Abstract
Mechanistic target of rapamycin complex 1 (mTORC1) responds to multiple distinct signals (growth factors, amino acids, stress, energy level) and coordinates cell growth and proliferation. The activity of mTORC1 is exquisitely regulated in response to distinct stimuli, however, the underlying molecular mechanisms are still not fully understood. The spatial compartmentalization of mTORC1 signaling has been suggested to be an important mechanism for mTORC1 to achieve the signal specificity and efficiency. To examine the spatial regulation of the activity of mTORC1 in live cells, we describe a protocol using a newly developed molecular tool, a genetically encoded fluorescence resonance energy transfer (FRET)-based mTORC1 activity reporter, TORCAR. When expressed in the cell, TORCAR acts as a surrogate substrate of mTORC1, and exhibits a change in FRET in response to phosphorylation by mTORC1. Genetically targeting TORCAR to specific subcellular locations further allows for the characterization of spatial compartmentalized mTORC1 signaling.
Keywords: mTOR, Kinase, Biosensor, Live-cell imaging, Fluorescence
INTRODUCTION
Mechanistic target of rapamycin (mTOR) is a highly conserved serine/threonine protein kinase that constitutes two structurally and functionally distinct complexes, mTOR complex 1 (mTORC1) and complex 2 (mTORC2). mTORC1 is acutely and allosterically inhibited by rapamycin, an antifungal microlide, whereas mTORC2 is inhibited by prolonged treatment of rapamycin. mTORC1 consists of mTOR, regulatory associated protein of mTOR (Raptor), and mammalian lethal with SEC13 protein 8 (mLST8), 40-kDa proline-rich Akt substrate (PRAS40), and DEP domain-containing mTOR-interacting protein (DEPTOR) (Laplante and Sabatini, 2012). The interaction of mTOR and Raptor is essential for the specificity of mTORC1 in recognizing and phosphorylating its protein substrates (Aylett et al., 2016; Hara et al., 2002; Kim et al., 2002), such as eukaryotic translation initiation factor 4E binding protein (4EBP1) (Hara et al., 1997), translational regulators ribosomal protein S6 kinase (S6K1) (Burnett et al., 1998), and Unc-51 like autophagy activating kinase 1 (Ulk1) (Kim et al., 2011). As a result of phosphorylation of downstream effectors, mTORC1 regulates a number of cellular anabolic and catabolic processes, such as promoting the biosynthesis of proteins and inhibiting autophagy (Dibble and Manning, 2013).
mTORC1 responds to many different extracellular and intracellular cues including nutrients, growth factors, stress, and cellular energy to coordinate cell growth and metabolism (Dibble and Cantley, 2015; Goberdhan et al., 2016). As a highly integrated signaling node, the activity of mTORC1 kinase complex is exquisitely regulated. Dysregulated mTORC1 activity is often associated with pathophysiological conditions such as cancer and type 2 diabetes (Dibble and Manning, 2013; Ilagan, 2016).
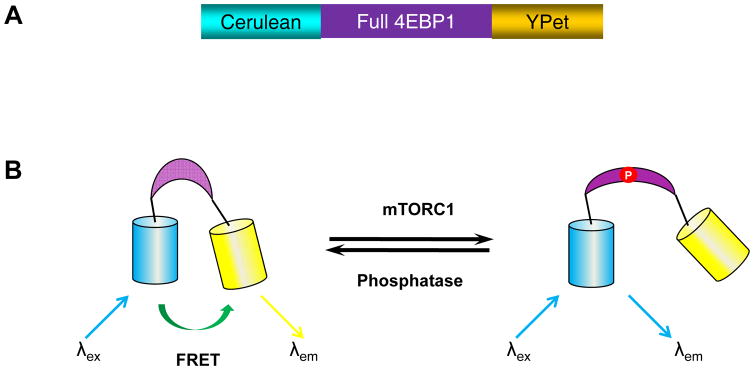
The utilization of genetically encoded fluorescence resonance energy transfer (FRET)-based mTORC1 activity reporter (TORCAR) enables us to detect the dynamic activity of mTORC1 in live cells with high spatiotemporal resolution. TORCAR was constructed using full length of 4EBP1, a well-studied mTORC1-specific substrate. 4EBP1 is sandwiched between a cyan fluorescent protein (cerulean) as a FRET donor and a yellow fluorescent protein (YPet) as a FRET acceptor (Figure 1A). When expressed in the cell, TORCAR acts as a surrogate substrate of mTORC1. Phosphorylation of TORCAR at two specific sites by mTORC1, Thr 37 and Thr 46 in the 4EBP1 region, induces a conformational change within the reporter, resulting in a decrease in FRET. The increase in the cyan-over-yellow (C/Y) emission ratio, which corresponds to a FRET decrease, can be used to indicate an increase in mTORC1 activity (Figure 1B). Below, we describe a general protocol of using TORCAR to monitor the mTORC1 activity in live cells, including specific, detailed procedures for the maintenance of NIH3T3 fibroblast cells, transfection of cells with TORCAR plasmid DNA, preparation of cells for imaging experiments, preparation of the imaging equipment, imaging of mTORC1 activity in live cells, and analysis of the acquired imaging data to quantify any observed changes in FRET.
Figure 1.
The design of TORCAR biosensor. (A) Domain structure of TORCAR construct. TORCAR is composed of cyan fluorescent protein (cerulean), full length 4EBP1, and yellow fluorescent protein (YPet) (B) General scheme depicting the TORCAR response. Active mTORC1 phosphorylates 4EBP1 in the TORCAR biosensor, which subsequently induced a conformational change that leads to an increase in the cyan-to-yellow emission ratio.
BASIC PROTOCOL
Monitoring mTORC1 activity using genetically encoded FRET-based biosensor TORCAR
The following protocol describes the use of a FRET-based biosensor TORCAR to track the activity of mTORC1 in NIH3T3 cells. First, cells are transfected with plasmid DNA encoding the TORCAR biosensor, and serum-starved overnight and amino acid starved for 2 hr. Then the microscope is prepared for the imaging experiment, followed by the acquisition of a time series of fluorescence intensity images. During imaging, the cells are stimulated by growth factors or amino acids to activate mTORC1, and the resulting changes in fluorescence are recorded. Finally, the images are analyzed to quantify the changes in mTORC1 activity.
Materials
NIH3T3 fibroblast cells plated on sterilized 35-mm glass-bottom dishes (see SUPPORT PROTOCOL)
Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco/BRL, Bethesda, MD) supplemented with 10 % calf serum (CS, ATCC 302030) and 1 % penicillin-streptomycin (Sigma-Aldrich) for use with NIH3T3 cells.
Solution of trypsin (0.25 %) and ethylenediamine tetraacetic acid (EDTA, 0.53 mM) (Invitrogen, Carlsbad, CA)
Opti-MEM I Reduced Serum Medium (Gibco)
Lipofectamine 2000 (Invitrogen)
Plasmid DNA of TORCAR and TORCAR targeted to subcellualr localization (Addgene)
HBSS* imaging buffer (see recipe)
Immersion oil Immersol 518F Fluorescence free (Carl Zeiss)
PDGF stock solution: 50 μg/ml PDGF-BB (Sigma-Aldrich) in H2O with 4 mM HCl and 0.1% BSA.
Torin1 stock solution: 1 mM torin1 (TOCRIS) in DMSO
Leucine O-Methyl ester (LeuOMe) stock solution: 3.25 M leucine O-methyl ester (NovaBiochem) in H2O
-
Inverted fluorescence microscope with appropriate objective, filters/mirrors, detector, and image acquisition software; for example:
Light source (e.g., XBO 75W xenon arc lamp, Carl Zeiss)
Zeiss Axiovert 200M microscope (Carl Zeiss)
40×/1.3-NA oil-immersion objective lens
Dichroic mirror, excitation filters (CFP, YFP), emission filters (CFP, YFP)
Cooled charge-coupled device (CCD) camera (e.g., MicroMAX BFT512, Roper Scientific)
Lambda 10-2 filter changer (Sutter Instruments)
METAFLUOR 6.2 imaging software (Universal Imaging)
PC to run microscope
Spreadsheet application (e.g., Microsoft Excel)
Cell Transfection and Starvation
-
1
NIH3T3 cells are seeded on 35-mm glass-bottom dish and grown to 40–60% confluency at 37 °C with 5% CO2 at the time of transfection.
-
2
Prepare DNA solution: in a microcentrifuge tube, prepare a 0.8 μg of TORCAR plasmid DNA and add Opti-MEM to bring the total volume to 50 μL. Gently mix the plasmid DNA with Opti-MEM and incubate at room temperature for 5 min. Opti-MEM should only be handled under sterile conditions, such as in a tissue culture hood.
-
3
Prepare Lipofectamine solution: in a separate tube, mix 2 μL Lipofectamine 2000 with Opti-MEM for a total volume of 50 μL and incubate at room temperature for 5 min.
-
4
Add all (50 μL) of the Lipofectamine solution dropwise to the DNA solution and pipet gently to mix. Incubate for 20 minutes at room temperature. Mix the Lipofectamine/DNA solution gently and do not vortex.
-
5
In a tissue culture hood, replace the medium in the 35-mm glass-bottom dish containing NIH3T3 cells with 2 ml of pre-warmed DMEM medium without serum. Add all (100 μL) of the Lipofectamine/DNA solution dropwise to the dish and rock the dish gently back and forth to evenly disperse the transfection reagents.
The volumes are given on a per-dish basis. Prepare tubes for each additional dish to be transfected. Optimize the amounts of components according to the Manufacturer’s instruction if necessary. Verify expression of reporter in the transfected cells 18–24 hr after transfection. For subcellular targeted TORCAR, it may be necessary to increase the DNA amount for transfection. -
6
Two hours before imaging, replace the serum-free DMEM with 2 ml of pre-warmed HBSS* imaging buffer (see recipe) for amino acid starvation, and incubate the dish in a CO2 independent incubator at 37 °C.
Prepare microscope for imaging
-
7
Turn on lamp, microscope, filter changer, camera, and computer. Load the METAFLUOR imaging software and an appropriate protocol for acquiring a time course of images for the FRET, CFP, and YFP channels. Use the imaging software to confirm the filter configuration and acquisition protocol are correct for the experiment. Filters should be verified using control experiments to make sure they are appropriate for imaging the desired fluorophores (e.g., CFP and YFP). The filter sets for individual channels are:
FRET — 420DF20 excitation filter, 450DRLP dichroic mirror, 535DF25 emission filter.
CFP — 420DF20 excitation filter, 450DRLP dichroic mirror, 475DF40 emission filter.
YFP — 495DF10 excitation filter, 515DRLP dichroic mirror, 535DF25 emission filter.
-
8
Aspirate the HBSS* imaging buffer from the side of the 35-mm glass-bottom imaging dish containing serum- and amino acid-starved NIH3T3 cells with minimal agitation to the cells. Add 1 mL of fresh HBSS* imaging buffer, gently rock the dish side to side and avoid pipetting directly over the cells.
-
9
Place a drop of immersion oil to the 40× objective and securely position the imaging dish onto the microscope stage. Focus the cells properly using the bright field setting, then switch to fluorescence mode and select cells exhibiting the appropriate healthy morphology and the desired expression level and cellular distribution of reporter.
Acquire images and data
-
10
Begin acquiring a time series of images for each channel (e.g., CFP, FRET, YFP). Use the imaging software to set the time lapse interval between each set of acquisitions, as well as the excitation exposure time for each individual channel. Images were typically taken every 25–30 seconds for TORCAR, and typical exposure times for FRET, CFP, and YFP are 500, 500, and 50 milliseconds, respectively, though these values can be adjusted based on expression levels and microscope components.
-
11
Stimulate mTORC1 activity. Acquire several sets of images in order to establish a baseline (generally 3 to 5 minutes, or until the baseline is flat). Take 300 to 500 μL of imaging buffer from the imaging dish and add to a microcentrifuge tube containing 1 μL of PDGF (50 μg/ml). Gently add this mixture back to the imaging dish (final concentrations: 50 ng/ml PDGF) by pipetting down to the side of the dish, careful not to disturb the cells. Pipet carefully to evenly mix the drug throughout the dish. Continue acquiring the image series to capture the stimulated response. To stimulate mTORC1 activity using leucine O-methyl ester (LeuOMe), the final concentration is 7.5 mM (2 μL of stock solution at 3.25 M).
-
12
(Optional) Inhibit mTORC1 activity. Take 300 to 500 μL of imaging buffer from the imaging dish and add to a microcentrifuge tube containing a small volume of inhibitors for mTOR kinase. Gently add this mixture back to the imaging dish (final concentration: 1 μM torin1) by pipetting down to the side of the dish, careful not to disturb the cells. Pipet carefully to evenly mix the drug throughout the dish. Continue acquiring the image series for several additional minutes to capture the response.
Analyze images and data
-
13
Using the imaging software, draw regions of interest (ROIs) for each fluorescent cell in the field, and choose a region where no transfected cells or no cells are present for background correction. Log the fluorescence intensities from each channel acquired during the experiment.
-
14Export the data to an excel file and calculate the FRET emission ratio (cyan-to-yellow) at each time point using the logged emission intensity data and the following equation.
-
15
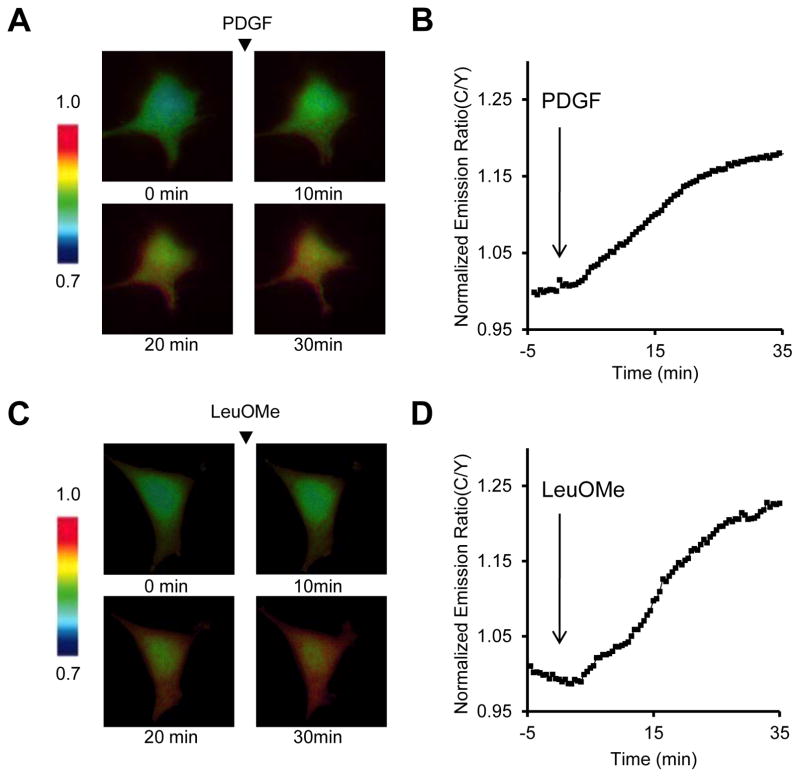
Set the time point immediately after addition of drug as time 0. Calculate the average of emission ratio before addition of drug, and normalize the emission ratio before addition of drug as 1. Generate a plot of the normalized cyan-to-yellow emission ratio versus time (Figure 2A and 2C). Pseudocolor images for each acquisition can also be generated using the METAFLUOR software to indicate the cyan-to-yellow emission ratio, where warmer colors represent high C/Y emission ratio and cooler colors low C/Y emission ratio (Figure 2B and 2D).
Figure 2.
TORCAR responses in live cells. Serum- and amino acid-starved NIH3T3 cells expressing TORCAR were stimulated with 50 ng/ml PDGF (A, B) or 7.5 mM LeuOMe (C, D). (A, C) Time courses of the cyan-to-yellow emission ratio of TORCAR upon stimulation. (B, D) Pseudo-colored images showing the FRET responses of TORCAR to PDGF (B) or LeuOMe (D) stimulation.
SUPPORT PROTOCOL
Maintenance of NIH3T3 cells in culture
The protocol below outlines the procedure for passaging NIH3T3 cells, both for prolonged maintenance and for use in imaging experiments.
Materials
NIH3T3 cells (American Type Culture Collection)
70% Ethanol (EtOH)
Dulbecco’s Phosphate Buffered Saline without Ca2+ or Mg2+ (DPBS, Gibco)
NIH3T3 cell culture medium (see recipe)
0.25% Trypsin/ethylenediamine tetraacetic acid (EDTA) solution (Gibco)
Tissue culture incubator
Tissue culture hood
Tissue culture flasks (e.g., T-25 cm2, BD Falcon)
35-mm glass-bottom imaging dishes (MatTEK)
Protocol steps
Maintain NIH3T3 cells in T-25 cm2 flasks in a humidified, 37 °C incubator with a 5% CO2 atmosphere. Passage cells whenever they reach 50 to 70% confluence by briefly examining the cells under a standard tissue culture microscope. The NIH3T3 cells should not be allowed to become completely confluent.
Before taking out the NIH3T3 cells from incubator for passaging, sterilize the tissue culture hood with 70% ethanol and have all the required supplies at hand, including pipets, pipet tips, flasks, media aliquots, dishes, etc., are sterile and available in the hood before beginning.
Examine the cells under a microscope for cell confluency and morphology. Carefully transfer the T-25 flask from the incubator to the hood.
Remove the culture medium from the flask. Gently wash two times with 2 mL of DPBS without calcium and magnesium to remove all traces of serum.
Pipet 300 μL of the trypsin/EDTA solution into the flask, and gently rock the flask back and forth several times. Let sit for 1 to 2 minutes, until 90% of cells begin to disperse (round up) by examining under a microscope.
Add 4.7 mL of fresh pre-warmed culture media to the flask and mix the cells by gently pipetting up and down. Avoid over-pipetting as this could damage the cells.
Perform a 1:6 split into a new T-25 flask to maintain the cells. Split cells every two or three days.
For imaging experiments, perform a 1:3 split of cells into the 35-mm glass-bottom dishes. Cells should reach 40–60% confluency after approximately 24 hours, and should be transfected at this confluency.
REAGENTS AND SOLUTIONS
All solutions should be made using water with 18.2 MΩ-cm resistivity, and prepared under sterile conditions in a tissue culture hood.
HBSS* imaging buffer
1× Hank’s Balanced Salt Solution (made from 10× HBSS (Gibco cat. 14065))
20 mM HEPES (Invitrogen)
2.0 g/L D-glucose (Sigma)
Adjust pH to 7.4, then filter sterilize using a 0.22 μm filter. Store a 50 mL aliquot at room temperature near the microscope room and store the remainder at 4 °C.
NIH3T3 cell culture medium
Dulbecco’s Modified Eagle’s Medium, 1 g/L glucose (Gibco 11885084)
10% calf serum (CS, ATCC 302030)
1% penicillin-streptomycin (Sigma-Aldrich)
Store culture media at 4 °C and warm to 37 °C before using with cells.
COMMENTARY
Background Information
Assays to measure the mTORC1 kinase activity
Multiple signal inputs, such as growth factors, amino acids, glucose and changes in energy status, stimulate the activity of mTORC1. To measure the mTORC1 kinase activity in vitro, a conventional biochemical approach is to incubate the immunoprecipitated mTORC1 complex with a purified protein substrate in the presence of ATP. Phosphorylation of the purified protein substrate by mTORC1 can then be assessed by different detection methods, such as radioactive detection (Hara et al., 1997; Wang et al., 2009) and chemiluminescent detection following immunoblotting (Sancak et al., 2007; Thoreen et al., 2009). One of the keys in this assay is the robust immunoprecipitation of the functional mTORC1 from cells using antibodies against subunits in the complex, the most commonly used being Raptor (Guertin et al., 2006; Sancak et al., 2007) or epitope tagged exogenous Raptor (Sancak et al., 2007). As the interaction between mTOR and Raptor is sensitive to Triton X-100 or NP-40, the mTORC1 complex is isolated using the zwitterionic detergent 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) from control or stimulated cells (Kim et al., 2002). Following incubation of the isolated mTORC1 with purified substrate and ATP, the mTORC1 activity can be measured, for example, as incorporation of 32P into a substrate by determining the 32P-radioactivity, or as the western blotting using antibodies against the phosphorylated substrates (Thoreen et al., 2009). This approach is useful in the determination of the functional status of mTORC1, quantification of the kinetics of substrate phosphorylation, and the comparison of in vitro activity in mutated complexes. Conventionally these assays are relatively time-consuming, and a loss of activity can occur during the immunoprecipitation process.
Direct detection of the phosphorylation level of mTORC1 substrates in cell lysates provides another method to measure the cellular mTORC1 activity, which also takes into account the potential role of endogenous phosphatase activity (Sancak et al., 2007). This approach depends on the availability of phosphorylation site-specific antibodies that recognize exclusively the phosphorylated substrates of mTORC1, including T37/46 in 4EBP1 (Gingras et al., 1999; Thoreen et al., 2009), T389 in S6K (Burnett et al., 1998), and S757 in Ulk1 (Kim et al., 2011).
Complementary to the in vitro kinase assay and phosphorylation specific antibody-based assay using lysed cells, a more recently developed approach involves a genetically encoded FRET-based reporter for quantification of endogenous mTORC1 activity in living cells (Zhou et al., 2015). This approach is based off the quantification of changes in FRET induced by a phosphorylation-dependent conformational change of the reporter TORCAR. In particular, TORCAR can be genetically targeted to specific subcellular location, such as plasma membrane, lysosomes, and nucleus. Subcellular targeted TORCAR allows detection of location-specific mTORC1 signaling in response to distinct upstream signals, such as growth factors and amino acids, and therefore, allowing a better understanding of the cellular regulation of mTORC1 signaling.
Critical Parameters
Starvation of cells for the imaging
This step focuses on starvation of cells for an optimized TORCAR response. As the activity of mTORC1 can be stimulated by growth factors and amino acids, the protocol described here is to starve cells for both serum and amino acids to lower the basal phosphorylation of the reporter before stimulation of mTORC1.
Criteria for selecting proper cells
Selecting proper cells is essential for successful imaging experiments. Unhealthy cells are likely to detach from the dish during imaging. The criteria for selecting cells include proper cell morphology, desired expression of reporter, and cellular distribution. First, cell morphology should be inspected to confirm the healthy status of the cells to be imaged. For example, healthy NIH3T3 cells should appear fibroblast-like and have elongated shapes, rather than being balled-up or rounded. Second, select cells with moderate fluorescence intensities. Too-high intensities due to high expression of the probe could raise the possibility of perturbation of endogenous signaling pathways, whereas too-low intensities may lead to reduced signal-to-noise ratios. Proper cellular distribution is examined by fluorescence intensity. For TORCAR without targeting, the yellow fluorescence and cyan fluorescence should be uniform and evenly distributed throughout the cells. For TORCAR with subcellular targeting, selected cells should exhibit well-localized fluorescence depending on the subcellular targeting. For example, co-staining with LysoTracker could be used to verify the specific expression of Lysosome-targeted TORCAR at the lysosomes.
Control experiments
It is important to confirm that changes in FRET ratio from TORCAR are due to the changes in mTORC1 activity. Specificity can be examined by pretreating cells with mTOR inhibitors, such as torin1 and torin2, or the allosteric inhibitor of mTORC1, rapamycin, before stimulating the cells. These treatments should block the FRET response. Addition of the inhibitors following the stimulation, should lead to a reverse of the TORCAR response. However, the reversal of the response may not be as robust as the blockade of response by pretreatment depending on the cellular phosphatase activities. Another negative control experiment is to use a reporter variant containing the T37A/T46A mutations (TORCAR-TA).
Troubleshooting
Table 1 describes some potential areas where problems may arise during the TORCAR experiment, along with possible causes and strategies for overcoming or avoiding these problems.
Table 1.
Possible Problems and Solutions
| Problem | Possible causes | Possible solutions |
|---|---|---|
| Dim cells | Poor transfection | Optimize transfection |
| Incorrect filter set | Verify microscope filter set | |
| No stable baseline | Changes in cell morphology | Optimize imaging conditions (e.g. temperature, imaging buffer) |
| Photobleaching | Reduce exposure time or decrease the illumination using neutral density filters | |
| Smaller/slower responses than expected | Incorrect drug addition | Mix well or use a perfusion system |
| Drug concentration too low | Verify amount of drug needed to full activate kinase | |
| Low expression of reporter | Optimize transfection | |
| High basal activity | Optimize the starvation conditions |
Anticipated Results
Treatment with PDGF should produce a increase in CFP fluorescence intensity, along with an decrease in the FRET channel.
Time Considerations
All of the steps outlined above can be completed over a period of 3 days. This includes passaging cells into 35-mm imaging dishes (~30 minutes, day 1), transfecting the cells (30 minutes to 1 hour, day 2), and acquiring and analyzing the data (1 hour per dish, day 3).
Significance Statement.
Mechanistic target of rapamycin complex 1 (mTORC1) integrates a variety of inputs such as growth factors and amino acids, to regulate cell metabolism and growth. As mTORC1 serves as a hub molecule regulating a number of processes, the activity of mTORC1 is exquisitely regulated. Spatial compartmentalization of mTORC1 signaling has been suggested to constitute an important mechanism to achieve signal specificity. Here we describe a protocol to track mTORC1 activity in living cells with high spatiotemporal resolution using a genetically encoded FRET-based mTORC1 activity reporter. This approach enables the detection of the mTORC1 activity in response to different stimuli, and will facilitate our understanding of the molecular mechanisms that regulate the mTORC1 activity in its native cellular environments.
Acknowledgments
This work is supported by National Institutes of Health R01 DK073368 and DP1 CA174423, and DK084171.
LITERATURE CITED
- Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. Architecture of human mTOR complex 1. Science (New York, NY. 2016;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends in cell biology. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nature cell biology. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goberdhan DC, Wilson C, Harris AL. Amino Acid Sensing by mTORC1: Intracellular Transporters Mark the Spot. Cell metabolism. 2016;23:580–589. doi: 10.1016/j.cmet.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Developmental cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. The Journal of biological chemistry. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- Ilagan E, Manning BD. Emerging Role of mTOR in the Response to Cancer Therapeutics. Trends in Cancer. 2016;2:241–251. doi: 10.1016/j.trecan.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Molecular cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence JC, Jr, Sturgill TW, Harris TE. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. The Journal of biological chemistry. 2009;284:14693–14697. doi: 10.1074/jbc.C109.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Clister TL, Lowry PR, Seldin MM, Wong GW, Zhang J. Dynamic Visualization of mTORC1 Activity in Living Cells. Cell reports. 2015;10:1767–1777. doi: 10.1016/j.celrep.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]