Abstract
Evasion of apoptosis is a hallmark of cancer, but the molecular circuitries of this process are not understood. Here we show that survivin, a member of the inhibitor of apoptosis gene family that is overexpressed in cancer, exists in a novel mitochondrial pool in tumor cells. In response to cell death stimulation, mitochondrial survivin is rapidly discharged in the cytosol, where it prevents caspase activation and inhibits apoptosis. Selective targeting of survivin to mitochondria enhances colony formation in soft agar, accelerates tumor growth in immunocompromised animals, and abolishes tumor cell apoptosis in vivo. Therefore, mitochondrial survivin orchestrates a novel pathway of apoptosis inhibition, which contributes to tumor progression.
Introduction
Deregulation of apoptosis is thought to invariably occur in human cancer, and to facilitate the acquisition of deleterious cancer traits (1), including loss of tumor suppressor genes, angiogenic changes, and immortalization (2). Among the regulators of apoptosis implicated in cancer cell survival, Bcl-2 family proteins (3) act at the mitochondrion (4) to control the release of apoptogenic proteins, notably cytochrome c and Smac (5), whereas members of the inhibitor of apoptosis (IAP) gene family function as endogenous inhibitors of caspases (6), the enzymatic effectors of apoptosis (7).
Survivin is a bifunctional IAP that has been implicated in protection from apoptosis and regulation of mitosis (8). Although an essential role of survivin at cell division has been linked to centrosomal function (9), metaphase and anaphase microtubule assembly (10, 11), and spindle checkpoint regulation (12), the mechanism by which survivin inhibits apoptosis has remained elusive (8). In particular, it has been debated whether survivin has a genuine function in apoptosis inhibition independently of its role as a master regulator of mitosis, and whether this is important in disease pathogenesis. Validating a cytoprotective mechanism of survivin has become a priority because of the dramatic exploitation of this pathway in human tumors (13), its frequent association with unfavorable disease outcome (14, 15), and the recent identification of molecular antagonists of survivin that are approaching clinical testing in cancer patients (8).
In this study, we asked whether subcellular compartmentalization of survivin could participate in cytoprotection, and whether this pathway has a direct role in tumor establishment and progression. We identified a novel pool of survivin that localizes to mitochondria and is rapidly released in the cytosol in response to apoptotic stimuli. In turn, mitochondrially released survivin suppresses caspase processing and enhances tumorigenesis in vivo.
Results
Identification of mitochondrial survivin.
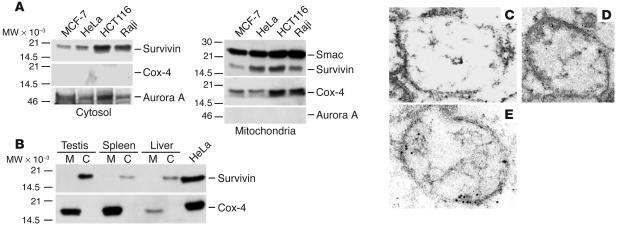
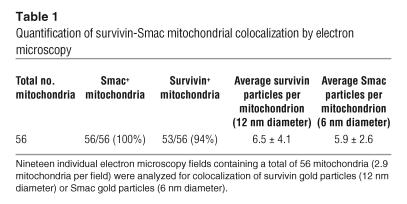
Because of its distribution among multiple subcellular compartments (16), we investigated whether survivin could also localize to mitochondria, an organelle with a central role in cell death pathways (4). A pool of survivin was found in purified cytosolic extracts of various tumor cell lines (Figure 1A), in agreement with previous observations (16). In addition, a pool of survivin was present in isolated mitochondrial fractions of tumor cells and colocalized with the reactivity of the mitochondrial proteins Smac and Cox-4, but not cytoplasmic Aurora A (Figure 1A). Next, we asked whether mitochondrial localization of survivin was a feature of transformed cells or was also observed in normal tissues. For these experiments, we prepared extracts from normal tissues known to express endogenous survivin and analyzed its distribution in cytosolic or mitochondrial fractions. Cytosolic extracts isolated from normal testis, liver, and spleen contained endogenous survivin, by immunoblotting (Figure 1B). In contrast, survivin was not detected in isolated mitochondrial fractions of the same tissues (Figure 1B). In immunoelectron microscopy experiments, analysis of MCF-7 cell pellets with a nonimmune IgG revealed negligible accumulation of gold particles (Figure 1C). In contrast, analysis of whole cell pellets (Figure 1D) or isolated mitochondrial fractions (Figure 1E) revealed staining for survivin in close proximity to the mitochondrial membrane(s), and colocalization with the reactivity of an antibody to Smac (Figure 1E). Quantification of 19 individual electron microscopy fields containing an average of 2.9 mitochondria each revealed that 94% of mitochondria examined exhibited dual localization of survivin and Smac, with comparable accumulation of gold particles per each marker (Table 1).
Figure 1.
Mitochondrial localization of survivin. (A) Survivin localization in tumor cell lines. Cytosolic (left panel) or mitochondrial (right panel) extracts from the indicated tumor cell lines were analyzed by immunoblotting. MW, molecular weight. (B) Subcellular fractionation of normal tissues. Mitochondrial (M) or cytosolic (C) fractions extracted from normal testis, spleen, and liver or unfractionated HeLa cell extracts were analyzed by immunoblotting. Cox-4 was used as a mitochondrial marker. (C–E) Immunoelectron microscopy. Mitochondrial pellets (C and E) isolated from MCF-7 cells or whole MCF-7 cell extracts (D) were stained with nonimmune IgG (C) or an antibody to survivin (D) followed by colloidal gold-conjugated secondary IgG. Isolated mitochondrial pellets (E) were simultaneously stained with antibodies to survivin (12 nm diameter gold particles) and Smac (6 nm diameter gold particles). Magnification, ×53,000 (C and D), ×104,000 (E).
Table 1.
Quantification of survivin-Smac mitochondrial colocalization by electron microscopy
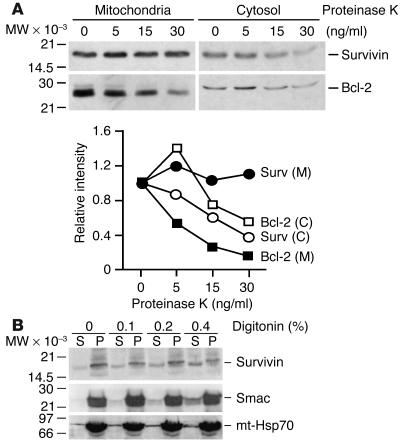
We next used a biochemical approach to map more precisely the submitochondrial topography of survivin. Treatment of mitochondrial pellets with proteinase K resulted in concentration-dependent loss of Bcl-2 (Figure 2A), which localizes to the outer mitochondrial membrane (3). In contrast, proteinase K treatment did not reduce survivin levels in mitochondria (Figure 2A). Conversely, proteinase K digested both Bcl-2 and survivin in isolated cytosolic extracts, confirming comparable susceptibility of both proteins to proteolysis (Figure 2A). We next permeabilized the outer mitochondrial membrane with digitonin (17) and looked for potential changes in survivin distribution. Digitonin treatment resulted in concentration-dependent relocalization of survivin from mitochondrial pellets into supernatants (Figure 2B). This was also associated with release of Smac (18) from mitochondrial pellets into supernatants, whereas the localization of nonextractable mitochondrial Hsp70 was unaffected by digitonin treatment (Figure 2B). Collectively, these data demonstrate that a novel pool of survivin localizes to mitochondria in tumor cells but not in normal tissues and is compartmentalized in the intermitochondrial membrane space.
Figure 2.
Topography of mitochondrial survivin. (A) Sensitivity to proteinase K. Mitochondrial or cytosolic fractions were treated with the indicated concentrations of proteinase K and analyzed by immunoblotting. Lower panel: Quantification of Bcl-2 or survivin proteolysis by proteinase K treatment. (B) Permeabilization of mitochondrial membrane. Mitochondrial fractions were treated with the indicated increasing concentrations of digitonin, and pellets (P) or supernatants (S) were analyzed by immunoblotting. mt-Hsp70, mitochondrial Hsp70.
Stress-response modulation of mitochondrial survivin.
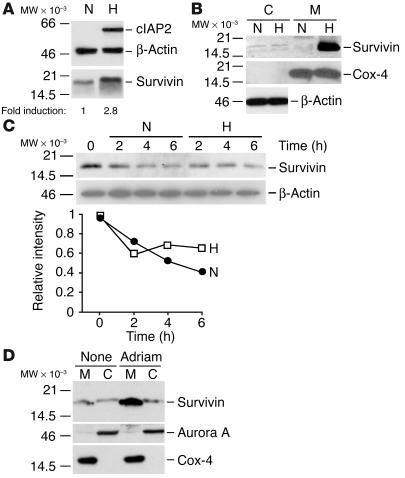
We next asked whether cellular stress stimuli influenced the expression and/or localization of the mitochondrial pool of survivin. Exposure of HeLa cells to hypoxia (∼1% O2) caused a dramatic upregulation of the IAP family protein cIAP2 (Figure 3A), in agreement with previous observations (19). Hypoxia also increased survivin levels approximately threefold, by immunoblotting (Figure 3A). Analysis of isolated subcellular fractions demonstrated that hypoxia-induced upregulation of survivin occurred exclusively in the mitochondrial pool with minimal changes in cytosolic survivin levels (Figure 3B). To begin to determine the mechanism(s) of survivin modulation by hypoxia, we examined changes in protein levels in cycloheximide block experiments. Cycloheximide treatment resulted in time-dependent turnover of endogenous survivin levels in HeLa cells, by immunoblotting (Figure 3C). Conversely, hypoxic HeLa cells exhibited stabilization of survivin levels over a 6-hour time interval, as compared with normoxic cultures (Figure 3C). In control experiments, no significant decrease of cell viability was observed 6 hours after hypoxia (data not shown). Finally, treatment of MCF-7 cells with nonapoptotic concentrations of the DNA-damaging agent adriamycin also resulted in selective expansion of mitochondrial survivin, with no significant changes in cytosolic survivin levels (Figure 3D).
Figure 3.
Modulation of mitochondrial survivin during the cellular stress response. (A) Regulation of survivin by hypoxia. HeLa cells were exposed to hypoxia, and analyzed by immunoblotting followed by densitometry. N, normoxic cultures; H, hypoxic cultures. (B) Subcellular fractionation. Cytosolic or mitochondrial fractions from HeLa cells were exposed to hypoxia, and analyzed by immunoblotting. (C) Cycloheximide block. Normoxic or hypoxic HeLa cells were treated with cycloheximide, and analyzed by immunoblotting at the indicated time intervals. Lower panel: β-Actin–normalized densitometric quantification of differential survivin stability in control versus hypoxic conditions. (D) Modulation by DNA damage. Untreated (None) or MCF-7 cells treated with nonapoptotic concentrations of adriamycin (Adriam) were fractionated in cytosolic and mitochondrial fractions, and analyzed by immunoblotting.
Loss of mitochondrial survivin influences cell death.
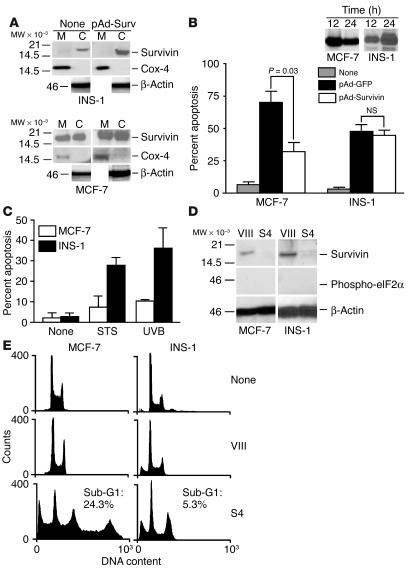
To begin to probe the functional role of mitochondrial survivin, we studied a rat insulinoma cell line, INS-1, in which survivin was found almost exclusively in the cytosol, but not in purified mitochondrial fractions (Figure 4A). When transduced with a replication-deficient adenovirus (pAd) encoding WT survivin (pAd-Survivin), INS-1 cells exhibited increased survivin expression in the cytosol, but not in mitochondria (Figure 4A). Conversely, MCF-7 cells expressed endogenous survivin in cytosolic and mitochondrial fractions, and both pools were coordinately increased by transduction with pAd-Survivin (Figure 4A). Transduction of MCF-7 cells with pAd-Survivin inhibited staurosporine-induced cell death by 65–70% (Figure 4B). In contrast, pAd-Survivin did not reverse apoptosis in INS-1 cells (Figure 4B). In control experiments, both INS-1 and MCF-7 cells expressed comparable levels of transduced survivin (Figure 4B, inset).
Figure 4.
Participation of mitochondrial survivin in cell death. (A) Defective mitochondrial import of survivin in INS-1 cells. INS-1 (upper panel) or MCF-7 (lower panel) cells were left untreated (None) or transduced with pAd-Survivin, and isolated mitochondrial or cytosolic fractions were analyzed by immunoblotting. (B) Cytoprotection. INS-1 or MCF-7 cells were transduced with pAd-GFP or pAd-Survivin, treated with staurosporine, and analyzed for hypodiploid DNA content. Data are the mean ± SEM of 3 independent experiments. Inset: Immunoblotting of survivin in transduced INS-1 or MCF-7 cells at the indicated time intervals. (C) Differential sensitivity to apoptosis. INS-1 or MCF-7 cells were treated with suboptimal concentrations of staurosporine (STS) or UVB and analyzed for nuclear morphology of apoptosis by DAPI staining. Data are the mean ± SD of 2 independent experiments. (D) RNAi knock-down. INS-1 or MCF-7 cells were transfected with dsRNA oligonucleotides to survivin (S4) or control (VIII), and analyzed by immunoblotting. (E) Cell cycle analysis. INS-1 or MCF-7 cells transfected with control or survivin dsRNA oligonucleotides were analyzed for cell cycle distribution by flow cytometry. The percentage of cells with hypodiploid (Sub-G1) DNA content is indicated.
To determine whether the lack of mitochondrial survivin resulted in increased sensitivity to apoptosis, we challenged INS-1 or MCF-7 cells with suboptimal doses of cell death stimuli. Concentrations of staurosporine (0.1 μM) or UVB (50 joules per square meter [J/m2]) that did not reduce the viability of MCF-7 cells resulted in extensive apoptosis of INS-1 cells (Figure 4C). In reciprocal experiments, we used RNA interference (RNAi) to acutely knock down survivin levels in INS-1 or MCF-7 cells and analyzed apoptosis and cell cycle progression (20). A survivin-derived double-stranded RNA (dsRNA) oligonucleotide (S4) ablated survivin levels in cytosolic and nuclear fractions of both MCF-7 and INS-1 cells, whereas a control dsRNA sequence (VIII) was without effect (Figure 4D and data not shown) (20). In control experiments, no phosphorylation of eIF2α was observed in RNAi-treated cells (Figure 4D). RNAi ablation of survivin in MCF-7 cells resulted in a dual phenotype of apoptosis and mitotic defects with polyploidy, by DNA content analysis and flow cytometry (Figure 4E), in agreement with previous observations (20). Although loss of survivin in INS-1 cells also caused mitotic defects predominantly seen as a G2/M block, this was not associated with significantly enhanced cell death, as compared with death of cells transfected with control dsRNA (VIII) (Figure 4E).
Mitochondrial survivin inhibits apoptosis.
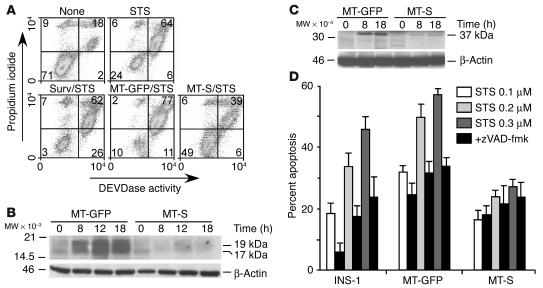
To further investigate a cytoprotective function of mitochondrial survivin, we engineered INS-1 cells to stably express WT survivin (Surv) or mitochondrially targeted survivin (MT-S), survivin targeted to mitochondria by the cytochrome c mitochondrial import sequence and containing GFP as a marker (INS-1/MT-S). By fluorescence microscopy, INS-1/MT-S cells exhibited perinuclear GFP expression consistent with mitochondrial localization (see below). Staurosporine treatment of parental INS-1 cells, or INS-1 cells stably transfected with mitochondrially targeted GFP (INS-1/MT-GFP), caused caspase-dependent cell death by multiparametric DEVDase activity (Figure 5A). Stable expression of Surv in INS-1 cells was unable to counteract staurosporine-induced apoptosis (Figure 5A). In contrast, mitochondrial targeting of survivin in INS-1/MT-S cells inhibited staurosporine-induced apoptosis and increased 5- to 16-fold the percentage of viable cells, by multiparametric flow cytometry (Figure 5A).
Figure 5.
Mitochondrial targeting of survivin in INS-1 cells inhibits apoptosis. (A) Cytoprotection in stable transfectants. Parental INS-1 cells or INS-1 cells stably transfected with Surv, MT-GFP, or MT-S were treated with staurosporine and analyzed for caspase-3 and caspase-7 activity (DEVDase activity, green fluorescence) and plasma membrane integrity (propidium iodide, red fluorescence), by multiparametric flow cytometry. The percentage of cells in each quadrant is indicated. (B) Caspase-3 cleavage. INS-1 cells expressing MT-GFP or MT-S were treated with staurosporine and analyzed at the indicated time intervals by immunoblotting. The position of active caspase-3 bands of 17 and 19 kDa is shown. (C) Caspase-9 cleavage. The experimental conditions were as in B except that samples were analyzed for generation of 37-kDa active caspase-9 fragments by immunoblotting. (D) Inhibition of caspase-dependent apoptosis. Parental INS-1 cells or INS-1 cells stably expressing MT-GFP or MT-S were treated with the indicated concentrations of staurosporine in the presence or absence of zVAD-fmk, and scored for nuclear morphology of apoptosis by DAPI staining. Data are the mean ± SEM of 3 independent experiments.
We next tested the effect of mitochondrially targeted survivin on proteolytic processing of effector and initiator caspases. Exposure of INS-1/MT-GFP cells to staurosporine resulted in time-dependent proteolytic cleavage of proform caspase-3 and caspase-9 to generate active fragments of 17 kDa and 19 kDa (Figure 5B), and 37 kDa (Figure 5C), respectively. Conversely, expression of mitochondrial survivin in stably transfected INS-1 (INS-1/MT-S) cells suppressed the generation of active caspase-3 (Figure 5B) and caspase-9 (Figure 5C) at the same time intervals examined. To further distinguish between caspase-dependent and -independent cell death modulated by mitochondrial survivin, we exposed differentially transfected INS-1 cells to staurosporine in the presence or absence of a broad-spectrum caspase inhibitor, z-Val-Ala-Asp (OMe)-fmk (zVAD-fmk). INS-1/MT-S cells were protected against apoptosis over a wide range of staurosporine concentrations, in a reaction that was not further enhanced by zVAD-fmk (Figure 5D). Conversely, staurosporine-induced apoptosis in INS-1 or INS-1/MT-GFP cells was inhibited by 50–75% by zVAD-fmk (Figure 5D), suggesting that mitochondrial survivin selectively protected against caspase-dependent cell death.
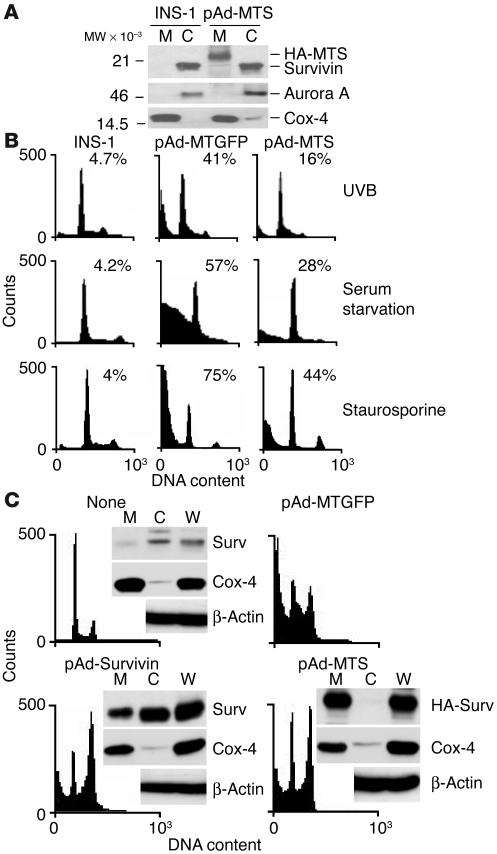
We next generated pAds encoding either MT-GFP (pAd-MTGFP) or hemagglutinin-tagged (HA) MT-S (pAd-MTS). Transduction of INS-1 cells with pAd-MTS resulted in selective accumulation of survivin in mitochondrial, but not cytosolic, extracts (Figure 6A). This was associated with strong inhibition of apoptosis induced by UVB, serum deprivation, or staurosporine, by hypodiploid DNA content and flow cytometry (Figure 6B). In contrast, transduction of INS-1 cells with pAd-MTGFP did not reverse apoptosis induced by the various cell death stimuli (Figure 6B). We next asked whether selective expansion of mitochondrial survivin was sufficient to inhibit apoptosis. Transduction of MCF-7 cells with pAd-Survivin resulted in increased expansion of both cytosolic and mitochondrial survivin, and inhibition of staurosporine-induced apoptosis (Figure 6C). By comparison, pAd-MTS resulted solely in expansion of mitochondrial, not cytosolic, survivin levels and was equally as effective as pAd-Survivin at suppressing staurosporine-induced apoptosis (Figure 6C). In control experiments, transduction of MCF-7 cells with pAd-MTGFP did not reduce staurosporine-mediated cell death (Figure 6C).
Figure 6.
Adenoviral targeting of survivin to mitochondria inhibits apoptosis. (A) Subcellular localization. INS-1 cells were infected with pAd-MTS, and isolated mitochondrial or cytosolic fractions were analyzed by immunoblotting. The position of endogenous or transduced (HA-MTS) survivin is indicated. (B) Cytoprotection by adenoviral transduction. INS-1 cells transduced with pAd-MTGFP or pAd-MTS were treated with UVB, exposed to serum starvation, or treated with staurosporine, and analyzed for DNA content by flow cytometry. Samples labeled as INS-1 were untreated cultures. The percentage of cells with hypodiploid (apoptotic) DNA content is indicated for each condition tested. (C) Mitochondrial survivin is sufficient for cytoprotection. MCF-7 cells were transduced with pAd-MTGFP, pAd-Survivin, or pAd-MTS, treated with staurosporine, and analyzed for hypodiploid DNA content and immunoblotting of whole cell extracts (W), mitochondrial, or cytosolic fractions. The percentages of cells with hypodiploid (apoptotic) DNA content were 47% (nontransduced cultures, data not shown), 40% (pAd-MTGFP), 25% (pAd-Survivin), and 29% (pAd-MTS).
Release of mitochondrial survivin during apoptosis antagonizes caspase-9 generation.
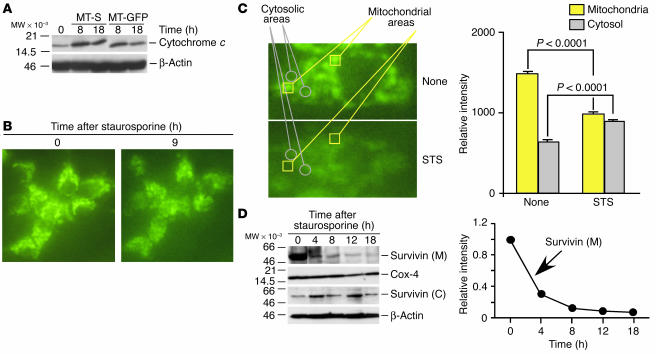
To begin to identify a mechanistic role of mitochondrial survivin in cytoprotection, we first monitored changes in permeability transition, i.e., the release of mitochondrial apoptogenic proteins following cell death stimulation (5). Stable expression of mitochondrially targeted survivin in INS-1/MT-S cells did not reduce the cytoplasmic accumulation of cytochrome c in response to staurosporine, as compared with cultures expressing mitochondrially targeted GFP (Figure 7A). Similar results were obtained with analysis of Smac in differentially transfected INS-1 cells following induction of apoptosis (data not shown). In contrast, treatment of INS-1/MT-S with staurosporine resulted in redistribution of the GFP signal from a punctate, perinuclear pattern observed in resting cells to a diffuse cytoplasmic localization (Figure 7B), suggestive of release of mitochondrial survivin. Fluorescence analysis of 264 individual stably transfected INS-1/MT-S cells revealed that staurosporine treatment resulted in translocation of the GFP signal from mitochondria to the cytosol in 93.3% ± 1.1% of the cell population examined. To further quantify the dynamics of mitochondrial release of survivin during apoptosis, single-cell fluorescence-analysis experiments were carried out. Determination of fluorescence intensity in individual areas corresponding to mitochondrial (Figure 7C, yellow squares) or cytosolic (Figure 7C, gray circles) regions demonstrated that staurosporine caused loss of mitochondrial GFP signal (1,475 ± 30.3 versus 980 ± 19, P < 0.0001, n = 16), which was associated with a parallel increase in cytosolic fluorescence (628 ± 21 versus 887 ± 14.4, P < 0.0001, n = 14).
Figure 7.
Mitochondrial release of survivin during apoptosis. (A) Effect on cytochrome c release. INS-1 cells stably transfected with MT-GFP or MT-S were treated with staurosporine, harvested at the indicated time intervals, and analyzed by immunoblotting. (B) Fluorescence microscopy. INS-1/MT-S cells were treated with staurosporine and analyzed for cytosolic redistribution of GFP. (C) Single-cell analysis. Areas corresponding to mitochondria (yellow squares) or cytosol (gray circles) were quantitatively analyzed for changes in fluorescence distribution before (None) or after staurosporine treatment (left panel). Right panel: Quantification of single-cell analysis. Changes in fluorescence intensity in individual mitochondrial or cytosolic areas were analyzed in the presence or absence of staurosporine treatment. Data represent an average of 16 individual determinations for mitochondrial areas and 14 individual determinations for cytosolic areas. (D) Time course of mitochondrial release of survivin during cell death. INS-1/MT-S cells were treated with staurosporine, and isolated mitochondrial or cytosolic fractions were analyzed at the indicated time intervals by immunoblotting. Right panel: Densitometric quantification of time-dependent depletion of mitochondrial survivin in response to staurosporine.
We next studied the time course of release of mitochondrial survivin during cell death in isolated cytosolic or mitochondrial fractions. In these experiments, staurosporine induced a rapid depletion of mitochondrial survivin, which was reduced by about 70% 4 hours after induction of apoptosis and was completely discharged by 12–18 hours (Figure 7D). This coincided with increased accumulation of survivin in isolated cytosolic extracts at the same time intervals (Figure 7D) and thus agrees with the fluorescence analysis reported above.
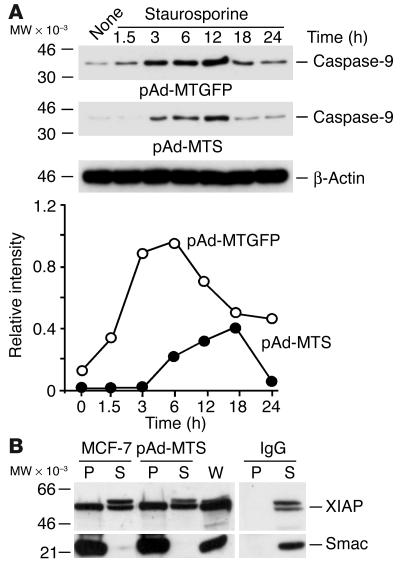
To determine the requirements of cytoprotection by mitochondrial survivin, we studied modulation of caspase-9 cleavage in transduced MCF-7 cells. Staurosporine treatment of MCF-7 cells transduced with pAd-MTGFP resulted in time-dependent cleavage of 46-kDa proform caspase-9 and generation of an active, 37-kDa caspase-9 fragment (Figure 8A). In contrast, transduction with pAd-MTS both delayed and attenuated the generation of active caspase-9 in staurosporine-treated MCF-7 cells (Figure 8A). Because survivin can associate with mitochondrial Smac (21), we asked whether cytoprotection by mitochondrial survivin involved binding and sequestration of Smac, which would be expected to relieve its inhibitory function on XIAP suppression of caspases (18). A specific interaction between survivin and Smac was demonstrated in vitro and in vivo (data not shown), in agreement with published data (21). However, transduction of MCF-7 cells with pAd-MTS did not affect the amount of Smac associated with XIAP during staurosporine-induced apoptosis, by coimmunoprecipitation and immunoblotting (Figure 8B). In control experiments, immune complexes precipitated by nonimmune IgG did not contain XIAP or Smac (Figure 8B).
Figure 8.
Mechanisms of cytoprotection by mitochondrial survivin. (A) Inhibition of caspase-9 processing. MCF-7 cells transduced with pAd-MTGFP or pAd-MTS were treated with staurosporine and analyzed for caspase-9 processing at the indicated time intervals, by immunoblotting. The position of approximately 37-kDa active caspase-9 is indicated. Lower panel: Quantification of caspase-9 generation by densitometry. (B) XIAP-Smac interaction. Control cultures or MCF-7 cells transduced with pAd-MTS were treated with staurosporine and immunoprecipitated with an antibody to Smac or control IgG, and pellets and supernatants were analyzed by immunoblotting with antibodies to XIAP or Smac.
Role of mitochondrial survivin in tumorigenesis.
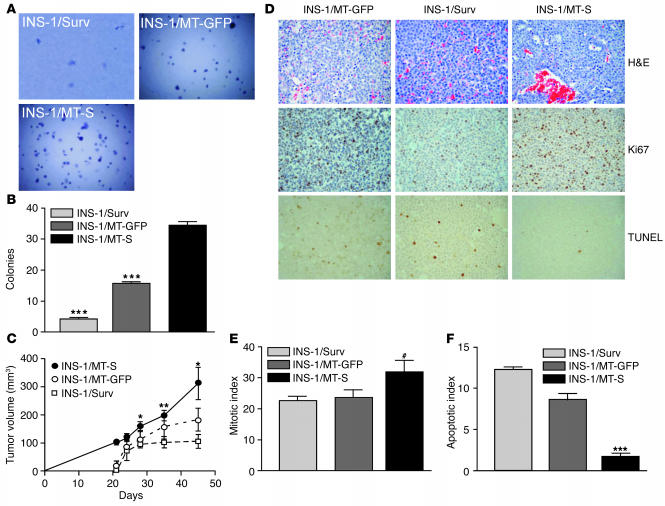
To determine whether the distinct subcellular pools of survivin were differentially involved in cellular transformation, we studied the ability of differentially transfected INS-1 cells to form colonies in soft agar, i.e., anchorage-independent cell growth. INS-1 cells expressing MT-GFP exhibited a limited degree of colony formation in soft agar over a 2-week interval (Figure 9, A and B). Expression of MT-S in INS-1 cells resulted in an approximately threefold increase in colony formation, as compared with expression of MT-GFP in INS-1 cells (Figure 9, A and B). In contrast, expression of cytosolic survivin that cannot be transported to mitochondria (INS-1/Surv) nearly completely abolished colony formation in soft agar (Figure 9, A and B).
Figure 9.
Mitochondrial survivin promotes tumorigenicity. (A) Colony formation in soft agar. Differentially transfected INS-1 cells were plated in semisolid medium and scored for colony formation by phase-contrast microscopy. (B) Quantification of colony formation in soft agar. The experimental conditions were as in A. Data are the mean ± SD of a representative experiment of at least 2 independent determinations. ***P = 0.0001. (C) Kinetics of tumor growth. Stably transfected INS-1 cells were injected subcutaneously in the flank of CB17 SCID/beige mice, and tumor volume was determined at the indicated time intervals. Statistical analysis compared growth of INS-1/Surv versus INS-1/MT-S tumors at day 28 (P = 0.024), day 35 (P = 0.0069), and day 45 (P = 0.015). *P < 0.05; **P < 0.01. (D) Histology. Tissue sections from the indicated INS-1 tumors were stained by H&E, Ki67 reactivity as a measure of cell proliferation, and TUNEL as a measure of apoptosis, in vivo. Magnification, ×400. (E) Mitotic index in vivo. The number of Ki67-positive (proliferating) cells was counted in 7 independent fields, each containing an average of 400 cells. #P = 0.031. (F) Apoptotic index in vivo. The number of TUNEL-positive (apoptotic) cells was counted in 7 independent fields, each containing an average of 400 cells.
We next injected differentially transfected INS-1 cells in immunocompromised mice and studied the kinetics of tumor formation. Tumors formed by INS-1/MT-GFP cells exhibited delayed onset and a flattened growth rate that did not significantly change during the last 3 weeks of examination (Figure 9C). Conversely, INS-1 cells expressing mitochondrially targeted survivin (INS-1/MT-S) gave rise to solid tumors characterized by early onset and steady exponential growth rate (Figure 9C). Stable expression of non-mitochondrially-targeted cytosolic survivin in INS-1 cells was associated with strong inhibition of tumor growth in immunocompromised animals (Figure 9C); this was similar to the findings regarding formation of colonies in soft agar (Figure 9, A and B). Histologically, tumors formed by INS-1 cells had neuroendocrine appearance and exhibited an extensive network of vascular sinusoids filled with blood (Figure 9D). Tumor cell proliferation as detected by Ki67 labeling (Figure 9D) was indistinguishable in tumors formed by INS-1 cells expressing mitochondrially targeted GFP or non-mitochondrially-targeted, cytosolic survivin (INS-1/Surv) (Figure 9E). Conversely, INS-1/MT-S tumors revealed a higher mitotic index (P = 0.031) as compared with INS-1/Surv tumors (Figure 9E). Quantification of internucleosomal DNA fragmentation by TUNEL staining (Figure 9D) revealed that expression of mitochondrially targeted survivin in INS-1/MT-S tumors resulted in nearly complete suppression of apoptosis in vivo (P = 0.0001), as compared with that in INS-1/Surv or INS-1/MT-GFP tumors (Figure 9F). When compared with INS-1/MT-GFP tumors, tumors expressing non-mitochondrially-targeted, INS-1/Surv resulted in a considerably increased (P = 0.0003) apoptotic index (Figure 9F), which may explain the reduction in colony formation (Figure 9, A and B) and the slower growth rate of these tumors in vivo (Figure 9C).
Discussion
In this study, we have uncovered a novel pathway of apoptosis inhibition centered on the compartmentalization of the IAP protein survivin to mitochondria. Dynamically expanded in response to cellular stress stimuli, and rapidly released in the cytosol following full-blown apoptotic stimulation, mitochondrial survivin exerts cytoprotection by preventing the activation of initiator caspase-9. This pathway has profound repercussions for tumor cell viability, as expression of mitochondrial survivin is sufficient to promote anchorage-independent cell growth, ablate tumor cell apoptosis in vivo, and sustain exponential tumor growth.
Since its discovery in 1997 (22), survivin has attracted attention as a unique member of the IAP gene family with a potential dual role in apoptosis inhibition and regulation of mitosis (8). The role of survivin in regulation of mitosis has recently become better understood, and linked to multiple spindle microtubule functions and mitotic checkpoints (8). In contrast, despite extensive experimental evidence in vitro, and in transgenic animals in vivo (8), the precise mechanism(s) by which survivin interferes with apoptosis has not been elucidated. The evidence presented here defines a novel antiapoptotic pathway mediated by survivin, functionally separable from its role in mitosis. Cytoprotection by survivin is more selective than that by other IAPs (6) and is specifically targeted at the initiation of mitochondrial apoptosis to prevent caspase-9 activation. This is consistent with the formation of a survivin–caspase-9 complex in vivo (23), the requirement of apoptosome components Apaf-1 and caspase-9 in the survivin pathway (24), and the ability of survivin to prevent incorporation of caspase-9 into a functional apoptosome (25). Although survivin may still oppose death receptor–initiated apoptosis, this is likely to occur in a cell type–specific fashion, in which mitochondrial amplification is required for effective execution of cell death (26).
Several independent lines of evidence demonstrated that survivin occurs in an abundant mitochondrial pool, localized to a digitonin-sensitive compartment in the inter–mitochondrial membrane space. The existence of a novel subcellular localization of survivin is consistent with the dynamic nature of this protein and with its distribution among multiple subcellular pools that are independently regulated, differentially posttranslationally modified, and uniquely immunoreactive (16). In mammalian cells, the mitochondrion has emerged as a central orchestrator of apoptosis (4, 5), by providing a specialized microenvironment for a multiplicity of cell death regulators (18, 27). A localization of certain IAPs to mitochondria has also been reported, and cIAP2 and XIAP were found in mitochondrial extracts of lung cancer cell lines, whereas cIAP1 became recruited to mitochondria in response to ionizing radiation (28). Although the mechanism(s) by which survivin, and possibly other IAPs, localize to mitochondria is currently not known, their lack of a recognizable mitochondrial import sequence suggests a potential role of an associated protein(s) with mitochondrial shuttling function. An attractive candidate for this pathway is the molecular chaperone Hsp90 (29), which participates in mitochondrial import of client proteins (30), associates with survivin (31) and other IAPs (our unpublished observations) in vivo, and exhibits an approximately 100-fold increased affinity for its client proteins in tumor cells (32), where survivin is abundantly found in mitochondria (this study). Conversely, survivin was not present in mitochondrial fractions of normal tissues, which suggests that its localization to mitochondria may be preferentially, or exclusively, associated with oncogenic transformation.
An intriguing feature of mitochondrial survivin is its dynamic expansion in response to cellular stress, which is in keeping with an emerging role of IAPs in the cellular adaptation to stress. Accordingly, exposure to chemotherapeutic drugs (33), exposure to heat shock (31), and exposure to hypoxia (19) have all been associated with increased IAP expression. For survivin, this appears to involve increased protein stability, and to result largely, if not exclusively, in an expansion of the mitochondrial pool. This may be envisioned as an adaptive response to environmental stresses, ideally suited to further elevate an antiapoptotic threshold in transformed cells.
As might have been predicted from its localization to the inter–mitochondrial membrane space, survivin, unlike Bcl-2 family proteins (3), did not affect the release of Smac or cytochrome c after cell death stimulation (4, 5). Rather, mitochondrial survivin exerted its cytoprotective function in the cytosol upon release from mitochondria during apoptosis. The kinetics of survivin release from mitochondria, with more than 70% of the protein discharged in the cytosol within 4 hours of apoptotic stimulation, closely mirror the time course of permeability transition (4, 5). This suggests a model in which loss of mitochondrial membrane integrity following cell death stimulation would result in the release not only of cell death amplifiers (18, 34, 35), but also of at least 1 cell death antagonist, i.e., survivin, ideally positioned to limit apoptosome-associated caspase activation. A critical feature of this pathway is that mitochondrial survivin is specifically competent to inhibit apoptosis, whereas overexpression of cytosolic survivin that cannot localize to mitochondria, i.e., in INS-1 cells, is unable to mediate cytoprotection. The idea that mitochondrial survivin could associate with Smac (21) and sequester it away from other IAP proteins, i.e., XIAP, in a mechanism that would be expected to remove its pro-apoptotic function (18), appears inconsistent with our findings that mitochondrial survivin does not reduce the association of Smac with XIAP during apoptosis. Conversely, it is possible that posttranslational modification(s) either within mitochondria or in cytosolic survivin may, respectively, enable or prevent its cytoprotective function. In this context, one attractive possibility consistent with recent results (36) is that mitochondrial shuttling of survivin promotes its ability to associate with XIAP, thus facilitating increased protein stability and synergistic suppression of caspase-9 (36). Further studies will test this possibility and delineate the structure-function requirements of cytoprotection by mitochondrial versus cytosolic survivin.
The biological context of the pathway described here is highlighted by the nearly ubiquitous overexpression of survivin in cancer, as opposed to normal tissues (22). In tumors, survivin is expressed at all cell cycle phases, thus making unlikely a sole role at mitosis, and it correlates with clinical phenotypes typically associated with apoptosis resistance, including low apoptotic index, resistance to therapy, and accelerated relapses (8). Nevertheless, it has been debated whether the high levels of survivin in cancer patients reflect exploitation of its mitotic function, somehow to override cell division checkpoints, or rather its second role in apoptosis inhibition. The data obtained here with differentially transfected INS-1 cells conclusively demonstrate that tumors selectively exploit the antiapoptotic function of survivin. Accordingly, targeting survivin to mitochondria was sufficient, alone, to enhance colony formation in soft agar, accelerate tumor growth in immunocompromised animals, and nearly completely ablate tumor cell apoptosis in vivo. Conversely, in stark contrast to the antiapoptotic gain afforded by mitochondrial survivin, increased expression of survivin that could not be transported to mitochondria, i.e., in INS-1 cells, not only did not contribute to tumorigenesis but actually severely impaired anchorage-independent cell growth, retarded tumor formation, and resulted in increased apoptosis in vivo. These data may be explained by an exaggerated function of survivin at mitosis, which is known to cause deregulated stability of spindle microtubules (10), and spindle checkpoint activation (37), leading to cell cycle delays and/or apoptosis. Because of its inhibition of apoptosome-associated caspase activity, survivin cytoprotection is ideally positioned to thwart cell death pathways typically activated in tumors, including oncogene-, chemotherapy-, or radiation therapy–mediated apoptosis (38, 39), and to accelerate and/or facilitate the acquisition of additional cancer traits (1), thus furthering tumor progression. This may explain the increased mitotic index of tumors expressing mitochondrial survivin, which may reflect oncogenic changes in key cell proliferation pathway(s).
In summary, the data presented here establish survivin as a bona fide antiapoptotic mediator of cancer progression and validate its importance as a therapeutic target (8). Because of its now established direct link to the apoptotic machinery, molecular antagonists of mitochondrial survivin may provide a desirable therapeutic window by selectively disabling tumor-associated cytoprotection without affecting the viability of normal tissues, where survivin is largely undetectable and its mitochondrial pool is not observed. Experimentally, this could be exemplified by the lack of toxicity of survivin ablation in INS-1 cells, as opposed to the robust apoptosis observed in MCF-7 cells (20). Because survivin is a short-lived protein at cell division, such an approach may not interfere with the mitotic function of survivin and may thus further limit toxicity for highly proliferating normal cellular compartments, which are typically compromised by conventional anticancer regimens. Therefore, these distinctive features of the survivin pathway provide a unique opportunity for rational anticancer treatment in humans (8).
Methods
Cell lines and antibodies.
Breast carcinoma MCF-7, cervical carcinoma HeLa, colorectal carcinoma HCT116, B lymphoma Raji, and human embryonic kidney HEK293T cells were obtained from the American Type Culture Collection. The rat insulinoma cell line INS-1 was the kind gift of R.S. Sherwin (Yale University School of Medicine, New Haven, Connecticut, USA). The antibodies to survivin (Novus Biologicals Inc.) were described previously (16). The following antibodies were also used: Aurora A (Novus Biologicals Inc.), Smac (1:1,000; ProSci Inc.), cytochrome c (1:1,000; BD Biosciences — Pharmingen), Cox-4 (1:5,000; BD Biosciences — Clontech), Bcl-2 (1:1,000; Santa Cruz Biotechnologies Inc.), XIAP (1:1,000; Transduction Laboratories), mitochondrial Hsp70 (Affinity BioReagents Inc.), and β-actin (1:5,000, clone AC-15, A5441; Sigma-Aldrich). An antibody to phosphorylated eIF2α was obtained from Cell Signaling Technology Inc.
Generation of transfected cell lines.
A human survivin cDNA containing BamHI sites was synthesized by PCR using the primers 5′-AGAGGTGGGATCCCGGCATG-3′ (forward) and 5′-AGCTCCGGATCCAGGCCTCTA-3′ (reverse). The cDNA was inserted in-frame into the BamHI sites of pECFP-Mito (BD Biosciences — Clontech) to construct pECFP-mt-Survivin. INS-1 cells were transfected with control vector or mitochondrially targeted survivin by LipofectAMINE (Invitrogen Corp.), and transferred to a 100-mm dish in selection medium containing 800 μg/ml G418 (GIBCO; Invitrogen Corp.). Colonies were picked 2–3 weeks later and expanded, and mitochondrial targeting of survivin in stable cell lines was demonstrated by fluorescence microscopy and immunoblotting of isolated mitochondrial fractions (see below). Experiments were conducted with 2 independent clonal cell lines with identical results.
Adenoviral vectors and cellular transduction.
The pAd vectors encoding Surv (pAd-Survivin) or GFP (pAd-GFP) have been described (40). pAd-MTS was established using the pAdEasy system, as described previously (40). To construct the shuttle vector, a cDNA encoding HA-survivin containing BamHI sites was synthesized by PCR using primers 5′-GGAGACCGGTGCTTATGTACCCAT-3′ (forward) and 5′-CATTTTATTAGGAAAGGACAGTGG-3′ (reverse). The cDNA was linearized and cloned in-frame into the BamHI sites of pECFP-Mito to construct pECFP-mt-HA-survivin. A NheI-NotI fragment was isolated from pECFP-mt-HA-survivin or pECFP-Mito and subcloned into pBluescript II KS(+) (Stratagene) to generate pBluescript-mtHAsurvivin or pBluescript-mtCFP, and a SalI-NotI fragment was subcloned into pAdTrack-CMV to generate pAd-MTS or pAd-MTCFP. Each shuttle vector was linearized with PmeI and electroporated in Escherichia coli BJ5183 for homologous recombination, and colonies were selected in 50 μg/ml kanamycin. Each pAd construct (4–8 μg) was digested with PacI and transfected in HEK293T cells by LipofectAMINE, and cultures were monitored for expression of GFP by fluorescence microscopy. High-titer viral stocks were generated as described previously (40) and purified by CsCl banding, and viral titer was quantified by green fluorescence–forming units in HEK293T cells infected with serial dilution of the viral stock. Cultures were transduced at a multiplicity of infection of 50 for 8 hours at 37°C in complete medium, washed with PBS, pH 7.4, and replenished with complete medium. The transduction efficiency (>90% of the cell population) was estimated from GFP fluorescence.
Preparation of mitochondrial and cytosolic extracts.
Mitochondrial and cytosolic fractions were extracted from MCF-7 cells (6 × 107 to 7 × 107) or INS-1 cells (7 × 107 to 8 × 107) using the ApoAlert Cell Fractionation Kit (BD Biosciences — Clontech), according to the supplier’s instructions. The isolated cytosolic fraction was further centrifuged at 10,000 g for 25 minutes at 4°C, and the supernatant was collected. Alternatively, mitochondrial extracts were prepared from MCF-7 or INS-1 cells (5 × 107) washed in TD buffer (135 mM NaCl, 5 mM KCl, 25 mM Tris-Cl, pH 7.6) and allowed to swell for 10 minutes in ice-cold hypotonic CaRSB buffer (10 mM NaCl, 1.5 mM CaCl2, 10 mM Tris-HCl, pH 7.5, plus protease inhibitors). Cells were Dounce-homogenized with 70 strokes, with addition of MS buffer (210 mM mannitol, 70 mM sucrose, 5 mM Tris, pH 7.6) to stabilize mitochondria (2 ml of 2.5× per 3 ml of homogenate). After removal of nuclear contaminants by centrifugation at 600 g for 15 minutes on ice, the supernatants were layered over a 1- to 2-M sucrose step gradient (10 mM Tris, 5 mM EDTA, pH 7.6, 2 mM DTT, plus protease inhibitors) and centrifuged at 110,000 g for 30 minutes at 4°C. Mitochondria were collected at the 1- to 1.5-M interphase by lateral suction through the tube, washed in 4 volumes of MS buffer at 9,400 g, and suspended in a final volume of 200 μl of MS buffer. The top layer, containing a cytosolic and a free protein fraction, was collected and used in parallel experiments. The purity of mitochondrial fractions was examined by immunoblotting with antibodies to mitochondrial Cox-4 or cytosolic Aurora A. In some experiments, testis, spleen, and liver isolated from 5-week-old FVB/N mice were fractionated in cytosolic and mitochondrial extracts using a Mitochondria Isolation Kit (Sigma-Aldrich), according to the manufacturer’s specifications.
Electron microscopy.
MCF-7 cell pellets or isolated mitochondrial fractions prepared by sucrose gradient ultracentrifugation were fixed in 3% formaldehyde and 0.1% glutaraldehyde (EM grade) for 10 minutes at 37°C. Samples were incubated in 50 mM NH4Cl in PBS for 60 minutes at 22°C to aminidate free aldehydes, dehydrated through a gradual series of ethanol to 100%, and transferred into a mixture of 50:50 (vol/vol) Lowicryl K4M resin/100% ethanol overnight at 22°C. Samples were transferred to aliquots of fresh resin (3 changes, 1 hour each) and applied to filling embedding capsules for 24 hours at 60°C. Thin sections were cut using a diamond knife on a Reichert-Jung Ultracut E ultramicrotome and placed on gold support grids for immunostaining. For labeling, sections were incubated in blocking solution (Zymed Laboratories Inc.) for 30 minutes at 22°C, rinsed, incubated with primary antibodies to survivin, Smac, or control IgG, rinsed again, and incubated with gold-conjugated secondary reagents (Smac, 6 nm diameter; survivin, 12 nm diameter) of appropriate specificities (1:20; Jackson ImmunoResearch Laboratories Inc.). After washes, samples were exposed to OsO4 vapor for 1 hour at 22°C, post-stained with uranyl acetate and lead citrate, and analyzed on a Philips CM10 electron microscope (Philips Electronics) at 80 kV. For quantification of mitochondrial colocalization of survivin and Smac, 19 individual electron microscopy fields containing an average of 2.9 mitochondria each were individually scored for accumulation of survivin- and Smac-associated gold particles.
Fluorescence microscopy.
INS-1 stable transfectants were plated onto 35-mm glass-bottom culture dishes with grid (MatTek Corp.). After 24 hours, cells were treated with staurosporine (Sigma-Aldrich), washed with PBS, and scanned with a manual/motorized reflected-fluorescence system (Olympus Corp.) at increasing time intervals after induction of apoptosis. Collected images were analyzed with IPLab version 3.5.4 software (Scanalytics Inc.). To quantify translocation of the GFP signal during apoptosis, 264 individual INS-1 cells stably transfected with mitochondrially targeted survivin (INS-1/MT-S) were scored for changes in GFP localization before or after staurosporine treatment. In single-cell analysis, regions corresponding to mitochondria or cytosol were analyzed for changes in fluorescence distribution before or after staurosporine treatment.
Mitochondrial sublocalization studies.
Mitochondrial or cytosolic extracts were incubated in MS buffer in the presence of 0–30 ng/ml proteinase K (Sigma-Aldrich) for 10 minutes at 0°C, and the reaction was stopped by addition of protease inhibitors. Modulation of survivin or Bcl-2 protein levels was monitored by immunoblotting. For differential extraction of mitochondrial proteins, freshly isolated mitochondrial pellets were incubated with increasing concentrations (0–4%) of digitonin (Sigma-Aldrich) for 15 minutes at 0°C. Pellets and supernatants were analyzed for differential redistribution of survivin or Smac proteins, by immunoblotting. Reactivity with nonextractable mitochondrial Hsp70 was used as control. The concentrations of digitonin used have been previously shown to extract proteins from the outer mitochondrial membrane (17).
Modulation of mitochondrial survivin during the cellular stress response.
For modulation of survivin expression and distribution during hypoxia, HeLa cells (2.5 × 105) were seeded in 35-mm-diameter tissue culture dishes. Cells were washed 3 times in PBS and incubated for 6 hours in KRB buffer (115 mM NaCl, 1 mM KH2PO4, 4 mM KCl, 1 mM MgSO4, 1.25 mM CaCl2, 27 mM NaHCO3) containing 1.2 U/ml Oxyrase (Oxyrase Inc.), in a GasPak 100 anaerobic chamber (BD). Whole cell extracts or isolated mitochondrial or cytosolic fractions were analyzed by immunoblotting. Control cells were incubated in KRB buffer alone. For cycloheximide block experiments, control or hypoxic HeLa cells were treated with cycloheximide (100 μg/ml), harvested after 2–6 hours, analyzed by immunoblotting, and quantified by β-actin–normalized densitometry. Alternatively, MCF-7 cells were treated with the chemotherapeutic drug adriamycin (100 nM), and isolated cytosolic or mitochondrial fractions were analyzed after 24 hours by immunoblotting.
Cell death analysis.
Subconfluent cultures of parental or transfected/transduced INS-1 or MCF-7 cells were exposed to staurosporine (0.1–1 μM), UVB irradiation (50–100 J/m2), or growth factor deprivation (0% FCS for 96 hours), harvested at increasing time intervals, and analyzed for hypodiploid DNA content by propidium iodide staining and flow cytometry. Alternatively, cells exposed to the various cell death stimuli were harvested, fixed in 4% paraformaldehyde containing 4% sucrose for 10 minutes at 22°C, and analyzed for nuclear morphology of apoptosis by staining with 6.5 μg/ml DAPI (Sigma-Aldrich) in 16% polyvinyl alcohol (Air Products and Chemicals Inc.) and 40% glycerol. In some experiments, the pan-caspase inhibitor zVAD-fmk (Enzyme Systems Products Inc.) was used (20 μM). For caspase activity, cells exposed to cell death stimuli were simultaneously analyzed for caspase activity and propidium iodide staining by multiparametric flow cytometry using the CaspaTag caspase-3 activity kit (Intergen Co.), or analyzed by immunoblotting using antibodies to activated caspase-3 or -9 (1:1,000; Transduction Laboratories). Acute ablation of survivin in MCF-7 or INS-1 cells was carried out by RNAi using the S4 dsRNA oligonucleotide GAGCCAAGAACAAAAUUGC, as described previously (20). Unrelated dsRNA oligonucleotide VIII was used as control (20). Cultures were analyzed for loss of survivin protein by immunoblotting, and analyzed for cell viability and mitotic defects by DNA content and flow cytometry. To determine the potential impact of mitochondrial survivin on the formation of a Smac-XIAP complex, MCF-7 cells were transduced with pAd-MTS, exposed to staurosporine, and immunoprecipitated with an antibody to Smac or control IgG. The immune complexes were probed with antibodies to XIAP or Smac, by immunoblotting.
Analysis of tumorigenesis.
For soft agar colony formation, 2 × 104 stable INS-1 transfectants were suspended in 1.5 ml of DMEM supplemented with 10% FBS and 0.35% bactoagar (BD) in 36-mm tissue culture plates containing 1.5 ml of 0.75% agarose in growth medium at the bottom layer. The plates were incubated at 37°C in a 5% CO2 incubator for 2 weeks. Colonies were stained with 0.005% crystal violet (Sigma-Aldrich), and counted using a dissecting microscope under high-power field. Experiments were conducted with 2 independent INS-1 clones with identical results. All experiments involving animals were approved by the institutional animal care and use committee. For xenograft tumor formation experiments, 2.5 × 106 INS-1 stable transfectants were injected into the flanks of 6- to 8-week-old female CB17 SCID/beige mice (Taconic; 3 animals per group, 2 tumors per animal). Tumor volume was measured in the 3 dimensions with a caliper at the indicated time intervals after injection. An independent experiment of tumor formation was repeated with 4 animals per group (2 tumors per animal) with identical kinetics of tumor growth.
Histology.
All tissue specimens were fixed in buffered formalin and embedded in paraffin. For tissue staining, sections were deparaffinized, rehydrated in water, and quenched for endogenous peroxidase. Epitope heat retrieval was carried out by steaming of the slides in 10% sodium citrate for 60 minutes. Processed slides were rinsed in PBS, and stained using standard avidin-biotin-peroxidase technique using the Histostain-Plus kit (Zymed Laboratories Inc.). After blocking, tissue sections were incubated overnight at 4°C with antibody Ki67 (1:25; DAKO Inc.), rinsed, and incubated with biotinylated anti-mouse IgG for 10 minutes at 22°C. Following addition of streptavidin-conjugated HRP, slides were incubated with 3′-3′-diamino-benzidine (DAB) as a chromogen and counterstained with hematoxylin. Negative control sections were processed as above without primary antibody and resulted in no detectable staining. A mitotic index was derived from the percentage of Ki67-positive cells counted under high-power field. For determination of internucleosomal DNA fragmentation by TUNEL staining, tissue sections were processed using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Chemicon International Inc.), according to the supplier’s specifications. An apoptotic index was calculated from the percentage of TUNEL-positive cells counted under high-power fields.
Statistical analysis.
Data were analyzed using the unpaired t test on a GraphPad Prism software package for Windows (GraphPad Software Inc.). A P value of 0.05 was considered statistically significant.
Acknowledgments
We thank Colin Duckett for critical reading of the manuscript, and Gregory Hendricks, Kevin Hendricks, and John Nunnari for electron microscopy. This work was supported by NIH grants CA78810, CA90917, and HL54131.
Footnotes
Nonstandard abbreviations used: dsRNA, double-stranded RNA; HA, hemagglutinin-tagged; IAP, inhibitor of apoptosis; J, joules; MT-GFP, mitochondrially targeted GFP; MT-S, mitochondrially targeted survivin; pAd, replication-deficient adenovirus; pAd-MTGFP, pAd encoding MT-GFP; pAd-MTS, pAd encoding HA MT-S; pAd-Survivin, pAd encoding WT survivin; RNAi, RNA interference; Surv, WT survivin; zVAD-fmk, z-Val-Ala-Asp (OMe)-fmk.Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 3.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 4.Zamzami N, Kroemer G. The mitochondrion in apoptosis: how Pandora’s box opens. Nat. Rev. Mol. Cell Biol. 2001;2:67–71. doi: 10.1038/35048073. [DOI] [PubMed] [Google Scholar]
- 5.Martinou JC, Green DR. Breaking the mitochondrial barrier. Nat. Rev. Mol. Cell Biol. 2001;2:63–67. doi: 10.1038/35048069. [DOI] [PubMed] [Google Scholar]
- 6.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 7.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 8.Altieri DC. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 9.Li F, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat. Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- 10.Giodini A, et al. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002;62:2462–2467. [PubMed] [Google Scholar]
- 11.Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- 12.Lens SM, Medema RH. The survivin/aurora B complex: its role in coordinating tension and attachment. Cell Cycle. 2003;2:507–510. doi: 10.4161/cc.2.6.559. [DOI] [PubMed] [Google Scholar]
- 13.Velculescu VE, et al. Analysis of human transcriptomes. Nat. Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 14.van ‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 15.van de Wetering M, et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 16.Fortugno P, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J. Cell Sci. 2002;115:575–585. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- 17.Gottlieb RA, Granville DJ. Analyzing mitochondrial changes during apoptosis. Methods. 2002;26:341–347. doi: 10.1016/S1046-2023(02)00040-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 19.Dong Z, et al. Up-regulation of apoptosis inhibitory protein IAP-2 by hypoxia. Hif-1-independent mechanisms. J. Biol. Chem. 2001;276:18702–18709. doi: 10.1074/jbc.M011774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltrami E, Plescia J, Wilkinson JC, Duckett CS, Altieri DC. Acute ablation of survivin uncovers p53-dependent mitotic checkpoint functions and control of mitochondrial apoptosis. J. Biol. Chem. 2004;279:2077–2084. doi: 10.1074/jbc.M309479200. [DOI] [PubMed] [Google Scholar]
- 21.Song Z, Yao X, Wu M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 2003;278:23130–23140. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- 22.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 23.O’Connor DS, et al. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanc-Brude OP, et al. Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin. Cancer Res. 2003;9:2683–2692. [PubMed] [Google Scholar]
- 25.Marusawa H, et al. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22:2729–2740. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scaffidi C, et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroemer G, Reed JC. Mitochondrial control of cell death. Nat. Med. 2000;6:513–519. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- 28.Ekedahl J, et al. Expression of inhibitor of apoptosis proteins in small- and non-small-cell lung carcinoma cells. Exp. Cell Res. 2002;279:277–290. doi: 10.1006/excr.2002.5608. [DOI] [PubMed] [Google Scholar]
- 29.Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 31.Fortugno P, et al. Regulation of survivin function by Hsp90. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13791–13796. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamal A, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 33.Chen JG, Yang CP, Cammer M, Horwitz SB. Gene expression and mitotic exit induced by microtubule-stabilizing drugs. Cancer Res. 2003;63:7891–7899. [PubMed] [Google Scholar]
- 34.Krajewski S, et al. Release of caspase-9 from mitochondria during neuronal apoptosis and cerebral ischemia. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5752–5757. doi: 10.1073/pnas.96.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Susin SA, et al. Mitochondrial release of caspase-2 and -9 during the apoptotic process. J. Exp. Med. 1999;189:381–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dohi T, et al. A IAP-IAP complex inhibits apoptosis. J. Biol. Chem. 2004;279:34087–34090. doi:10.1074/jbc.C400236200. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 37.Lens SM, et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003;22:2934–2947. doi: 10.1093/emboj/cdg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debatin KM, Poncet D, Kroemer G. Chemotherapy: targeting the mitochondrial cell death pathway. Oncogene. 2002;21:8786–8803. doi: 10.1038/sj.onc.1206039. [DOI] [PubMed] [Google Scholar]
- 39.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 40.Mesri M, Wall NR, Li J, Kim RW, Altieri DC. Cancer gene therapy using a survivin mutant adenovirus. J. Clin. Invest. 2001;108:981–990. doi:10.1172/JCI200112983. doi: 10.1172/JCI12983. [DOI] [PMC free article] [PubMed] [Google Scholar]