α-actinin-2 (Figure 2A, middle), whereas the adjacent amino acids 284–345 of atrogin-1 are required for binding to calcineurin A (Figure 2A, bottom). To further confirm the region of atrogin-1 required for association with calcineurin A, a full-length calcineurin A GST fusion was tested in GST pull-down assays with a series of atrogin-1 deletion mutants expressed in vivo in COS-7 cells. Amino acids 284–345 of atrogin-1 were also required for calcineurin A binding in this assay (Figure 2B). These data, summarized in Figure 2C, indicate that topologically adjacent domains of atrogin-1 are required for α-actinin-2 and calcineurin A binding.
Figure 2.
Mapping the interaction domains of atrogin-1, α-actinin-2, and calcineurin A. (A) The residues of atrogin-1 required for binding to α-actinin-2 and calcineurin A were determined with GST pull-down assays. GST–atrogin-1 fusion proteins were purified and analyzed for expression (top). The ability of the truncated atrogin-1 fusion proteins to bind to calcineurin A (expressed in COS-7 cells as a GFP fusion) and α-actinin-2 (expressed as an HA fusion) was analyzed by blotting with antibodies against HA (middle) and GFP (bottom). (B) GST–calcineurin A was affinity-purified and analyzed by blotting with antibody against GST (top). The pull-down (middle) and input (bottom) fractions of COS-7 cell extracts expressing the indicated Myc-tagged atrogin-1 truncations were immunoblotted with antibodies against Myc. (C) Schematic representation of atrogin-1 truncations that interact with α-actinin-2 and calcineurin A. F-box, F-box domain; NLS, nuclear location sequence; PDZ, PDZ domain. (D) The region of calcineurin A involved in binding to atrogin-1 was analyzed in pull-down assays. GST–calcineurin A fusion proteins were affinity purified and analyzed by blotting with GST antibody (top). The ability of the various calcineurin A fusion proteins to bind to atrogin-1 expressed as a Myc-tagged fusion in COS-7 cells was analyzed by blotting with Myc antibody (bottom). (E) Schematic representations of calcineurin A residues that bind to atrogin-1. CnB, calcineurin B–binding domain; CaM, calmodulin-binding domain; AID, autoinhibitory domain.
To identify the sequences in calcineurin A required for interaction with atrogin-1, a series of truncations of calcineurin A expressed as GST fusions (Figure 2D, top) was used in pull-down assays with Myc–atrogin-1 expressed in COS-7 cells. The region of calcineurin A containing amino acids 287–337 was required for binding to atrogin-1 (Figure 2D, bottom). These data (summarized in Figure 2E) indicate that the atrogin-1–interacting domain of calcineurin A resides within the C-terminal portion of its catalytic domain and is distinct from the binding site for other known calcineurin-interacting proteins.
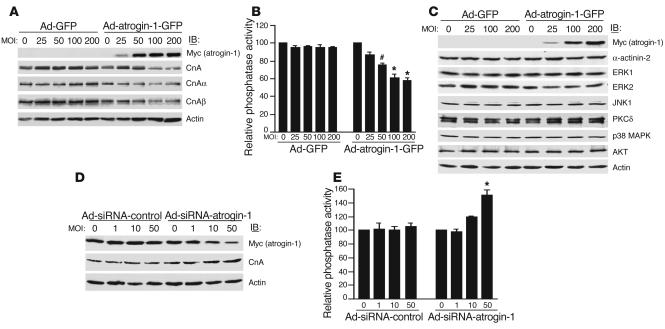
Atrogin-1 reduces endogenous calcineurin A protein levels and decreases calcineurin A activity in neonatal cardiomyocytes.
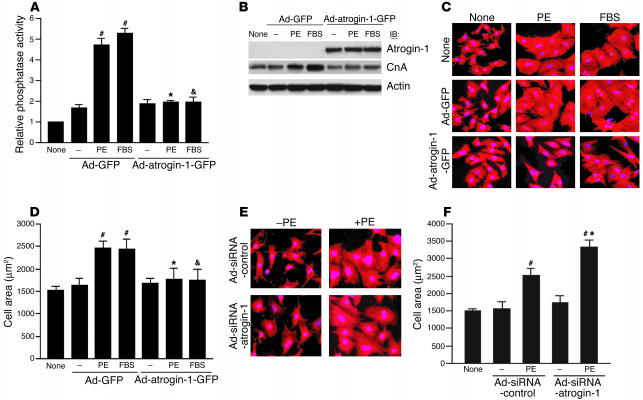
Because atrogin-1 is a putative ubiquitin ligase component, we tested the ability of atrogin-1 to regulate the level of endogenous calcineurin A protein in cardiomyocytes infected with adenoviruses Ad-GFP and Ad-atrogin-1-GFP after FBS stimulation. There are 3 known genes that encode catalytic calcineurin A proteins, calcineurin Aα, calcineurin Aβ, and calcineurin Aγ. Infection with Ad-atrogin-1-GFP caused a dose-dependent reduction in the level of total endogenous calcineurin A, including both calcineurin Aα and calcineurin Aβ (Figure 3A), demonstrating that atrogin-1 reduces calcineurin A protein levels in cardiomyocytes. To establish the cellular consequences of the effects of atrogin-1 on endogenous calcineurin A protein, we measured calcineurin phosphatase activity in cardiomyocytes infected with either Ad-GFP or Ad-atrogin-1-GFP after FBS stimulation. Calcineurin phosphatase activity was significantly decreased in a dose-dependent manner in Ad-atrogin-1-GFP–infected compared with Ad-GFP–infected cardiomyocytes (Figure 3B; P < 0.05 or 0.001). To determine whether atrogin-1 might influence endogenous proteins in other signaling pathways, cardiomyocytes were infected with adenoviruses Ad-GFP and Ad-atrogin-1-GFP after FBS stimulation. Immunoblotting indicated that atrogin-1 does not appear to have a major role in regulating the levels of other endogenous proteins, including α-actinin-2, ERK, JNK, PKCδ, p38 MAPK, and Akt (Figure 3C); this demonstrates at least some degree of specificity for the effect of atrogin-1 on calcineurin A.
Figure 3.
Atrogin-1 regulates endogenous calcineurin A levels and activity. (A) Cardiomyocytes were infected with increasing multiplicities of infection (MOI) of Ad-atrogin-1-GFP and Ad-GFP with FBS stimulation for 36 hours. The levels of expressed atrogin-1 and endogenous calcineurin A were determined by immunoblotting with anti-Myc or –calcineurin A antibodies, respectively. An antibody against β-actin was used to assess protein loading. A representative blot is shown for each condition. (B) Calcineurin A phosphatase activity was measured with 5 μg of cell extracts. The data are from 3 independent experiments in duplicate. *P < 0.05, #P < 0.001 vs. Ad-GFP. (C) Adenovirus-infected cardiomyocytes were tested for expression of endogenous α-actinin-2, ERK1, ERK2, JNK1, PKCδ, p38 MAPK, and Akt by immunoblotting with the indicated antibodies. An antibody against β-actin was used to assess protein loading. (D) Cardiomyocytes were infected with increasing MOI of Ad-siRNA-atrogin-1 and Ad-siRNA-control with FBS stimulation for 36 hours, and the levels of expressed atrogin-1 and endogenous calcineurin A were determined by Western blotting. (E) Calcineurin A phosphatase activity was measured in cardiomyocytes infected with Ad-siRNA-atrogin-1 and Ad-siRNA-control. The data are from 3 independent experiments in duplicate. *P < 0.001 vs. Ad-siRNA-control.
In addition, we tested whether endogenous atrogin-1 is critical in regulating the level of calcineurin A in cardiomyocytes using an adenoviral small interfering RNA (siRNA) strategy. siRNA targeted to unique sequences in atrogin-1 effectively reduced atrogin-1 protein levels by 80% in FBS-treated cardiomyocytes compared with an adenovirus expressing a control siRNA, and suppression of atrogin-1 protein was accompanied by increased calcineurin A expression (Figure 3D). Similarly, downregulation of atrogin-1 resulted in a significant increase in calcineurin phosphatase activity, whereas the control siRNA had no effect (Figure 3E). Together, these data demonstrate that atrogin-1 inhibits endogenous calcineurin A activity by reducing calcineurin A levels in FBS-stimulated cardiomyocytes.
Atrogin-1 represses calcineurin A–dependent transcriptional responses in neonatal cardiomyocytes.
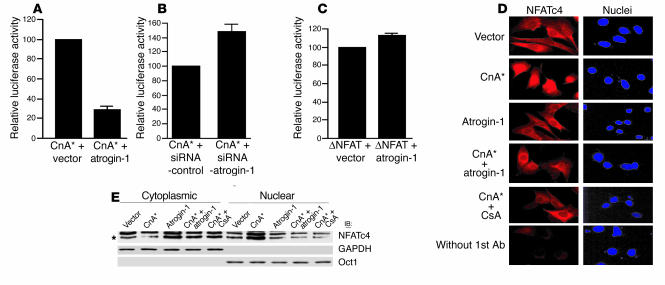
Nuclear translocation and activation of target genes by NFAT depends on calcineurin signaling in cardiomyocytes (4), which led us to test the effects of atrogin-1 on calcineurin-NFAT function. Cardiomyocytes were cotransfected with reporter construct pIL2-Luc (containing an NFAT-binding site from the IL-2 promoter upstream of the luciferase gene) and plasmids expressing activated calcineurin A alone or in combination with atrogin-1. Atrogin-1 potently repressed calcineurin-dependent transactivation (Figure 4A). In contrast, transfection of a plasmid expressing atrogin-1 siRNA enhanced calcineurin-dependent transactivation by more than 45% (Figure 4B). Interestingly, atrogin-1 had no effect on pIL2-Luc transactivation by constitutively activated NFAT (ΔNFAT, Figure 4C), which suggests that atrogin-1 does not impair NFAT-dependent transactivation directly and instead exerts its effects at the level of calcineurin A activity.
Figure 4.
Atrogin-1 blocks calcineurin-dependent transcriptional responses and nuclear translocation of NFATc4 but does not inhibit a constitutively active form of NFAT in cardiomyocytes. (A) The calcineurin-dependent transcriptional response was measured in cardiomyocytes by cotransfection of a pIL2-Luc reporter plasmid and active calcineurin A (CnA*) together with atrogin-1 or the empty expression vector as control, and luciferase activity was measured. (B) A pIL2-Luc reporter plasmid and active calcineurin A together with plasmids expressing siRNA-atrogin-1 or siRNA-control were transfected in cardiomyocytes, and luciferase activity was measured. (C) An NFAT-dependent pIL2-Luc reporter plasmid, an expression vector encoding active NFAT (ΔNFAT), and atrogin-1 or the empty expression vector as control were transfected in cardiomyocytes, and luciferase activity was measured. (D) Cardiomyocytes transfected with the indicated plasmids were immunostained 36 hours after transfection with an antibody against NFATc4 (red), and nuclei were counterstained with DAPI (blue). (E) Cardiomyocytes transfected with the indicated plasmids were separated into cytoplasmic and nuclear fractions. Equal amounts of protein were analyzed by immunoblotting with NFATc4, GAPDH, and Oct1 antibodies. GAPDH and Oct1 served as cytoplasmic and nuclear markers, respectively. A representative blot is shown for each condition. CsA, cyclosporin A.
To extend this observation, we tested whether atrogin-1 alters nuclear translocation of NFAT, which occurs rapidly after dephosphorylation by calcineurin and is required for its activity. Neonatal cardiomyocytes were immunostained 36 hours after plasmid transfection with an antibody against NFATc4 (Figure 4D, red), and nuclei were counterstained with DAPI (Figure 4D, blue). In cells transfected with vector alone, NFATc4 was localized to the cytoplasm. Following stimulation with activated calcineurin A, NFATc4 translocated to the nucleus, as has been described previously (15, 16). Cotransfection with activated calcineurin A and atrogin-1 together completely blocked nuclear translocation of NFATc4, which was similar to the effects of the calcineurin A inhibitor cyclosporin A on NFAT mobilization. We measured transfected NFATc4 translocation quantitatively in cardiomyocytes by Western blot analysis to confirm that atrogin-1 selectively blocked calcineurin-dependent NFAT nuclear translocation (Figure 4E). These experiments indicate that atrogin-1 can inhibit calcineurin-dependent nuclear translocation of NFAT in cardiomyocytes, without having direct effects on NFAT or on other pathways that affect NFAT activity.
Atrogin-1 inhibits calcineurin-dependent cardiomyocyte hypertrophy.
Previous studies have shown that calcineurin is activated by hypertrophic agonists in cultured cardiomyocytes (4, 8). To determine directly whether atrogin-1 regulates the agonist-induced calcineurin signaling pathway that mediates cardiac hypertrophy, cardiomyocytes were infected with Ad-GFP or Ad-atrogin-1-GFP and subsequently treated with phenylephrine (PE) or FBS for 36 hours. Calcineurin phosphatase activity was increased approximately 5-fold by stimulation with PE and FBS in cardiomyocytes infected with Ad-GFP (Figure 5A). In comparison, Ad-atrogin-1-GFP infection completely blocked the increases in calcineurin activity. In addition, Western blotting demonstrated that Ad-atrogin-1-GFP infection reduced basal calcineurin A protein levels and repressed agonist-induced induction of calcineurin A protein in response to PE and FBS stimulation (Figure 5B).
Figure 5.
Ad-atrogin-1-GFP infection blocks agonist-induced cardiomyocyte hypertrophy. (A) Cardiomyocytes were infected with Ad-GFP or Ad-atrogin-1-GFP and were stimulated with PE (100 μM) or FBS for 36 hours. Calcineurin phosphatase activity was measured with 5 μg of cell extracts. (B) Cardiomyocytes were cultured and infected with adenoviruses as described above. The levels of expressed atrogin-1 and endogenous calcineurin A protein were determined by immunoblotting with anti-Myc or –calcineurin A antibodies, respectively. (C) Cardiomyocytes were cultured and infected with the indicated adenoviruses. Cells were stained with α-actinin antibody (red), and nuclei were stained with DAPI (blue). A representative field is shown for each condition. Magnification, ×200. (D) Quantitation of cell surface area in C (100–120 random cells measured in each group). #P < 0.001 vs. serum-free control; *P < 0.001 vs. Ad-GFP + PE; &P < 0.001 vs. Ad-GFP + FBS. (E) Cardiomyocytes were infected with Ad-siRNA-control or Ad-siRNA-atrogin-1 and were treated with PE for 36 hours. Cells were stained with α-actinin antibody. A representative field is shown for each condition. Magnification, ×200. (F) Quantitation of cell surface area (100–120 random cells measured in each group). #P < 0.001 vs. serum-free control; *P < 0.001 vs. Ad-siRNA-control + PE.
To investigate the effects of atrogin-1 on the morphologic changes induced by hypertrophic agents, cardiomyocytes were infected with Ad-GFP, Ad-atrogin-1-GFP, or no virus and stimulated with PE or FBS for 36 hours, fixed, and stained with anti–sarcomeric α-actinin antibody. Whereas either medium alone (Figure 5C, top) or infection with Ad-GFP (Figure 5C, middle) had no effect on agonist-induced hypertrophy, cardiomyocytes infected with Ad-atrogin-1-GFP repressed PE- or FBS-induced hypertrophy (Figure 5C, bottom). Measurement of cell surface area indicated that cells infected with Ad-atrogin-1-GFP did not increase in size after PE or FBS stimulation (Figure 5D). Furthermore, infection of cardiomyocytes with Ad-siRNA-atrogin-1 enhanced PE-induced cardiomyocyte hypertrophy (Figure 5E). Cell surface area was increased by 1.4-fold in Ad-siRNA-atrogin-1–infected cardiomyocytes compared with cells infected with Ad-siRNA-control (Figure 5F).
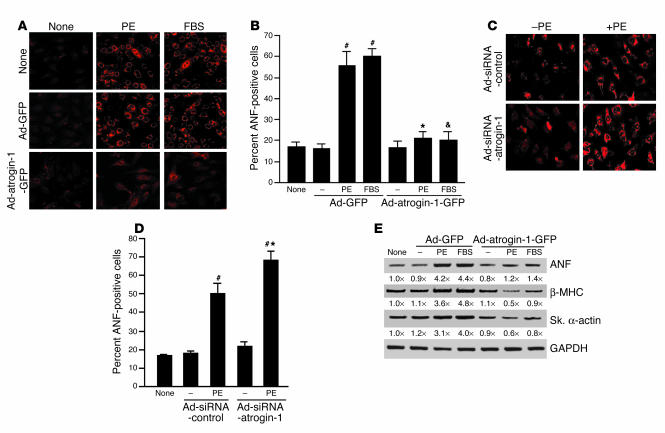
The preceding experiments suggest that inhibition of calcineurin activity by atrogin-1 is sufficient to inhibit agonist-induced hypertrophy in cardiomyocytes. To confirm that the effects of atrogin-1 on cell size reflect inhibition of the hypertrophic fetal gene expression profile in neonatal cardiomyocytes, cells were infected with either Ad-GFP or Ad-atrogin-1-GFP, stimulated by agonists, and immunostained with antibody against atrial natriuretic factor (ANF). Cardiomyocytes infected with Ad-atrogin-1-GFP did not appreciably increase ANF protein in response to stimulation by PE and FBS (Figure 6A, bottom), whereas ANF was abundantly expressed in cells conditioned with media alone or Ad-GFP (Figure 6A, top and middle). Quantitative analysis indicated that the number of ANF-positive cells was not increased above base line after PE and FBS treatment in cells treated with Ad-atrogin-1-GFP (Figure 6B). Conversely, cardiomyocytes infected with Ad-siRNA-atrogin-1 demonstrated a modest but statistically significant increase in ANF positivity in response to stimulation by PE (Figure 6, C and D). In addition, RT-PCR analysis demonstrated that atrogin-1 prevented induction of the mRNA for hypertrophy markers ANF, β–myosin heavy chain (β-MHC), and skeletal α-actin (Figure 6E). Taken together, these data indicate that atrogin-1 blocks calcineurin A–dependent, agonist-induced hypertrophy and prevents hypertrophic gene induction in vitro, which is consistent with its ability to inhibit calcineurin A activity and decrease calcineurin A protein levels.
Figure 6.
Ad-atrogin-1-GFP inhibits fetal gene expression in cardiomyocytes. (A) Cardiomyocytes were infected and were stimulated as indicated. Cells were fixed and stained with antibody against ANF (perinuclear red signal). A representative field is shown for each condition. Magnification, ×200. (B) Percentage of ANF-positive cells (100–120 random cells measured in each group). #P < 0.001 vs. serum-free control; *P < 0.001 vs. Ad-GFP + PE; &P < 0.001 vs. Ad-GFP + FBS. (C) Cardiomyocytes were infected with Ad-siRNA-control or Ad-siRNA-atrogin-1 and were stimulated with PE (100 μM) for 36 hours. Cells were fixed and stained with antibody against ANF. Magnification, ×200. (D) Percentage of ANF-positive cells (100–120 random cells measured in each group). #P < 0.001 vs. serum-free control; *P < 0.001 vs. Ad-siRNA-control + PE. (E) Cardiomyocytes were infected with adenoviruses and stimulated as indicated. The transcripts of ANF, β-MHC, and skeletal α-actin (Sk. α-actin) were measured by quantitative RT-PCR. ANF, skeletal α-actin, and β-MHC levels are shown relative to those in untreated, uninfected control cells and normalized to the level of GAPDH below each panel. A representative experiment is shown for each condition.
The SCFatrogin-1 complex has an E3 ubiquitin ligase activity and promotes ubiquitination of calcineurin A in vitro.
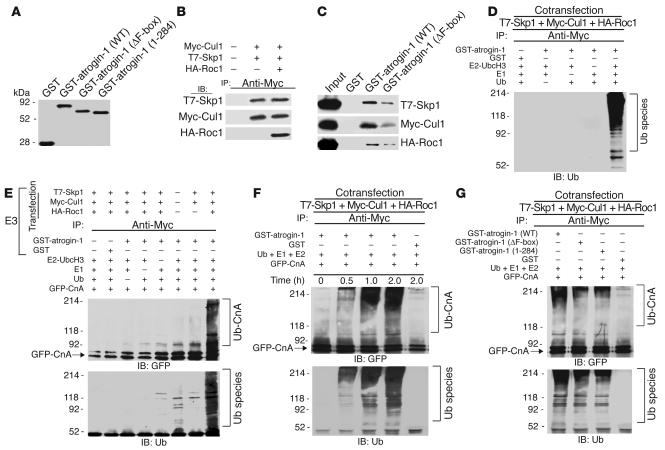
Atrogin-1 contains an F-box domain (amino acids 228–267), which is characteristic of proteins that are components of SCF ubiquitin ligase complexes (11, 12, 17). F-box proteins are adaptors that associate with Skp1, Cul1, and Roc1 of the SCF complex through the F-box and simultaneously directly bind and recruit proteins to be ubiquitinated (17). F-box proteins therefore provide substrate specificity to SCF complexes, and they are also the rate-limiting components for substrate ubiquitination. To determine whether atrogin-1 associates with other components of the SCF complex, we purified GST–atrogin-1 fusion proteins in bacteria (WT, lacking the F-box, or lacking the calcineurin-binding domain; Figure 7A) and tested formation of the Skp1-Cul1-Roc1 complex by coimmunoprecipitation of epitope-tagged proteins. Under these conditions, Myc-tagged Cul1 immunoprecipitated T7-tagged Skp1 and HA-tagged Roc1 (Figure 7B). GST pull-down assays demonstrated that Skp1, Cul1, and Roc1 proteins present in cell extracts bound efficiently to immobilized GST–atrogin-1 but not GST alone, and only weakly to a fusion protein lacking the F-box (Figure 7C). These observations extend previous reports (11, 12) and confirm that atrogin-1 incorporates into SCF complexes in an F-box–dependent fashion.
Figure 7.
Atrogin-1 participates in an SCFatrogin-1 complex that ubiquitinates calcineurin A in vitro. (A) GST–atrogin-1 (WT), atrogin-1 ΔF-box, and atrogin-1 1–284 were purified from bacteria. The purity of each purified protein was verified by SDS-PAGE and Coomassie staining. (B) Association of Skp1 and Roc1 with Cul1 was confirmed by cotransfection with T7-Skp1, Myc-Cul1, and HA-Roc1 in COS-7 cells. Equal amounts of cell extract were immunoprecipitated with Myc antibody and analyzed by immuoblotting with antibodies against T7 (Skp1, top), Myc (Cul1, middle), and HA (Roc1, bottom), respectively. (C) Interaction of atrogin-1 with Skp1, Cul1, and Roc1 was analyzed in in vitro GST pull-down assays. The ability of Skp1 (top), Cul1 (middle), and Roc1 (bottom) to bind GST, GST–atrogin-1, or GST–atrogin-1 ΔF-box was analyzed by immunoblotting. (D) The SCFatrogin-1 complex was evaluated for ubiquitin ligase activity in the presence of recombinant E1, UbcH3/CDC34 (E2 UbcH3), and ubiquitin (Ub) as indicated. (E) The SCFatrogin-1 complex was evaluated for its capacity to ubiquitinate purified GFP-tagged calcineurin A. After the in vitro ubiquitylation reaction, the samples were analyzed by immunoblotting with an anti-ubiquitin antibody to identify ubiquitinated products or with an anti-GFP antibody directed to calcineurin A to reveal ubiquitinated calcineurin A species. (F) Purified GFP-tagged calcineurin A was incubated with SCFatrogin-1 complex for the indicated times, and calcineurin A ubiquitination was examined by anti-GFP (top) or anti-ubiquitin (bottom) antibodies. (G) Purified GFP-tagged calcineurin A was incubated with SCFatrogin-1 complex containing atrogin-1 WT, the ΔF-box mutant, or the 1–284 mutant, and calcineurin A ubiquitination was examined by immunoblotting.
Ubiquitin ligases interact functionally with specific E2 ubiquitin-conjugating enzymes to facilitate substrate recognition and ubiquitin transfer (18). We tested the ability of the SCFatrogin-1 complex to assemble polyubiquitin chains in the presence of a panel of E2 ubiquitin-conjugating enzymes. We found that UbcH3/CDC34 was able to interact efficiently with the SCFatrogin-1 complex. However, other ubiquitin-conjugating enzymes — including UbcH5, UbcH4, E2-20, and E2-25 — were ineffective in facilitating SCFatrogin-1 complex–mediated polyubiquitin chain formation (data not shown). To determine whether atrogin-1–containing SCFatrogin-1 complexes have intrinsic ubiquitin ligase activity, we performed an in vitro ubiquitination assay with the immunoprecipitated Skp1-Cul1-Roc1 complex, purified GST–atrogin-1 or GST alone, recombinant E1, E2 (UbcH3/CDC34), and ubiquitin. SCFatrogin-1 complex–dependent multiubiquitin chain assembly (an established marker of ubiquitin ligase activity [ref. 18]) was readily detected and dependent on the presence of atrogin-1 and E1 and E2 enzymes (Figure 7D). Thus, atrogin-1 assembles a functional ubiquitin ligase complex in vitro.
To examine directly whether the SCFatrogin-1 complex can promote calcineurin A ubiquitination in vitro, we reconstituted a reaction using the immunoprecipitated Skp1-Cul1-Roc1 complex, GST–atrogin-1, and recombinant E1 and UbcH3. Reactions were performed with calcineurin A expressed as a GFP fusion in COS-7 cells. Ubiquitination of calcineurin A was evaluated by immunoblotting. A high–molecular weight species indicative of polyubiquitin chain assembly was detected in the presence of the SCFatrogin-1 complex, ubiquitin, E1, and UbcH3; Roc1 (the RING finger component of the reaction) dramatically enhanced the ubiquitination of calcineurin A (Figure 7E). Omitting ubiquitin, E1, UbcH3, or GST–atrogin-1 abolished formation of the polyubiquitin chain, and GFP alone was not efficiently ubiquitinated under these conditions (data not shown), which is consistent with previous observations that unmodified GFP is not a substrate for the ubiquitination machinery (19). More importantly, omission of the SCF complex also prevented calcineurin A polyubiquitination. Thus, the observed product in our assays represents the polyubiquitinated form of calcineurin A. These data suggest that an SCF complex containing atrogin-1 as an F-box component catalyzes polyubiquitination of calcineurin A.
Since efficient transfer of ubiquitin to calcineurin A requires the direct interaction of calcineurin A with the UbcH3/SCFatrogin-1 machinery, atrogin-1 mutants lacking the F-box or the calcineurin-binding site should be resistant to ubiquitination. To test this hypothesis, we engineered 2 mutant forms of atrogin-1, containing either a deletion of the entire F-box (atrogin-1 ΔF-box) or atrogin-1 1–284, which lacks the calcineurin A–binding site. In the presence of the SCFatrogin-1 complex, atrogin-1 dramatically enhanced the ubiquitination of calcineurin A in a time-dependent manner (Figure 7F). Moreover, deletion of the F-box or the calcineurin A–binding site markedly attenuated atrogin-1–mediated enhancement of calcineurin A polyubiquitination (Figure 7G, compare second and third lanes with first lane). Collectively, these data indicate that atrogin-1 functions as a limiting component of an F-box–dependent E3 ubiquitin ligase and that the F-box domain and calcineurin A–binding site (amino acids 284–345) of atrogin-1 are both required for calcineurin A ubiquitination.
Atrogin-1 transgenic mice are resistant to pressure overload–induced cardiac hypertrophy.
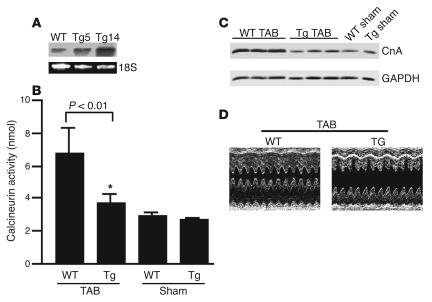
To determine whether atrogin-1 is also capable of repressing calcineurin A activity and cardiac hypertrophy in vivo, we created transgenic mice that overexpress atrogin-1 in the heart under the control of the α-myosin heavy chain (α-MHC) promoter (data not shown) (20). Two independently derived atrogin-1 founders (Tg5 and Tg14) contained approximately 3 and 15 copies, respectively, of the transgene as assessed by Southern blot analysis (data not shown). Northern blot analysis demonstrated that expression of the atrogin-1 transgene was restricted to the heart (data not shown), and expression of atrogin-1 in Tg5 and Tg14 was increased 4.4- and 12.2-fold, respectively, compared with that in nontransgenic mice (Figure 8A). (Because the transgenic lines showed a similar phenotype, we will only describe data from line Tg14.) Transgenic mice were phenotypically indistinguishable from their WT littermates, and none of the transgenic lines was characterized by premature lethality. There was no significant difference in heart weight/body weight ratios between WT and atrogin-1 transgenic mice, and echocardiography failed to reveal any abnormalities in cardiac function in transgenic mice up to 8 months of age (data not shown). This is not surprising, since calcineurin inhibitors such as cyclosporin A do not have significant effects on base-line cardiac function (1).
Figure 8.
Effects of overexpressed atrogin-1 on calcineurin A activity, protein level, and cardiac function. (A) Northern blot analysis of transgene expression. Ten micrograms of total RNA from heart of Tg5 and Tg14 (second and third lanes) and WT (first lane) was analyzed by Northern blotting with a cDNA probe corresponding to a 1.1-kb fragment containing the mouse atrogin-1 coding sequences. The bottom panel corresponds to the ethidium bromide staining of ribosomal 18S RNA of the same RNA sample separated on an identical gel. (B) Eight-week-old mice were subjected to thoracic aortic banding (TAB) or to sham surgery. Fourteen days later, mice were sacrificed, the hearts were freshly isolated from atrogin-1 transgenic (Tg) and nontransgenic control (WT) mice, and calcineurin A activity was determined (n = 7). *P < 0.001 vs. WT. (C) Western blot analysis of calcineurin A and GAPDH protein levels from hearts of WT and transgenic mice was measured 14 days after thoracic aortic banding or sham surgery. Numbers indicate the expression level of calcineurin A in aortic-banded WT and Tg hearts relative to sham-operated WT hearts, normalized to GAPDH. (D) M-mode echocardiographic analysis of hearts from Tg and WT mice.
We wondered whether these mice might be sensitized to hypertrophic stimuli based on our observations in vitro, as would be predicted if atrogin-1 is a suppressor of calcineurin A activity. Transgenic mice and age- and sex-matched WT littermates were subjected to aortic banding to induce pressure overload (or sham surgery) for 14 days. In response to aortic banding, calcineurin A activity was elevated about 3-fold in WT mice, but cardiac calcineurin A activity was only minimally increased in hearts from atrogin-1 transgenic mice (Figure 8B). Similarly, calcineurin A protein levels were increased by pressure overload in WT mice but were reduced by 70% in atrogin-1 transgenic mice (Figure 8C). These data confirm our in vitro observations and indicate that atrogin-1 inhibits calcineurin A activity and reduces its protein levels in vivo.
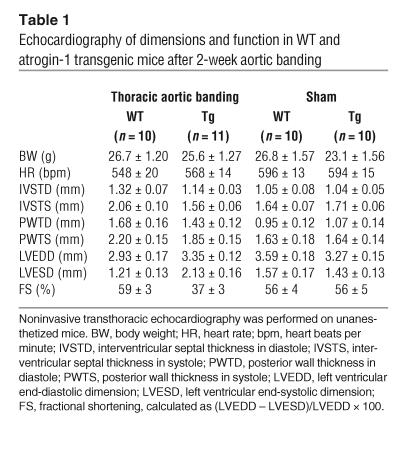
To assess cardiac function accurately, transthoracic echocardiography was performed. In the aortic-banded group, atrogin-1 transgenic mice developed left ventricular dilation, characterized by increased left ventricular end-systolic dimensions (Figure 8D; Table 1). In contrast, WT mice had reduced left ventricular dimensions and increased septal thickness 14 days after banding. Furthermore, atrogin-1 transgenic mice had decreased interventricular septal thickness in systole compared with nontransgenic mice and had a dramatic reduction in ejection fraction, as estimated by fractional shortening (37% versus 59% in WT mice) after aortic banding. No differences in echocardiographic parameters were detected in sham-treated mice in either group. These measurements indicate that atrogin-1 transgenic mice display left ventricular dilatation and reduced cardiac function rather than the expected ventricular hypertrophy after aortic banding.
Table 1.
Echocardiography of dimensions and function in WT and atrogin-1 transgenic mice after 2-week aortic banding
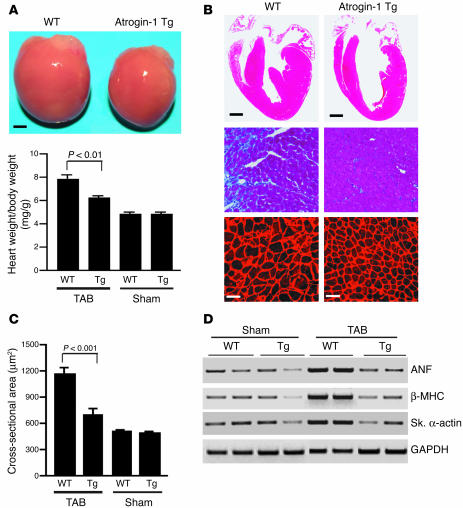
On gross postmortem examination, atrogin-1 transgenic mice did not develop cardiac hypertrophy after banding (Figure 9A, top) and had a 25% reduction in heart weight/body weight ratios compared with age-matched aortic-banded WT mice (Figure 9A, bottom). There were no significant differences in heart size or heart weight/body weight ratios between transgenic mice and nontransgenic mice after sham surgery. Histologic analysis of aortic-banded transgenic hearts revealed that the walls of the septum and left ventricular chambers were much thinner than in nontransgenic hearts (Figure 9B, top). Masson’s trichrome staining of heart sections from transgenic mice indicated reduced interstitial cell fibrosis (Figure 9B, middle), which is reminiscent of the reduction in collagen deposition observed in MCIP1-overexpressing hearts (21). Histologic sections were also stained with wheat germ agglutinin–TRITC to determine cross-sectional areas of cardiac myocytes (Figure 9B, bottom). Myocyte cross-sectional area was not significantly different between transgenic and nontransgenic mice in sham groups; however, the cross-sectional area of cardiac myocytes in nontransgenic hearts was significantly increased, whereas myocytes in transgenic hearts increased very little compared with those in sham-treated mice, with the result that the cross-sectional area of cardiac myocytes in aortic-banded transgenic hearts was 41% less than in controls (Figure 9C).
Figure 9.
Atrogin-1 blunts the hypertrophic response to pressure overload. (A) Representative heart sizes and heart weight/body weight ratios of atrogin-1 transgenic mice were compared with those of nontransgenic mice. Eight-week-old mice were subjected to thoracic aortic banding (TAB) or sham surgery. Fourteen days later, animals were sacrificed, the hearts were freshly isolated from transgenic (Tg) and nontransgenic (WT) mice, and heart weight/body weight ratios were determined (n = 7). Scale bar: 1 mm. (B) Representative macroscopic histologic analysis of H&E-stained hearts from indicated mice after 2 weeks of aortic banding is shown (top panels). Scale bar: 1 mm. Histologic sections were also stained with Masson’s trichrome to detect interstitial cell fibrosis in hearts (middle panels; magnification, ×200), and with wheat germ agglutinin–TRITC conjugate to determine cell size (bottom panels; scale bars: 50 μm). (C) Quantitation of myocyte cross-sectional areas from the indicated groups (n = 200 cells per section). (D) Analysis of hypertrophic markers. Total RNA was isolated from hearts of mice of the indicated genotype, and expression of transcripts for ANF, β-MHC, skeletal α-actin, and GAPDH was determined by slot blot analysis. A representative analysis is shown.
We also examined changes in fetal gene expression to determine whether atrogin-1 transgenic hearts have a stress-associated expression pattern. RT-PCR analysis was performed to quantify mRNA levels of hypertrophic genes from hearts of transgenic and nontransgenic mice after banding for 2 weeks (Figure 9D). Levels of cardiac ANF, β-MHC, and skeletal α-actin mRNA, which are typically induced during hypertrophy, were decreased by more than 55% in hearts from atrogin-1 transgenic mice compared with nontransgenic hearts. Collectively, these data indicate that atrogin-1 acts as a suppressor of the molecular program for cardiac hypertrophy in vivo, at least in part by suppressing calcineurin A protein expression and thus inhibiting calcineurin A activity. In response to pressure overload, transgenic hearts expressing elevated levels of atrogin-1 fail to increase wall thickness, thereby increasing wall stress and causing cardiac chamber enlargement.
Discussion
The principal finding of the present study is that the F-box protein atrogin-1 is capable of binding the calcineurin A–α-actinin-2 complex and inhibiting calcineurin A activity. Atrogin-1, which is structurally distinct from other proteins that inhibit calcineurin, is specifically expressed in heart and skeletal muscle and recruits components of the SCF ubiquitin ligase complex to promote ubiquitination of calcineurin. Consistent with these observations, transgenic overexpression of atrogin-1 in the heart attenuates induction of calcineurin A activity and protein level and blunts cardiac hypertrophy and the fetal gene expression program. Taken together, these observations demonstrate a new mechanism whereby calcineurin A activity may be regulated in a muscle-specific fashion to manipulate the response to hypertrophic stimuli and modulate cardiac function.
Atrogin-1 and inhibition of cardiomyocyte hypertrophy.
The present studies indicate that atrogin-1 attenuates the cardiac hypertrophic response with selectivity at the level of calcineurin signaling, insofar as atrogin-1 does not affect the ability of cardiomyocytes to respond to hypertrophic signaling events downstream of calcineurin (Figure 4C). Previous studies have demonstrated that the calcineurin-antagonist drugs cyclosporin A and FK506 can block hypertrophic responses in cultured cardiomyocytes and in animal models (reviewed in ref. 1). Similarly, endogenous calcineurin inhibitors including AKAP79, Cabin/Cain, and MCIP1 attenuate pathologic hypertrophy when overexpressed in cultured neonatal cardiomyocytes and in the heart (1, 21–23). Although mice with cardiac-specific overexpression of calcineurin-inhibitory proteins are in some cases protected from adverse effects of stress signaling and heart failure progression (21, 24, 25), these protein inhibitors are not all cardiac-specific in their endogenous expression, and some also may have other functions. For example, the calcineurin-inhibitory domain of Cabin/Cain directly inhibits the transcriptional activity of myocyte enhancer factor-2, suggesting an alternative mechanism whereby Cabin/Cain might inhibit cardiac hypertrophy (26). In addition, recent studies have identified dual effects of MCIP1 on calcineurin activity. Overexpression of MCIP1 can suppress calcineurin activity through direct interaction with the catalytic domain of the enzyme. However, mice lacking MCIP1 have either diminished or enhanced hypertrophic responses depending on the stimulus (22), and a 4-aminopyridine derivative that induces MCIP1 expression paradoxically stimulates cardiac hypertrophy in a calcineurin-dependent fashion (27). Atrogin-1 therefore represents a unique cardiac-specific mechanism for regulating calcineurin activity (although we do not discount the possibility that other hypertrophy-associated atrogin-1 substrates exist). By virtue of its ability to target calcineurin for degradation, atrogin-1 provides a rapid method to potently attenuate calcineurin activity and hypertrophic signaling both in vitro and in vivo.
Calcineurin-binding domains have been identified in Cabin/Cain and MCIP1, as well as in NFATs. All of these proteins bind calcineurin through a PXIXIT (or closely related) motif that interacts with the C-terminus of the catalytic domain of the enzyme (5). Interestingly, the calcineurin-binding domain of atrogin-1 (Figure 2) does not contain this motif and instead interacts through a cysteine/histidine-rich domain (residues 284–345). The presence of sequences with the potential for highly ordered structure is typical of protein recognition motifs in other F-box proteins. Interestingly, we observe that the effects of atrogin-1 on calcineurin activity and hypertrophy both in vitro and in vivo are most marked under conditions in which hypertrophy is stimulated, and atrogin-1 has only modest or no effects under physiologic conditions. This suggests that the activity of atrogin-1 is enhanced by hypertrophic stimulation. This may occur via agonist-induced posttranslational modifications, which are generally known to affect the affinity of F-box proteins for their substrates. Although the present studies do not directly address the factors that regulate association of atrogin-1 and calcineurin, signals such as phosphorylation may also regulate the stability of calcineurin under physiologic conditions (28).
Atrogin-1 and calcineurin regulation.
Atrogin-1 localizes to the Z-disc (Figure 1), which plays a pivotal role in diverse aspects of cardiac muscle structure and function. One of the major components of the Z-disc is α-actinin, which maintains the structural integrity of the sarcomere by cross-linking actin and titin molecules (29). Mutations in several proteins associated with α-actinin and/or the Z-disc, including muscle LIM protein (29), have been implicated in the development of cardiomyopathy. Furthermore, α-actinin interacts with several signaling molecules, such as protein kinase N (30), G protein–coupled receptor kinases (31), phospholipase D2 (32), and calcineurin (5), indicating that Z-disc proteins also link signaling pathways in the heart. The colocalization of atrogin-1 with calcineurin A and α-actinin-2 at the Z-disc satisfies a necessary condition to allow atrogin-1 to regulate calcineurin signaling via direct interactions.
Ubiquitin-dependent degradation requires a cascade of 3 enzymes that catalyze ubiquitin activation, conjugation, and ligation. Among these ubiquitination enzymes, the ubiquitin ligases provide the crucial elements of specificity that direct the formation of polyubiquitin chains and targeting of substrates (13). One of the best-characterized ligases is the SCF complex, in which Skp1 associates with the N-terminal domain of Cul1 and with the C-terminal domain of Roc1/Rbx1/Hrt1, a small RING finger protein (33). Through Skp1, Cul1 interacts with an F-box protein that in turn binds its substrate (34–36). As the substrate-specific component of these complexes, F-box–containing proteins are generally rate-limiting for protein degradation. To our knowledge, our studies provide the first evidence that atrogin-1 is capable of interacting with calcineurin A and inhibiting calcineurin-dependent transcriptional activity. Moreover, the SCFatrogin-1 complex has ubiquitin ligase activity and can target calcineurin A for ubiquitination and degradation. Ubiquitination via the SCFatrogin-1 complex therefore represents an additional mechanism for regulating calcineurin A protein levels and activity in cardiomyocytes. Our data (Figures 3 and 4) and those of others (37) indicate that changes in calcineurin activity are closely correlated with changes in calcineurin protein levels, and the SCFatrogin-1 complex provides one plausible mechanism to coordinate cues that determine calcineurin activity at this level.
Regulation of atrogin-1 activity.
Most of what is known about the regulation of atrogin-1 is based on studies in skeletal muscle. Atrogin-1, like the RING finger protein MuRF1, is highly inducible at the mRNA level by denervation and disuse (11, 12). Whether similar stimuli regulate atrogin-1 mRNA in the heart is not known, but it would make sense that factors that regulate cardiomyocyte size would be upstream in the pathway of atrogin-1 expression.
Recent studies implicate Foxo transcription factors as critical regulators of atrogin-1 expression in skeletal muscle (38, 39). In its hypophosphorylated, nuclear-localizing state, Foxo3 is a potent transcriptional activator of the atrogin-1 promoter. However, Foxo proteins can be phosphorylated by the IGF-1/PI3K/Akt pathway, which results in transcriptional inactivation of Foxo3. Because IGF-1 is a potent stimulus for skeletal muscle hypertrophy, coordinate regulation of atrogin-1 by this pathway provides a mechanism to determine skeletal muscle cell size through the balance of protein synthesis and degradation. Since IGF-1 also regulates some types of hypertrophy of the heart, it is logical to speculate that this same pathway may regulate cardiac atrogin-1 expression. Some data indicate that calcineurin- and IGF-1–dependent hypertrophy occurs via a distinct signaling pathway and reflects differences between pathologic and physiologic hypertrophy (23). Targeted degradation of calcineurin by the SCFatrogin-1 complex may therefore represent a common pathway for reversing hypertrophy of the heart, even if the prohypertrophic arms of the calcineurin and IGF-1 pathways are independent of one another.
Atrogin-1 and hypertrophy-induced damage.
Hearts from transgenic mice overexpressing atrogin-1 are functionally normal under physiologic conditions but fail to develop hypertrophy and instead undergo dilatation in response to pressure overload (Figures 8 and 9). It is remarkable that increasing atrogin-1 levels under physiologic conditions has no adverse consequences on cardiac function; this may indicate that signaling events that occur during cardiac stress are permissive for atrogin-1 activity. Although generally it is well accepted that inhibition of calcineurin-NFAT signaling blocks the hypertrophic response, the precise response has varied in different experimental systems. Pharmacologic inhibition of calcineurin activity with cyclosporin A at doses that are used clinically for immunosuppression has little effect on the hypertrophic response, whereas doses above those used in the clinical range inhibit pressure-induced hypertrophy without adversely compromising cardiac function (at least over a period of several weeks) (40). However, cyclosporin treatment also induces myocyte apoptosis after pressure overload in vivo (41), as does direct molecular inhibition of NFAT activity in vitro (42). Thus, our observations are generally consistent with the most severe phenotypes observed after calcineurin inhibition. That atrogin-1 both inhibits hypertrophy and induces stress-dependent left ventricular dysfunction may indicate that targeted degradation is an especially efficient means to inhibit calcineurin activity. It is also possible that the phenotypes of atrogin-1 transgenic mice are further modified by other substrates for the SCFatrogin-1 complex, or through other effects of atrogin-1 that are independent of its ubiquitin ligase activity. Ubiquitin ligases often control the ubiquitination of multiple substrates (43), so further work will be required to uncover additional substrates for atrogin-1, which may represent other arms in the cardiac hypertrophic signaling cascade.
Clinical implications.
To our knowledge, the present studies provide the first evidence of a pathophysiologic role for atrogin-1 in the heart and identify the first substrate for its ubiquitin ligase activity. While cardiac hypertrophy is thought to initially benefit the heart by maintaining left ventricular function in the setting of increased afterload, hypertrophy ultimately causes diastolic dysfunction of the heart and is a leading predictor of the development of sudden death and heart failure (44). In contrast, the clinical reversal of hypertrophy is a powerful indicator of event-free survival in patients at otherwise high cardiovascular risk (45). There are growing lines of evidence to indicate that pharmacologic or genetic overexpression of calcineurin inhibitors attenuates the development of cardiac hypertrophy. Whether inhibition of calcineurin activity is sufficient to blunt long-term changes in the heart in response to hypertrophic stimuli remains unclear, and further studies will be necessary to determine whether and how salutary effects can be distinguished from hemodynamically maladaptive responses. Nevertheless, a better understanding of how proteins such as atrogin-1 — a new repressor of calcineurin activity with tissue-specific expression — impact on cardiac pathophysiology may open new doors to specific therapies that affect cardiac size and function.
Methods
Yeast 2-hybrid analyses.
The C-terminus of atrogin-1 (nucleotides encoding amino acids 272–355) was PCR-amplified from a full-length mouse atrogin-1 cDNA clone (AF441120). This fragment, fused to the GAL4 DNA-binding domain in pGBKT7 vector, was used as bait in a 2-hybrid screen of a human heart Matchmaker cDNA library (BD Biosciences — Clontech). From this screen, we identified a cDNA encoding α-actinin-2, and that was characterized by sequencing using the BLAST algorithm.
Plasmid construction and antibodies.
The mouse atrogin-1 cDNA was isolated from a mouse cardiac cDNA library by PCR. The full-length and truncated forms of mouse atrogin-1 and calcineurin A were generated by PCR. The F-box deletion of atrogin-1 was constructed by megaprimer PCR amplification to delete amino acids 228–267 (46). The isolated PCR products were subcloned into mammalian expression plasmid pCMV-TBs or GST fusion protein expression plasmid pGEX-KG. A luciferase reporter plasmid, pIL2-Luc (containing an NFAT-responsive element, generously provided by Jeff Molkentin, Children’s Hospital Medical Center, Cincinnati, Ohio, USA), the β-gal reporter plasmid pCMV-lacZ, the mammalian calcineurin A and NFATc4 expression constructs, and plasmid pTB701 HA-tagged α-actinin-2 (a gift from Ju Chen, UCSD, La Jolla, California, USA) have been described previously (47–49). siRNA sequences (siRNA–atrogin-1, 5′-CTACGTAGTAAGGCTGTTG-3′; siRNA-control, 5′-GTGCGTTGCTAGTACCAAC-3′) were incorporated into 64-bp self-annealing oligonucleotides and cloned into pSIREN-RetroQ-ZsGreen (BD Biosciences — Clontech). The following primary and secondary antibodies were used: anti-HA (12CA5; Roche Diagnostics Corp.); anti-Xpress (Invitrogen Corp.); anti–sarcomeric α-actinin (EA-53), anti-GST (both from Sigma-Aldrich); anti–calcineurin A (clone 29; Transduction Laboratories); anti-Myc (9E10), anti-CnAα, anti-CnAβ (C-20), anti-NFATc4 (H-74), anti-Oct1 (C-21), anti–Akt1/2 (H-136), anti-JNK1 (C-17; all from Santa Cruz); anti-PKCδ (Upstate Group Inc.); anti-ANF (IHC9103; Peninsula Laboratories Inc.); anti–p38 MAPK, anti-ERK (both from Cell Signaling Technology Inc.); anti-ubiquitin, anti-GAPDH (both from Chemicon International Inc.); and anti-mouse– or anti-rabbit–conjugated antibodies (both from Invitrogen Corp.). Anti–atrogin-1 antibody was raised in rabbits against synthetic peptides corresponding to the N-terminal 13 amino acids of mouse atrogin-1 (SKTKTQYFHQEKW) and purified by affinity chromatography.
Cell culture and adenoviral infection.
COS-7 cells were cultured in DMEM supplemented with 10% FBS. COS-7 cells were transiently transfected using FuGENE (Roche Diagnostics Corp.) as previously described (18). Neonatal rat cardiomyocytes were isolated by enzymatic disassociation of 1- to 2-day-old neonatal rat hearts with the Neonatal Cardiomyocyte Isolation System (Worthington Biochemical Corp.). Eighteen hours after plating, cells were infected with adenovirus expressing atrogin-1 for 2 hours and subsequently cultured for 24 hours before stimulation. To induce the hypertrophic response, PE or FBS (1%) was added to cultures in serum-free media for 36 hours. When indicated, cyclosporin A (Sigma-Aldrich) was present at 1 μM. Recombinant adenoviruses expressing GFP alone (Ad-GFP), Myc-tagged atrogin-1 and GFP (Ad-atrogin-1-GFP), siRNA-control, or siRNA-atrogin-1 driven by the cytomegalovirus promoter were generated using the AdEasy system (MP Biomedicals Inc.) as described previously (50).
Immunocytochemistry.
Neonatal rat cardiomyocytes were processed for immunofluorescence as described previously (4). The visualization of cardiomyocyte size, sarcomeric organization, perinuclear ANF expression, and NFATc4 translocation was performed by immunostaining with appropriate primary and secondary antibodies. Fluorescent images were collected on an epifluorescence microscope (Eclipse E800; Nikon Inc.). Quantitation of cardiomyocyte cell surface area was performed on digitized images using NIH Image software. One hundred to one hundred twenty cardiomyocytes in 15–20 fields were examined in 3 independent experiments. To identify nuclei in some experiments, fixed cells were incubated in DAPI stain (10 μg/ml) for 5 minutes at room temperature before the final wash steps.
Immunoprecipitation and immunoblotting.
COS-7 cells were cotransfected with expression vectors for Xpress-tagged atrogin-1, HA–α-actinin-2, and/or GFP–calcineurin A using FuGENE 6 (Roche Diagnostics Corp.). Immunoprecipitation was performed as described previously (51). Tagged proteins were immunoprecipitated for 1–2 hours at 4°C, using protein A/G agarose and 2 μg of the appropriate antibody (anti-Xpress, anti-HA, anti-GFP). The beads were washed and fractionated by 10% SDS-PAGE followed by immunoblotting as previously described (51).
GST pull-down assays.
Purified GST-tagged proteins were prepared as previously described (51). Five micrograms of each purified GST fusion protein or GST protein alone was added to 500 μg of cell extracts from COS-7 cells, and incubated in GST lysis buffer (20 mM Tris-HCl, pH 7.4, 0.5 mM DTT, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, plus protease inhibitors) for 1 hour at 4°C. After addition of 25 μl of glutathione–Sepharose 4B, the binding reaction was continued for an additional 45 minutes at 4°C. The bound beads were washed 4 times with the same lysis buffer and analyzed by SDS-PAGE.
In vitro ubiquitination reactions.
Skp1, Cul1, and Roc1 immunocomplexes were precipitated from transfected COS-7 cells with 2 μg of anti-Myc antibody as described previously (52). Individual immunocomplexes were immobilized with Myc antibody on agarose G beads and washed 3 times with lysis buffer containing 50 mM Tris-HCl (pH 7.4), 50 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM DTT, 10 mM NaF, and 10% glycerol. Washed immunocomplexes were added to a ubiquitination reaction (final volume 30 μl) containing 50 mM Tris-HCl (pH 7.4), 5 mM MgCl, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 60 ng of E1, 600 ng of UbcH3, 1 μg of purified GST–atrogin-1, 1 μg purified calcineurin A, and 10 μg of ubiquitin. Reactions were incubated at 30°C for 2 hours unless otherwise indicated, terminated by boiling for 5 minutes with SDS–sample buffer containing 0.1 M DTT, and resolved by SDS-PAGE, followed by immunoblotting with appropriate antibodies.
Reporter gene assays.
NFAT-driven luciferase activity assay was performed using the Luciferase Assay System (Promega Corp.). Briefly, neonatal cardiomyocytes were transiently transfected with expression vectors carrying atrogin-1 and luciferase reporter constructs (5 μg) using LipofectAMINE PLUS (Invitrogen Corp.). Cells were harvested 40 hours after transfection and analyzed for luciferase activity. The data represent the mean ± SEM of 3 independent experiments in duplicate and are corrected for differences in transfection efficiency by cotransfection with pCMV-β-gal and normalization for β-gal activity.
Calcineurin phosphatase activity assay.
Endogenous calcineurin phosphatase activity was measured with the QuantiZyme Assay System (BIOMOL Research Laboratories Inc.). Cardiomyocytes were infected with Ad-atrogin-1-GFP or Ad-GFP for 24 hours and treated with PE or FBS. Calcineurin phosphatase activity was measured colorimetrically by detection of free phosphate released from the calcineurin-specific RII phosphopeptide using the BIOMOL green reagent (BIOMOL Research Laboratories Inc.). Assays were performed in duplicate at each time point in 3 separate experiments.
Generation of transgenic mice.
Transgenic mice expressing atrogin-1 in the heart were created by cloning of a cDNA fragment encoding full-length mouse atrogin-1 downstream of the murine α-MHC promoter (gift of Jeffrey Robbins, Children’s Hospital Medical Center, Cincinnati, Ohio, USA). After purification, the resulting construct was used for pronuclear microinjection. At 3 weeks of age, a 1-cm portion of tail was removed from offspring for DNA analysis. The potentially transgenic mice were screened twice by PCR using the following primers: forward, 5′-CATATGGGATGGGATATAAAGGGGCTGGAGCAC-3′; reverse, 5′-CCACTTTTCTTGATGGAAGTACTGAGTTTTGGT-3′. Founders were confirmed by Southern blot analysis of tail DNA. EcoRI- or BamHI-digested genomic DNA was transferred to nitrocellulose membranes and hybridized to a 1.1-kb radiolabeled fragment containing exons 2 and 3 from the 5′ untranslated region of the murine α-MHC gene and a portion of atrogin-1 or a 1.1-kb fragment of atrogin-1 cDNA. In all experiments performed in this study, age- and sex-matched nontransgenic littermate mice were used for comparison with the atrogin-1 transgenic mice.
RNA analysis.
Total RNA was purified from cultured cells or intact hearts with Trizol (Invitrogen Corp.). For RT-PCR, hypertrophic marker gene analysis of ANF, β-MHC, skeletal α-actin, and GAPDH (as a control) was performed with primers designed to detect mouse gene products as described previously (53). Northern blot was performed as previously described (51). Blots were hybridized with a [γ-32P]ATP-labeled oligonucleotide probe or a [α-32P]dCTP-labeled fragment of the mouse atrogin-1 cDNA.
Transverse aortic constriction surgery and echocardiography.
Transverse thoracic aortic constriction was performed as described previously (54). Eight-week-old WT and atrogin-1 transgenic mice were anesthetized with isoflurane. A 7.0 nylon suture ligature was tied against a 27-gauge needle at the transverse aorta to produce a 65–70% constriction following removal of the needle. Doppler analysis was performed to ensure that physiologic constriction of the aorta was induced. Fourteen days after surgery, echocardiography was performed with a Vevo 660 ultrasound system (VisualSonics Inc.) equipped with a 30-MHz transducer. All procedures were approved by and performed in accordance with the University of North Carolina Institutional Animal Care and Use Committee.
Histology and microscopy.
Histologic analyses of tissues were performed according to standard protocols (37). Hearts from WT and transgenic mice were collected at the indicated times, fixed overnight in 10% formalin, and embedded in paraffin. Serial 5-μm heart sections from each group were analyzed. Samples were stained with H&E for routine histologic examination, Masson’s trichrome for collagen, and wheat germ agglutinin–TRITC conjugate to identify sarcolemmal membranes so that myofiber diameter could be quantified.
Statistical analysis.
Data are presented as means ± SEM. Differences between groups were evaluated for statistical significance using Student’s t test. P values less than 0.05 were regarded as significant.
Acknowledgments
This study was supported by grants from the NIH (HL65619, HL072347). C. Patterson is an Established Investigator of the American Heart Association and a Burroughs Wellcome Fund Clinical Scientist in Translational Research. We thank Yue Xiong, Jeff Molkentin, Jeffrey Robbins, Yoshitaka Ono, and Manabu Furukawa for their generous gifts of plasmids and antibodies, and Yaxu Wu and Wei-Bing Xing for assistance with luciferase measurement.
Footnotes
Nonstandard abbreviations used: AKAP79, A kinase–anchoring protein 79; ANF, atrial natriuretic factor; GST, glutathione-S-transferase; HA, hemagglutinin; MCIP1, modulatory calcineurin–interacting protein 1; MHC, myosin heavy chain; NFAT, nuclear factor of activated T cells; PE, phenylephrine; siRNA, small interfering RNA.Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Molkentin JD. Calcineurin and beyond: cardiac hypertrophic signaling. Circ. Res. 2000;87:731–738. doi: 10.1161/01.res.87.9.731. [DOI] [PubMed] [Google Scholar]
- 2.Vakili BA, Okin PM, Devereux RB. Prognostic implications of left ventricular hypertrophy. Am. Heart J. 2001;141:334–341. doi: 10.1067/mhj.2001.113218. [DOI] [PubMed] [Google Scholar]
- 3.Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–558. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 4.Molkentin JD, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14632–14637. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bueno OF, et al. Impaired cardiac hypertrophic response in Calcineurin Abeta-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4586–4591. doi: 10.1073/pnas.072647999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Windt LJ, et al. Targeted inhibition of calcineurin attenuates cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3322–3327. doi: 10.1073/pnas.031371998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taigen T, De Windt LJ, Lim HW, Molkentin JD. Targeted inhibition of calcineurin prevents agonist-induced cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1196–1201. doi: 10.1073/pnas.97.3.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coghlan VM, et al. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 10.Vega RB, Yang J, Rothermel BA, Bassel-Duby R, Williams RS. Multiple domains of MCIP1 contribute to inhibition of calcineurin activity. J. Biol. Chem. 2002;277:30401–30407. doi: 10.1074/jbc.M200123200. [DOI] [PubMed] [Google Scholar]
- 11.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 12.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson PK, et al. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 14.Jackson PK, Eldridge AG. The SCF ubiquitin ligase: an extended look. Mol. Cell. 2002;9:923–925. doi: 10.1016/s1097-2765(02)00538-5. [DOI] [PubMed] [Google Scholar]
- 15.Delling U, et al. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol. Cell. Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rooij E, et al. Requirement of nuclear factor of activated T-cells in calcineurin-mediated cardiomyocyte hypertrophy. J. Biol. Chem. 2002;277:48617–48626. doi: 10.1074/jbc.M206532200. [DOI] [PubMed] [Google Scholar]
- 17.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 19.Lindsten K, Menendez-Benito V, Masucci MG, Dantuma NP. A transgenic mouse model of the ubiquitin/proteasome system. Nat. Biotechnol. 2003;21:897–902. doi: 10.1038/nbt851. [DOI] [PubMed] [Google Scholar]
- 20.Subramaniam A, et al. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- 21.van Rooij E, et al. MCIP1 overexpression suppresses left ventricular remodeling and sustains cardiac function after myocardial infarction. Circ. Res. 2004;94:e18–e26. doi: 10.1161/01.RES.0000118597.54416.00. [DOI] [PubMed] [Google Scholar]
- 22.Vega RB, Bassel-Duby R, Olson EN. Control of cardiac growth and function by calcineurin signaling. J. Biol. Chem. 2003;278:36981–36984. doi: 10.1074/jbc.R300023200. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins BJ, et al. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ. Res. 2004;94:110–118. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 24.Esposito G, et al. Genetic alterations that inhibit in vivo pressure-overload hypertrophy prevent cardiac dysfunction despite increased wall stress. Circulation. 2002;105:85–92. doi: 10.1161/hc0102.101365. [DOI] [PubMed] [Google Scholar]
- 25.Hill JA, et al. Targeted inhibition of calcineurin in pressure-overload cardiac hypertrophy. Preservation of systolic function. J. Biol. Chem. 2002;277:10251–10255. doi: 10.1074/jbc.M110722200. [DOI] [PubMed] [Google Scholar]
- 26.Youn HD, Sun L, Prywes R, Liu JO. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286:790–793. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 27.Bush E, et al. A small molecular activator of cardiac hypertrophy uncovered in a chemical screen for modifiers of the calcineurin signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2870–2875. doi: 10.1073/pnas.0308723101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto Y, Soderling TR. Regulation of calcineurin by phosphorylation. Identification of the regulatory site phosphorylated by Ca2+/calmodulin-dependent protein kinase II and protein kinase C. J. Biol. Chem. 1989;264:16524–16529. [PubMed] [Google Scholar]
- 29.Knoll R, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 30.Mukai H, et al. Interaction of PKN with alpha-actinin. J. Biol. Chem. 1997;272:4740–4746. doi: 10.1074/jbc.272.8.4740. [DOI] [PubMed] [Google Scholar]
- 31.Freeman JL, Pitcher JA, Li X, Bennett V, Lefkowitz RJ. α-Actinin is a potent regulator of G protein-coupled receptor kinase activity and substrate specificity in vitro. FEBS Lett. 2000;473:280–284. doi: 10.1016/s0014-5793(00)01543-x. [DOI] [PubMed] [Google Scholar]
- 32.Park JP, et al. Amplification of the MLL region in acute myeloid leukemia. Cancer Genet. Cytogenet. 2000;121:198–205. doi: 10.1016/s0165-4608(00)00256-9. [DOI] [PubMed] [Google Scholar]
- 33.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 34.Bai C, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 35.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 36.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 37.Lim HW, et al. Calcineurin expression, activation, and function in cardiac pressure-overload hypertrophy. Circulation. 2000;101:2431–2437. doi: 10.1161/01.cir.101.20.2431. [DOI] [PubMed] [Google Scholar]
- 38.Stitt TN, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 39.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- 41.Yang G, et al. Cyclosporine reduces left ventricular mass with chronic aortic banding in mice, which could be due to apoptosis and fibrosis. J. Mol. Cell. Cardiol. 2001;33:1505–1514. doi: 10.1006/jmcc.2001.1413. [DOI] [PubMed] [Google Scholar]
- 42.Pu WT, Ma Q, Izumo S. NFAT transcription factors are critical survival factors that inhibit cardiomyocyte apoptosis during phenylephrine stimulation in vitro. Circ. Res. 2003;92:725–731. doi: 10.1161/01.RES.0000069211.82346.46. [DOI] [PubMed] [Google Scholar]
- 43.Koepp D, Harper J, Elledge S. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 44.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Engl. J. Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 45.Verdecchia P, et al. Prognostic significance of serial changes in left ventricular mass in essential hypertension. Circulation. 1998;97:48–54. doi: 10.1161/01.cir.97.1.48. [DOI] [PubMed] [Google Scholar]
- 46.Burke E, Barik S. Megaprimer PCR: application in mutagenesis and gene fusion. Methods Mol. Biol. 2003;226:525–532. doi: 10.1385/1-59259-384-4:525. [DOI] [PubMed] [Google Scholar]
- 47.Chin ER, et al. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 49.Rothermel B, et al. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- 50.Dai Q, et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ballinger CA, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohta T, Xiong Y. Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases. Cancer Res. 2001;61:1347–1353. [PubMed] [Google Scholar]
- 53.Jones WK, et al. Ablation of the murine alpha myosin heavy chain gene leads to dosage effects and functional deficits in the heart. J. Clin. Invest. 1996;98:1906–1917. doi: 10.1172/JCI118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill JA, et al. Cardiac hypertrophy is not a required compensatory response to short-term pressure overload. Circulation. 2000;101:2863–2869. doi: 10.1161/01.cir.101.24.2863. [DOI] [PubMed] [Google Scholar]