Abstract
Peptide deformylase activity was thought to be limited to ribosomal protein synthesis in prokaryotes, where new peptides are initiated with an N-formylated methionine. We describe here a new human peptide deformylase (Homo sapiens PDF, or HsPDF) that is localized to the mitochondria. HsPDF is capable of removing formyl groups from N-terminal methionines of newly synthesized mitochondrial proteins, an activity previously not thought to be necessary in mammalian cells. We show that actinonin, a peptidomimetic antibiotic that inhibits HsPDF, also inhibits the proliferation of 16 human cancer cell lines. We designed and synthesized 33 chemical analogs of actinonin; all of the molecules with potent activity against HsPDF also inhibited tumor cell growth, and vice versa, confirming target specificity. Small interfering RNA inhibition of HsPDF protein expression was also antiproliferative. Actinonin treatment of cells led to a tumor-specific mitochondrial membrane depolarization and ATP depletion in a time- and dose-dependent manner; removal of actinonin led to a recovery of the membrane potential consistent with indirect effects on the electron transport chain. In animal models, oral or parenteral actinonin was well tolerated and inhibited human prostate cancer and lung cancer growth. We conclude that HsPDF is a new human mitochondrial enzyme that may provide a novel selective target for anticancer therapy by use of actinonin-based antibiotics.
Introduction
The N-terminal methionine excision (NME) pathway is an essential mechanism in all organisms (1, 2). Although N-formyl groups modify the first methionine of all newly synthesized proteins in the cytoplasm of prokaryotes and organelles of eukaryotes, this formyl-methionine is often not retained and is cleaved from the mature protein as a part of a posttranslational modification that may be linked to the N-end rule governing the half-lives of proteins (3). Two enzyme families are involved in this NME pathway, peptide deformylase (PDF) and methionine aminopeptidase (MAP). PDF removes all N-formyl groups and unmasks the amino group of the first methionine, which is a prerequisite for the subsequent action of MAP (2).
Although most of the initial work characterizing PDFs has focused on E. coli, there is an increasing interest in PDF as a target for developing therapies against other pathogens. These include Thermus thermophilus and Bacillus stearothermophilus (4), Staphylococcus aureus (5), Haemophilus influenzae, Streptococcus pneumoniae (6), Leptospira interrogans (7), and Mycobacterium tuberculosis (8). In fact, numerous PDF inhibitors have been shown to have in vitro activities against several of these pathogens, and it has been speculated that these same molecules could be used as broad-spectrum antibiotics against a variety of infectious diseases (9).
The development of antimicrobial agents targeting PDF could encounter a potential hurdle. While PDF was originally thought to be a prokaryotic enzyme, recent genome-database searches have revealed eukaryotic PDF-like sequences in parasites, plants, and mammals (10), and recent studies have shown that these eukaryotic PDFs are active in vitro and in vivo (11–15). There are 3 classes of PDFs based on structural and sequence analyses (16, 17). Type 1 is divided into 2 subclasses: PDF1a includes plant and mammalian mitochondrial PDFs, whereas PDF1b includes enzymes found in Gram-negative bacteria, some Gram-positive bacteria, and plants. The eukaryotic PDF1b enzymes are targeted to both plastids and mitochondria (10). Type 2 and type 3 PDFs are found only in Gram-positive bacteria; however, type 3 PDFs have no associated deformylase activity.
The first eukaryotic PDF to be identified was in the higher plant Arabidopsis thaliana. The 2 deformylase-like genes in this plant code for functional eukaryotic PDFs (10). These enzymes appear to be localized in the mitochondria and plastids (10, 12), 2 organelles of prokaryotic origin. Another eukaryotic PDF was later identified and characterized in Plasmodium falciparum, the primary parasite responsible for malaria in humans (11). This catalytically active deformylase (PfPDF) is inhibited by actinonin and other known PDF inhibitors, which suggests PfPDF as a potential target for antimalarial therapies (11, 18).
The human Pdf gene contains 2 exons on chromosome 16, and the gene product is homologous to other characterized PDFs, albeit with some significant differences. The human Pdf mRNA has been reported to be expressed at the same level in all types of human tissues (10). More recently, we and other investigators have shown that a recombinantly expressed human PDF (Homo sapiens PDF, or HsPDF) is active in vitro (13–15). Therefore, the data suggest that N-terminal protein processing is evolutionarily conserved and may be important in the chloroplasts or mitochondria of at least some eukaryotic organisms, although its role in mammals is controversial (8, 14).
Previous work in our laboratory showed that actinonin, a naturally occurring antibiotic derived from a Streptomyces species (19) that inhibits aminopeptidase N (APN, or CD13), has antiproliferative effects on human leukemia and lymphoma cells in vitro and antitumor activity in a syngeneic AKR leukemia mouse model (20, 21). However, the antiproliferative effects were seen in APN-negative cells, and therefore the activity could not be mediated by APN inhibition. Therefore, we sought another target of actinonin that might explain its selective anticancer activity. Actinonin’s potent inhibition of bacterial, plant, and parasitic PDFs (11, 12, 22) prompted our investigation of the possibility that HsPDF may be a new cancer cell target if it were a functionally important human mitochondrial enzyme. We show here that HsPDF is a functional mitochondrial enzyme necessary for cell growth and proliferation. Moreover, we designed and synthesized a class of new inhibitors, based on actinonin, which selectively inhibit the growth of a broad variety of human cancer cells in vitro and human tumors in vivo in mice. We also propose a model for actinonin’s mechanism of action. These findings have implications for the development of various therapeutic strategies for bacteria, mycobacteria, parasites, or cancer that target this enzyme.
Results
HsPDF activity against human mitochondrial substrates.
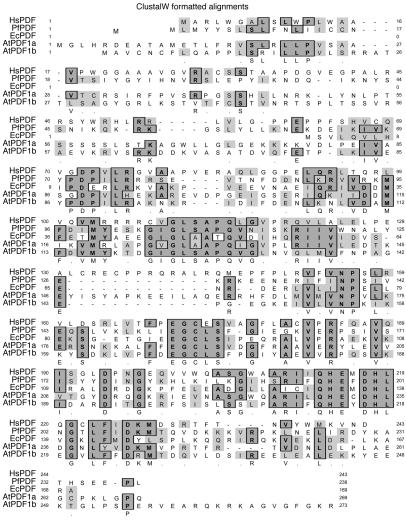
The ClustalW (23) sequence alignment (Figure 1) of amino acid residues of HsPDF compared with other forms of PDF shows that the human protein has significant identity (30–40%) to the other catalytically active proteins. In addition, the critical metal-binding residues (C172, H214, H218) and the catalytic residue (E215) are conserved (18, 24, 25). We have previously described the expression cloning and purification of HsPDF for enzymatic studies (13). N-terminal truncation allowed expression of a soluble, active enzyme. All enzymatic analyses reported here were conducted with the cobalt-substituted N-terminal truncation mutant extended with a C-terminus 6x-histidine tag for purification.
Figure 1.
ClustalW alignment of the deduced HsPDF amino acid sequence with bacterial, plant, and parasitic PDF sequences from the National Center for Biotechnology Information (NCBI) database using the MacVector program (Accelrys Inc.). Hs, Homo sapiens (NP_071736); Pf, P. falciparum (NP_704619); Ec, E. coli (NP_417745); At, A. thaliana (AAG33973 and AAG33980).
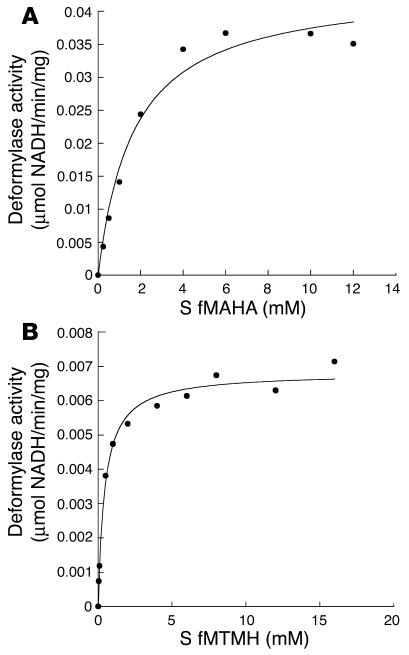
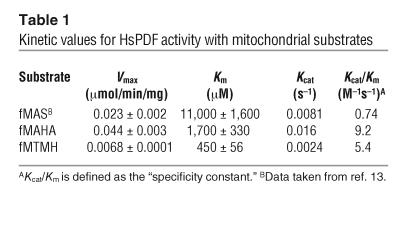
We examined the suitability of different formylated peptides as potential substrates (Figure 2, A and B). HsPDF was able to deformylate 2 formylated peptides, formyl-Met-Ala-His-Ala (fMAHA) and formyl-Met-Thr-Met-His (fMTMH), that were designed to mimic the N-terminal amino acid sequence of 2 different human mitochondrially encoded proteins, cytochrome c oxidase II and NADH dehydrogenase subunit V, respectively. The Kcat/Km values (Table 1) for these mitochondrial peptides are 5- to 10-fold higher than those for the generic substrate formyl-Met-Ala-Ser (fMAS), which does not correlate with any mitochondrially encoded proteins. This increase in Kcat/Km, sometimes referred to as the “specificity constant,” suggests a higher catalytic efficiency toward the mitochondrial peptides. Interestingly, HsPDF was not active against the bacterial peptide formyl-Met-Leu-Phe (fMLP/fMLF) (13).
Figure 2.
Kinetic analysis of HsPDF activity against mitochondrial peptides. (A) HsPDF activity was measured as a function of substrate (S) fMAHA using the formate dehydrogenase–coupled PDF assay. (B) HsPDF activity was measured as a function of substrate fMTMH using the formate dehydrogenase–coupled PDF assay. All determinations are representative of at least 2 independent experiments. Kinetic curves were generated by the KaleidaGraph computer program (Synergy Software). Kinetic values derived from these curves are displayed in Table 1.
Table 1.
Kinetic values for HsPDF activity with mitochondrial substrates
Localization of HsPDF in the mitochondria of living cells.
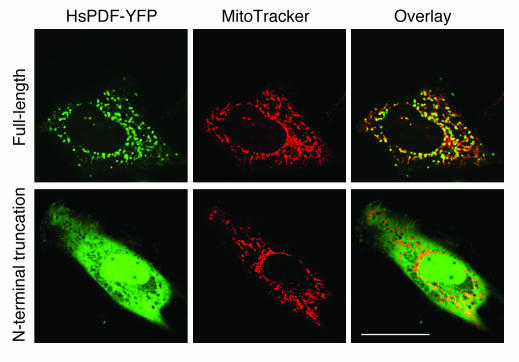
The full-length amino acid sequence of HsPDF contains an N-terminal sequence that is predicted to serve as a mitochondrial targeting motif (11). To test the hypothesis that HsPDF is targeted to the mitochondria, we transfected HeLa cells with HsPDF–yellow fluorescent protein (HsPDF-YFP) fusion constructs. Live-cell confocal microscopy showed that the full-length HsPDF colocalizes with mitochondria that are stained with MitoTracker Red (Invitrogen Corp.) (Figure 3). As a control, we tagged with YFP an N-terminal truncation mutant of HsPDF (AAs 64–244) that lacks the mitochondrial signal sequence. The truncated HsPDF-YFP fusion protein failed to colocalize with the mitochondrial marker and was instead homogenously distributed in the cytosol and nucleoplasm, a pattern indistinguishable from that of YFP expressed alone.
Figure 3.
The N-terminus targets HsPDF to the mitochondria. HeLa cells were transfected with full-length or N-terminally truncated HsPDF (AAs 64–244) tagged at the C-terminus with YFP and imaged alive by laser scanning confocal microscopy 24 hours after transfection and 30 minutes after addition of MitoTracker Red CMXRos to the media. Scale bar: 10 μm.
Actinonin inhibits cell growth in various human tumor cell lines.
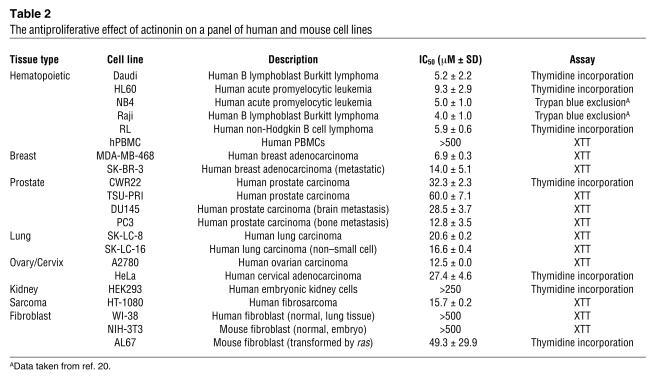
We have previously shown that actinonin has antiproliferative effects on both HL60 leukemia cells and Daudi lymphoma cells, and that antiproliferative activity does not correlate with APN expression. Therefore, we now hypothesized that HsPDF is a target of actinonin in tumor cells. As HsPDF is expressed diversely in human tissues (10, 26), we investigated whether actinonin inhibits the growth and proliferation of a variety of human tumor cell lines. Actinonin had antiproliferative activity against 16 of 17 human tumor cell lines tested, representing 9 different cell lineages (Table 2). Only HEK293 renal cells were resistant. In comparison, a number of “normal” cell lines were found to be resistant to actinonin, as well. These were the WI-38 and NIH-3T3 cell lines (human and mouse normal fibroblast cell lines, respectively) and normal human PBMCs (hPBMCs). The actinonin-sensitive mouse fibroblast cell line AL67 differs from the NIH-3T3 parental cells in that it has been transformed by ras. We have previously reported that actinonin may have some myelosuppressive activities, as evidenced by decreased colony formation of human bone marrow cells (20). However, the dose required for 50% growth inhibition of these normal cells was more than 5 times the IC50 required for the tumorigenic hematopoietic cell lines tested here.
Table 2.
The antiproliferative effect of actinonin on a panel of human and mouse cell lines
Actinonin analogs reveal a link between HsPDF inhibition and antiproliferative activity.
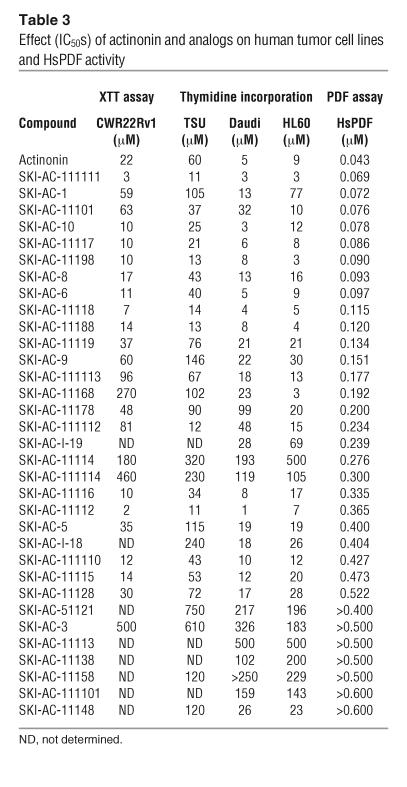
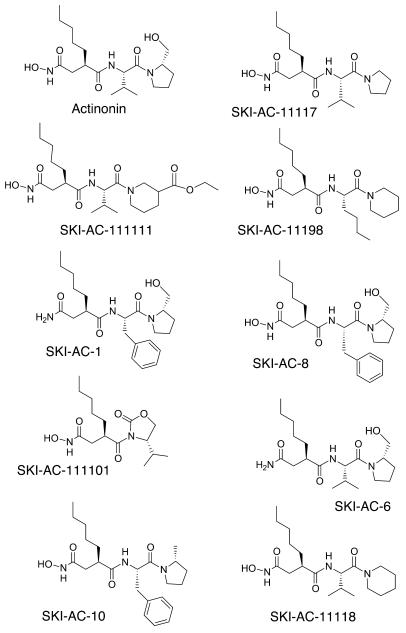
Thirty-three analogs of actinonin were synthesized to determine whether there was a correlation between inhibition of HsPDF activity and human tumor cell growth and to find more potent and specific compounds. We screened actinonin and its analogs for both HsPDF enzyme inhibition and human tumor cell growth inhibition as measured by XTT (tetrazolium salt) metabolism and tritiated thymidine incorporation (Table 3 and Figure 4). The cell lines used for the screen were CWR22Rv1 and TSU-PRI (human prostate cancer), Daudi (human Burkitt lymphoma), and HL60 (human acute myeloid leukemia). The 2 prostate cancer cell lines (APN-negative) were chosen as representative solid tumor cell lines, with and without androgen-resistance characteristics. As actinonin can also inhibit APN, 2 hematopoietic cell lines were chosen, 1 with and 1 without APN expression. All 9 compounds that inhibited HsPDF with an IC50 of less than 0.1 μM were also potent antiproliferatives (IC50 < 50 μM on at least 1 cell line). Of the 16 compounds that inhibited HsPDF with an IC50 of 0.2 μM or less, all were also antiproliferatives. Moreover, 6 of the 7 compounds with no measurable HsPDF inhibition exhibited no inhibition of cell growth. These data show that HsPDF inhibition is consistently associated with antiproliferative activity against human tumor cells. Conversely, lack of activity against HsPDF is generally associated with inability to inhibit the growth of tumor cells. The observation that 9 compounds displaying weak HsPDF inhibition were antiproliferative suggests that these compounds inhibit cell growth via HsPDF-independent mechanisms.
Table 3.
Effect (IC50s) of actinonin and analogs on human tumor cell lines and HsPDF activity
Figure 4.
Chemical structures of actinonin and selected analogs with the lowest IC50 against HsPDF (see Table 3).
Knockdown of HsPDF by small interfering RNA leads to inhibition of cell proliferation.
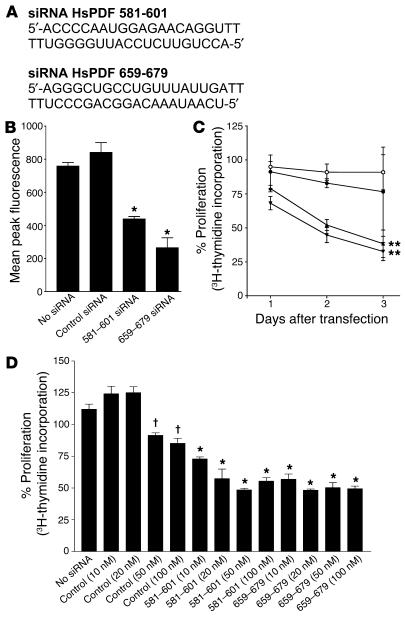
If the antitumor effect of actinonin is mediated by inhibition of HsPDF, then cell growth must depend to some extent on HsPDF activity. To test this hypothesis, we examined the effect of silencing the human Pdf gene on tumor cell growth. Transfection of HeLa cells with small interfering RNA (siRNA) duplexes that specifically target human Pdf mRNA (Figure 5A) led to a significant decrease in both Pdf mRNA (data not shown) and protein expression (Figure 5B) as well as a decrease in cell proliferation as evidenced by the tritiated thymidine incorporation assay (Figure 5C). Control siRNAs (nonspecific duplexes) had no effect on mRNA levels, protein expression, or proliferation. The target-specific siRNAs did not reduce expression of the fluorescent protein control (data not shown). Kinetic analysis revealed that the substantial decrease of both protein expression and cell proliferation was not observed until 48 hours after transfection, consistent with the requirement for catabolism of preformed protein to observe the functional effects of gene silencing. The siRNA sequences that showed significant inhibition of cell proliferation targeted the region between bases 580 and 680. Several ineffective siRNA sequences targeted other regions of the mRNA, consistent with the general observation that siRNA is effective for some but not all sequences within a message.
Figure 5.
siRNAs designed to target HsPDF inhibit protein expression and tumor cell proliferation. (A) siRNAs were designed to target HsPDF. The 2 siRNA sequences that were effective were HsPDF 581–601 and HsPDF 659–679. The numbers refer to the nucleotide number relative to the start codon of human Pdf mRNA. (B) HeLa cells stably expressing HsPDF-YFP fusion protein were transfected with 100 nM of control or HsPDF-specific siRNAs. Mean peak fluorescence determined 48 hours after transfection shows that the target-specific siRNAs significantly reduced protein expression (*P < 0.001). (C) HeLa cells were transfected with 100 nM of control or HsPDF-specific siRNAs. Cell proliferation was determined by tritiated thymidine incorporation over a period of 3 days following transfection. The control siRNA group (filled circles) showed no significant changes (P > 0.05) in cell proliferation when compared with the no-siRNA control group (open circles). siRNA HsPDF 581–601 (upright triangles) and siRNA HsPDF 659–679 (inverted triangles) caused significant decreases (**P < 0.05) in cell proliferation 1 and 2 days after transfection. (D) HeLa cells were transfected with 10, 20, 50, and 100 nM of control or HsPDF-specific siRNAs. Cell proliferation was determined by tritiated thymidine incorporation 2 days after transfection. The HsPDF-specific siRNAs significantly inhibited cell proliferation (*P < 0.001) at all 4 doses when compared with both the no-siRNA group and the corresponding dose of control siRNA, while significant inhibition ( P < 0.001) was seen only at the higher doses (50 and 100 nM) of the control group.
In order to address concerns regarding off-target and concentration-dependent effects on mammalian gene expression (27–29), we also tested a range of doses for our effective siRNAs. It has been reported that a characteristic feature of nonspecific effects on gene expression is dependence on siRNA concentration and that nonspecific effects occur at siRNA concentrations of 100 nM but not at 20 nM (29). We found that the 2 effective siRNAs were able to significantly inhibit tumor cell proliferation at a dose as low as 10 nM (Figure 5D), which is well below the concentration for nonspecific effects reported in the literature. Moreover, there was a concentration-dependent effect only with the control siRNAs, not with the target-specific siRNAs. These data confirm that HsPDF is important for cancer cell growth and survival.
Thus, the antiproliferative effects of actinonin and numerous analogs on a wide variety of human tumor cell lines, in combination with the requirement of HsPDF for cell growth, provide compelling evidence that HsPDF is a critical target of actinonin and that the inhibition of this protein in the mitochondria leads to cell death in tumor cells.
Actinonin causes selective mitochondrial membrane depolarization.
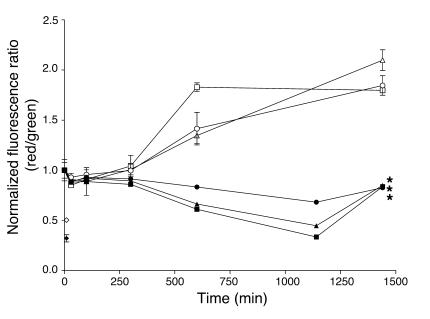
The evidence that actinonin targets HsPDF, a mitochondrial protein, suggests that actinonin may mediate its antiproliferative action via disruption of mitochondrial function. A key early indicator of mitochondrial toxicity is the loss of potential across the mitochondrial membrane (30). We monitored mitochondrial membrane potential by using a fluorescent dye, JC-1 (described in Methods), which exhibits a membrane potential-dependent accumulation in mitochondria. JC-1 emits at 525 nm (green) in monomeric form; in the presence of high mitochondrial membrane potential, JC-1 forms red (590 nm) fluorescent aggregates. Thus, mitochondrial depolarization can be quantitated by a decrease in the red/green fluorescence intensity ratio. We show that actinonin treatment of RL cells (human B cell lymphoma line) causes a significant time- and dose-dependent depolarization of the mitochondrial membrane (Figure 6). The magnitude of the depolarization at the highest dose (100 μg/ml) is equal to that of the positive control, carbonyl cyanide m-chlorophenylhydrazone (CCCP), a proton ionophore that irreversibly uncouples respiration (31). The kinetics of the actinonin effect were far slower than for CCCP, which directly disrupts the electron transport chain; this is consistent with an expected indirect effect of actinonin on this pathway. In addition, when cells were treated with actinonin for 24 hours and then removed from treatment, the mitochondrial membrane potential recovered to the cells’ normal resting potential after approximately 8 hours (data not shown). This actinonin-dependent depolarization and subsequent recovery with actinonin wash-out was also seen with HeLa (cervical adenocarcinoma) and HL60 (myeloid leukemia) cell lines. In contrast, normal peripheral blood lymphocytes incubated with actinonin at the same 3 doses did not exhibit a depolarization over time (Figure 6).
Figure 6.
Actinonin selectively causes mitochondrial membrane depolarization in a time- and dose-dependent manner. RL lymphoma cells (bottom 3 curves) were incubated with actinonin at 10 μg/ml (filled circles), 20 μg/ml (filled triangles), or 100 μg/ml (filled squares), treated with JC-1 dye, and analyzed by flow cytometry. Actinonin treatment led to a time- and dose-dependent depolarization of the mitochondrial membrane, as evidenced by the significant decrease (*P < 0.05) in the fluorescent red/green ratio. CCCP (filled diamond) was used as a positive control and showed depolarization after 10 minutes of treatment. Normal peripheral blood lymphocytes (top 3 curves) were incubated with actinonin at 10 μg/ml (open circles), 20 μg/ml (open triangles), or 100 μg/ml (open squares), treated with JC-1 dye, and analyzed by flow cytometry. There was no time- or dose-dependent depolarization of the mitochondrial membrane with actinonin treatment, although the CCCP control (open diamond) was effective at 10 minutes. All graphed values (mean ± SD) represent a normalized fluorescence ratio (red/green) calculated by division of the ratio for each time point by the ratio at time 0 (RL cells at t0, 1.853 ± 0.079; normal cells at t0, 0.349 ± 0.106).
We then hypothesized that ATP depletion might be a possible consequence of actinonin-induced mitochondrial dysfunction. To further examine this question, we measured ATP levels in Daudi cells treated with varying doses of actinonin (5, 10, and 20 μg/ml) and found that it depleted ATP levels in a time- and dose-dependent manner. At the highest dose of 20 μg/ml, actinonin depleted ATP levels by 56.0% at 12 hours, 67.3% at 24 hours, and 95.8% at 36 hours. We also conducted some preliminary experiments to examine actinonin-induced apoptosis. We previously published a report (20) that actinonin induces a low level of apoptosis (approximately 10%) in non–APN-expressing cells, based on the TUNEL assay. We confirmed this low level of apoptosis using annexin V staining and found no more than 7% annexin V–positive, propidium iodide–negative Daudi cells in response to the same dose of actinonin.
Actinonin inhibits human tumor xenograft growth in mice.
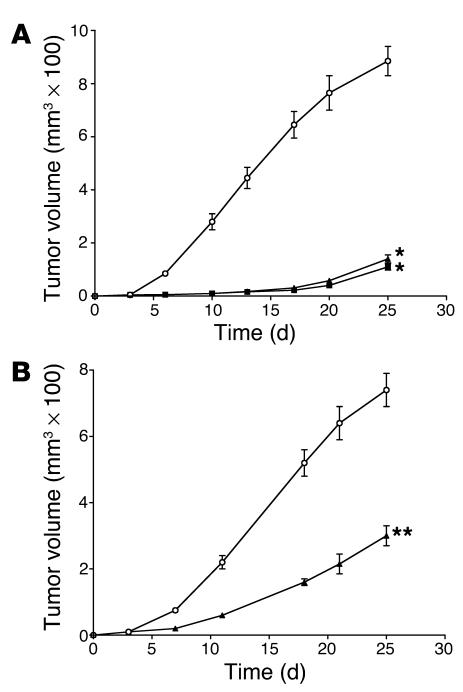
Although previous work has shown that actinonin was effective in a syngeneic AKR leukemia mouse model, the broad anticancer activity seen here in vitro prompted an investigation of whether actinonin would be effective against mouse xenograft models of human solid tumors. Actinonin has been safely administered to mice as an antibiotic at doses up to 400 mg/kg (19). Therefore, it does not appear to have significant toxicity to normal tissues, despite its antitumor activity in vitro. Remarkably, actinonin exhibited significant antitumor activity when given i.p. or orally in a CWR22 human prostate tumor xenograft model in nude mice (Figure 7A). During treatment, the animals showed no signs of clinical toxicity. The 250-mg/kg i.p. regimen and the 500 mg/kg oral regimen were similarly effective at inhibiting tumor growth; this suggests that actinonin is orally bioavailable. Similar antitumor activity was seen in 2 other human tumor xenograft models in nude mice bearing A549 human non–small cell lung cancer (Figure 7B) and PC3 human prostate cancer (data not shown). Therefore, it appears that actinonin has significant activity in vivo against human tumors with tolerable toxicity.
Figure 7.
Actinonin inhibits human tumor growth in xenograft mouse models. (A) Athymic nude mice bearing CWR22 human prostate tumor xenografts were treated with vehicle control (open circles), actinonin at 250 mg/kg i.p. (filled triangles), or actinonin at 500 mg/kg orally (filled squares) twice a day for 2 weeks (excluding weekends) as soon as s.c. tumors were palpable. Tumor size was measured every 3–5 days with calipers. Actinonin administered either i.p. or orally significantly inhibited the growth of the human prostate tumor xenografts (*P < 0.005). (B) Athymic nude mice bearing A549 human non–small cell lung cancer xenografts were treated with vehicle control (open circles) or actinonin at 150 mg/kg i.p. (filled triangles) once a day for 2 weeks (excluding weekends) as soon as s.c. tumors were palpable. Tumor size was measured every 3–5 days with calipers. Actinonin significantly inhibited the growth of the human lung cancer xenograft (**P < 0.01).
Discussion
The biochemistry of protein deformylation in various non-mammalian organisms is of considerable interest because of the potential for use of the enzymes involved as targets for therapeutic interventions. Bacterial PDF has been studied extensively and is currently considered a target for the development of antibacterial drugs (16, 17, 32–38). The goal of this work was to determine whether the newly discovered human mitochondrial deformylase was the target of actinonin, and whether actinonin had broad cancer therapeutic activity. APN was originally considered the relevant target of actinonin in mammalian cells (39, 40). However, previous work in our laboratory (20, 21) demonstrated that the antileukemic activity of actinonin, a peptidomimetic antibiotic, is not mediated by APN. This finding, in combination with actinonin’s known bactericidal effects that are due to its inhibition of PDF (5, 6, 22), led us to examine the possibility that HsPDF may be an important enzyme in human cells and, thus, the potential target of actinonin in tumor cells. Therefore, we also explored the possibility that a functional human peptide-deformylase protein might be a potential target for novel cancer therapeutics.
We show here that HsPDF is expressed in human mitochondria and capable of selectively deformylating model human mitochondrial N-methionyl formylated proteins. Moreover, we show that actinonin and numerous newly designed chemical analogs, which are all capable of potent HsPDF inhibition, all result in potent antiproliferative effects on tumor cells. In addition, siRNA reduction of HsPDF mRNA and protein results in reduction in tumor cell proliferation. Finally, we demonstrate that actinonin is an active and tolerable anticancer agent in human solid tumor xenograft models in mice.
Genome-database searches had revealed that a human Pdf homolog exists (10) and that the deduced amino acid sequence exhibits significant sequence similarities to E. coli PDF (EcPDF), including key catalytic residues. The encoded human protein, HsPDF, was found to be enzymatically active in the presence of various N-formylated peptides (13–15). Our results confirmed that HsPDF is enzymatically active for generic N-formylated peptides and, furthermore, has even greater activity for peptides that mimic the N-terminus of human mitochondrially encoded proteins. We also showed that HsPDF is localized in the mitochondria by virtue of an N-terminal targeting sequence.
The complete function and relevance of HsPDF in human cells remain to be determined. Recently, Nguyen et al. (14) suggested that HsPDF is likely to be an evolutionary remnant without any functional role in protein formylation/deformylation, based on HsPDF’s weak catalytic activity. In contrast, Serero et al. (15) conclude that human mitochondria have a functional human PDF that is involved in the conserved NME pathway. Our data support the conclusion that HsPDF is a functional enzyme located in the mitochondria. We extend this to show that HsPDF is important for tumor cell growth and survival and is the target of a new class of small molecules.
These conclusions are consistent with data from plant and bacterial systems. Plant and parasitic PDFs have been shown to be expressed in organelles such as plastids and mitochondria (10–12, 41, 42). HsPDF is nuclear-encoded but predicted to be localized in the mitochondria (11, 43). This prediction was based on the realization that the only nascent proteins requiring deformylation in eukaryotic cells are those synthesized on mitochondrial ribosomes. We have now confirmed this mitochondrial localization. Moreover, HsPDF’s higher specificity toward 2 formylated peptides (fMAHA and fMTMH) designed to mimic the N-terminal sequence of 2 human mitochondrially encoded proteins supports the idea that HsPDF evolved to deformylate proteins that are synthesized in mammalian mitochondria. This subset of cellular proteins is well defined; the mitochondrial genome codes for only 13 polypeptides that are subunits of the various complexes that make up the respiratory chain (44, 45). Thus, inhibition of HsPDF in the mitochondria could presumably lead to disruption of protein complexes in the mitochondrial respiratory chain and possibly even cell death. This hypothesis is consistent with evidence of an analogous role for plant PDF’s deformylation of chloroplast-encoded proteins, which are crucial components of photosystem II (42). Actinonin, which causes bleaching of the plant, inhibits plant PDF and therefore leads to destabilization of specific proteins of photosystem II. The antibacterial and antiplant activities of actinonin would lead to the supposition that actinonin is inhibiting the growth of tumor cells via inhibition of HsPDF. Here we provide evidence to support this hypothesis. Numerous analogs that potently inhibit HsPDF enzymatic activity all potently inhibit cell growth. Nearly all analogs that have no anti-PDF activity are not antiproliferative. In addition, siRNA knockdown of HsPDF confirms that its presence is necessary for cell proliferation.
It has been previously reported that aminopeptidase inhibitors, including actinonin and bestatin, inhibit cell proliferation of leukemia cells via suppression of serine phosphorylation of both MAPK and GSK-3β (46). However, this mechanistic description refers to cellular events in response to APN inhibition and is limited to APN-expressing cells. Furthermore, bestatin does not inhibit HsPDF (data not shown), which suggests that actinonin’s inhibition of HsPDF and subsequent inhibition of APN-negative cell proliferation rely on a different mechanism. We believe that actinonin provides an interesting mechanistic model for its effects because it is inhibiting posttranslational modifications of proteins. HsPDF is thought to be involved in the deformylation and processing of mitochondrially encoded proteins. These proteins make up the subunits of 4 of the 5 complexes in the electron transport chain. When HsPDF is inhibited by actinonin, there would be an accumulation of unprocessed proteins that could lead to 2 possible outcomes. The reduction in properly assembled electron transport chain complexes would lead to a reduction in proton gradient, and consequently reduced ATP synthesis. This loss of the proton gradient would result in a depolarization of the mitochondrial membrane, and we have reported here that actinonin causes not only mitochondrial membrane depolarization but also ATP depletion. In addition, the accumulation of unfolded proteins in the mitochondria may induce a mitochondrial-specific stress response, which reportedly results in an increase in the level of a transcription factor called C/EBP homology protein (CHOP) (47). Preliminary gene-chip analysis (data not shown) suggests that actinonin upregulates the expression of CHOP in human cancer cells, and CHOP has been implicated in programmed cell death (48). Under either mechanism, prolonged actinonin inhibition of HsPDF leading to membrane depolarization and/or a mitochondrial stress response would ultimately lead to cell death.
The mitochondria play a pivotal role in cell death and apoptosis (49–51). These organelles produce the bulk of the cell’s ATP, which is an endogenous inhibitor of the permeability transition pore complex (PTPC) (52, 53). ATP depletion, which could occur if the mitochondrial respiratory chain was inhibited, might facilitate PTPC opening and mitochondrial membrane permeabilization. Thus, it is conceivable that inhibition of HsPDF could lead to inhibition of the respiratory chain and subsequent ATP depletion and thus facilitate mitochondrial membrane permeabilization, a critical event for apoptosis. Actinonin has already been shown to induce G1 arrest and apoptosis in leukemia and lymphoma cell lines (20). However, the low level of apoptosis seen in non–APN-expressing cells does not support a substantial role for apoptosis as the primary mechanism by which actinonin kills cells.
Interestingly, we have also shown that withdrawal of actinonin after a 24-hour incubation leads to a recovery of the mitochondrial membrane depolarization reported here. Thus, if HsPDF activity is transiently inhibited by actinonin, the unfolded proteins should still be available as substrates for HsPDF when actinonin is removed, leading to the recovery of a functional electron transport chain. This effect is in contrast to that seen with uncouplers of respiration such as CCCP, which irreversibly uncouples respiration.
Despite the inhibition of cell-free HsPDF and antiproliferative activity against all but 1 human tumor cell line tested (n = 17), actinonin has been shown to be safe at high doses in mice (19), and we show that it has only slight antiproliferative effects on normal cells in culture as well. Remarkably, actinonin had significant antitumor effects in human xenograft mouse models of prostate and lung cancer, as both an oral and a parenteral agent. In addition, preliminary results with SKI-AC-111111 suggest that the analog is not only a more potent antiproliferative agent in vitro compared with actinonin but is also approximately 3 times more potent as an in vivo antitumor agent in a human prostate tumor xenograft model (data not shown). However, the mechanisms for the apparently selective anticancer in vivo activity are not clear. There has been recent attention to mitochondria as potential targets of anticancer therapy. Evidence suggests that the mitochondria of tumor cells are different enough from normal cells (44, 54) that tumor cells may be more sensitive to mitochondrial insult. MKT-077, a cationic rhodacyanine dye, is an example of a drug that is selectively toxic to carcinoma cells (55), and one explanation for its mechanism of action is the inhibition of mitochondrial respiration. Recently, Fantin et al. (56) showed that a mitochondriotoxic small molecule can selectively inhibit tumor cell growth. Thus, actinonin’s inhibition of HsPDF, resulting in mitochondrial disruption, could have a similar tumor specificity. We show here that actinonin does not cause mitochondrial membrane depolarization of normal circulating lymphocytes; this indicates tumor cell selectivity. The cause of this is under investigation. Human bone marrow colony growth is reduced by actinonin in vitro (20); however, in the cancer therapeutic studies in mice described here, effective antitumor doses of actinonin did not give rise to obvious toxic side effects.
Therefore, we conclude that HsPDF is a new human mitochondrial enzyme that may provide a novel and tumor-selective target for the development of anticancer therapies.
Methods
Reagents.
N-formylated peptide substrates used in the deformylase assay were formyl-Met-Ala-Ser (fMAS), formyl-Met-Ala-His-Ala (fMAHA), and formyl-Met-Thr-Met-His (fMTMH). All substrates were either purchased from or custom synthesized by Bachem Bioscience Inc. Actinonin and CCCP were purchased from Sigma-Aldrich. JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide) was purchased from Invitrogen Corp. Analogs of actinonin were synthesized by the Organic Synthesis Core Facility at Memorial Sloan-Kettering Cancer Center.
PDF assay.
The spectrophotometric assay is based on the method described by Lazennec and Meinnel (57) and is a formate dehydrogenase–coupled assay for PDF activity. Our measurements were conducted at 25°C in polystyrene cuvettes containing 50 mM HEPES (pH 7.4), 10 mM NaCl, 0.2 mg/ml BSA, 2.4 mM NAD+ (Roche Applied Science), 1 U formate dehydrogenase (Sigma-Aldrich), and 0–32 mM N-formylated peptide (Bachem Bioscience Inc.). The reaction was initiated with the addition of 20–100 μg HsPDF enzyme. The rate of NADH production was measured by monitoring of the increase in absorbance at 340 nm using a Spectronic Genesys 2 spectrophotometer (Spectronic Instruments).
Localization of HsPDF in living cells.
The full coding sequence or a truncation of the nucleotides encoding the first 63 amino acids of the human Pdf cDNA was amplified by PCR and cloned in-frame into pEYFP-N1 (BD Biosciences — Clontech). HeLa cells (American Type Culture Collection) were maintained in 5% CO2 in DMEM supplemented with 4 mM L-glutamine, 4.5 g/l glucose, and 10% FBS (cellgro; Mediatech Inc.). HeLa cells to be examined by fluorescence microscopy were plated at 2 × 105 per plate into 35-mm dishes containing a glass coverslip–covered 15-mm cutout (MatTek Corp.) and transfected the next day using LipofectAMINE 2000 (Invitrogen Corp.) according to the manufacturer’s instructions. To stain mitochondria, MitoTracker Red CMXRos (Invitrogen Corp.) was added to cell cultures 30 minutes prior to imaging. Living cells were imaged with a Zeiss 510 inverted laser scanning confocal microscope (LSM; Carl Zeiss Inc.). Representative images were processed with Adobe Photoshop 7.0 (Adobe Systems Inc.).
siRNA preparation and transfection.
Twenty-four hours before transfection, HeLa cells were trypsinized, diluted in fresh media without antibiotics, and transferred to 96-well plates at a density of 10,000 cells per well. On the day of transfection, cells (50–70% confluent) were washed with PBS and the media was replaced with Opti-MEM I reduced-serum medium (Invitrogen Corp.). Transfections of siRNA were carried out using Oligofectamine (Invitrogen Corp.) according to the manufacturer’s instructions. Target-specific siRNA duplexes were designed using a proprietary algorithm of the Sloan-Kettering Institute siRNA Core Facility and provided at a concentration of 10 μM. The siRNAs specific to human Pdf (National Center for Biotechnology Information [NCBI] accession no. NM_022341) were positioned at 581–601 and 659–679 relative to the start codon and were compared with sequences in the human-genome database to confirm that no other genes were targeted. A nonspecific duplex control pool (Dharmacon Inc.) was used as a control and was composed of 4 duplexes containing 33% GC content. All experiments were carried out twice, in triplicate each time. HeLa cell growth was determined via tritiated thymidine incorporation on days 1, 2, and 3 after transfection.
Determination of HsPDF mRNA levels and protein expression.
To confirm knockdown of HsPDF mRNA, HeLa cell RNA was harvested 24, 48, and 72 hours after siRNA transfection. DNase-treated total RNA (5 μg) was used to prepare cDNA for subsequent real-time PCR. The ThermoScript RT-PCR System (Invitrogen Corp.) was used according to the manufacturer’s instructions. The cDNA samples were diluted 1:5 with 5 mM Tris-HCl (pH 8.5), and 3 μl was used for a 25-μl reaction in a MicroAmp optical 96-well reaction plate on an ABI PRISM 7700 sequence detection system (Applied Biosystems). The master mixes contained 2× TaqMan Universal Master Mix, RNase/DNase–free water, and the respective primers and probe. The HsPDF master mix contained primers at 700 mM and HsPDF probe at 200 mM. The β-actin master mix (housekeeping reference gene) contained primers at 500 mM and β-actin probe at 150 mM. The sequences for primers (Gene Link Inc.) and probes (Applied Biosystems) are as follows: PDF forward primer 5′-GGGCAGCCCGCATCA-3′, PDF reverse primer 5′-TGCTGTCCATTTTGTCAATAAACA-3′, β-actin forward primer 5′-CTGGCACCCAGCACAATG-3′, β-actin reverse primer 5′-GCCGATCCACACGGAGTACT-3′, PDF probe 6FAM-CAGCACGAGATGGACCACCTGCAG-TAMRA, and β-actin probe 6FAM-TCAAGATCATTGCTCCTCCTGAGCGC-TAMRA. For final calculations, each sample’s cycle threshold value was subtracted from its respective β-actin value.
For HsPDF protein expression analysis, we used HeLa cells stably expressing the same full-length HsPDF-YFP construct used in the confocal imaging studies. As a control, we used HeLa cells stably expressing a fluorescent vector control. Twenty-four and 48 hours after siRNA transfection, the cells were analyzed by flow cytometry (Cytomics FC 500; Beckman Coulter Inc.) and mean peak fluorescence was determined by a flow cytometry analysis program (FlowJo 4.5; Tree Star Inc.).
The data regarding siRNA knockdown of mRNA, protein, and proliferation are represented as the mean ± SD and were analyzed for statistical significance by an ANOVA with a Newman-Keuls post-test using Prism 3.0 (GraphPad Software).
Cell proliferation assays.
A variety of human and mouse cell lines were used to assess the antiproliferative activity of actinonin. Human tumor cell lines included hematopoietic (Daudi, HL60, NB4, Raji, RL), breast cancer (MDA-MB-468, SK-BR-3), prostate cancer (CWR22Rv1, TSU-PRI, DU145, PC3), lung cancer (SK-LC-8, SK-LC-16), ovarian cancer (A2780), cervical cancer (HeLa), and other (HEK293, HT-1080). We also used human PBMCs (hPBMCs), human fibroblasts (WI-38), mouse fibroblasts (NIH-3T3), and a mouse fibroblast cell line, NIH-3T3, transformed with ras (AL67). To quantitate the effect of actinonin on cancer cell proliferation, 2 different assays were used. We have shown previously that assays of cell viability, such as XTT metabolism and trypan blue exclusion, directly correlate with tritiated thymidine incorporation results from cells treated with actinonin. Assay I was the tritiated thymidine incorporation assay, in which an aliquot of 200 μl of cells (10,000 cells per well) was plated and incubated at 37°C in 96-well plates in the presence or absence of actinonin. Serial dilutions of actinonin were made in complete media. After 1–5 days of incubation, 50 μl of 10 μCi/ml tritiated 3H-thymidine (PerkinElmer Inc.) was added to each well and allowed to incorporate for 5 hours. Plates were frozen at –80°C overnight, and cells were harvested onto filtermats (PerkinElmer Inc.) using a semiautomatic harvester (Skatron Instruments). Filtermats were counted in a 1205 Betaplate liquid scintillation counter (PerkinElmer Inc.). Assay II was the XTT assay, in which cells in log-phase growth were plated in a 96-well plate at a certain density. Actinonin was added to the plate at various concentrations made from serial dilutions in complete media. When the cells in the control wells reached confluence, 50 μl of the tetrazolium salt XTT (dissolved at 1 mg/ml in 37°C serum-free medium) was added with phenazine methosulphate (an electron transfer reagent) to each well of the plate, and the cells were incubated again for 2–4 hours at 37°C. Absorbances were read at a dual wavelength of 450/630 nm using a plate reader.
Determination of mitochondrial membrane depolarization.
RL cells (human B cell lymphoma) were cultured in RPMI supplemented with 20% FBS, 10 mM HEPES, 1 mM sodium pyruvate, 4.5 g/l glucose, 1.5 g/l NaHCO3, 2 mM L-glutamine, and 1% penicillin/streptomycin. Cells were incubated with varying concentrations of actinonin (0–100 μg/ml) at a density of 1 × 106 cells/ml. Maximal dissipation of mitochondrial membrane potential was determined by incubation with the proton ionophore CCCP at a final concentration of 100 nM for 10 minutes prior to data acquisition. Mitochondrial mass and membrane potential were assessed by incubation of cells with JC-1 for 5 minutes at 37°C. Cells were washed 3 times in PBS and resuspended in media supplemented with actinonin at the original concentration. Flow cytometry was performed using a FACSCalibur (BD) running CellQuest software. Fifty thousand events were acquired in list mode for each data point and analyzed with FlowJo software. Mitochondrial mass was measured in FL-1 (“green,” mean excitation emission at 525 nm). Negative mitochondrial membrane potential was measured in FL-2 (“red,” mean excitation emission at 590 nm). Mitochondrial membrane potential was expressed as a ratio of the negative membrane potential to the mass (red/green ratio). All measurements were performed in triplicate and are expressed as the mean ± SD. Statistical significance was determined by an ANOVA with a Newman-Keuls post-test using Prism 3.0.
Measurement of ATP depletion.
Daudi (human B cell lymphoma) cells were cultured in RPMI media supplemented with 1 mM L-glutamine and 10% FBS. Cells were treated with 3 doses of actinonin (5, 10, and 20 μg/ml) for varying periods of time (1–36 hours). At each time point, cells were counted and resuspended to a final concentration of 100,000 cells/ml. ATP was quantitated in 1,000 cells using an ATP assay kit (EMD Biosciences Inc.) following the manufacturer’s instructions. Immediate luminescence detection over 1 minute was performed with a Berthold Lumat LB 9501 luminometer (Berthold Technologies). ATP determinations were calculated using an ATP standard curve, and results were expressed as grams ATP per 1,000 cells.
Annexin V staining.
Daudi (human B cell lymphoma) cells were cultured in RPMI media supplemented with 1 mM L-glutamine and 10% FBS. Cells were treated with 3 doses of actinonin (5, 10, and 20 μg/ml) for varying periods of time (1–96 hours). At each time point, 1 × 106 cells were rinsed with binding buffer and stained with annexin V and propidium iodide (control) using the BD ApoAlert kit (BD Biosciences — Clontech) according to the manufacturer’s instructions. Cells were analyzed by flow cytometry (Cytomics FC 500), and the percentage annexin V–positive, propidium iodide–negative, was determined by a flow cytometry analysis program (FlowJo 4.5).
Human tumor xenograft models in mice.
Eight- to ten-week-old athymic NCr-nu mice were inoculated s.c. in the flank with minced CWR22 (prostate), A549 (lung), or PC3 (prostate) tumor cells mixed with Matrigel (BD). Once the tumors became palpable, the mice were randomized into control and treatment groups with 3–5 animals per group. Actinonin was administered i.p. or orally daily for 2 weeks except for weekends. The doses of actinonin chosen for these studies were based on a preliminary toxicity study in which weight loss was the limiting criterion for calculation of the maximal tolerated dose. Mice in the control group were given vehicle alone. Tumors were measured every 3–4 days with calipers. Tumor volumes were calculated by the formula 4/3 × π × [(larger diameter + smaller diameter) / 4]3. The data are represented as the mean ± SD and were analyzed for statistical significance by nonparametric Wilcoxon rank-sum test using SAS 8.2 (SAS Institute Inc.). All animal studies were conducted with the approval of the Institutional Animal Care and Use Committee at Sloan-Kettering Institute.
Screening of actinonin analogs for HsPDF inhibition and antiproliferative activity.
All actinonin analogs were dissolved in 10–50% DMSO, and subsequent dilution in the enzymatic and antiproliferation assays brought the final DMSO concentration to below 1%. In addition, DMSO controls were used to calculate percentage inhibition by analogs. CWR22Rv1 and TSU-PRI cancer cell lines were used to assess the antiproliferative efficacy of analogs in the XTT assay. Details of the XTT assay have been described in a previous section. Daudi and HL60 cancer cell lines were used in the tritiated thymidine incorporation assay, as described above. HsPDF inhibition by analogs was determined with the formate dehydrogenase–coupled PDF assay as described in a previous section. IC50 values represent the concentration at which the compound inhibits 50% of cell proliferation or enzymatic activity when compared with controls.
Acknowledgments
We thank Yang Xu and David Ron for their helpful discussions concerning this work, Ouathek Ouerfelli for the design and synthesis of the siRNAs, Bryan Soper and Barry Kappel for expert assistance in expression cloning and flow cytometry, respectively, and the Memorial Sloan-Kettering Cancer Center Flow Cytometry Core Facility. This work was supported by NIH training grant CA062948 NIH grant R01 CA55349, the Doris Duke Charitable Foundation, William H. Goodwin and Alice Goodwin, the Commonwealth Cancer Foundation for Research, and the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center. D.A. Scheinberg is a Doris Duke Distinguished Clinical Scientist.
Footnotes
Nonstandard abbreviations used: APN, aminopeptidase N; CCCP, carbonyl cyanide m-chlorophenylhydrazone; fMAHA, formyl-Met-Ala-His-Ala; fMAS, formyl-Met-Ala-Ser; fMTMH, formyl-Met-Thr-Met-His; NME, N-terminal methionine excision; PDF, peptide deformylase; siRNA, small interfering RNA; YFP, yellow fluorescent protein.Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Chang SY, McGary EC, Chang S. Methionine aminopeptidase gene of Escherichia coli is essential for cell growth. J. Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solbiati J, Chapman-Smith A, Miller JL, Miller CG, Cronan JE., Jr Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J. Mol. Biol. 1999;290:607–614. doi: 10.1006/jmbi.1999.2913. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ragusa S, Blanquet S, Meinnel T. Control of peptide deformylase activity by metal cations. J. Mol. Biol. 1998;280:515–523. doi: 10.1006/jmbi.1998.1883. [DOI] [PubMed] [Google Scholar]
- 5.Margolis PS, et al. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob. Agents Chemother. 2000;44:1825–1831. doi: 10.1128/aac.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margolis P, et al. Resistance of Streptococcus pneumoniae to deformylase inhibitors is due to mutations in defB. Antimicrob. Agents Chemother. 2001;45:2432–2435. doi: 10.1128/AAC.45.9.2432-2435.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Chen Z, Gong W. Enzymatic properties of a new peptide deformylase from pathogenic bacterium Leptospira interrogans. Biochem. Biophys. Res. Commun. 2002;295:884–889. doi: 10.1016/s0006-291x(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 8.Cynamon MH, Alvirez-Freites E, Yeo AE. BB-3497, a peptide deformylase inhibitor, is active against Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2004;53:403–405. doi: 10.1093/jac/dkh054. [DOI] [PubMed] [Google Scholar]
- 9.Meinnel T. Peptide deformylase of eukaryotic protists: a target for new antiparasitic agents? Parasitol. Today. 2000;16:165–168. doi: 10.1016/s0169-4758(99)01627-0. [DOI] [PubMed] [Google Scholar]
- 10.Giglione C, Serero A, Pierre M, Boisson B, Meinnel T. Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 2000;19:5916–5929. doi: 10.1093/emboj/19.21.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracchi-Ricard V, et al. Characterization of an eukaryotic peptide deformylase from Plasmodium falciparum. Arch. Biochem. Biophys. 2001;396:162–170. doi: 10.1006/abbi.2001.2631. [DOI] [PubMed] [Google Scholar]
- 12.Dirk LM, Williams MA, Houtz RL. Eukaryotic peptide deformylases. Nuclear-encoded and chloroplast-targeted enzymes in Arabidopsis. Plant Physiol. 2001;127:97–107. doi: 10.1104/pp.127.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MD, et al. A new human peptide deformylase inhibitable by actinonin. Biochem. Biophys. Res. Commun. 2003;312:309–315. doi: 10.1016/j.bbrc.2003.10.123. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen KT, et al. Characterization of a human peptide deformylase: implications for antibacterial drug design. Biochemistry. 2003;42:9952–9958. doi: 10.1021/bi0346446. [DOI] [PubMed] [Google Scholar]
- 15.Serero A, Giglione C, Sardini A, Martinez-Sanz J, Meinnel T. An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway. J. Biol. Chem. 2003;278:52953–52963. doi: 10.1074/jbc.M309770200. [DOI] [PubMed] [Google Scholar]
- 16.Guilloteau JP, et al. The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents. J. Mol. Biol. 2002;320:951–962. doi: 10.1016/s0022-2836(02)00549-1. [DOI] [PubMed] [Google Scholar]
- 17.Giglione C, Pierre M, Meinnel T. Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents. Mol. Microbiol. 2000;36:1197–1205. doi: 10.1046/j.1365-2958.2000.01908.x. [DOI] [PubMed] [Google Scholar]
- 18.Kumar A, et al. Crystals of peptide deformylase from Plasmodium falciparum reveal critical characteristics of the active site for drug design. Structure (Camb.). 2002;10:357–367. doi: 10.1016/s0969-2126(02)00719-0. [DOI] [PubMed] [Google Scholar]
- 19.Gordon JJ, Kelly BK, Miller GA. Actinonin: an antibiotic substance produced by an actinomycete. Nature. 1962;195:701–702. doi: 10.1038/195701b0. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Lai LT, Gabrilove JL, Scheinberg DA. Antitumor activity of actinonin in vitro and in vivo. Clin. Cancer Res. 1998;4:171–176. [PubMed] [Google Scholar]
- 21.Xu Y, Scheinberg DA. Elimination of human leukemia by monoclonal antibodies in an athymic nude mouse leukemia model. Clin. Cancer Res. 1995;1:1179–1187. [PubMed] [Google Scholar]
- 22.Chen DZ, et al. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry. 2000;39:1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinnel T, Blanquet S, Dardel F. A new subclass of the zinc metalloproteases superfamily revealed by the solution structure of peptide deformylase. J. Mol. Biol. 1996;262:375–386. doi: 10.1006/jmbi.1996.0521. [DOI] [PubMed] [Google Scholar]
- 25.Chan MK, et al. Crystal structure of the Escherichia coli peptide deformylase. Biochemistry. 1997;36:13904–13909. doi: 10.1021/bi9711543. [DOI] [PubMed] [Google Scholar]
- 26.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 27.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 28.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semizarov D, et al. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elmore SP, Nishimura Y, Qian T, Herman B, Lemasters JJ. Discrimination of depolarized from polarized mitochondria by confocal fluorescence resonance energy transfer. Arch. Biochem. Biophys. 2004;422:145–152. doi: 10.1016/j.abb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Nieminen AL, et al. Protection by acidotic pH and fructose against lethal injury to rat hepatocytes from mitochondrial inhibitors, ionophores and oxidant chemicals. Biochem. Biophys. Res. Commun. 1990;167:600–606. doi: 10.1016/0006-291x(90)92067-a. [DOI] [PubMed] [Google Scholar]
- 32.Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao B, et al. Structural basis for the design of antibiotics targeting peptide deformylase. Biochemistry. 1999;38:4712–4719. doi: 10.1021/bi982594c. [DOI] [PubMed] [Google Scholar]
- 34.Durand DJ, Gordon Green B, O’Connell JF, Grant SK. Peptide aldehyde inhibitors of bacterial peptide deformylases. Arch. Biochem. Biophys. 1999;367:297–302. doi: 10.1006/abbi.1999.1274. [DOI] [PubMed] [Google Scholar]
- 35.Clements JM, et al. Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor. Antimicrob. Agents Chemother. 2001;45:563–570. doi: 10.1128/AAC.45.2.563-570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackbarth CJ, et al. N-alkyl urea hydroxamic acids as a new class of peptide deformylase inhibitors with antibacterial activity. Antimicrob. Agents Chemother. 2002;46:2752–2764. doi: 10.1128/AAC.46.9.2752-2764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta MK, Mishra P, Prathipati P, Saxena AK. 2D-QSAR in hydroxamic acid derivatives as peptide deformylase inhibitors and antibacterial agents. Bioorg. Med. Chem. 2002;10:3713–3716. doi: 10.1016/s0968-0896(02)00421-2. [DOI] [PubMed] [Google Scholar]
- 38.Jones RN, Rhomberg PR. Comparative spectrum and activity of NVP-PDF386 (VRC4887), a new peptide deformylase inhibitor. J. Antimicrob. Chemother. 2003;51:157–161. doi: 10.1093/jac/dkg055. [DOI] [PubMed] [Google Scholar]
- 39.Umezawa H, et al. Production of actinonin, an inhibitor of aminopeptidase M, by actinomycetes. J. Antibiot. 1985;38:1629–1630. doi: 10.7164/antibiotics.38.1629. [DOI] [PubMed] [Google Scholar]
- 40.Tieku S, Hooper NM. Inhibition of aminopeptidases N, A and W. A re-evaluation of the actions of bestatin and inhibitors of angiotensin converting enzyme. Biochem. Pharmacol. 1992;44:1725–1730. doi: 10.1016/0006-2952(92)90065-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giglione C, Meinnel T. Organellar peptide deformylases: universality of the N-terminal methionine cleavage mechanism. Trends Plant Sci. 2001;6:566–572. doi: 10.1016/s1360-1385(01)02151-3. [DOI] [PubMed] [Google Scholar]
- 42.Giglione C, Vallon O, Meinnel T. Control of protein life-span by N-terminal methionine excision. EMBO J. 2003;22:13–23. doi: 10.1093/emboj/cdg007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emanuelsson O, von Heijne G. Prediction of organellar targeting signals. . Biochim. Biophys. Acta. 2001; 1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- 44.Penta JS, Johnson FM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutat. Res. 2001;488:119–133. doi: 10.1016/s1383-5742(01)00053-9. [DOI] [PubMed] [Google Scholar]
- 45.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. . Biochim. Biophys. Acta. 1999; 1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 46.Sawafuji K, et al. Aminopeptidase inhibitors inhibit proliferation and induce apoptosis of K562 and STI571-resistant K562 cell lines through the MAPK and GSK-3beta pathways. Leuk. Lymphoma. 2003;44:1987–1996. doi: 10.1080/1042819031000122033. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Q, et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinszner H, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costantini P, Jacotot E, Decaudin D, Kroemer G. Mitochondrion as a novel target of anticancer chemotherapy. J. Natl. Cancer Inst. 2000;92:1042–1053. doi: 10.1093/jnci/92.13.1042. [DOI] [PubMed] [Google Scholar]
- 50.Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 51.Nieminen AL. Apoptosis and necrosis in health and disease: role of mitochondria. Int. Rev. Cytol. 2003;224:29–55. doi: 10.1016/s0074-7696(05)24002-0. [DOI] [PubMed] [Google Scholar]
- 52.Bernardi P. The permeability transition pore. Control points of a cyclosporin A-sensitive mitochondrial channel involved in cell death. . Biochim. Biophys. Acta. 1996; 1275:5–9. doi: 10.1016/0005-2728(96)00041-2. [DOI] [PubMed] [Google Scholar]
- 53.Zoratti M, Szabo I. The mitochondrial permeability transition. . Biochim. Biophys. Acta. 1995; 1241:139–176. doi: 10.1016/0304-4157(95)00003-a. [DOI] [PubMed] [Google Scholar]
- 54.Cavalli LR, Liang BC. Mutagenesis, tumorigenicity, and apoptosis: are the mitochondria involved? Mutat. Res. 1998;398:19–26. doi: 10.1016/s0027-5107(97)00223-6. [DOI] [PubMed] [Google Scholar]
- 55.Modica-Napolitano JS, et al. Selective damage to carcinoma mitochondria by the rhodacyanine MKT-077. Cancer Res. 1996;56:544–550. [PubMed] [Google Scholar]
- 56.Fantin VR, Berardi MJ, Scorrano L, Korsmeyer SJ, Leder P. A novel mitochondriotoxic small molecule that selectively inhibits tumor cell growth. Cancer Cell. 2002;2:29–42. doi: 10.1016/s1535-6108(02)00082-x. [DOI] [PubMed] [Google Scholar]
- 57.Lazennec C, Meinnel T. Formate dehydrogenase-coupled spectrophotometric assay of peptide deformylase. Anal. Biochem. 1997;244:180–182. doi: 10.1006/abio.1996.9910. [DOI] [PubMed] [Google Scholar]