Abstract
Previous analyses have identified related cytoplasmic Lsm1-7p and nuclear Lsm2-8p complexes. Here we report that mature heat shock and MET mRNAs that are trapped in the nucleus due to a block in mRNA export were strongly stabilized in strains lacking Lsm6p or the nucleus-specific Lsm8p protein but not by the absence of the cytoplasmic Lsm1p. These nucleus-restricted mRNAs remain polyadenylated until their degradation, indicating that nuclear mRNA degradation does not involve the incremental deadenylation that is a key feature of cytoplasmic turnover. Lsm8p can be UV cross-linked to nuclear poly(A)+ RNA, indicating that an Lsm2-8p complex interacts directly with nucleus-restricted mRNA. Analysis of pre-mRNAs that contain intronic snoRNAs indicates that their 5′ degradation is specifically inhibited in strains lacking any of the Lsm2-8p proteins but Lsm1p. Nucleus-restricted mRNAs and pre-mRNA degradation intermediates that accumulate in lsm mutants remain 5′ capped. We conclude that the Lsm2-8p complex normally targets nuclear RNA substrates for decapping.
Sm-like (Lsm) proteins have been identified in all kingdoms of life and participate in numerous RNA processing and degradation pathways. The Sm and Lsm complexes are all likely to form similar structures with seven member rings (or six in the case of Escherichia coli Hfq) with a central hole, through which the RNA may pass (1, 2, 14, 50, 75, 82). In the nucleus, an Lsm2-8p complex associates with the U6 snRNA and is important for U6 accumulation, snRNP biogenesis, and pre-mRNA splicing (1, 27, 49, 57, 64, 67). This complex is also implicated in the processing of pre-tRNAs, pre-small nucleolar RNAs (snoRNAs), and pre-rRNAs (40, 41, 42). In contrast to the Lsm2-8p proteins, which have nuclear roles, a homologous Lsm1-7p complex functions in cytoplasmic mRNA degradation. The Lsm1-7p complex functions in both the 5′ and 3′ degradation of mRNAs, promoting mRNA decapping and 5′ degradation, probably via interactions with the decapping enzymes and Xrn1p (10, 12, 70, 71), and also protects the 3′ ends from premature degradation (29).
Control of mRNA stability is a key step in the regulation of gene expression, regulating both the amount of mRNA that accumulates following transcription and the time for which the mRNA remains functional. In yeast, a major pathway of cytoplasmic mRNA degradation involves deadenylation-dependent removal of the 5′ cap by the decapping complex Dcp1p/Dcp2p followed by 5′→3′ degradation by the exonuclease Xrn1p (8, 17, 32, 44, 52, 53). In an alternative pathway, deadenylated mRNAs are degraded 3′→5′ by the cytoplasmic exosome (5).
Pathways that degrade mRNA precursors in the nucleus have also been identified (11, 16, 30, 74; reviewed in references 35, 37, and 51), with different pathways apparently utilizing the same RNA degradation machinery. Unspliced nuclear pre-mRNAs are very rapidly degraded, predominantly 3′→5′, by the nuclear exosome. In contrast, 5′→3′ degradation by the exonuclease Rat1p plays a minor role in this pathway (11). The nuclear exosome also functions in the rapid degradation of pre-mRNAs that have failed to undergo correct 3′ cleavage (74), but in this pathway there is no evidence for significant 5′→3′ degradation. Pre-mRNAs that lack poly(A) tails or carry aberrant tails are restricted to the nucleus (20, 30, 36) and are probably also degraded by the nuclear exosome. Finally, mRNAs that are restricted to the nucleus due to a defect in nuclear-cytoplasmic transport are stabilized by deletion of the nuclear exosome component Rrp6p and by deletion of Rai1p, which functions together with Rat1p in rRNA processing (16). These results indicate that nucleus-restricted mRNAs can be degraded both 3′→5′ and 5′→3′.
The 5′→3′ degradation of nuclear pre-mRNAs presumably requires their decapping prior to exonuclease digestion. In the cytoplasm, decapping is not a default activity but is highly regulated and integrated with mRNA deadenylation and translational status. Efficient decapping requires the translation factor Pat1p/Mrt1p and the cytoplasmic Lsm1-7p complex, several members of which can interact with Dcp1p, Dcp2p, and Xrn1p (12, 24, 25, 28, 70, 71, 76).
Here we report that the decapping and 5′ degradation of both unspliced pre-mRNAs and otherwise mature mRNAs that are restricted to the nucleus due to a defect in mRNA export require the nuclear Lsm2-8p complex.
MATERIALS AND METHODS
Strains.
The transformation procedure was performed as described previously (26). Saccharomyces cerevisiae strains used in this work are listed in Table 1. Strains YJK23, YJK24, YJK26, and YJK27 were constructed by PCR-based gene disruption of LSM6 (YJK23, YJK26, and YJK27) or LSM1 (YJK24) in strains FYEF95, YCB81, and YCB63, respectively, by using plasmid pTL54 as a PCR template (43). Disruption was confirmed by PCR analysis. Strain YJK46 was generated by one-step PCR using pFA6a-kanMX6-pGAL1-3HA as a template (47). The phenotypes of respective constructs were confirmed by Northern hybridization. Strains YJK49 and YJK51 were constructed by a one-step PCR procedure as described previously (59). Correct integration was confirmed by PCR analysis, and the expression of Lsm8p-TAP was tested by Western blotting. Strain BSY557 was cotransformed with plasmids pBS1181 and pBS1184; this was followed by the loss of plasmid pBS1191 by growth on medium containing 5-fluoroorotic acid as described previously (48). Strains BMA64, AEMY19 (lsm6Δ), and AEMY46 (GAL-lsm8) were transformed with a multicopy plasmid (pYX172) carrying the SNR6 gene, which encodes the U6 snRNA (49).
TABLE 1.
Yeast strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| AEMY19 | MATα ade2-1 his3Δ200 leu2-3,112 trp1Δ1 ura3-1 LSM6::HIS3 | 49 |
| AEMY22 | MATα ade2-1 his3Δ200 leu2-3,112 trp1Δ1 ura3-1 LSM7::HIS3 | 49 |
| AEMY24 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM1::TRP1 | 49 |
| AEMY31 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM3::TRP1 [pBM125-GAL1-HA-LSM3] | 49 |
| AEMY33 | MATα ade2-1 his3Δ200 leu2-3,112 trp1Δ1 ura3-1 LSM2::HIS3 [pBM125-GAL1-LSM2-HA] | 49 |
| AEMY46 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM8::TRP1 [pBM125-GAL1-HA-LSM8] | 49 |
| AEMY47 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM5::TRP1 [pBM125-GAL1-HA-LSM5] | 49 |
| MCY4 | MATaade1-101 his3-Δ1 trp1-289 ura3-52 LEU2-GAL1-LSM4 | 15 |
| BMA64 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ ura3-1 | F. Lacroute |
| D270 | MATaade2 his3 leu2 trp2 ura3 | 78 |
| D271 | MATα ade2 his3 leu2 trp2 ura3 | 78 |
| YCBA63 | As D271 but HIS3-GAL::ProtA-RRP41 | 74 |
| YCBA81 | As D271 but HIS5sp-GAL-3HA-DOB1 | 74 |
| FYEF95 | MATaura3-Δ851 trp-Δ1Δ63 leu2Δ1 Δnup145 [ARS-CEN-LEU2-ProtA-NUP145N] | 69 |
| BSY557 | MATaade2 arg4 leu2-3, 112 trp1-289 ura3-52 SNR6::ADE2 pBS1191 (CEN-URA3-SNR6) | 48 |
| YJK23 | As FYEF95 but LSM6::Kl URA | This work |
| YJK24 | As FYEF95 but LSM1::Kl URA | This work |
| YJK26 | As YCB81 but LSM6::Kl URA3 | This work |
| YJK27 | As YCB63 but LSM6::Kl URA3 | This work |
| YJK46 | As FYEF95 but kanMX6-GAL1-LSM8 | This work |
| YJK49 | As FYEF95 but Lsm8-TAP | This work |
| YJK51 | As D270 but Lsm8-TAP | This work |
RNA extraction and Northern hybridization.
For depletion of the essential Lsm proteins, cells were harvested at intervals following the shift from RSG medium (2% galactose, 2% sucrose, 2% raffinose) or yeast-peptone-Gal medium containing 2% galactose to yeast-peptone-dextrose (YPD) medium containing 2% glucose. Otherwise, strains were grown in YPD medium. The lsm1Δ, lsm6Δ, lsm7Δ, lsm2(Ts), and lsm5(Ts) strains were pregrown at 23°C and transferred to 37°C. For the analysis of heat shock (HS) mRNAs, cells were grown at 23°C, transferred to 42°C for 15 min, and further grown at 37°C. Strains AEMY46 and YJK46 were pregrown in RSG medium and transferred to YPD medium for 18 h prior to the heat shock. The transcriptional shutdown of GAL or MET mRNAs was performed by addition of glucose, to achieve a 4% final concentration, to cell cultures pregrown to mid-log phase (optical density at 600 nm [OD600], ∼0.4) on galactose or by addition of methionine, to achieve a 2 mM final concentration, to cell cultures pregrown in minimal medium lacking methionine, respectively. The cultures were shifted to 37°C for 30 min to achieve a loss-of-function phenotype prior to transcriptional shutdown. Strains AEMY46 and YJK46 were pregrown in RSG medium and transferred to minimal galactose medium lacking methionine for 18 h prior to the transcriptional shutdown. For depletion of U6 snRNA, cells were pregrown in selective minimal medium with 2% lactate, 2% glycerol, and 0.05% glucose to mid-log phase and then supplemented with 2% galactose as described previously (48).
RNA extraction and Northern hybridization were carried out as described previously (9, 73). Small RNAs were separated on a 6% acrylamide gel containing 8.3 M urea and transferred to a Hybond N+ membrane by electrotransfer. High-molecular-weight RNAs were analyzed on 2% agarose gels and transferred by capillary elution.
Quantification of Northern blots was performed using a Storm 860 PhosphorImager and ImageQuant software (Molecular Dynamics). Half-lives of MET mRNAs were determined by plotting the mRNA levels versus the time on a semilog plot.
For RNA hybridization the following oligonucleotides were used: 008 (18S), 5′-CATGGCTTAATCTTTGAGAC; 205 (U18), 5′-GTCAGATACTGTGATAGTC; 206 (U18-3′), 5′-GCTCTGTGCTATCGTC; 250 (scR1), 5′-ATCCCGGCCGCCTCCATCAC; 255 (snR38), 5′-GAGAGGTTACCTATTATTACCCATTCAGACAGGGATAACTG; 261 (U6), 5′-AAAACGAAATAAATTCTTTGTAAAAC; 270 (U2), 5′-TGAAGAAACCATGAGCGAAGAAA; 400 (ACT1), 5′-TCTTGGTCTACCGACGATAGATGGGAAGACAGCA; 418 (PGK1), 5′-ATTCCAAAGAAGCACCACCACC; 470 (RPS3), 5′-GACACCGTCAGCGACTAG; 472 (MET3), 5′-GCTCTCACAGTCAACTCCCTGTGGGCTCTATGCATT; 473 (MET6), 5′-GCGTTGGCAGGCAAGTCCAAAACTAAGACAGGTTC; 474 (SSA3), 5′-CCAAGCCATAAGCAATTGCTGCTGCAGTGGGTTCATTG; 475 (SSA1-RH), 5′-GCTGGCAGTGGTGTTGCTGTC; 476 (SSA1-3′), 5′-CACCTGGAGCACCACCAGCTTGGTACAACTTAGACAT; 477 (SSA4-RH), 5′-CGGTGGAGGCCGCTTGCGAAGC; 478 (SSA4-3′), 5′-CTCCAGCACCCGGAACTGGGCCTGCTCCTGGGGCAC; 483 (TEF4), 5′-CCACCGACCAAGTTGTTGGACATGAAAGTCAAGGTAAG; 484 (EFB1), 5′-GCAACCATTTCTTCCAAATTGGTTTCATCATCCCATGG; 485 (MET3-3′), 5′-TGCTTGCCAACAACAGAGTCCTTGTCATCTTCCC; 486 (MET3-RH), 5′-GTCCACTACCAGAACCAATG; 763 (SSA4), 5′-GTGGTACGCCTCTTGGAGCGGGTGGAAACCGCTC; and 765 (SSA1), 5′-AATTTGTCACCAGCTTCAGAAATGGTGTTCTTCAAAGAG.
Immunoprecipitation.
Immunoprecipitation of 7-methyl guanosine (m7G)-capped or trimethyl guanosine (TMG)-capped RNAs was performed as described previously (39). Twenty micrograms of RNA was incubated on a rotating wheel in buffer A (150 mM potassium acetate [pH 7.5], 20 mM Tris acetate; 5 mM magnesium acetate) for 2 h at 4°C with cap-specific serum (R1131) or monoclonal antibody (Ab) against both m7G and trimethyl guanosine (TMG) cap structures (H20) (both kindly provided by R. Lührmann, MPI, Göttingen, Germany) bound to protein A-Sepharose. After washing the pellet in buffer A, bound RNAs were eluted with 10 mM m7G(5′)ppp(5′)G (Pharmacia) in buffer A. The RNAs were extracted with guanidium thiocyanate and phenol-chloroform and ethanol precipitated.
RNase H treatment.
RNase H cleavage and deadenylation were performed essentially as described previously (54). For deadenylation, samples of 20 μg of total RNA were annealed with 400 ng of oligo(dT) at 68°C for 10 min and digested with 1.5 U of RNase H at 30°C for 1 h. RNase H cleavage of SSA4 and SSA1 heat shock mRNAs or MET3 mRNA was carried out in similar conditions except that 40 ng of oligonucleotide complementary to the specific mRNA sequence was annealed with the RNA sample.
UV cross-linking of poly(A)+ RNPs.
Cross-linking and purification of poly(A)+ RNPs were performed exactly as described (6, 61) from 1.2 liters of cells (strains YJK49 and YJK51). Strain YJK51 was grown at 30°C to an OD600 of 0.75. Strain YJK49 was pregrown at 23°C to an OD600 of 0.6 and transferred to 37°C for 30 min to achieve a loss-of-function phenotype. Recovered RNPs were treated with micrococcal nuclease (Roche) and RNase A (Sigma) as described previously and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blot analysis was performed using anti-protein A Ab to detect ProtA-Nup145N and Lsm8-TAP, C4-1.2 Ab against Npl3p (62) followed by horseradish peroxidase-conjugated donkey anti-rabbit Ab, and monoclonal A66 Ab against Nop1p (7) followed by horseradish peroxidase-conjugated sheep anti-mouse Ab.
RESULTS
A difficulty in analyzing the nuclear degradation of mRNAs is distinguishing it from normal cytoplasmic mRNA turnover. Here we have analyzed two distinctly different classes of RNA that are degraded within the nucleus: nucleus-restricted mRNAs that fail to be exported due to truncation of the nuclear pore complex component Nup145p and unspliced pre-mRNAs, which are normally very rapidly degraded within the nucleus.
Depletion of Lsm proteins stabilizes mRNAs trapped in the nucleus.
Strains carrying the nup145N allele, which lacks the C-terminal domain of the nucleoporin Nup145p, are temperature sensitive lethal and show rapid accumulation of nuclear poly(A)+ RNA after transfer to the nonpermissive temperature of 37°C (18, 69).
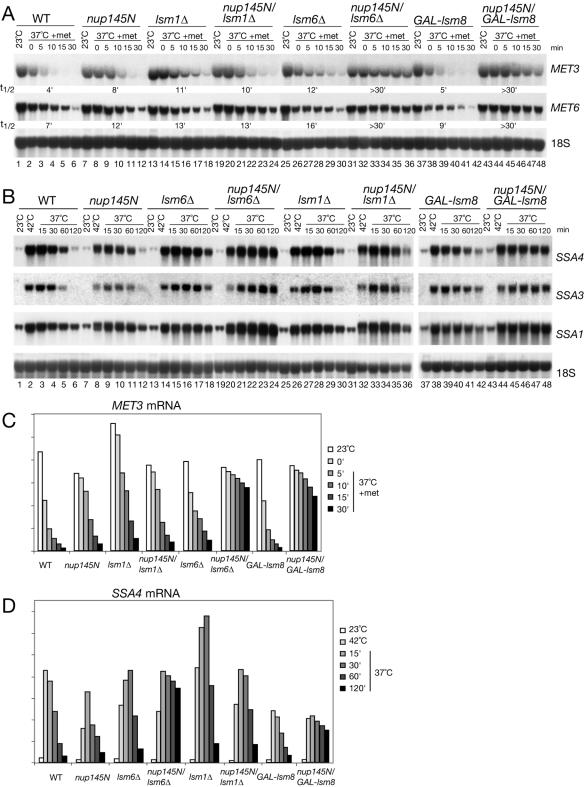
To analyze the decay of the repressible MET3 and MET6 mRNAs, transcription was inhibited by addition of methionine 15 min after transfer to 37°C (Fig. 1A and C). Both mRNAs had approximately twofold-longer half-lives in the nup145N strain (Fig. 1A, lanes 7 to 12) than in the wild type (Fig. 1A, lanes 1 to 6), indicating that their nuclear decay was slower than the normal cytoplasmic turnover. This is in marked contrast to the very rapid decay previously observed for unspliced or 3′-unprocessed nuclear RNAs (11, 16, 30, 74). Decay of MET3 and MET6 was substantially slower in the nup145N strain that also lacked Lsm6p (Fig. 1A, lanes 31 to 36) or was depleted for the nucleus-specific component Lsm8p (Fig. 1A, lanes 43 to 48). In contrast, the absence of the cytoplasmic Lsm1p from the nup145N strain conferred no clear stabilization of the MET mRNAs (Fig. 1A), confirming their nuclear degradation. As expected, the absence of only Lsm1p (Fig. 1A, lanes 13 to 18) or Lsm6p (Fig. 1A, lanes 25 to 30) from the otherwise wild-type strain stabilized the MET mRNAs, due to their roles in cytoplasmic mRNA turnover (10, 12, 70), whereas depletion of Lsm8p alone conferred little stabilization (Fig. 1A, lanes 37 to 42). These data were quantified by PhosphorImager analysis, and those for MET3 are presented in Fig. 1C. Similar results were observed during transcription shutdown of repressible GAL1, GAL7, and GAL10 mRNAs in nup145N/lsm6Δ, nup145N/GAL::lsm8, and nup145N/lsm1Δ mutants following transfer to glucose medium at 37°C (data not shown).
FIG. 1.
Transcriptional shutdown of transiently expressed mRNAs. Northern analysis of MET (A) and heat shock (B) mRNAs. (A) Strains were pregrown in minimal medium lacking methionine at 23°C (23°C lanes), shifted to 37°C for 30 min (37°C 0-min lane), and supplemented with methionine to a 2 mM final concentration. The GAL::lsm8 and nup145N/GAL::lsm8 strains (lanes 37 to 48) were pregrown on galactose and transferred to minimal glucose medium lacking methionine for 18 h before the shift to 37°C. mRNA half-lives (t1/2, indicated in minutes) are shown below each panel. (B) Strains were pregrown at 23°C (23°C lanes), shifted to 42°C for 15 min (42°C lanes), and then transferred to 37°C (37°C lanes) for the times indicated. The GAL::lsm8 and nup145N/GAL::lsm8 strains (lanes 37 to 48) were pregrown on galactose and transferred to glucose for 18 h before the heat shock. RNA was separated on 2% agarose gels and hybridized with oligonucleotide probes complementary for the mRNAs indicated on the right. The level of 18S rRNA (probe 008) was used as a loading control. (C and D) Graphic representation of mRNA levels during transcriptional shutdown for MET3 (C) and SSA4 (D) and mRNAs. Values for each RNA were standardized relative to those for the loading control (18S rRNA). RNA levels were calculated based on PhosphorImager quantification of Northern hybridization data from panels A and B. WT, wild type.
In wild-type strains, brief (15-min) transfer to 42°C induces a pulse of HS transcripts, which decay during subsequent incubation at 37°C, permitting a pseudo pulse-chase analysis (shown for three HS mRNAs, SSA1, SSA3, and SSA4, in Fig. 1B, lanes 1 to 6; quantified data are shown in Fig. 1D) (63, 68). HS mRNAs are efficiently retained in the nuclei of nup145N strains and other mRNA export mutants, as shown by in situ hybridization and a corresponding failure to synthesize HS proteins at the nonpermissive temperature (33, 72; L. Milligan and D. Tollervey, data in preparation). Decay of the HS mRNAs in the nup145N strain was similar to or slower than the cytoplasmic turnover in the wild type (Fig. 1B, compare lanes 1 to 6 and lanes 7 to 12). For the nup145N and lsmΔ strains the level of HS mRNAs shows a transient increase after the shift to 37°C before decaying, in contrast to the WT. These distinct kinetics prevent a direct comparison of HS mRNA decay rates in different strains. However, it is clear that in the nup145N/lsm6Δ and nup145N/GAL::lsm8 strains the HS mRNAs were more significantly stabilized than the nup145N single mutant strain (Fig. 1B, compare lanes 19 to 24 and 43 to 48 to lanes 7 to 12). In contrast, the nup145N/lsm1Δ (Fig. 1B, lanes 31 to 36) did not show significantly increased stabilization. These data are presented for SSA4 in Fig. 1D.
From these analyses we conclude that Lsm6p and Lsm8p are required for efficient degradation of nucleus-restricted mRNAs, whereas Lsm1p is not needed for this activity. This conclusion strongly suggests the involvement of the Lsm2-8p complex in nuclear mRNA degradation.
Nucleus-restricted heat shock mRNAs stabilized in lsm mutants are polyadenylated.
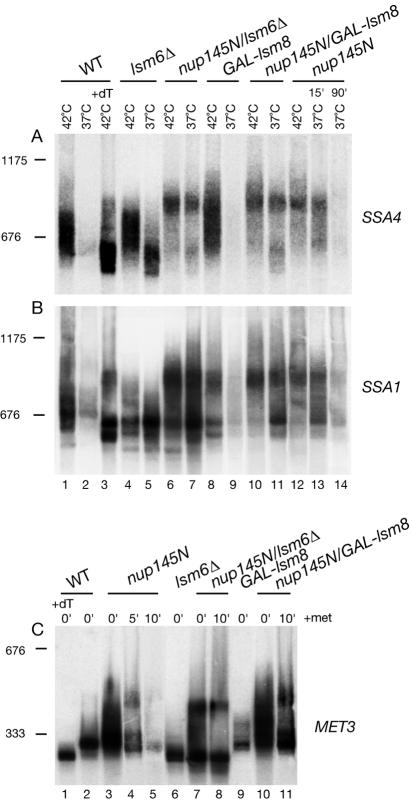
In the cytoplasm, mRNAs undergo distributive, incremental deadenylation prior to complete degradation (52, 53). To determine whether this is also the case for nucleus-restricted mRNAs, the poly(A) tails were analyzed for the SSA1, SSA4 and MET3 mRNA. To allow resolution of the poly(A) tail, the mRNAs were specifically cleaved by using a complementary oligonucleotide and RNase H, prior to Northern analysis (see Materials and Methods).
In the wild-type strain, after HS induction for 15 min at 42°C, strong signals were seen for each HS mRNA with heterogeneity that corresponds to the poly(A) tail length distribution resulting from cytoplasmic deadenylation (Fig. 2A and B, lanes 1). Following deadenylation by addition of oligo(dT) to the RNase H reaction (Fig. 2A and B, lanes 3) the size of the mRNA is reduced, although some heterogeneity is still observed for both SSA1 and SSA4. This may represent intrinsic 3′-end heterogeneity, rather than incomplete deadenylation, since it was not seen for MET3 under equivalent conditions (Fig. 2C, lane 1). In the wild-type strain following transfer to 37°C for 90 min (Fig. 2A and B, lanes 2) most of the HS mRNA is degraded, and the residual mRNAs are largely oligoadenylated. In the lsm6-Δ strain more deadenylated HS mRNA remains after 90 min at 37°C (Fig. 2A and B, lanes 5), presumably due to the inhibition of cytoplasmic 5′ degradation as previously shown for other mRNAs (10, 12, 29). In contrast, HS mRNAs in the Lsm8p-depleted single mutant strains undergo normal deadenylation at 42°C (Fig. 2A and B, lanes 8) and are lost after 90 min at 37°C (Fig. 2A and B, lanes 9).
FIG. 2.
Heat shock mRNAs stabilized in lsm mutants are polyadenylated. Strains were grown, and total RNA was prepared as described for Fig. 3. (A and B) Strains were pregrown at 23°C, shifted to 42°C for 15 min (42°C lanes), and then transferred to 37°C for 15 or 90 min (37°C lanes). (C) Strains pregrown in minimal medium lacking methionine at 23°C were shifted to 37°C for 30 min (0-min lane), supplemented with methionine to a 2 mM final concentration, and further incubated for 5 or 10 min (5- and 10-min lanes). GAL::lsm8 strains were pregrown on galactose and transferred to glucose medium for 18 h before a temperature shift. RNA samples were annealed with oligonucleotide complementary to either SSA4 (A, oligonucleotide 477), SSA1 (B, oligonucleotide 475), heat shock mRNAs, or MET3 (C, oligonucleotide 486) and treated with RNase H. RNA from the wild type (WT) was also treated with oligo(dT) (lane marked +dT) to visualize deadenylated species. Samples were separated on a 6% acrylamide gel and hybridized with anti-SSA4-3′ probe (A, oligonucleotide 478), anti-SSA1-3′ probe (B, oligonucleotide 476), or anti-MET3-3′ probe (C, oligonucleotide 485). The positions of migration of scR1 (676 nucleotides) and U2 snRNA (1,175 nucleotides) determined by hybridization of the same filters are indicated on the left as size markers.
In the nup145N strain after 15 min at 42°C (Fig. 2A and B, lanes 12), the mRNAs are fully adenylated, consistent with their nuclear localization. In fact, each of the mRNAs tested was mildly hyperadenylated, as previously reported for other mutations that inhibit nuclear export of mRNAs (31, 36). After a further 15 min at 37°C (Fig. 2A and B, lanes 13), most mRNA remains hyperadenylated, although a deadenylated pool is visible. After 90 min at 37°C the mRNA is largely degraded, but a hyperadenylated population remains detectable (Fig. 2A and B, lanes 14). In the nup145N strain lacking Lsm6p (Fig. 2A and B, lanes 7) or depleted of Lsm8p (Fig. 2A and B, lanes 11), substantial HS mRNA remains after 90 min at 37°C. The majority of this RNA remains hyperadenylated, although a fully deadenylated population is also clearly stabilized in the nup145N/GAL::lsm8 and nup145N/lsm6Δ strains, in contrast to the GAL::lsm8 and lsm6Δ single mutants. Unexpectedly, hyperadenylated SSA1 and SSA4 were also detected in the GAL::lsm8 single mutant strain (Fig. 2A and B, lanes 8). We speculate that this finding represents a fraction of the HS mRNA population that would normally have been rapidly degraded within the nucleus.
The poly(A) tail status of the MET3 mRNA was tested before (0-min lanes) or after (5- and 10-min lanes) transcriptional shutdown by addition of methionine in cells inhibited for mRNA export. In all nup145N strains, hyperadenylation of mRNA in the nucleus was observed (Fig. 2C, compare lanes 3, 7, and 10 to lane 2). In the lsm6Δ single mutant, the cytoplasmic mRNA is deadenylated [compare lane 6 to lane 1 where RNA was deadenylated in the presence of oligo(dT)]. After transcription inhibition, MET3 mRNA in the nup145N strain shows hyperadenylated and deadenylated populations (Fig. 2C, lane 4), both of which are apparently stabilized by the absence of Lsm6p (Fig. 2C, lane 8) or depletion of Lsm8p (Fig. 2C, lane 11). The basis of the difference in migration of the smallest forms in the Lsm6p- and Lsm8p-depleted strains is unclear.
We conclude that a fraction of the nucleus-restricted mRNAs present in the nup145N strain become fully deadenylated, but they do not appear to undergo the slow, incremental deadenylation of the entire mRNA population that is a major characteristic of cytoplasmic mRNA degradation. Moreover, depletion of Lsm6p or Lsm8p did not preferentially stabilize deadenylated forms of the mRNAs, in contrast to the effects of mutation of Lsm1p to 7p on cytoplasmic mRNAs. We cannot, however, exclude the possibility that some of the deadenylated species are mRNAs that have escaped to the cytoplasm.
5′ fragments of unspliced pre-mRNAs are stabilized in lsm mutants.
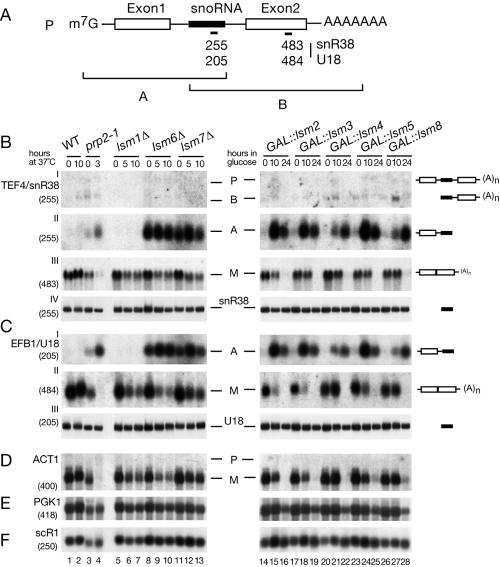
Many snoRNAs are encoded within pre-mRNA introns (Fig. 3A) and are released by exonuclease digestion of the excised intron following splicing or during degradation of the unspliced pre-mRNA (4, 55, 58, 77, 79). The presence of the intronic snoRNA inhibits the activity of degradative exonucleases on the unspliced pre-mRNA, allowing the contribution of 5′ and 3′ degradation pathways to be distinguished (11). To demonstrate whether Lsm proteins are involved in the 5′ or 3′ degradation pathway, the accumulation of specific degradation intermediates for pre-mRNAs containing intron-encoded snoRNAs was assessed in strains lacking each of the Lsm proteins. GAL-regulated alleles of LSM2 to LSM5 and LSM8 (42, 49) were analyzed by transferring the strains from permissive RSG medium (0-h samples) to repressive glucose medium. Strains lacking Lsm1p, Lsm6p, or Lsm7p are temperature sensitive for growth (70), as are nuclear localization signal-containing fusions between Lsm2p and Lsm5p and the DNA-binding domain of Gal4p [referred to here as lsm2(ts) and lsm5(ts)], constructed for use in two-hybrid analyses (24). These strains were grown in glucose medium at 23°C (0-h samples) and transferred to the nonpermissive temperature of 37°C.
FIG. 3.
5′-unprocessed fragments of pre-mRNAs containing intronic snoRNAs accumulate in lsm mutants. (A) Schematic representation of degradation intermediates for a pre-mRNA containing an intronic snoRNA. The locations of numbered oligonucleotide probes are shown below. P, full-length precursor; A, product of 3′→5′ degradation that extends from the 5′ end of the transcript to the end of the snoRNA; B, product of 5′→3′ degradation that extends from the 5′ end of the snoRNA to the 3′ end of the transcript (see Fig. 3 in reference 11). (B to F) Analysis of pre-mRNA degradation in GAL::lsm and lsm-Δ strains. Strains carrying GAL-regulated constructs (GAL::lsm) (lanes 14 to 28) were grown in permissive RSG medium (0 h) and transferred to repressive, glucose medium at 30°C for the times indicated. Strains in which Lsm1p, Lsm6p, or Lsm7p was deleted (lanes 5 to 13), the temperature-sensitive prp2-1 strain (lanes 3 to 4) and the wild-type (WT) strain (lanes 1 to 2) were pregrown at 23°C (0 h) and transferred to 37°C for the times indicated. RNA was separated on 2% agarose gels or on 6% polyacrylamide gel (BIV and CIII) and hybridized with the oligonucleotide probes shown in parentheses. The names of RNA species are on the left or between two columns (BIV and CIII); the positions of pre-mRNAs (P), degradation intermediates (A, A′, and B), and mature mRNAs (M) are indicated, and schematic representations are shown on the right. The level of scR1 RNA is shown as a control for loading.
Figure 3 shows the analysis of snoRNA-containing transcripts, TEF4 (the host gene for snR38) (Fig. 3B), and EFB1 (the host gene for U18) (Fig. 3C); BEL1 (the host gene for U24 snoRNA) was also analyzed and gave similar results (data not shown). The spliced but non-snoRNA-containing ACT1 (Fig. 3D) and nonspliced PGK1 mRNAs (Fig. 3E) were used as controls. Mutation of the splicing factor Prp2p led to loss of the mature mRNAs (M) accompanied by mild accumulation of the primary transcript (P), the 3′-processed pre-mRNA fragment (termed A) and the 5′-processed pre-mRNA fragment (termed B) (Fig. 3A). In the prp2-1 strain more of the A form is visible than the B form because 3′ degradation by the exosome is more active than 5′ degradation (11).
The absence of Lsm6p or Lsm7p resulted in dramatic accumulation of the A forms (i.e., the 5′-unprocessed species) of all tested snoRNA-containing pre-mRNAs (Fig. 3B and C, lanes 8 to 13; see Table 2 for PhosphorImager quantification). This was seen even in cells grown at the permissive temperature of 23°C (Fig. 3B and C, lanes 8 and 11), when pre-mRNA splicing is not clearly defective (Fig. 3D, ACT1 mRNA). Previous analyses indicated that a fraction of the unspliced pre-mRNA population is degraded in wild-type cells (11), and this degradation is presumably the origin of the A fragments that are stabilized in the lsm6Δ and lsm7Δ strains at a permissive temperature. In strains genetically depleted of Lsm2-5p or the nucleus-specific Lsm8p, strong accumulation of the A forms of TEF4/snR38 and EFB1/U18 was seen 10 h after transfer to glucose medium (Fig. 3B and C, lanes 14 to 28). At this time point splicing is reduced but not blocked, as shown by the continued synthesis of mature ACT1 mRNA (Fig. 3D). In contrast, strains lacking the cytoplasmic Lsm1p showed no clear accumulation of any pre-mRNA tested (Fig. 3BI and II and CI, lanes 5 to 7). The levels of mature snoRNAs, the RNA component of the signal recognition particle (scR1) and the intronless PGK1 mRNA were little affected in lsm mutants (Fig. 3BIV, CIII, and D to F).
TABLE 2.
Accumulation of pre-mRNA degradation intermediates in prp2-1 and lsm6-Δ strains based on PhosphorImager quantification of Northern hybridization data from Fig. 3
| Mutant and temp | Accumulation of indicated RNA speciesa
|
|
|---|---|---|
| snR38-A | U18-A | |
| WT, 23°C | 1 | 1 |
| prp2-1, 23°C | 3.6 | 4.2 |
| prp2-1, 37°C | 7.1 | 11.7 |
| lsm6-Δ, 23°C | 30 | 13.5 |
| lsm6-Δ, 37°C | 43.6 | 19.3 |
All values are standardized to the level of scR1 RNA used as a loading control. Numbers expressed are relative to the level in the wild type (wt), which was arbitrarily set at 1.
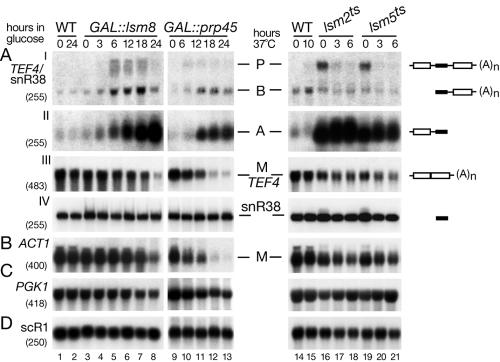
The kinetics of pre-mRNA accumulation were further examined in the GAL::lsm8 strain at earlier time points after transfer to glucose medium (Fig. 4, lanes 1 to 8). Accumulation of the A form of TEF4/snR38 was seen 3 h after transfer to glucose medium, well before any detectable growth defect (49) and prior to visible depletion of the TEF4 mRNA. GAL-regulated alleles of known pre-mRNA splicing factors Prp45p (Fig. 4, lanes 9 to 13) (3) and Syf3p (data not shown) (60) were also analyzed. Consistent with a splicing defect, depletion of Prp45p strongly reduced the levels of mature TEF4 and ACT1 mRNAs (Fig. 4AIII and B). However, despite the stronger splicing defect, accumulation of the 5′-unprocessed intermediate (A form) was substantially lower than that which occurred on depletion of Lsm8p (Fig. 4AII).
FIG. 4.
Analysis of pre-mRNA degradation in GAL::lsm8, GAL::prp45, lsm2(Ts), and lsm5(Ts) strains. Strains were grown, and RNA was prepared as described for Fig. 3. RNA was separated on 2% agarose gels or on 6% polyacrylamide gel (AIV) and hybridized with the oligonucleotide probes shown in parentheses. The names of RNA species are on the left or between two columns (AIII, AIV); the positions of pre-mRNAs (P), degradation intermediates (A and B), and mature mRNAs (M) are indicated, and schematic representations are shown on the right.
In the lsm2(ts) and lsm5(ts) strains, strong, nonconditional accumulation of the A form of TEF4/snR38 was seen with little inhibition of pre-mRNA splicing (Fig. 4, lanes 16 to 21). In addition, some accumulation of the primary transcript was seen at 23°C (Fig. 4A, lanes 16 and 19), which was lost following transfer to 37°C. Analysis of pre-tRNA processing also showed defects in these strains at 23°C, which were lost at 37°C (42). This is in contrast to cytoplasmic mRNA degradation, which is more defective at 37°C (70), indicating that the nuclear and cytoplasmic defects show distinct temperature dependence, possibly related to the presence of strong nuclear localization signal elements in these fusion constructs.
We conclude that the Lsm2-8p proteins are all required specifically for the 5′→3′ degradation of unspliced nuclear pre-mRNAs.
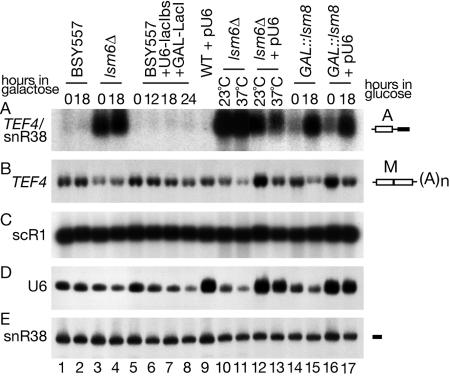
The Lsm2-8p complex is associated with the U6 snRNA, and depletion of Lsm proteins, particularly Lsm6p, leads to a reduced abundance of U6 (49, 64). Moreover, overexpression of U6 has been reported to suppress some of the RNA processing phenotypes associated with Lsm depletion (22). To assess the involvement of U6 in nuclear RNA turnover, we analyzed the degradation of the TEF4/snR38 pre-mRNA in a strain in which U6 transcription can be repressed by inducible LacI binding (Fig. 5, lanes 5 to 8) (48). Reduction in the level of U6 to an extent comparable to that of the lsm6Δ strain did not lead to any accumulation of the 5′-unprocessed pre-mRNA (Fig. 5A). Overexpression of U6 from a multicopy plasmid (pU6/pYX172) (49) caused some reduction in the accumulation of the A form of the TEF4/snR38 pre-mRNA in the lsm6Δ strain (Fig. 5A, compare lanes 10 to 11 with lanes 12 to 13) but had no effect in a strain depleted of Lsm8p (Fig. 5A, compare lanes 15 and 17).
FIG. 5.
Alterations in U6 abundance are not responsible for the defects in nuclear pre-mRNA degradation. RNAs were extracted from strain BSY557 carrying the U6 gene under a promoter containing a LacI binding site and a LacI gene under the GAL1 promoter (BSY557+U6-lacIbs+GAL-lacI; lanes 5 to 8), from its isogenic wild-type (WT) strain (BSY557, lanes 1 to 2) and from the lsm6-Δ strain (lanes 2 to 3) grown in minimal medium with 2% lactate, 2% glycerol, and 0.05% glucose (0 h) and supplemented with 2% galactose. The lsm6Δ strain (lanes 10 to 11), the GAL-lsm8 strain (lanes 14 to 15), BMA64+(pU6 (lane 9), the lsm6Δ+pU6 strain (lanes 12 to 13), and the GAL-lsm8+pU6 strain (lanes 16 to 17) expressing U6 from a high-copy-number 2μm plasmid were grown as described for Fig. 3. RNA was separated on 2% agarose gel (A to C) or on 6% polyacrylamide gel (D and E). The names of RNA species are on the left, and schematic representations of the 5′-unprocessed degradation intermediate (A) and mature mRNA (M) are shown on the right.
We conclude that alterations in the abundance of U6 snRNA do not play a major role in the inhibition of nuclear mRNA degradation in the lsm mutants.
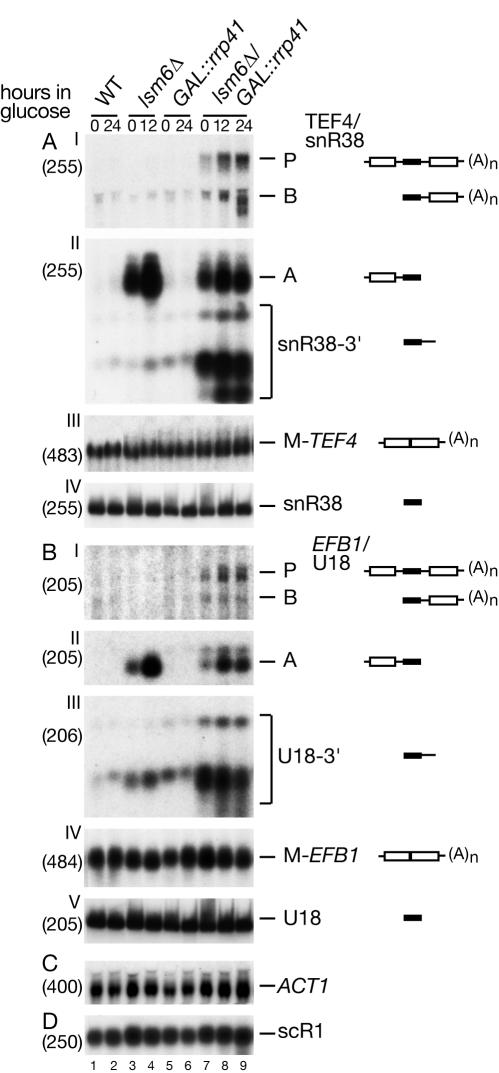
Pre-mRNAs are subject to 3′ degradation by the exosome complex of 3′→5′ exonucleases, so the combination of reduced exosome activity with the absence of an Lsm protein might be predicted to lead to accumulation of the pre-mRNA primary transcripts. To test this prediction, the lsm6Δ mutation was combined with a GAL-regulated allele of the exosome component Rrp41p (Fig. 6) and the putative RNA helicase Mtr4p, an essential cofactor for the nuclear exosome (data not shown). Accumulation of the TEF4, EFB1, and BEL1 primary transcripts was seen in both double mutant strains, while accumulation of snR38-A was reduced compared to that of the lsm6Δ single mutant, consistent with the role of Lsm6p in 5′ degradation (Fig. 6A and B and data not shown). Exosome mutations also lead to accumulation of shorter 3′-extended forms of snR38, U18, and U24 snoRNAs (4, 77). The 3′-extended snR38, U18, and U24 were much more abundant in the GAL::rrp41/lsm6Δ strain than in the GAL::rrp41 single mutant strain (Fig. 6A and B and data not shown). This stabilization may result from synergistic inhibition of 3′ processing. The Lsm2-8p complex has been implicated in the 3′ maturation of the U3 snoRNA (41) as well as other small stable RNAs (40, 42, 49, 56).
FIG. 6.
Inactivation of the nuclear 3′→5′ degradation pathway in lsm6Δ strains. Northern blot analysis of the lsm6-Δ strain deficient in exosome-mediated degradation. Strains were pregrown in galactose medium at 23°C (0 h) and transferred to glucose medium for the times indicated. RNA was separated on 2% agarose gels or on 6% polyacrylamide gel (AIV, BV) and hybridized with the oligonucleotide probes shown in parentheses. The names of RNA species and the positions of pre-mRNAs (P), degradation intermediates (A and B), and 3′-extended snoRNAs are indicated, and schematic representations are shown on the right. WT, wild type.
From the above we conclude that strains lacking any of the proteins Lsm2p to Lsm8p are defective in pre-mRNA 5′ degradation, whereas Lsm1p is not required for nuclear degradation.
Nucleus-restricted mRNAs stabilized in lsm mutants are 5′ capped.
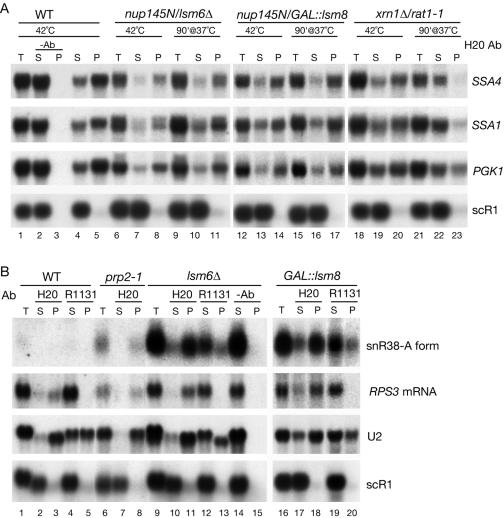
The 5′ degradation of nuclear mRNAs and premRNAs presumably requires decapping, since the only known 5′ exonucleases, Rat1p and Xrn1p, are unable to digest capped mRNAs (32, 67). To determine whether depletion of Lsm2-8p inhibits nuclear mRNA decapping or stabilizes decapped RNAs, we determined the cap status of the stabilized RNAs by immunoprecipitation with anti-cap antibodies (monoclonal Ab H20, generously provided by R. Lührmann).
The cap status of nucleus-restricted SSA1 and SSA4 mRNAs (Fig. 7A) was assessed by precipitation with H20 during induction at 42°C (42°C lanes) and after incubation at 37°C for 90 min (37°C lanes). In the nup145N strains either lacking Lsm6p (Fig. 7A, lanes 6 to 11) or depleted of Lsm8p (Fig. 7A, lanes 12 to 17), the efficiency of precipitation of SSA1 and SSA4 mRNA was similar at 42°C (lanes 8 and 14) or following transfer to 37°C for 90 min (lanes 11 and 17), showing that the nucleus-restricted, stabilized mRNAs remain predominantly capped. PhosphorImager quantification of these data is presented in Table 3. In a strain lacking the 5′→3′ exonucleases Rat1p and Xrn1p, SSA4 and SSA1 were predominately capped shortly after synthesis at 42°C (Fig. 7A, lane 20) but were much more weakly precipitated after incubation at 37°C for 90 min (Fig. 7A, lane 23). This is consistent with the accumulation of decapped cytoplasmic mRNAs in the absence of the 5′ exonucleases. The stable PGK1 mRNA, which is largely cytoplasmic and fully capped during heat shock, was precipitated with an efficiency similar to that of SSA1 and SSA4, whereas the noncapped scR1 RNA was not precipitated.
FIG. 7.
mRNAs stabilized in lsm mutants are 5′ capped. (A) Immunoprecipitation of total RNAs with monoclonal anti-cap antibody (H20). RNAs were extracted from the wild type (WT, lanes 1 to 5), nup145N/lsm6-Δ (lanes 6 to 11), and xrn1-Δ/rat1-1 (lanes 18 to 23) strains pregrown in glucose at 23°C, shifted to 42°C for 15 min (42°C lanes), and then transferred to 37°C for 90 min (37°C lanes) or from the nup145N/GAL::lsm8 strain pregrown at the permissive RSG medium and transferred to the repressive glucose medium for 18 h prior to the heat shock and shift to 37°C for 90 min. RNAs were immunoprecipitated with H20 or R1131 antibodies or mock treated (lanes marked −Ab) as outlined in Materials and Methods, recovered from the lysate (T), supernatant (S), and immunoprecipitate (P), separated on 2% agarose gels, and analyzed by Northern hybridization using probes complementary to the RNA species indicated on the right. (B) Immunoprecipitation of total RNAs with TMa cap-specific antibody (R1131) or monoclonal antibody against both m7G and m3G cap structures (H20). RNAs were extracted from the wild-type strain BMA64 (WT, lanes 1 to 5) and the lsm6Δ strain (lanes 9 to 15) grown on glucose at 23°C, from the prp2-1 strain (lanes 6 to 8) pregrown at 23°C and transferred to 37°C for 3 h, and from the GAL::lsm8 strain pregrown in permissive RSG medium and transferred to the repressive glucose medium for 18 h.
TABLE 3.
Efficiency of immunoprecipitation by H20 cap antibodies based on PhosphorImager quantification of Northern hybridization data from Fig. 6A
| Parameter | Efficiency (%) in indicated strain or mutanta
|
|||||||
|---|---|---|---|---|---|---|---|---|
| WT (Ab−) | WT | nup145N/lsm6-Δ | nup145N/lsm6-Δ | nup145N/GAL-lsm8 | nup145N/GAL-lsm8 | xrn1-Δ/rat1-1 | xrn1-Δ/rat1-1 | |
| Assay condition(s) | 42°C | 42°C | 42°C | 90 min, 37°C | 42°C | 90 min, 37°C | 42°C | 90 min, 37°C |
| mRNAs | ||||||||
| SSA4 | 0.7 | 72 | 70 | 70 | 69 | 71 | 62 | 18 |
| SSA1 | 0.7 | 63 | 64 | 66 | 61 | 64 | 55 | 20 |
| PGK1 | 0.55 | 72 | 70 | 71 | 68 | 69 | 58 | 36 |
| scR1 | 0.6 | 5.3 | 4.8 | 4.7 | 4.3 | 4 | 5.8 | 6.1 |
Supernatant plus pellet equals 100%. WT, wild type.
We wanted to determine whether stabilized A forms of the snoRNA-containing pre-mRNAs carried m7G caps or the 2,2,7 TMG cap structures found on the mature snoRNAs. For this we compared monoclonal Ab H20, which recognizes both m7G and TMG cap structures, with Ab R1131, which is specific for TMG caps (R. Lührmann) (Fig. 7B). In the wild-type strain, the m7G-capped RPS3 mRNA was precipitated by H20 but not by R1131, whereas the TMG-capped U2 snRNA was precipitated by both antibodies (Fig. 7B, lanes 1 to 5). In the prp2-1, lsm6Δ, and Lsm8p-depleted strains, the A form of TEF4/snR38 was efficiently precipitated by H20. This shows that this RNA is capped but does not reveal the nature of the cap structure. In the lsm mutant strains the A form of TEF4/snR38 was also precipitated by the TMG-specific antibody, R1131, but with slightly lower efficiency than the U2 snRNA. PhosphorImager quantification of these data is presented in Table 4. The stabilized 5′ fragment of the snoRNA/mRNA therefore largely undergoes cap trimethylation in strains lacking Lsm6p or Lsm8p. This may render these fragments more resistant to nuclear decapping and contribute to their very high levels in strains lacking Lsm2-8p. We cannot exclude that the 5′ fragments having trimethylated caps and containing snoRNA sequences are 5′ extended but otherwise functional snoRNAs.
TABLE 4.
Efficiency of immunoprecipitation by R1131 or H20 cap antibodies based on PhosphorImager quantification of Northern hybridization data from Fig. 6B.
| Parameter | Efficiency (%) in indicated strain or mutanta
|
|||||||
|---|---|---|---|---|---|---|---|---|
| WT | WT | prp2-1 | lsm6Δ | lsm6Δ | lsm6Δ | GAL::lsm8 | GAL::lsm8 | |
| Ab | H20 | R1131 | H20 | H20 | R1131 | None | H20 | R1131 |
| mRNAs | ||||||||
| snR38-A | ND | ND | 72.6 | 73 | 33 | 3.1 | 68 | 27 |
| RPS3 | 64 | 4.6 | 69 | 76 | 4.4 | 2.9 | 71 | 2.7 |
| U2 | 78.3 | 44 | 81 | 80 | 43 | 1.6 | 73 | 33 |
| scR1 | 6.9 | 6.2 | 7.4 | 8.6 | 5.8 | 2.1 | 6.8 | 6.2 |
Supernatant plus pellet equals 100%. ND, not detected; WT, wild type.
We conclude that nuclear mRNAs are not decapped in strains lacking Lsm6p or Lsm8p. It is very likely that the inhibition of nuclear decapping activity is responsible for the stabilization of nucleus-restricted mRNAs and the 5′ fragments of nuclear pre-mRNAs in the strains lacking Lsm2p to Lsm8p.
Lsm8p is associated with nucleus-restricted mRNAs.
Previous analyses reported that components of the Lsm1-7p complex, but not Lsm8p, coimmunoprecipitate cytoplasmic mRNAs (70, 71). To determine whether the Lsm2-8p complex is directly associated with the nucleus-restricted mRNAs, we tested whether Lsm8p can be UV cross-linked to poly(A)+ RNA in the nup145N strain at the nonpermissive temperature, when it is predominantly nuclear (18).
We compared UV-irradiated or nonirradiated wild-type cells expressing Lsm8p-TAP (Fig. 8, lanes 1 to 3) to nup145N cells expressing Lsm8p-TAP (Fig. 8, lanes 4 to 6). After irradiation, RNPs cross-linked to poly(A)+ RNA were isolated from cell lysates by double passage over an oligo(dT) cellulose column followed by low-salt elution (see Materials and Methods). Proteins recovered from the total lysate and from the oligo(dT) eluate were analyzed by Western blotting. As previously reported (70, 71), Lsm8p was not detectably recovered in association with poly(A)+ RNA in the absence of a block in mRNA export (Fig. 8, lane 3). Lsm8p was, however, clearly cross-linked to poly(A)+ RNA in the nup145N strain (Fig. 8, lane 6), although the protein had apparently undergone some degradation during incubation.
FIG. 8.
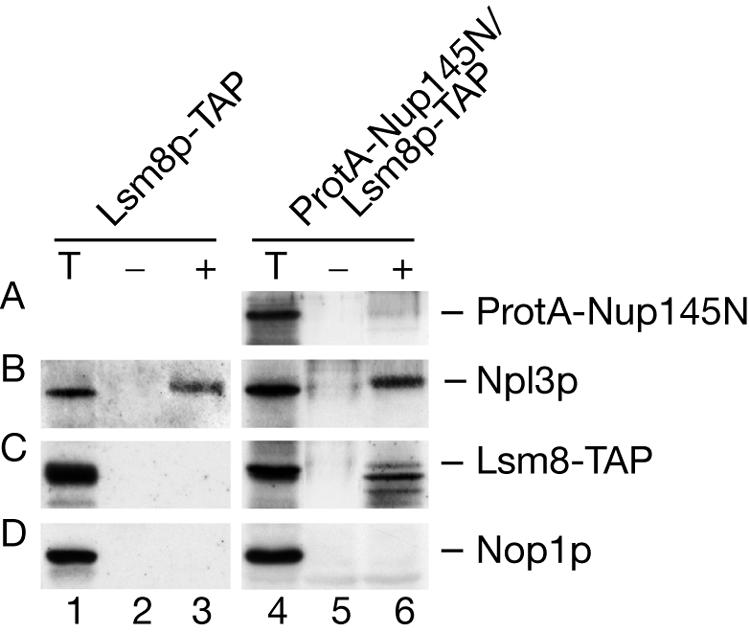
Lsm8p can be cross-linked to polyadenylated nuclear RNAs. A Western blot of proteins UV cross-linked to poly(A)+ RNAs is shown. The Lsm8-TAP strain was grown at 30°C to an OD600 of 0.75, and the nup145N/Lsm8-TAP strain was pregrown at 23°C to an OD600 of 0.6 and transferred to 37°C for 30 min to block mRNA export. UV cross-linking and purification of poly(A)+ RNPs on oligo(dT)-cellulose was performed as described in Materials and Methods. Western analysis was performed on total lysate (T) and on RNP complexes recovered from the nonirradiated (−) and irradiated (+) cells resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis following RNase digestion of the bound material. Approximately 90-fold more material was loaded for the eluate fractions compared to the total fractions. Western blot analysis was decorated with anti-protein A Ab to detect ProtA-Nup145N (A), Lsm8-TAP (C), anti-Npl3 Ab (B), and anti-Nop1p Ab (D).
As a positive control, the Western blots were decorated with antibodies against Npl3p, a well-characterized poly(A)+ RNA binding protein that shuttles between the nucleus and the cytoplasm (23, 61, 65, 81). Cross-linking of Npl3p was observed in the presence or absence of mRNA export, with efficiency similar to that of Lsm8p in the nup145N strain. Nop1p is a nucleolar protein that does not associate with poly(A)+ RNA (61) and was used as a negative control. The nup145N construct also carries a protein A tag on the residual Nup145Np N-terminal fragment (69). Since this region does not include the RNA binding domain of Nup145p (18, 21, 69), it is not expected to efficiently associate with poly(A)+ RNA, and only weak cross-linking was observed (Fig. 8, lanes 4 to 6).
We conclude that Lsm8p-TAP can be cross-linked to nucleus-restricted poly(A)+ RNA but not to cytoplasmic mRNA, suggesting a direct role for the Lsm2-8p complex in nuclear mRNA metabolism. These observations also confirm that the Lsm complex is involved in a deadenylation-independent degradation pathway.
DISCUSSION
The Lsm1-7p complex plays an important role in cytoplasmic mRNA turnover, promoting mRNA decapping and subsequent 5′ degradation and preventing inappropriate 3′ degradation (29, 70). Here we show that the related Lsm2-8p complex functions in the nucleus to promote decapping and 5′ degradation of nuclear mRNAs and pre-mRNAs. The stabilization of capped forms of two quite different classes of nuclear RNA, nucleus-restricted mRNAs in strains defective for mRNA export and the 5′ fragments of unspliced pre-mRNAs that contain intronic snoRNAs, was observed. Other aspects of the nuclear degradation of these RNAs are quite different. Unspliced pre-mRNAs undergo very rapid degradation, predominately 3′→5′, by the exosome complex (11). In contrast, nucleus-restricted mRNAs are much more slowly degraded, with a substantial contribution from 5′→3′ degradation (16).
In the cytoplasm, the translational status of the mRNA plays a key role in establishing decapping rates. However, despite reports of the localization of translation factors and translation activity in the nuclei of metazoan cells (13, 34; reviewed in reference 80), we think it unlikely that the decapping rates of yeast nuclear RNAs are determined by translation. Rather, we envisage that the RNP structure of the pre-mRNA is assessed and can trigger this activity. The hnRNP or other protein(s) involved remains to be identified.
The striking increase in 5′-unprocessed pre-mRNAs seen in the lsm6Δ or lsm7Δ strains growing at a permissive temperature was not accompanied by any clear defect in pre-mRNA splicing. However, a previous analysis concluded that a significant fraction of the unspliced nuclear pre-mRNA population is degraded even in wild-type yeast cells (11). The 5′ fragments of the snoRNA-containing pre-mRNAs stabilized in these lsm mutants are very likely to have been generated in this way. In contrast to strains defective only in exosome components, where increased pre-mRNA levels are observed (11), inactivation of Lsm proteins does not clearly stabilize unspliced full-length pre-mRNAs, indicating that these are rapidly degraded 3′→5′ by the exosome. Reducing or inhibiting exosome activity in cells lacking Lsm6p leads to only modest accumulation of primary transcripts. Moreover, pre-mRNAs that had been 5′ degraded to the end of the snoRNA were also detected in the Lsm exosome double mutants. This indicates that depletion of Lsm2-8p delays but does not fully block 5′ degradation. Similar observations have been made for the role of the Lsm1-7p in the 5′ degradation of cytoplasmic mRNAs (70).
In nup116Δ strains nucleus-restricted mRNAs are reported to be stabilized by the absence of components of the nuclear cap-binding complex (CBC) (16). This stabilization might result from replacement of CBC by eIF4E, which normally binds to cytoplasmic mRNA cap structures but can also be detected in the nucleus in yeast and mammalian cells (19, 45, 46). Alternatively, the mammalian poly(A) binding protein PABP has been shown to bind to the cap structure of polyadenylated mRNAs (38), although this activity has not yet been reported for the yeast protein. In either case, we speculate that these interactions may be less readily displaced by the Lsm2-8p complex or other components of the nuclear decapping machinery than is CBC.
Nuclear mRNA and pre-mRNA degradation involves a set of enzymes and cofactors that strikingly mirror the cytoplasmic mRNA degradation machinery. Nuclear 3′ degradation involves the nuclear exosome complex together with the putative RNA helicase Mtr4p/Dob1p (11, 16, 30, 74), while cytoplasmic 3′ turnover involves the cytoplasmic exosome complex and a related helicase, Ski2p (5). Nuclear 5′ decapping is stimulated by Lsm2-8p, and subsequent 5′ degradation probably involves the predominantly nuclear 5′→3′ exonuclease Rat1p (11, 16), while the Lsm1-7p complex (10, 12, 70, 71) and the homologous Xrn1p exonuclease (32, 52, 53) play the same roles in the cytoplasm.
The clearest differences between nuclear and cytoplasmic degradation appear to be in the role of deadenylation. A key feature of mRNA turnover in the cytoplasm is continuous, incremental deadenylation that is normally rate limiting and gives rise to the characteristic, even distribution of heterogeneous mRNA lengths. However, this does not appear to be the case for nucleus-restricted mRNAs. In the nup145N strains hyperadenylation was seen for all mRNAs tested, as previously observed for other mutants defective in mRNA export (31, 36). These hyperadenylated mRNAs were strongly stabilized by depletion of Lsm6p or Lsm8p. In addition, a population of nuclear mRNAs that were largely or fully deadenylated was stabilized, with relatively few RNAs of intermediate size. This suggests that prior to 3′ degradation of the RNA body, nuclear mRNAs are subject to a distinct deadenylation activity. This appears to show processive activity, fully deadenylating the RNA once bound, in contrast to the distributive activity of cytoplasmic deadenylation.
Acknowledgments
We thank Phil Mitchell for critical reading of the manuscript, Reinhard Lührmann for the R1131 and H20 sera, John Aris for the A66 antibodies, Bertrand Séraphin for strain BSY557 and plasmids pBS1181 and pBS1184, and Emmanuelle Fabre for providing strain FYEF95.
C.B.-A. was the recipient of a Marie Curie fellowship from the EU. This work was supported by the Wellcome Trust.
REFERENCES
- 1.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Lührmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achsel, T., H. Stark, and R. Lührmann. 2001. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. USA 98:3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albers, A., A. Diment, M. Muraru, C. S. Russell, and J. Beggs. 2002. Identification and characterisation of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9:138-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, J. S., and R. Parker. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson, J. T., S. M. Wilson, K. V. Datar, and M. S. Swanson. 1993. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 13:2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aris, P., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 9.Beltrame, M., and D. Tollervey. 1992. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeck, R., B. Lapeyre, C. E. Brown, and A. B. Sachs. 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 12.Bouveret, E., G. Rigaut, A. Shevchenko, M. Wilm, and B. Séraphin. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19:1661-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brogna, S., T. A. Sato, and M. Rosbash. 2002. Ribosome components are associated with sites of transcription. Mol. Cell 10:93-104. [PubMed] [Google Scholar]
- 14.Collins, B. M., S. J. Harrop, G. D. Kornfeld, I. W. Dawes, P. M. Curmi, and B. C. Mabbutt. 2001. Crystal structure of a heptameric Sm-like protein complex from Archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. 309:915-923. [DOI] [PubMed] [Google Scholar]
- 15.Cooper, M., L. H. Johnston, and J. Beggs. 1995. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 14:2066-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, B., J. S. Butler, and F. Sherman. 2003. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 23:5502-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 18.Dockendorff, T. C., C. V. Heath, A. L. Goldstein, C. A. Snay, and C. N. Cole. 1997. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol. Cell. Biol. 17:2347-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dostie, J., M. Ferraiuolo, A. Pause, S. A. Adam, and N. Sonenberg. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 19:3142-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dower, K., and M. Rosbash. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabre, E., W. Boelens, C. Wimmer, I. Mattaj, and E. C. Hurt. 1994. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell 78:275-289. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez, C. F., B. K. Pannone, X. Chen, G. Fuchs, and S. L. Wolin. 2004. An Lsm2-Lsm7 complex in Saccharomyces cerevisiae associates with the small nucleolar RNA snR5. Mol. Biol. Cell. 15:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flach, J., M. Bossie, J. Vogel, A. Corbett, T. Jinks, D. A. Willins, and P. A. Silver. 1994. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol. Cell. Biol. 14:8399-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromont-Racine, M., A. E. Mayes, A. Brunet-Simon, J. C. Rain, A. Colley, I. Dix, L. Decourty, N. Joly, F. Ricard, J. D. Beggs, and P. Legrain. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavin, A.-C., M. R. Bösche, O. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A.-M. Michon, C.-M. Cruciat, M. Remor, C. Höfert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M.-A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Séraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 26.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficient transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottschalk, A., G. Neubauer, J. Banroques, M. Mann, R. Lührmann, and P. Fabrizio. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18:4535-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatfield, L., C. A. Beelman, A. Stevens, and R. Parker. 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5830-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, W., and R. Parker. 2001. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158:1445-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 31.Hilleren, P., and R. Parker. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu, C. L., and A. Stevens. 1993. Yeast cells lacking 5′-3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurt, E., K. Strasser, A. Segref, S. Bailer, N. Schlaich, C. Presutti, D. Tollervey, and R. Jansen. 2000. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem. 275:8361-8368. [DOI] [PubMed] [Google Scholar]
- 34.Iborra, F. J., D. A. Jackson, and P. R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139-1142. [DOI] [PubMed] [Google Scholar]
- 35.Jensen, T. H., K. Dower, D. Libri, and M. Rosbash. 2003. Early formation of mRNP: license for export or quality control? Mol. Cell 11:1129-1138. [DOI] [PubMed] [Google Scholar]
- 36.Jensen, T. H., K. Patricio, T. McCarthy, and M. Rosbash. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7:887-898. [DOI] [PubMed] [Google Scholar]
- 37.Jensen, T. H., and M. Rosbash. 2003. Co-transcriptional monitoring of mRNP formation. Nat. Struct. Biol. 10:10-12. [DOI] [PubMed] [Google Scholar]
- 38.Khanna, R., and M. Kiledjian. 2004. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 23:1968-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kufel, J., C. Allmang, G. Chanfreau, E. Petfalski, D. L. J. Lafontaine, and D. Tollervey. 2000. Precursors to the U3 snoRNA lack snoRNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20:5415-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kufel, J., C. Allmang, E. Petfalski, J. Beggs, and D. Tollervey. 2003. Lsm proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem. 278:2147-2156. [DOI] [PubMed] [Google Scholar]
- 41.Kufel, J., C. Allmang, L. Verdone, J. Beggs, and D. Tollervey. 2003. A complex pathway for 3′ processing of the yeast U3 snoRNA. Nucleic Acids Res. 31:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kufel, J., C. Allmang, L. Verdone, J. Beggs, and D. Tollervey. 2002. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell. Biol. 22:5248-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lafontaine, D., and D. Tollervey. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaGrandeur, T. E., and R. Parker. 1998. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang, V., N. I. Zanchin, H. Lunsdorf, M. Tuite, and J. E. McCarthy. 1994. Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA, and consequences of its overproduction. J. Biol. Chem. 269:6117-6123. [PubMed] [Google Scholar]
- 46.Lejbkowicz, F., C. Goyer, A. Darveau, S. Neron, R. Lemieux, and N. Sonenberg. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA 89:9612-9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 48.Luukkonen, B. G., and B. Seraphin. 1998. Construction of an in vivo-regulated U6 snRNA transcription unit as a tool to study U6 function. RNA 4:231-238. [PMC free article] [PubMed] [Google Scholar]
- 49.Mayes, A. E., L. Verdone, P. Legrain, and J. D. Beggs. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 51.Moore, M. J. 2002. Nuclear RNA turnover. Cell 108:431-434. [DOI] [PubMed] [Google Scholar]
- 52.Muhlrad, D., C. J. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′Æ 3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 53.Muhlrad, D., C. J. Decker, and R. Parker. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15:2145-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muhlrad, D., and R. Parker. 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6:2100-2111. [DOI] [PubMed] [Google Scholar]
- 55.Ooi, S. L., D. Samarsky, M. Fournier, and J. D. Boeke. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4:1096-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pannone, B. K., S. Do Kim, D. A. Noe, and S. L. Wolin. 2001. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics 158:187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pannone, B. K., D. Xue, and S. L. Wolin. 1998. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 17:7442-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petfalski, E., T. Dandekar, Y. Henry, and D. Tollervey. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 60.Russell, C. S., S. Ben-Yehuda, I. Dix, M. Kupiec, and J. D. Beggs. 2000. Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA 6:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russell, I., and D. Tollervey. 1995. Yeast Nop3p has structural and functional similarities to mammalian pre-mRNA binding proteins. Eur. J. Cell Biol. 66:293-301. [PubMed] [Google Scholar]
- 62.Russell, I. D., and D. Tollervey. 1992. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J. Cell Biol. 119:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saavedra, C., K. S. Tung, D. C. Amberg, A. K. Hopper, and C. N. Cole. 1996. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 10:1608-1620. [DOI] [PubMed] [Google Scholar]
- 64.Salgado-Garrido, J., E. Bragado-Nilsson, S. Kandels-Lewis, and B. Seraphin. 1999. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 18:3451-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singleton, D. R., S. Chen, M. Hitomi, C. Kumagai, and A. M. Tartakoff. 1995. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J. Cell Sci. 108:265-272. [DOI] [PubMed] [Google Scholar]
- 66.Stevens, A., and T. L. Poole. 1995. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem. 270:16063-16069. [DOI] [PubMed] [Google Scholar]
- 67.Stevens, S. W., and J. Abelson. 1999. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. USA 96:7226-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stutz, F., J. Kantor, D. Zhang, T. McCarthy, M. Neville, and M. Rosbash. 1997. The yeast nucleoporin Rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 11:2857-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teixeira, M. T., S. Siniossoglou, S. Podtelejnikov, J. C. Benichou, M. Mann, B. Dujon, E. Hurt, and E. Fabre. 1997. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 16:5086-5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tharun, S., W. He, A. E. Mayes, P. Lennertz, J. D. Beggs, and R. Parker. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404:515-518. [DOI] [PubMed] [Google Scholar]
- 71.Tharun, S., and R. Parker. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8:1075-1083. [DOI] [PubMed] [Google Scholar]
- 72.Thomsen, R., D. Libri, J. Boulay, M. Rosbash, and T. H. Jensen. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9:1049-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Torchet, C., C. Bousquet-Antonelli, L. Milligan, E. Thompson, J. Kufel, and D. Tollervey. 2002. Processing of 3′ extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 9:1285-1296. [DOI] [PubMed] [Google Scholar]
- 75.Törö, I., S. Thore, C. Mayer, J. Basquin, B. Séraphin, and D. Suck. 2001. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 20:2293-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockson, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 77.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Venema, J., and D. Tollervey. 1996. RRP5 is required for the formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15:5701-5714. [PMC free article] [PubMed] [Google Scholar]
- 79.Villa, T., F. Ceradini, C. Presutti, and I. Bozzoni. 1998. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol. 18:3376-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilkinson, M. F., and A. B. Shyu. 2002. RNA surveillance by nuclear scanning? Nat. Cell Biol. 4:E144-E147. [DOI] [PubMed] [Google Scholar]
- 81.Wilson, S. M., K. V. Datar, M. R. Paddy, J. R. Swedlow, and M. S. Swanson. 1994. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol. 127:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]