Abstract
A system-wide understanding of biological processes requires a comprehensive knowledge of the proteins in the biological system. The eosinophil is a type of granulocytic leukocyte specified early in hematopoietic differentiation that participates in barrier defense, innate immunity, and allergic disease. The proteome of the eosinophil is largely unannotated with under 500 proteins identified. We now report a map of the nonstimulated peripheral blood eosinophil proteome assembled using two-dimensional liquid chromatography coupled with high-resolution mass spectrometry. Our analysis yielded 100,892 unique peptides mapping to 7,086 protein groups representing 6,813 genes as well as 4,802 site-specific phosphorylation events. We account for the contribution of platelets that routinely contaminate purified eosinophils and report the variability in the eosinophil proteome among five individuals and proteomic changes accompanying acute activation of eosinophils by interleukin-5. Our deep coverage and quantitative analyses fill an important gap in the existing maps of the human proteome and will enable the strategic use of proteomics to study eosinophils in human diseases.
Keywords: immunology, eosinophil, quantitative proteomics
Graphical abstract
INTRODUCTION
Acquiring deep proteomic coverage through mass spectrometry (MS)-based analysis can aid understanding of system-wide processes, confirm expression of transcripts, inform experiments of specific proteins and protein complexes of particular interest, and generate hypotheses about pathways altered in disease. Spurred by the improvement of mass spectrometers and increasingly streamlined sample preparation, recent proteomic studies dive deep into biological samples ranging from cell lines such as HeLa cells to the wide variety of tissues and cells characterized in the two draft maps of the human proteome.1–3 These two maps provide extensive coverage of a variety of tissues, organs, and cell types, but both exclude many hematopoietic cells, such as eosinophils.
Eosinophils are a specific type of granular leukocyte critical to inflammatory and immunoregulatory responses such as tissue homeostasis and inflammation of allergic diseases.4,5 Allergic rhinitis and asthma, for example, are characterized by the presence and activation of eosinophils.6,7 In hypereosinophilic syndrome and eosinophilic gastrointestinal diseases, eosinophils participate in the pathogenesis of disease through several mechanisms, including the release of a unique set of granule components, secretion of cytokines, and regulation of T cells.4,6 Many characteristics of eosinophils are well-defined, such as their differentiation and maturation in bone marrow before moving into the blood.4,6,8,9 However, other molecular mechanisms and signaling events important for eosinophil function remain unclear or unknown and will require novel approaches to identify. An NIH taskforce report on eosinophil-associated diseases recently highlighted this critical need to better understand eosinophil immunobiology.10 To help fulfill that need and advance further study, a comprehensive analysis of the eosinophil proteome would create a guiding map of cellular contents, identify pathways and biological functions unique to or enriched in eosinophils, and impact future research of allergies, asthma, and other diseases affected by eosinophils.
The most recent and in-depth eosinophil proteome study, based on subcellular fractionation and two-dimensional electrophoresis, identified 426 unique nonredundant proteins, over half of which were not previously observed in eosinophils.11 Although this work provided an initial characterization of the eosinophil proteome, advances in mass spectrometry instrumentation, sample preparation, chromatography, and informatics now offer vastly deeper proteomic coverage at much greater speed. These advances enable a “shotgun” approach aimed at analyzing proteolytic fragments encompassing the whole proteome. Compare, for example, a 2001 proteomics result to a 2014 achievement: 1,483 yeast proteins identified in 68 h of mass spectral analysis12 versus 3,977 yeast proteins identified in just over an hour. This stark difference shows how improved instrumentation allows deep coverage of the proteome within a practical time frame.13
Given the medical relevance of eosinophils, it is imperative to bring recent technological developments in proteomics to bear on these cells. We have assembled a comprehensive map of the eosinophil proteome using mass spectrometry and an optimized sample preparation workflow, including high pH reversed phase fractionation prior to liquid chromatography–mass spectrometry (LC-MS/MS). We provide a resource of 7,086 proteins and over 4,800 sites of protein phosphorylation identified from purified eosinophils. We estimate protein cellular abundance from these data. In addition, we have utilized isobaric labeling to assess the variability in eosinophil proteins among five individuals and the proteomic changes that accompany acute activation of eosinophils by interleukin-5 (IL5). Finally, we account for the contribution of platelets, which associate with and thus routinely contaminate purified eosinophils, to the determined proteomes.
MATERIALS AND METHODS
The University of Wisconsin-Madison Health Sciences Institutional Review Board approved this study. Informed written consent was obtained from each subject before participation.
Purification of Eosinophils and Platelets
Eosinophils were purified from 200 mL of heparinized blood of donors with allergic rhinitis or allergic asthma by Percoll centrifugation (density of 1.090 g/mL) and negative selection. Such donors had eosinophil counts in the high normal range of 200–500 per μL that allowed reproducible purification of 20–50 million eosinophils per purification. The gradient separated eosinophilic and neutrophilic granulocytes from mononuclear cells such as NK cells and B cells. Negative selection with the AutoMACS system (Miltenyi, Auburn, CA) used a cocktail of anti-CD16, anti-CD14, anti-CD3, and antiglycophorin A coupled magnetic beads (Figure 1A).14,15 Purity of eosinophils compared to other leukocytes was ≥98% as determined by Wright–Giemsa staining followed by microscopic scoring of 500 cells. Viability was ≥98% as assessed by trypan blue staining. To assess the impact of platelet contamination, we purified an additional sample of eosinophils from a single donor in which negative selection was accomplished with the antibody cocktail described above or with the cocktail plus an antibody to CD61, the β3 subunit of the major platelet integrin. Purified eosinophils, received on ice in 2% calf serum, were resuspended in RPMI at 2.5 × 106 cells/ml, incubated at 37 °C for 1 h,14 and then centrifuged at 1200 rpm (260g in a Sorvall Technospin R centrifuge, Du Pont, Wilmington, DE) for 10 min at 4 °C. The supernatant was aspirated, and the cell pellet was immediately frozen by immersing the tube in N2(l). The sample was then stored at −80 °C. Platelets were purified from citrated blood (from a donor with allergic asthma) by differential centrifugation and washing as described.16
Figure 1.
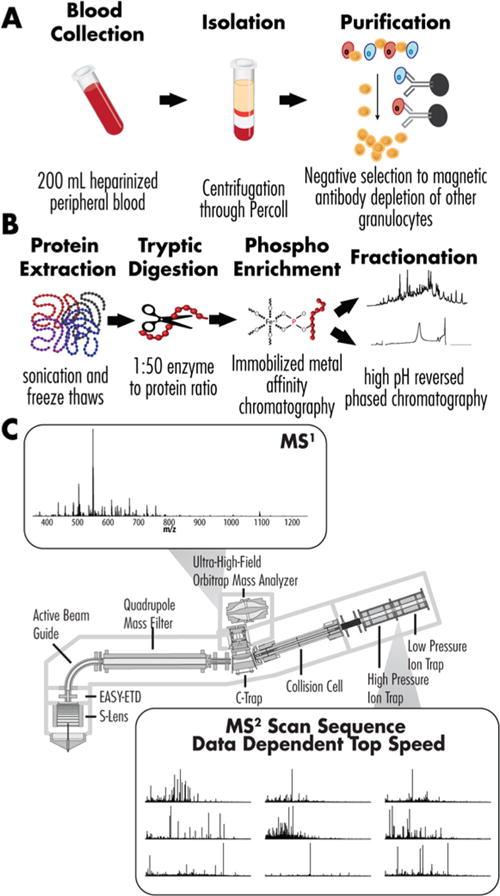
Overview of the analytical strategy. (A) Peripheral blood was donated from volunteers with eosinophil counts between 250 and 500/ul. Granulocytes were isolated via centrifugation and purified using negative selection with magnetic beads bearing antibodies CD3, CD14, CD16, and glycophorin-A. (B) The combined eosinophils were lysed via probe sonication, digested with trypsin, and enriched for phosphorylation (immobilized metal affinity chromatography). The enriched and nonenriched samples were fractionated using high pH reversed phase chromatography. (C) Fractions were run on a Q-OT-qIT mass spectrometer (Orbitrap Fusion) using a data-dependent top speed. MS1 were collected in the Orbitrap. MS2 scans were collected in the ion trap for peptide fractions and in the Orbitrap for phosphopeptide fractions.
Acute Activation of Eosinophils
Eosinophils were purified from five different donors with approximately 20 million cells each. After incubation at 37 °C for 1 h, purified eosinophils from each donor were split in half, and each half was treated for 5 min with saline alone or IL5 at 20 ng/mL as described,14 after which the cell pellets were collected by centrifugation and snap frozen.
Eosinophil Lysis and Digestion
Pellets from three donors were solubilized on ice in 8 M urea, 40 mM Tris, pH 8, 30 sodium chloride, 6 mM sodium pyrophosphate, 2 mM magnesium chloride, 1 mM calcium chloride, phosSTOP phosphatase inhibitor (Roche), and 2x mini EDTA-free protease inhibitor (Roche). The combined 75 million purified eosinophils were lysed via sonication and freeze–thaw cycles yielding 1.26 mg of protein for digestion. Protein concentration was determined by BCA assay (Thermo Pierce). Disulfide bonds were reduced at 58 °C for 45 min with 5 mM DTT and carbamidomethylated using 15 mM iodoacetamide for 45 min in the dark at room temperature. The samples were then quenched with 5 mM DTT for 15 min at room temperature and then diluted with 50 mM Tris, pH 8, and 5 mM calcium chloride to reach a final concentration of 1.5 M urea. The sample was digested with trypsin (Promega) (enzyme/protein 1:50) at room temperature on a rocker overnight. The sample was acidified to 0.5% TFA prior to desalting on a C18 solid-phase extraction column (Waters), and the desalted sample was dried to completion.
Phosphopeptide Enrichment
Peptides were enriched for the phosphoryl moiety using IMAC with magnetic beads (Qiagen). The beads were washed three times with water and incubated with 40 mM EDTA for 1 h with shaking. The beads were then washed three times with water and incubated with 100 mM iron chloride (FeCl3) for 45 min with shaking. Afterward, the beads were washed four times with 80% acetonitrile (ACN) and 0.15% trifluoroacetic acid (TFA) in water. The desalted eosinophil peptides were resuspended in 80% ACN and 0.15% TFA and incubated with the beads for 30 min with shaking. The flow-through was saved and combined with the first of three washes with 80% ACN and 0.15% TFA in water. Phosphopeptides were eluted with 100 μL of 50% ACN and 0.7% ammonium hydroxide. The sample was immediately acidified with 50 μL of 5% formic acid (FA), frozen, then dried to completion.
Isobaric Labeling
Peptides from the ten different samples (five donors, one stimulated and one resting from each donor) were labeled with one of the ten tandem mass tags (TMT). Tags were resuspended in 50 uL of 100% ACN. Samples were resuspended in 100 uL of 100 mM tetraethylammonium bromide (TEAB). The tag was added to samples, and they were labeled on a shaker for 2 h. The reaction was quenched with 0.8 uL of 50% hydroxylamine, and the samples were mixed in equal amounts.
High pH Reversed Phase Chromatography
Peptides (IMAC flow through) and phosphopeptides were separately fractionated by high pH reversed phase chromatography using a 250 mm by 4.6 mm column packed with 5 μm C18 particles (Gemini, Phenomenex) on a Surveyor LC quaternary pump (Thermo Scientific). The samples were resuspended in high pH reversed phase buffer, injected, and separated over a 60 min gradient (buffer A: 20 mM ammonium formate, pH 10; buffer B: 20 mM ammonium formate, pH 10, in 80% ACN). Flow rate was held at 0.8 mL/min except during re-equilibration when it was increased to 1 mL/min. Fractions were collected each min from 10 to 36 min and combined into 20 fractions for the peptides and 10 fractions for the phosphopeptides, which were frozen and dried to completion. The combined TMT-labeled sample was also fractionated using the same method. The eosinophil, platelet-depleted eosinophil, and platelet samples were fractionated using the same gradient but concatenated to 16 fractions.
LC-MS/MS Analysis
Fractions were resuspended in 0.2% FA (approximately 1 ug/uL), and 1 ug was loaded onto a 50 cm silica capillary column (75 μm i.d., 360 μm o.d) packed with 1.7 μm, 130 Å pore size bridged ethylene hybrid C18 particles (Waters). Each fraction was separated over a 100 min analysis on an Acquity nanoHPLC (Waters), including time for re-equilibration. Buffer A was water with 0.2% FA and 5% DMSO, and buffer B was ACN with 0.2% FA. The flow rate was 0.300 μL/min. Eluted peptide cations were analyzed with an Orbitrap Fusion (Q-OT-qIT) mass spectrometer for the global analysis and TMT-labeled samples.
Full MS scans (350–1250 m/z) were acquired in the Orbitrap at 60K resolution, and MS2 scans were collected using data-dependent top speed mode with a cycle time of 3 s and priority for the most intense precursors. After a peptide precursor was sampled by MS/MS, the precursor with a 10 ppm tolerance was excluded from repetitive sampling for 15 s. For MS/MS for the protein analysis, precursors were isolated in the quadrupole with an isolation window of 0.7 m/z, fragmented by higher energy collisional dissociation (HCD) with a normalized collision energy of 30, and analyzed in the ion trap. The automatic gain control (AGC) target was set to 5 × 105 for MS scans and 104 for MS/MS scans, and the maximum injection times were 100 and 25 ms, respectively. For phosphopeptide analysis, each of the ten fractions was run in a technical duplicate. For MS/MS, precursors were isolated in the quadrupole within a window of 3 m/z, fragmented as above, and analyzed in the Orbitrap with 60K resolution. The maximum injection times and AGC targets were the same as for the MS scans but increased to 200 ms and 5 × 104 for the MS/MS scans, respectively.
For the TMT-labeled eosinophil sample, high-resolution MS/MS scans (60,000 K) were collected to differentiate between the closely spaced TMT reporter ions. As before, the instrument was operated in data-dependent top speed mode with a cycle time of 3 s and priority for most intense precursors. For MS/MS, precursors were isolated in the quadrupole with an isolation window of 1 m/z, fragmented by HCD with a normalized collision energy of 37, and analyzed in the Orbitrap with a first mass of 105 m/z. The AGC was set to 5 × 105 for MS and MS/MS scans. The maximum injection times were 100 ms for MS scans and 60 ms for MS/MS scans.
The peptides of eosinophils purified without or with the extra depletion of platelets or of purified platelets were analyzed on an Orbitrap Elite (Thermo Scientific). The fractions were resuspended in 0.2% FA as before but separated on a Dionex nanoHPLC over a 120 min gradient including trapping time. The instrument was operated in data-dependent top 20 mode using collisional-induced dissociation (CID) with a normalized collision energy of 35. The AGC was set to 1 × 106 for MS and to 5 × 105 for MS/MS. The maximum injection times were 50 ms for MS scans and 50 ms for MS/MS scans.
All raw files analyzed are available on Chorus under Eosinophil Proteome project 1015, as described in Supporting Information.
Data Analysis
Raw data were processed using MaxQuant (Version 1.5.0.25).17 Searches were performed against a target-decoy database of reviewed proteins plus isoforms (Uniprot (human), www.uniprot.org, April 4, 2014) and the Andromeda search algorithm.18 Searches were conducted using 6 ppm MS1 precursor tolerance and 0.35 Da product mass tolerance for the peptide analysis and 0.015 Da product mass tolerance for the phosphopeptide analysis. Minimum Andromeda score was zero for unmodified peptides and 40 for modified peptides as default MaxQuant values. A maximum of two missed tryptic cleavages was allowed. Searches were performed with a fixed modification for carbamidomethylation of cysteine residues and a variable modification for the oxidation of methionine. For the phosphopeptide analysis, phosphorylation (serine, threonine, and tyrosine) was added as a variable modification. Peptides and proteins were reduced using a 1% peptide spectrum match (PSM) false discovery rate (FDR) and a 1% protein FDR determined by the target-decoy method.19 Proteins were identified and quantified based on at least one unique peptide. Intensity-based absolute quantitation (iBAQ) values were obtained for the protein runs through the default label-free quantitation settings in MaxQuant.20,21 Data are summarized in Supplemental Tables 1 and 2. The data for the eosinophils, platelet-depleted eosinophils, and platelet comparison were also analyzed in MaxQuant using the same settings as before but with MaxLFQ enabled using the default settings.
The TMT-labeled sample data was analyzed using our in-house software suite COMPASS.22 The product mass tolerance was 0.015 Da, and a maximum of three missed tryptic cleavages was allowed. Searches were performed with fixed modifications for carbamidomethylation of cysteine residues, TMT on lysine residues and the N-terminus, and variable modifications for the oxidation of methionine and TMT on tyrosine residues. Peptides and proteins were reduced using a 1% PSM FDR and a 1% protein FDR determined by the target-decoy method.23 TMT quantitation was performed in TagQuant, part of COMPASS, and was corrected for tag impurities.22 Data analysis was then continued in Excel. Statistical significance was determined using a two-tailed and equal variance t test. Hierarchical clustering was performed in Perseus using Euclidean distance and average linkage with the data preprocessed with k-means.24
RESULTS AND DISCUSSION
To map the eosinophil proteome, we combined 75 million purified eosinophils from three donors. The yields of eosinophils in the blood varied from donor to donor, and thus, the contributions of individual donors to the pool were not equivalent. After tryptic digestion, the combined sample, 1.26 mg of protein, provided sufficient material for analysis of the global proteome and phosphoproteome (Figure 1) by enabling IMAC followed by prefractionation via high pH reversed phase chromatography into 20 fractions for the peptide sample and 10 fractions for the phosphopeptide fraction. The resulting detection of lower abundance proteins deepened our proteomic coverage from the LC-MS/MS analyses. Combining 20 HPLC-MS/MS peptide analyses (Figure 1C) totaling 1,522,655 MS/MS scans, 264,899 peptide spectral matches were obtained, corresponding to 100,892 unique peptide sequences and 7,086 proteins (Supplemental Table 1) with an FDR of less than 1%. The median sequence coverage was 28.8% (mean sequence coverage = 32.6%), and 94% of the proteins were identified with at least two unique peptides. Following protein grouping, 3,184 proteins were unambiguously identified as a single splice isoform; other proteins were grouped as nondistinguishable proteins or several isoforms. The 451 proteins that were identified by only one peptide can be found in Sheet 2 of Supplemental Table 1. Our data correlated well with previously published results, covering 416 of the 426 proteins reported.11 Of the ten proteins not observed, four were unreviewed Uniprot entries or otherwise not included in our database. From the phosphopeptide analysis, 4,802 sites of phosphorylation were identified and localized to a single amino acid with probability of greater than 75% (Supplemental Table 2). The distribution of serine, threonine, and tyrosine phosphorylation was 87, 12, and 1%, respectively, which is similar to previous global phosphorylation studies of other biological samples.25,26
To provide estimates of relative abundance of identified proteins, we used intensity-based absolute quantification (iBAQ), which sums the peptide intensities of all identified peptides for a protein and then divides each by the number of theoretically observable peptides based on an in silico protein digest.21 With this approach, we determined that the 15 most abundant proteins comprise 25% of the proteome (Figure 2A, Table 1). Of the 7,086 proteins, the most abundant 385, or a little over 5%, make up 75% of the eosinophil proteome mass (Figure 2A). The quantified proteome followed a sigmoidal distribution over seven orders-of-magnitude with a median Log2 intensity of 25.2 (Figure 2B). As a point of comparison, 25% of the HeLa cell proteome comprises the 40 most abundant proteins (0.5%), and 75% of the proteome mass is attributed to the 600 most abundant proteins (7.4%).27 The distribution of iBAQ intensity values across ranked protein percent (Figure 2B) follows the same distribution observed in HeLa cells.27 Our experimentally derived protein expression values correlate with the intensities of spots on two-dimensional gels presented by Straub et al. with actin being the most predominant protein.11
Figure 2.
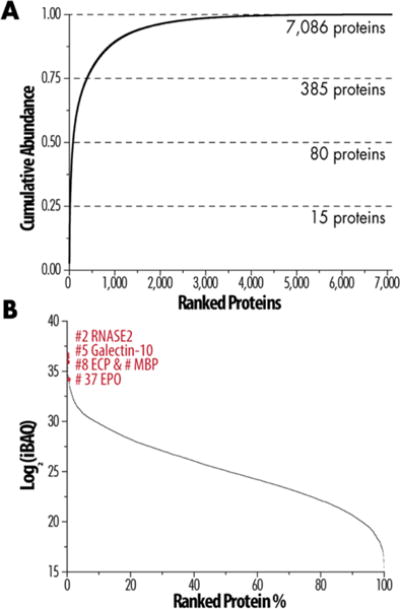
Quantitative analysis to estimate protein abundance. (A) Cumulative protein mass of ranked proteins from highest to lowest abundance. (B) Expression of all quantified proteins over 7 orders of magnitude with several important proteins identified by their numerical rank.
Table 1.
Top 15 Most Abundant Proteins in Eosinophils
| rank | gene name | protein name | peptides | differentiating peptides | sequence coverage [%] | iBAQ |
|---|---|---|---|---|---|---|
| 1 | ACTB | Actin, cytoplasmic 1; Actin, cytoplasmic 1, N-terminally processed | 62 | 1 | 85% | 1.246 × 1011 |
| 2 | RNASE2 | Nonsecretory ribonuclease | 10 | 10 | 41% | 1.107 × 1011 |
| 3 | HIST1H4A | Histone H4 | 21 | 21 | 65% | 1.105 × 1011 |
| 4 | HIST1H2AJ; HIST1H2AH; HIST1H2AG; H2AFJ; HIST2H2AC; HIST2H2AA3; HIST1H2AA | Histone H2A type 1-J; Histone H2A type 1-H; Histone H2A type 1; Histone H2A. J; Histone H2A type 2-C; Histone H2A type 2-A; Histone H2A type 1-A | 12 | 0 | 50% | 9.8 × 1010 |
| 5 | CLC | Charcot–Leyden crystal protein (galectin-10) | 21 | 21 | 99% | 8.991 × 1010 |
| 6 | HIST1H2BH; HIST2H2BF | Histone H2B type 1-H; Histone H2B type 2-F | 28 | 0 | 83% | 7.833 × 1010 |
| 7 | PFN1 | Profilin-1 | 26 | 21 | 79% | 7.35 × 1010 |
| 8 | PRG2 | Bone marrow proteoglycan 2; Eosinophil granule major basic protein | 15 | 15 | 59% | 6.587 × 1010 |
| 9 | RNASE3 | Eosinophil cationic protein | 22 | 22 | 80% | 6.359 × 1010 |
| 10 | CFL1 | Cofilin-1 | 39 | 26 | 99% | 4.895 × 1010 |
| 11 | S100A9 | Protein S100-A9 | 19 | 19 | 91% | 4.482 × 1010 |
| 12 | PRG3 | Bone marrow proteoglycan 3 | 20 | 20 | 76% | 4.331 × 1010 |
| 13 | ANXA1 | Annexin A1 | 72 | 72 | 93.90% | 4.26 × 1010 |
| 14 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 54 | 6 | 98.50% | 4.246 × 1010 |
| 15 | HIST1H3A; HIST3H3; H3F3C | Histone H3.1; Histone H3.1t; Histone H3.3C | 22 | 2 | 81.60% | 3.704 × 1010 |
Selected Eosinophil Proteins of Interest
Among the most abundant 37 proteins are all of the six major granule proteins known to play important roles in human eosinophils. Charcot–Leyden crystal protein (CLC, galectin-10) was the fifth most abundant protein (Table 1) with 21 unique peptides and 99.3% coverage. A mannose-binding lectin with obscure function, CLC is the principal component of the primary granule of eosinophils into which it is packaged despite lacking a signal sequence.28 Eosinophil-derived neurotoxin (EDN, RNASE2) and eosinophil cationic protein (ECP, RNASE3) were the second and ninth most abundant proteins identified, respectively, by ten unique peptides that mapped 41% of the sequence and 22 unique peptides covering 80% of the sequence. The sequences of EDN and ECP are 67% identical, and both are members of the RNase family.29 EDN and ECP are located in the secondary granule of eosinophils and are released during activation.6 EDN has antiviral and RNase activities,29 and ECP has cytotoxic and noncytotoxic activities, such as suppression of T cell proliferative responses.6 Major basic protein (MBP), another secondary granule protein responsible for both cytotoxic and noncytotoxic activities,4 was the eighth most abundant protein with 15 unique peptides and 59% sequence coverage.
The last of the major secondary granule proteins is eosinophil peroxidase (EPX), which was the 37th most abundant protein with 78.2% sequence coverage from 80 unique peptides (Supplemental Table 1). The content of EPX has been estimated to be 90 × 106 per eosinophil,30 but because of its larger size, it is estimated to make up approximately 25% of the total protein mass of the granules.6 EPX and PRG3 are the only granule proteins considered unique to eosinophils.29 The eosinophil granule proteins have been linked to human diseases due to their toxicity to tissues and other cells.29 For example, MBP disrupts the lipid bilayer of cells and interrupts enzyme activity.31 Also, asthma patients have elevated levels of circulating MBP in biological fluids, presumably due to degranulation.29 The ability to quantify the major eosinophil granule proteins, such as PRG3, through proteomic analysis provides an additional tool to look for evidence of prior degranulation in eosinophils isolated from these fluids.
We also identified four sialic acid-binding immunoglobulin-type lectins (Siglecs): CD33 (or Siglec-3), SIGLEC7, SIGLEC8, and SIGLEC10 (Table 2). The ranking is similar to the expected expression based primarily on flow cytometry7 with CD33 being the 4,596th and SIGLEC8 the 930th most abundant proteins, respectively. A variety of other receptors important for eosinophil function were found, including receptors for lipid mediators, adhesion receptors, and Fc receptors.4 CC-chemokine receptor 3 (CCR3), the 4,316th most abundant protein (Supplemental Table 1), plays a major role in the biology of the eosinophil as the receptor for eotaxins.4 Antagonism of CCR3 is a current field of research,32 as are preclinical studies for SIGLEC8-targeted treatments.32
Table 2.
Sialic Acid-Binding Immunoglobulin-Type Lectins
| rank | protein | peptides | differentiating peptides | % sequence coverage | iBAQ |
|---|---|---|---|---|---|
| 930 | SIGLEC8 | 19 | 19 | 47.5% | 6.33 × 1008 |
| 1166 | SIGLEC10 canonical and isoforms 3, 4, 5, 6, 7, 9 | 27 | 8 | 57.2% | 4.40 × 1008 |
| 4596 | CD33 | 3 | 3 | 15.8% | 1.37 × 1007 |
| 4747 | SIGLEC7 | 8 | 8 | 23.8% | 1.19 × 1007 |
| 6635 | SIGLEC10 isoform 8 | 19 | 1 | 45.6% | 8.43 × 1005 |
A large number of chemokines and cytokines have been attributed to eosinophils based largely on immunoassays.33 Of these, we detected only four: TGFA, TGFB1, CCL5, and CCL23 (Supplemental Table 1). We also detected CSF1, CCL18, CCL24, CXCL12, IL18, TNFSF13, and TNFSF14. CCL5 and IL18 were the most abundant. Of the seven modulatory proteins for which we could not find literature associating the proteins with eosinophils, five were identified in bronchoalveolar lavage fluid of patients with idiopathic pulmonary fibrosis in a focused proteomic screen that employed multiple reaction monitoring.34 In addition, IL18 was recently also identified immunologically in eosinophils.35 Proteomic analysis clearly should play an important role in future studies of eosinophil effectors that modulate tissue regeneration and host defenses by eosinophils.
The most abundant protein was cytoplasmic actin-1 (ACTB) with 62 distinct peptides and 85% coverage of the coding sequence, including one unique peptide that differentiated ACTB from other actins (Table 1). However, our proteomic data allows us to identify other members of the actin family present in eosinophils. An actin, either skeletal muscle cell cytoplasmic actin (ACTA1) or cardiac actin (ACTC1), was present at ∼20% the abundance of ACTB and identified by 35 peptides, two being unique to ACTC1 or ACTA1, with 50% coverage. Cytoplasmic actin-2 (ACTG1), identified by 62 different peptides covering 85% of coding sequence, including one peptide that differentiated ACTG from other actins, was present at 0.005% the abundance of ACTB. Two other top 15 proteins, prolifin-1 (PFN1) and cofilin-1 (CFL1), are modulators of actin dynamics.36,37 Actin is crucial to the role of eosinophils because they are dynamic cells that upon activation, such as by IL5, undergo reorganization of the cytoskeleton, organelles, adhesion receptors, and signaling molecules.14
Acute Activation of Eosinophils by IL5
We sought to investigate the proteome and phosphoproteome upon acute activation with IL5, which is known to cause global reorganization of eosinophils.14 Using a multiplexed proteomic strategy, we compared eosinophils from five unique donors with allergic asthma. Half of the purified eosinophils from a donor were left unstimulated, and half were acutely activated for 5 min. The number of identified proteins (4,446) and phosphosites (1,819) was less than in the label-free analysis described above. Of the identified proteins, 651 were quantified by only one peptide (Supplemental Table 3, Sheet 2). A short activation time of 5 min induces maximal polarization and activation of MAPK1/3, STAT1, and STAT5;14 however, as expected, we detected minimal changes in the proteome with only five proteins measuring a fold change greater than two and a p-value of less than 0.01 (Figure 3A). Examination of the phosphoproteome, however, revealed a dynamic cellular response with 220 significantly changed phosphoisoforms (Figure 3B). When we examined the motifs of the upregulated phosphosites, we identified many phosphorylation sites mapping back to the MAPK and CAMKII motifs (Figure 3C). Although the upregulation of MAPK was expected based on earlier findings, our analysis provides a broader view of what phosphosites are changed (Supplemental Table 3), which will inform pathway analysis and identify new end effectors. Among the sites showing the highest change in modification were those in vimentin (VIM), which relocalizes in eosinophils upon IL5-induced shape change,14 and of residues in two proteins, PTPN1138 and LCP1,39 for which phosphorylation is critical to activation of eosinophils by IL5 family cytokines. However, other highly modified sites were in proteins that currently lack function in eosinophils and warrant further study. These proteins include PPP1R9B, which has been characterized in other cells including platelets,40 and NHSL-2, which is largely uncharacterized.
Figure 3.
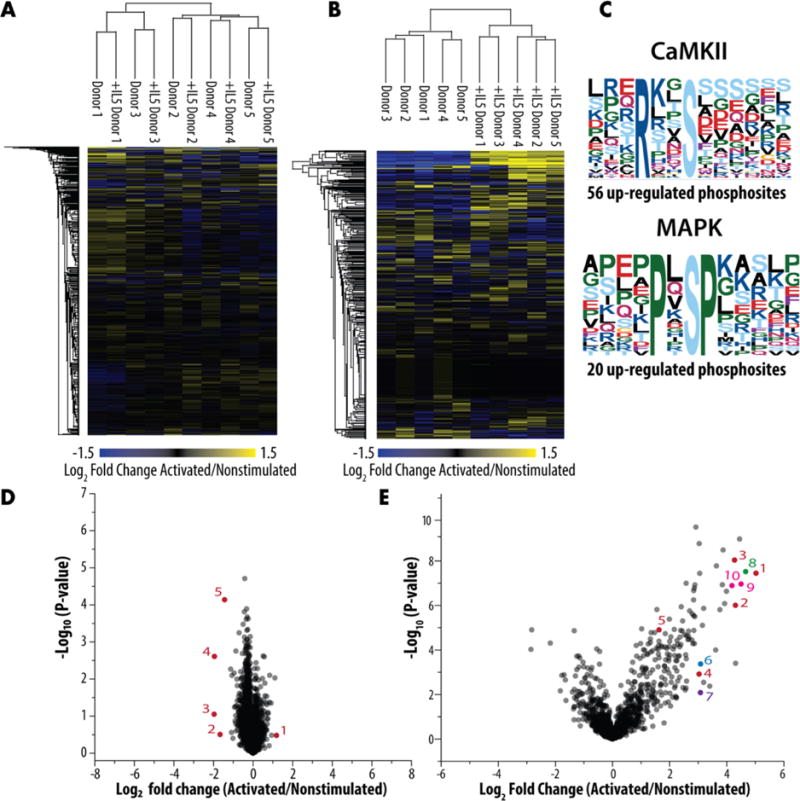
Changes in the proteome and phosphoproteome upon acute activation. Heat maps of the (A) proteome and (B) phosphoproteome based on the multiplexed analysis of acutely activated and unstimulated eosinophils. (C) Phosphoproteome motifs. (D) Volcano plot of fold-change versus significance of change at the protein level with significantly changing proteins annotated in red: (1) isoform 2 of proteasome assembly chaperone 4, (2) uncharacterized protein C18orf25, (3) putative uncharacterized protein LOC100996504, (4) Cannabinoid receptor 2, and (5) isoform 2 of copper-transporting ATPase 1. (E) Volcano plot of fold-change versus significance of change for phosphoisoforms with ones of interest highlighted in red (vimentin (VIM)): (1) Y11, (2) S8, (3) S7, (4) S47, (5) S39 and (blue) tyrosine-protein phosphatase nonreceptor type 11 (PTPN11), (6) Y546 and (purple) Plastin-1 (LCP1), (7) S5 and (green) PPP1R9B protein (PPP1R9B), (8) S94 and (pink) NHS-like protein 2 (NHSL2), (9) S393, and (10) T392/S393.
Individual Variation
Isolating the quantitative data from the five donor samples that were left unstimulated in the isobaric labeling experiment provides an opportunity to explore the extent of individual variation within the eosinophil proteome. Eosinophils demonstrate a low level of individual variation, as shown in Figure 4, by the distribution of coefficient of variation (CV). Some exceptions of proteins with more variation include seven histocompatibility antigen isoforms (HLA proteins) with a CV greater than 1. The knowledge of minimal individual variation is not only important for designing future proteomic studies but also useful for planning immunostaining and any other eosinophil biology studies using purified eosinophils from different donors. Individual variation is particularly relevant as precision medicine and targeted treatments become a focus in the medical community.
Figure 4.
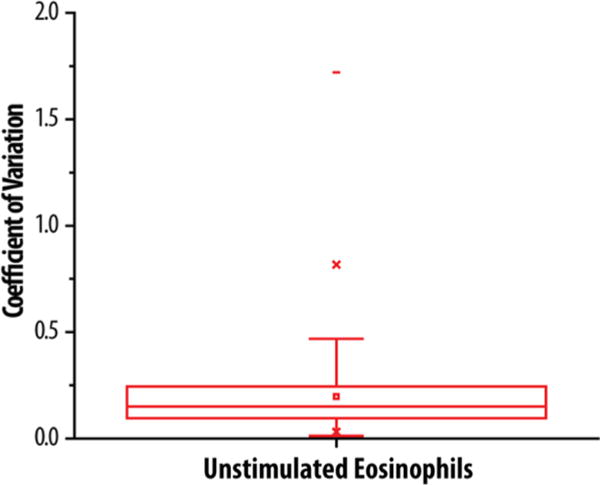
Shown is a box plot of the 1SD and 2SD of the coefficients of variation limited to no variation among proteins from purified eosinophils collected from 5 different donors. Proteins with a high coefficient of variation include seven histocompatibility antigen isoforms with a CV greater than 1.
Sample Purity
Eosinophils constitute less than 0.002% of the volume of blood, 0.005% of all blood cells, and 10% of white blood cells. Our purification was designed to minimize contamination from blood plasma proteins, red blood cells, lymphocytes, monocytes, neutrophils, and other granulocytes while allowing good (∼50%) yields of eosinophils. The eosinophils used in all proteomic studies were purified by centrifugal separation on Percoll and negative selection to remove neutrophils and monocytes with antibodies bound to magnetic beads to at least 98% purity with respect to other nucleated cells as assessed by Wright–Giemsa staining (Figure 1A). Albumin (ALB), which represents ∼50% of plasma proteins and ∼15% of protein in blood, was among the top 100 most abundant proteins; less abundant plasma proteins, such as fibronectin, were absent. Alpha (HBA1) and beta (HBB) globin subunits of red cell hemoglobin, which represent ∼70% of protein in blood, were also in the top 100 and of nearly identical abundance (Supplemental Table 1). Other red cell-specific proteins, such as erythrocyte membrane protein 4.1 (EPB41) or glycophorin A, were present in trace amounts or absent.
Normal reference values for a differential white blood count are typically 40–70% for neutrophils, 22–44% for lymphocytes, 4–11% for eosinophils, and 0–3% for basophils.41 Lymphocytes are mononuclear cells that include T, B, and NK cells. Because lymphocytes are separated from granulocytes in the Percoll gradient, and staining of 500 cells to differentiate among leukocytes regularly demonstrated that the eosinophils were >98% pure, we did not perform additional selection with anti-CD19 and anti-CD56. For each of the samples pooled for global proteomic analysis, no neutrophils and <1% lymphocytes were found. The IGHD B-cell receptor and the GRAH granzyme of NK cells were not detected in the lysates of purified eosinophils. Because neutrophils were found in three of the five samples for the TMT quantitative experiment, we compared the global proteome of neutrophils and eosinophils. Global proteomics of neutrophils has been limited to only 1,000–2,500 proteins in recent studies.42,43 Overlap between the global proteome of eosinophils and the largest reported neutrophil proteome was greater than 80%.42
The most troublesome protein contaminants were from platelets, which adhere to a proportion of eosinophils.44–46 We would expect many proteins to be shared between the two cell types; for example, CD9 is an established marker for both eosinophils7 and platelets.47 In prior immunostaining studies, we demonstrated that αIIb-integrin (ITGA2B) and thrombospondin-1 (THBS1) are present in the occasional platelet associated with eosinophils but not eosinophils themselves;44,45 these were identified as the 977th and 1566th most abundant proteins, respectively. Other identified proteins almost certainly originating from platelets include platelet factor 4 (PF4 V1) and β-thromboglobulin (also called pro platelet basic protein, PPBP).48 These were present in abundances that were 4–6% the abundance of eosinophil major basic protein (PRG2).
To examine overlap between eosinophils and platelets, we did an experiment in which half of the granulocytes were subjected to our usual negative selection cocktail and half were subjected to negative selection in which the cocktail included an antibody to CD61 to remove platelets and platelet–eosinophil complexes. CD61 is the β3 subunit of the major platelet integrin. To demonstrate depletion of platelets independently, we carried out flow cytometry, which demonstrated less cells reactive with anti-CD41 (αIIb), the partner of β3 (Figure 5A), and Western immunoblotting, which demonstrated 4-fold less THBS1 (Figure 5B). We analyzed the platelet proteome by the same workflow and employed a label-free quantitation strategy to provide relative quantification among eosinophils purified by the usual method, platelet-depleted eosinophils, and platelets. Supplemental Table 4 shows the relative quantification across identified proteins, demonstrating the relative amount of overlapping proteins between cell types and the reduction in platelet contamination proteins. Figure 6 plots the ratios of proteins found in all three samples. A number of proteins likely to originate from platelets were significantly increased in platelets and eosinophils purified without platelet depletion compared to the platelet-depleted eosinophils. These data can serve as a guide to assess the platelet contamination that routinely occurs during purification of eosinophils and demonstrates that the addition of antibodies to deplete platelets, although unlikely to result in 100% depletion, is warranted in future studies of the eosinophil proteome. In troublesome cases, immunostaining can serve as a facile method to sort out which proteins are unique to platelets and eosinophils.
Figure 5.
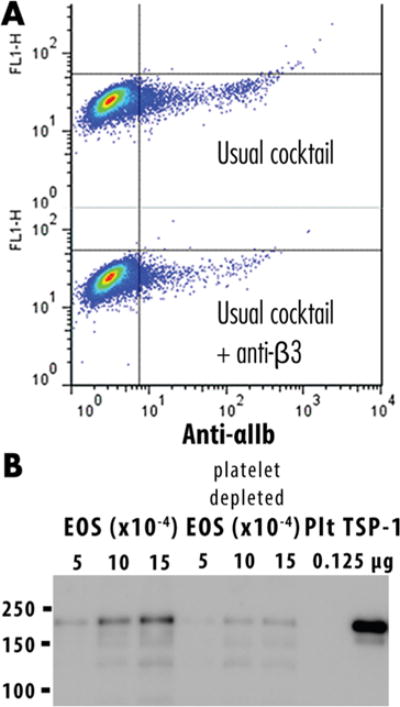
Depletion of platelet contamination. (A) Addition of the anti-CD61 (β3 integrin) purification step yielded eosinophils that were less reactive with anti-CD41 αIIb integrin, the partner of β3 integrin, as shown by flow cytometry. (B) Immunoblotting demonstrates that the platelet-depleted eosinophils contain less THBS1.
Figure 6.
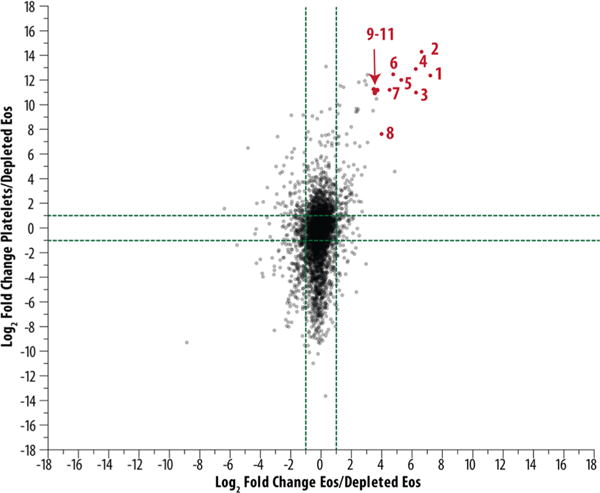
Depletion of platelet contamination as shown through mass spectrometry. Label-free quantitation was used to demonstrate that proteins shown in red were upregulated in eosinophils purified with the usual cocktail and platelets versus platelet-depleted eosinophils. The proteins in red are likely candidates of platelet contamination, including (1) serum deprivation-response protein, (2) tubulin β1 chain, (3) platelet factor 4, (4) fibrinogen β chain, (5) integrin αIIb, (6) coagulation factor XIII A chain, (7) platelet basic protein, (8) tubulin α8 chain, and (9–11) integrin β3, platelet glycoprotein Ib α chain, and isoform γ A of fibrinogen γ chain.
CONCLUSIONS
Here we have provided a comprehensive list of proteins in eosinophils that can be used to generate hypotheses for further study, described changes in the phosphoproteome upon activation, and evaluated the level of platelet contamination in purified eosinophils. We have only touched upon the information contained in the Supplementary Tables. This information represents a new starting point for future studies on the functions and localizations of proteins in the biology of eosinophils at baseline and in different states of activation in vitro after stimulation by different mediators. The list of proteins and their phosphorylation status provides insight into splice isoforms, phosphorylation events that change upon activation, and relative abundances. For example, the proteomic data have already informed further study of the different splice isoforms of STAT3 from evidence of the STAT3 deletion 701 isoform in the proteomics data.49 The sample preparation and further depletion of platelets indicate how studies of all types can be improved to limit impact from the platelets adhering to eosinophils. The publicly accessible data will provide a basis for eosinophil biologists to conduct future translational and clinical research on the similarities and differences in the proteome of eosinophils in different anatomic compartments and activation states in vivo; in different eosinophil-associated disorders such as asthma, eosinophilic gastrointestinal diseases, and hyper-eosinophilic syndrome; after different interventions and among different subjects; and on the possible associations of differences with disease activity, which are all currently areas of great interest.
Supplementary Material
Acknowledgments
This study was possible because of purified blood eosinophils provided by Paul Fichtinger of the Eosinophil Core Laboratory, whom we thank profusely. We are also indebted to Frances Fogerty for preparation of cells, Doug Annis for immunoblotting studies and advice on platelet purification, and Timothy Rhoads for critical reading of the manuscript. This work was supported in part by a Program Project Grant on the Role of Eosinophils in Airway Inflammation and Remodeling, P01HL088585 (N.N.J.), NIGMS R01 GM080148 (J.J.C.), and a seed grant from the University of Wisconsin Carbone Cancer Center to D.F.M. and J.J.C. E.M.W. gratefully acknowledges support from National Institutes of Health-funded Research Training in Hematology (T32 HL007899).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00006.
Summary of proteins identified in global analysis with proteins ordered from most to least abundant (XLSX) Summary of phosphosites identified in global analysis with proteins ordered alphabetically by protein abbreviation (XLSX)
Summary of proteins and phosphosites quantified in comparative analysis of unstimulated and acutely activated eosinophils with entries ordered according to fold differences (XLSX)
Summary of label-free quantification of eosinophils, platelets, and platelet-depleted eosinophils with entries ordered according to fold change (XLSX)
- Platelet Contamination: https://chorusproject.org/anonymous/download/experiment/2ae5a0c2ee6444b98031c17f5e6720e2
Author Contributions
E.M.W. designed and performed research, analyzed data, and wrote the manuscript. M.W.J. designed and performed research and wrote the manuscript. M.S.W., J.J.C, A.S.H., and D.F.M designed experiments and wrote the manuscript. E.A.S. was responsible for purification of eosinophils and wrote the manuscript. N.N.J. and S.K.M. arranged for study material.
Notes
The authors declare no competing financial interest.
References
- 1.Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell. 2013;49(4):583–90. doi: 10.1016/j.molcel.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LD, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TS, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509(7502):575–81. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–7. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13(1):9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wynn TA. Type 2 cytokines: mechanisms and therapeutic strategies. Nat Rev Immunol. 2015;15(5):271–82. doi: 10.1038/nri3831. [DOI] [PubMed] [Google Scholar]
- 6.Konikoff MR, Blanchard C, Kirby C, Buckmeier BK, Cohen MB, Heubi JE, Putnam PE, Rothenberg ME. Potential of blood eosinophils, eosinophil-derived neurotoxin, and eotaxin-3 as biomarkers of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2006;4(11):1328–36. doi: 10.1016/j.cgh.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Lee JJ, Rosenberg HF. Eosinophils in health and disease. 1st. Elsevier; London: 2013. p. 654. [Google Scholar]
- 8.Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med. 2009;206(1):183–93. doi: 10.1084/jem.20081756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouffi C, Kartashov AV, Schollaert KL, Chen X, Bacon WC, Weirauch MT, Barski A, Fulkerson PC. Transcription Factor Repertoire of Homeostatic Eosinophilopoiesis. J Immunol. 2015;195(6):2683–95. doi: 10.4049/jimmunol.1500510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bochner BS, Book W, Busse WW, Butterfield J, Furuta GT, Gleich GJ, Klion AD, Lee JJ, Leiferman KM, Minnicozzi M, Moqbel R, Rothenberg ME, Schwartz LB, Simon HU, Wechsler ME, Weller PF. Workshop report from the National Institutes of Health Taskforce on the Research Needs of Eosinophil-Associated Diseases (TREAD) J Allergy Clin Immunol. 2012;130(3):587–96. doi: 10.1016/j.jaci.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straub C, Pazdrak K, Young TW, Stafford SJ, Wu Z, Wiktorowicz JE, Haag AM, English RD, Soman KV, Kurosky A. Toward the proteome of the human peripheral blood eosinophil. Proteomics: Clin Appl. 2009;3(10):1151–1173. doi: 10.1002/prca.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 13.Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. The one hour yeast proteome. Mol Cell Proteomics. 2014;13(1):339–47. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han ST, Mosher DF. IL-5 induces suspended eosinophils to undergo unique global reorganization associated with priming. Am J Respir Cell Mol Biol. 2014;50(3):654–64. doi: 10.1165/rcmb.2013-0181OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson MW, Annis DS, Mosher DF. alpha(M)beta(2) integrin-mediated adhesion and motility of IL-5-stimulated eosinophils on periostin. Am J Respir Cell Mol Biol. 2013;48(4):503–10. doi: 10.1165/rcmb.2012-0150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, Geiger J, Sickmann A, Zahedi RP. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012;120(15):e73–82. doi: 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 18.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 19.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 20.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics. 2014;13:2513. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 22.Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011;11(6):1064–74. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J, Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinf. 2012;13(Suppl 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chua JC, Douglass JA, Gillman A, O’Hehir RE, Meeusen EN. Galectin-10, a potential biomarker of eosinophilic airway inflammation. PLoS One. 2012;7(8):e42549. doi: 10.1371/journal.pone.0042549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acharya KR, Ackerman SJ. Eosinophil granule proteins: form and function. J Biol Chem. 2014;289(25):17406–17415. doi: 10.1074/jbc.R113.546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abu-Ghazaleh RI, Dunnette SL, Loegering DA, Checkel JL, Kita H, Thomas LL, Gleich GJ. Eosinophil granule proteins in peripheral blood granulocytes. J Leukocyte Biol. 1992;52(6):611–8. doi: 10.1002/jlb.52.6.611. [DOI] [PubMed] [Google Scholar]
- 31.Kay AB. Allergy and allergic diseases. 2nd. Wiley-Blackwell; West Sussex; Hoboken, NJ: 2008. [Google Scholar]
- 32.Fulkerson PC, Rothenberg ME. Targeting eosinophils in allergy, inflammation and beyond. Nat Rev Drug Discovery. 2013;12(2):117–29. doi: 10.1038/nrd3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davoine F, Lacy P. Eosinophil cytokines, chemokines, and growth factors: emerging roles in immunity. Front Immunol. 2014;5:570. doi: 10.3389/fimmu.2014.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster MW, Morrison LD, Todd JL, Snyder LD, Thompson JW, Soderblom EJ, Plonk K, Weinhold KJ, Townsend R, Minnich A, Moseley MA. Quantitative proteomics of bronchoalveolar lavage fluid in idiopathic pulmonary fibrosis. J Proteome Res. 2015;14(2):1238–49. doi: 10.1021/pr501149m. [DOI] [PubMed] [Google Scholar]
- 35.Gatault S, Delbeke M, Driss V, Sarazin A, Dendooven A, Kahn JE, Lefevre G, Capron M. IL-18 Is Involved in Eosinophil-Mediated Tumoricidal Activity against a Colon Carcinoma Cell Line by Upregulating LFA-1 and ICAM-1. J Immunol. 2015;195(5):2483–92. doi: 10.4049/jimmunol.1402914. [DOI] [PubMed] [Google Scholar]
- 36.Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18(1):11–7. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18(1):26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Pazdrak K, Olszewska-Pazdrak B, Stafford S, Garofalo RP, Alam R. Lyn, Jak2, and Raf-1 kinases are critical for the antiapoptotic effect of interleukin 5, whereas only Raf-1 kinase is essential for eosinophil activation and degranulation. J Exp Med. 1998;188(3):421–9. doi: 10.1084/jem.188.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pazdrak K, Young TW, Straub C, Stafford S, Kurosky A. Priming of eosinophils by GM-CSF is mediated by protein kinase CbetaII-phosphorylated L-plastin. J Immunol. 2011;186(11):6485–96. doi: 10.4049/jimmunol.1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma P, Ou K, Sinnamon AJ, Jiang H, Siderovski DP, Brass LF. Modulating platelet reactivity through control of RGS18 availability. Blood. 2015;126(24):2611–20. doi: 10.1182/blood-2015-04-640037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kratz A, Ferraro M, Sluss PM, Lewandrowski KB. Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Laboratory reference values. N Engl J Med. 2004;351(15):1548–63. doi: 10.1056/NEJMcpc049016. [DOI] [PubMed] [Google Scholar]
- 42.Zhou JY, Krovvidi RK, Gao Y, Gao H, Petritis BO, De AK, Miller-Graziano CL, Bankey PE, Petyuk VA, Nicora CD, Clauss TR, Moore RJ, Shi T, Brown JN, Kaushal A, Xiao W, Davis RW, Maier RV, Tompkins RG, Qian WJ, Camp DG, 2nd, Smith RD. Trauma-associated human neutrophil alterations revealed by comparative proteomics profiling. Proteomics: Clin Appl. 2013;7(7–8):571–83. doi: 10.1002/prca.201200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLeish KR, Merchant ML, Klein JB, Ward RA. Technical note: proteomic approaches to fundamental questions about neutrophil biology. J Leukocyte Biol. 2013;94(4):683–692. doi: 10.1189/jlb.1112591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johansson MW, Han ST, Gunderson KA, Busse WW, Jarjour NN, Mosher DF. Platelet activation, P-selectin, and eosinophil beta1-integrin activation in asthma. Am J Respir Crit Care Med. 2012;185(5):498–507. doi: 10.1164/rccm.201109-1712OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson MW, Mosher DF. Activation of beta1 integrins on blood eosinophils by P-selectin. Am J Respir Cell Mol Biol. 2011;45(4):889–97. doi: 10.1165/rcmb.2010-0402OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Page C, Pitchford S. Platelets and allergic inflammation. Clin Exp Allergy. 2014;44(7):901–13. doi: 10.1111/cea.12322. [DOI] [PubMed] [Google Scholar]
- 47.Kishimoto T. Leucocyte typing VI: white cell differentiation antigens: proceedings of the sixth international workshop and conference held in Kobe, Japan, 10–14 November 1996. xxxiv. Garland Pub; New York: 1998. p. 1342. [Google Scholar]
- 48.McRedmond JP, Park SD, Reilly DF, Coppinger JA, Maguire PB, Shields DC, Fitzgerald DJ. Integration of proteomics and genomics in platelets: a profile of platelet proteins and platelet-specific genes. Mol Cell Proteomics. 2004;3(2):133–144. doi: 10.1074/mcp.M300063-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Turton KB, Annis DS, Rui L, Esnault S, Mosher DF. Ratios of Four STAT3 Splice Variants in Human Eosinophils and Diffuse Large B Cell Lymphoma Cells. PLoS One. 2015;10(5):e0127243. doi: 10.1371/journal.pone.0127243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


