Abstract
La protein binds precursors to 5S rRNA, tRNAs, and other transcripts that contain 3′ UUU-OH and also promotes their maturation in the nucleus. Separate from this function, human La has been shown to positively modulate the translation of mRNAs that contain complex 5′ regulatory motifs that direct internal initiation of translation. Nonphosphorylated La (npLa) inhibits pre-tRNA processing, while phosphorylation of human La serine-366 (S366) promotes pre-tRNA processing. npLa was found specifically associated with a class of mRNAs that have unusually short 5′ untranslated regions comprised of terminal oligopyrimidine (5′TOP) tracts and that encode ribosomal proteins and translation elongation factors. Although La S366 represents a CK2 phosphorylation site, there was no evidence that CK2 phosphorylates it in vivo. We used the CK2-specific inhibitor, 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB), and antisense-mediated knockdown to demonstrate that CK2 is responsible for La S366 phosphorylation in vivo. Hypophosphorylation was not associated with significant change in total La levels or proteolytic cleavage. Quantitative reverse transcription-PCR revealed increased association of the 5′TOP-mRNA encoding ribosomal protein L37 (rpL37) with La after TBB treatment. Transfection revealed more rpL37 mRNA associated with nonphosphorylatable La A366 than with La S366, concomitant with La A366-specific shift of a fraction of L37 mRNA off polysomes. The data indicate that CK2 phosphorylates La S366 in vivo, that this limits 5′TOP mRNA binding, and that increasing npLa leads to greater association with potentially negative effects on TOP mRNA translation. Consistent with data that indicate that phosphorylation reverses negative effects of npLa on tRNA production, the present data suggest that CK2 phosphorylation of La can affect production of the translational machinery.
La is an abundant RNA-binding phosphoprotein that interacts with different classes of RNAs (34, 55). Although most La resides in the nucleoplasm, the amount in the nucleolus and cytoplasm can vary (33, 34, 55). La interacts with some of its ligands by recognition of the UUU-OH ends that terminate nascent RNA polymerase (pol) III transcripts, including precursors to 5S rRNA and tRNAs, as well as a limited number of pol II small nuclear and small nucleolar RNA (snoRNA) intermediates (30, 32, 56). For RNAs of this class, La has been shown to stabilize and retain them in the nucleus during their maturation (33, 34, 55). Accordingly, disruption of the nuclear retention of La leads to dysfunctional processing of its associated pre-tRNAs (21).
Analyses of yeast and human La proteins revealed that the conserved N-terminal domain, comprised of two different RNA binding motifs (2, 12) in conjunction with trafficking signals, is sufficient for its nuclear function in vivo (21, 23, 53, 57). It is therefore intriguing that metazoan La proteins also contain an extended C-terminal domain (CTD) that includes an atypical RNA recognition motif (RRM) (25) and a short basic motif (SBM) that contribute to RNA 5′-end recognition distinct from UUU-OH binding (Fig. 1A) (27). For human La, phosphoserine-366 inhibits this mode of RNA binding, and this phosphorylation is required to prevent La from interfering with tRNA production (14, 33, 34). Specifically, full-length nonphosphorylated La (npLa) binds pre-tRNAs in a manner that inhibits their 5′ processing, whereas the S366 phosphoprotein (pLa) facilitates tRNA production (14, 23). Accordingly, whereas CTD-truncated human La (hLa) proteins are active for tRNA production, full-length npLa is inhibitory while full-length pLa facilitates tRNA production in vivo (23). Presumably, inhibition of pre-tRNA production by npLa is avoided in vivo by the large excess of pLa in the nucleoplasm, whereas a substantial fraction of npLa is cytoplasmic and is associated with specific mRNAs (22). Although the function of the hLa CTD is uncertain, it may be more related to mRNA than tRNA activities (14, 22, 27).
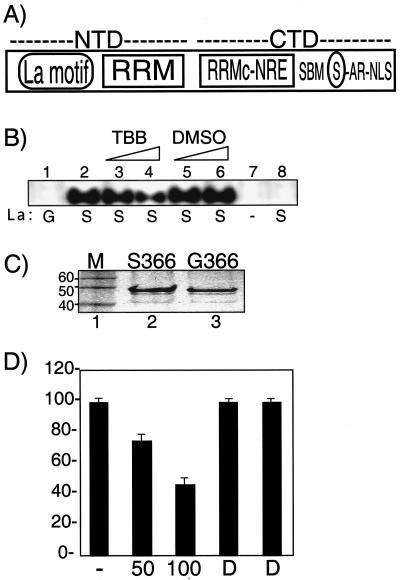
FIG. 1.
TBB inhibits CK2-mediated phosphorylation of La on S366 in vitro. (A) Schematic drawing of the human La protein, comprised of N-terminal and C-terminal domains (labeled NTD and CTD, respectively), as described in the text. Note that the nuclear retention element (NRE) is an integral part of the structure of the atypical RRM of the CTD (2). The acidic region (AR) and nuclear localization signal (NLS) are indicated. The encircled S represents serine-366. (B) A total of 0.5 μg of purified, recombinant, wild-type La S366 (lanes 2 to 6 and lane 8) or an equal amount of the point mutant La G366 (lane 1) were incubated with [γ-32P]GTP and recombinant CK2 (lanes 1 to 7) as part of a standard kinase reaction mixture that contained 500 μM GTP and 50 or 100 μM TBB dissolved in DMSO (lanes 3 and 4) or equivalent amounts of DMSO alone (lanes 5 and 6). Additional control reactions lacked La (lane 7) or CK2 (lane 8). Products were separated by denaturing polyacrylamide gel electrophoresis and were analyzed by autoradiography. (C) Recombinant La proteins were separated by sodium dodecyl sulfate-12% PAGE and stained with Coomassie blue. M, molecular size marker, indicated in kilodaltons to the left. (D) Quantitation of triplicate CK2 assays performed as described for panel A. The amount of 32P-La from reactions with no addition (lane −), 50 or 100 μM TBB (lanes 50 and 100), and DMSO alone (lanes D), as indicated on the x axis. Results are expressed as the percentage of the radioactive counts in the no-addition and DMSO reactions; error bars reflect the range of three experiments.
Ample evidence indicates that La can promote the translation of certain specific mRNAs which lack 3′ UUU-OH but bear complex 5′ regulatory motifs. hLa has been shown to promote translation of mRNAs that bear internal ribosome entry sites (IRES) (3, 6, 19, 24, 28, 35, 43, 47; reviewed in reference 55). Association of La with an Mdm2 mRNA isoform that contains two upstream open reading frames promotes Mdm2 translation with a consequent decrease in p53 and increased leukemia progression (52). La can also have negative effects on translation, depending on the 5′ untranslated region (5′UTR) context and the mechanism of translation initiation (50). La also binds the 5′-terminal oligopyrimidine (5′TOP) motifs found on a family of mRNAs that encode ribosomal proteins and translation factors (9, 11, 22). Regulation of 5′TOP-containing mRNAs controls ribosome biogenesis and, therefore, the capacity for translation (36). The translational efficiency of 5′TOP mRNAs changes in response to nutritional status and other signals, although the trans-acting factors and pathways involved remain incompletely understood (36). In addition, some small snoRNAs that participate in rRNA biogenesis are contained in introns of 5′TOP pre-mRNAs (48). In general, TOP-containing mRNAs have remarkably short (e.g., 30 nucleotides) 5′UTRs with no upstream AUGs (36). However, while some reports describe a positive effect of Xenopus La (9, 11, 38), others described an inhibitory effect of human La on TOP-mRNA translation (58). In any case, it is interesting that although yeasts do not have recognizable 5′TOP motifs per se, Lhp1p, the yeast La homolog, has been noted to associate with ribosomal protein mRNAs as well as with some snoRNA sequences (20). Because La participates in the metabolism of tRNAs, 5S rRNA, snoRNAs, and 5′TOP mRNAs that encode ribosomal proteins, a potential role for it in the coordination of the production of the translation machinery is fitting (27).
Consistent with the data reviewed above, npLa rather than pLa was found associated with 5′TOP mRNA in HeLa cells (22), suggesting that La phosphorylation may determine the extent to which it associates with TOP mRNAs. Consistent with this, only a fraction of L37 5′TOP mRNA was found associated with npLa in HeLa cells, suggesting that npLa may be limiting for L37 mRNA binding (22). The differential phosphorylation of hLa in the context of TOP mRNA metabolism had not been examined previously. Likewise, several aspects of La phosphorylation in vivo remain unclear.
Efforts to elucidate pathways that affect La-5′TOP mRNA interactions should include identification of the kinase that phosphorylates La S366 in vivo. Notably, La S366 comprises a very good consensus site for protein kinase CK2 which can efficiently phosphorylate this site in vitro, La S366 is found phosphorylated in vivo, and CK2 copurifies with pLa (15). Thus, although it seems likely that La S366 would be phosphorylated by CK2 in vivo, appropriate evidence of this has been absent, in part due to the lack of CK2-specific inhibitors. CK2 is unusual in its use of GTP and ATP as phosphoryl donors, a conserved feature that may account for the unique properties of its nucleotide binding site (13, 16, 18, 41, 42). These features both distinguish CK2 among protein kinases and allow development of a unique CK2 inhibitor, 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB), which has been well documented as very highly specific for CK2 (46, 51).
We report that TBB inhibits phosphorylation of human La S366 in vivo, as does antisense-mediated knockdown of CK2. The fraction of npLa increases after TBB treatment, and this is associated with a comparable increase in the rpL37 mRNA bound to La. Transfection resulted in more rpL37 mRNA associated with La A366 than with La S366, with shift of a fraction of L37 mRNA off of polysomes. The data indicate that CK2 phosphorylates La S366 in vivo and suggest that this limits TOP mRNA binding by La. Prior data indicated that pLa promotes pre-tRNA production while npLa inhibits it. Consistent with a role for La in production of the translational machinery, the present data suggest that increased levels of npLa may also be inhibitory to 5′TOP mRNA translation in human cells, in agreement with previous work (58).
MATERIALS AND METHODS
Tissue culture, drug treatments, and extract preparation.
Human embryonic kidney (HEK 293) cells were obtained from American Type Culture Collection (Manassas, Va.) and grown at 37°C with 5% CO2 in complete DMEM (Dulbecco's modified Eagle's medium plus 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 0.25 μg of amphotericin B/ml [Fungizomer, Life Technologies, Inc.]). TBB was a generous gift of L. Pinna (University of Padua, Italy); it was dissolved in dimethyl sulfoxide (DMSO) (<0.5% of the medium). 6-Dimethylaminopurine (6-DMAP) was from Fisher Scientific and was dissolved in distilled H2O. All cell treatments were conducted in triplicate, and control cells were treated with equal amounts of the solvent DMSO. Cell growth and viability were assessed using a hemacytometer and the trypan blue exclusion assay; only cells with viability of >85% were used for further analysis. At the end of the incubation period, cells were collected by manual scraping of the culture flask, circumventing the use of trypsin protease for cell detachment. Whole-cell extracts (WCE) were made by Dounce homogenization of cells (washed three times in PBS) in NET.5 buffer (150 mM NaCl, 2 mM EDTA, 50 mM Tris-Cl [pH 7.5], 0.5% Nonidet P-40) containing fresh phenylmethylsulfonyl fluoride (PMSF) (2 mM) and a combination of protease inhibitor cocktails obtained from Sigma (#P-1860) and Upstate Biotechnology (#20-116) as well as the RNase inhibitor VRC, obtained from GibcoBRL (#15552014). A pilot experiment determined that exogenous phosphatase inhibitors being present during extract preparation does not alter the relative amounts of pLa and npLa isolated (data not shown).
One-dimensional isoelectric focusing (IEF) and immunoblotting were performed as described previously (15). Immunoblotting used anti-La antibody (Go; at 1:5,000; gift of J. Keene and D. Kenan) and 125I-protein A (Amersham) as described previously (21, 23) unless otherwise indicated. Anti-phosphoserine-366 La (AbP) was previously described (22).
In vitro phosphorylation of recombinant La proteins by CK2.
His-tagged La S366 and the point mutant La G366 were purified using a B-PER 6× His fusion protein purification kit (Pierce) (15). A standard CK2 kinase assay (15) using human CK2 (500,000 U/ml; Calbiochem, La Jolla, Calif.) and 0.5 μg of purified La was performed with master mix technology such that each reaction mixture contained 0.5 mM GTP, 10 mM MgCl2, 10 mM Tris-Cl (pH 7.5), 1.5 μl of [γ-32P]GTP (10 uCi/μl; New England Nuclear), and 25 U of CK2 (kinase master mix). Reactions were started by adding equal aliquots of the master mix to equal amounts of the La substrates and were stopped with 2× lithium dodecyl sulfate (LDS) loading buffer containing fresh beta-mercaptoethanol and were loaded onto a 4 to 20% polyacrylamide LDS NuPage-Novex gel (Invitrogen). Quantitation was by Fuji PhosphorImager and Image Gauge software.
CK2-mediated, in vitro phosphorylation of native npLa S366 isolated from cells was performed as described above, except that the substrate was native cellular La in WCE and 2 μl of CK2 was used. For the calf intestinal phosphatase (CIP) and mock CIP reactions, the extracts were incubated with 10 μl of CIP (10 U/μl; New England Biolabs) for 30 min at 37°C and subsequently were supplemented with a phosphatase inhibitor cocktail from Sigma (#P-2850), which efficiently inhibits CIP under these conditions (data not shown), prior to the CK2 reaction. WCE (1.4 ml) was made from 175 cm2 of control or TBB-treated 293 cells. For each reaction mixture, 15 μl of WCE was added to 15 μl of kinase master mix (described above), and the mixture was incubated at 30°C for 45 min. The volume was then increased to 200 μl with NET-2 (150 mM NaCl, 2 mM EDTA, 50 mM Tris [pH 7.5], 0.05% Nonidet P40) and incubated at 4°C for 3 h with prewashed protein A-Sepharose that had been conjugated to 5 μl of anti-La or nonimmune immunoglobulin G. Immunoprecipitation was then carried out in a total volume of 200 μl of NET-2, washed 5× with 1 ml of NET-2, eluted with an equal volume of 2× loading buffer containing fresh beta-mercaptoethanol, and examined by LDS-polyacrylamide gel electrophoresis (LDS-PAGE) (NuPage Novex) autoradiography and chemiluminescent immunoblotting. The blot was incubated for 1 h with anti-La in phosphate-buffered saline (PBS) with 0.1% Tween-20 and 5% bovine serum albumin followed by incubation with horseradish peroxidase-conjugated rabbit anti-human immunoglobulin G (1/1,000) and was processed using an ECL kit (Amersham).
Coimmunoprecipitation of La-associated RNA was performed as described above, with 25 μl of protein A-Sepharose conjugated to 3 μl of anti-green fluorescent protein (GFP) antibody (#8372-2; BD Biosciences) or 5 μl of anti-La incubated with 200 μl of WCE in a total volume of 400 μl.
Transient transfection was mediated by 70 μl of Lipofectamine 2000 (#11668-027; Invitrogen) and 30 μg of total DNA (10 μg of pEGFP-hLaA366, pEGFP-hLaS366, or pEGFP plasmid DNA plus 20 μg of carrier pcDNA3) mixed with 1.75 ml of OPTI-MEM I (Invitrogen). After 20 min, another 15.75 ml of OPTI-MEM I was added and applied to freshly washed 293 cells in 100- by 20-mm tissue culture dishes. After overnight incubation at 37°C in 5% CO2 the transfection medium was replaced with complete DMEM. Cells were harvested 24 h after the transfection, and 500 μl of WCE was made from each dish. For mock transfection OPTI-MEM I alone was used instead of DNA. The pGFP-hLaS366 construct was previously described (21); pGFP-hLaA366 was created using the same strategy, subcloning of the SalI/BamHI fragment of pRep4-hLaA366 (23) into the corresponding sites in pEGFP-C1 (Clontech).
pCl-neo-hLaA366 is the construct used to express the nonfusion protein, hLaA366. The EcoRI/MluI fragment from pEGFP-hLaA366 was cloned into the corresponding sites in pCl-neo (Promega). pCl-neo-hLaS366 was created by site-directed mutagenesis of pCl-neo-hLaA366 using the appropriate oligonucleotide and QuikChange mutagenesis (Stratagene).
Standard reverse transcription-PCR (RT-PCR) was performed using gene-specific primers positioned in adjoining exons so that the RT-PCR products reflect spliced mRNA as described previously (22). Conditions were developed in the dynamic range for each assay (data not shown) to reflect quantitative differences in the samples. Negative control reactions that heat inactivated the reverse transcriptase prior to PCR cycling or included water instead of RNA were performed with each analysis to monitor quality control (data not shown).
Quantitative real-time RT-PCR was performed with the same primers as those for standard RT-PCR and used a LightCycler and Fast Start RNA Master SYBR Green I reagents (Roche Diagnostics) with an experimental run protocol as follows: RT reaction was at 61°C for 20 min, followed by denaturation at 95°C for 30 s and then repeated cycles of 95°C for 2 s, 60°C for 5 s, and 72°C for 10 s with a single fluorescence measurement. The melting curve program was 60 to 95°C with a heating rate of 0.1°C per s with continuous fluorescence measurement and a final cooling step to 40°C. Negative control experiments that omitted the RT step or used water instead of RNA yielded no products (data not shown). Integrity of all reactions was confirmed by demonstration of product melting curves with a melting point of ∼83°C and negligible nonspecific products. In addition, gel electrophoresis and ethidium bromide staining of aliquots of real-time RT-PCR products revealed the correct-sized bands as the only products (data not shown).
Monitoring proteolytic cleavage of hypophosphorylated La and apoptosis.
The percentage of cells undergoing early stages of apoptosis were identified with an annexin V-fluorescein isothiocyanate-conjugated apoptosis detection kit (49) (BD Biosciences Clontech, Palo Alto, Calif.) according to the manufacturer's protocol, using a Becton Dickinson FACSCalibur system and CellQuest v.3.1 software (San Jose, Calif.). Apoptosis was also monitored by the presence or absence of poly(ADP-ribose) polymerase (PARP; 116 kDa) and its p85 kDa caspase cleavage fragment, using a p85 PARP-specific antibody for immunoblotting. Anti-PARP was from Roche (#1835238) and was used at 1:2,000; anti-p85 PARP p85 fragment was from Promega (#G7341) and was used at 1:300. Both were incubated with the blots overnight followed by incubation with 125I-protein A (Amersham).
Polysome mRNA profile analysis.
Extract from 35-ml cultures (5 × 107 cells) was used for each sucrose gradient. One hour prior to harvest, cycloheximide (#C-7698; Sigma) was added to 50 μg/ml. Cells were washed three times with 1× PBS containing cycloheximide and was pelleted at 4,000 × g at 4°C for 5 min. The pellet was resuspended in 200 μl of TMK100 lysis buffer (10 mM Tris-HCl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 1% Triton X-100 in diethylpyrocarbonate-treated water). Protease inhibitor cocktail 1/100 V (#P-1860; Sigma), inhibitor cocktail 1/100 V (#20-116; Upstate Biotechnology), 0.2 M PMSF in ethanol (EtOH) and 1/100 V (#93482; Sigma), and 2 mM dithiothreitol (DTT) were added fresh just before use. The mixture was pipetted up and down five times. Cell debris was removed by sedimentation in a microcentrifuge at 10,000 × g at 4°C for 5 min. The supernatant was layered onto a 14- by 89-mm 4.5 to 45% (wt/vol) sucrose gradient that contained gradient salt buffer (20 mM Tris-HCl [pH 7.5], 150 mM KCl, 10 mM MgCl2). The sample was sedimented at 39,000 rpm at 4°C in an SW41 rotor for 150 min. Fractions (0.6 ml) were collected into 1.5-ml tubes. One milliliter of 100% EtOH was added to 0.4 ml of each fraction, the samples were vortexed, and they were put at −80°C to precipitate RNA. Pellets were collected by sedimentation, washed in 75% EtOH, resuspended in 0.4 ml of Tris-EDTA (TE; pH 8.0), and transferred to 1.5-ml tubes. The RNA was again precipitated by addition of 50 μl of 3 M sodium acetate (pH 5.2) and 1 ml of 100% EtOH. Each pellet was washed in 75% EtOH and resuspended in 25 μl of water. Equal volumes (5 μl) of all samples were analyzed by RT-PCR.
Antisense-mediated CK2 knockdown.
We used a published method to knock down CK2β levels in vivo (26). Oligonucleotides (sense, 5′-GACGTGAAGATGAGCAGCTC-3′; antisense, 5′-GAGCTGCTCATCTTCACGTC-3′) were incubated with cells prior to preparation of cell extract, as indicated in the figure legend. Anti-CK2β was purchased from Calbiochem (#218712). Anti-β-actin was from Sigma (#A-5316).
RESULTS
TBB inhibits CK2-mediated phosphorylation of human La S366 in vitro.
The majority of S366 of La is found phosphorylated in vivo (7, 15), and recombinant La can be phosphorylated on S366 by CK2 in vitro (14, 15, 23). A schematic drawing of the human La protein is shown in Fig. 1A. To examine the effects of TBB on CK2 phosphorylation of La in vitro, reaction conditions used previously, including 500 μM GTP, [γ-32P]GTP, and purified recombinant CK2 (rCK2), were employed (14, 15, 23) (Fig. 1B). The purified recombinant proteins, La S366 and La G366, are shown in Fig. 1C. La S366 was readily phosphorylated by CK2 (Fig. 1B, lane 2), while an equal amount of La G366 was not (lane 1). Omission of either La S366 or rCK2 from the reaction abolished the signal (lanes 7 and 8), establishing that CK2 phosphorylates La specifically on S366 in this assay. TBB led to a dose-dependent inhibition of La phosphorylation (lanes 3 and 4) while addition of solvent alone (DMSO) had negligible effects (lanes 5 and 6). Quantitation of three experiments as summarized in Fig. 1D demonstrate that TBB inhibits CK2-mediated phosphorylation of S366 of La in vitro. Because 100 μM TBB decreased phosphorylation to less than 50% when GTP was present at 500 μM, these results support the conclusion that TBB is a potent inhibitor of CK2 (46).
TBB inhibits phosphorylation of La antigen in vivo.
Although Jurkat cells are quite sensitive to the apoptosis-inducing effect of TBB (45), direct comparisons revealed that human embryonic kidney (293) cells were significantly less sensitive (data not shown). Because dose-response pilot experiments revealed that 50 μM TBB for 24 h led to La hypophosphorylation with limited cell toxicity (data not shown), these conditions were used subsequently.
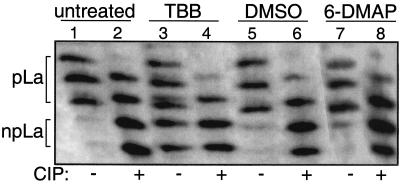
One-dimensional IEF followed by immunoblotting with anti-La can monitor the relative amounts of native npLa and pLa isoforms in cellular extract (23). Analysis is assisted by treating an aliquot of the extract with CIP, which converts pLa to npLa, whereas mock or no treatment reflects the distribution of npLa and pLa as they exist in the extract (Fig. 2, compare lanes 1 and 2). pLa and npLa each separate into multiple bands due to additional modifications as described previously (23). In agreement with previous studies of HeLa cells, the 293 cells contained mostly pLa with substantially less npLa (Fig. 2, lane 1) (22, 23). The CIP-treated extracts reveal a significant amount of npLa (even-numbered lanes) which served as mobility markers and as internal controls. Extracts from untreated 293 cells as well as cells incubated with 50 μM TBB, solvent (DMSO) alone, or a commonly used inhibitor of other kinases, 6-DMAP, were compared (Fig. 2, odd-numbered lanes). Untreated, DMSO-treated, and 6-DMAP-treated cells contained mostly pLa with much less npLa (lanes 1, 5, and 7). In contrast to this, cells treated with TBB contained a substantial amount of npLa in addition to pLa (lane 3). Additional IEF analyses revealed that TBB caused a dose- and time-dependent increase in npLa (data not shown). Quantitation revealed that pLa represents ∼85% while npLa represents ∼15% of total La and that TBB led to a more than twofold increase in the nonphosphorylated fraction of La in cells (data not shown; see below).
FIG. 2.
TBB increases hypophosphorylated La isoforms in vivo. Results are from one-dimensional IEF followed by immunoblotting with anti-La serum. WCEs were prepared from untreated 293 cells (lanes 1 and 2) or 293 cells incubated for 24 h in 50 μM TBB (lanes 3 and 4), solvent alone (DMSO; lanes 5 and 6), or 6-dimethyl-aminopurine (6-DMAP; lanes 7 and 8). The extracts were treated in vitro with CIP (+) or were mock-treated (−) as indicated below the lanes. Lane 1 reflects a substantially larger amount of pLa compared to that of npLa as they exist in untreated extract; pLa and npLa each separate into multiple bands due to acidic modifications that are found at positions other than S366, as described previously (23).
Although the previously documented high selectivity of TBB (46) would suggest that the increase in npLa might be due to direct inhibition of CK2-mediated phosphorylation of La in vivo, we note that IEF can reveal the distribution of La isoforms but does not unequivocally identify the hypophosphorylated site. Therefore, to examine if S366 was hypophosphorylated in TBB-treated cells, the La S366 site-specific rCK2-mediated in vitro kinase assay was used on La protein isolated from cells after TBB treatment as described below.
TBB inhibits phosphorylation of S366 in vivo.
It was previously shown that La protein that already contains phosphate on S366 is not phosphorylated by [γ-32P]GTP and rCK2 in vitro and that the magnitude of the 32P-La signal obtained from the in vitro reaction increases with an increase in the amount of npLa (see Fig. 5B in reference 15). This suggested that rCK2 exogenously added to extract would phosphorylate the cell-derived npLa on S366 in vitro and that this would reveal the degree of hypophosphorylation of S366. Therefore, we used the rCK2-dependent in vitro assay to examine cellular pLa and npLa levels. After [γ-32P]GTP-mediated phosphorylation by rCK2 in vitro, La protein was isolated by immunoprecipitation (IP) and was analyzed by gel autoradiography (Fig. 3A) and also by chemiluminescence immunoblotting to assess the specific activity of the isolated La protein (Fig. 3B).
FIG. 5.
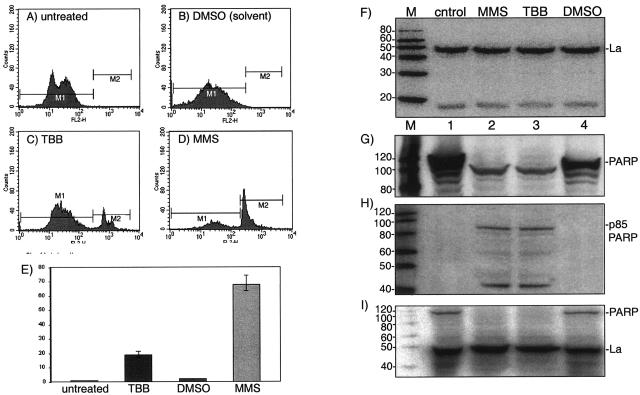
TBB treatment leads to apoptosis in HEK293 cells with no hypophosphorylation-associated cleavage of La. Fluorescence-activated cell sorting (FACS) analysis of apoptosis using annexin-V staining of untreated (A), solvent-treated (B), 50 μM TBB-treated (C), and MMS-treated (D) 293 cells as indicated. (E) Quantitation summary of three experiments. The y axis is the percentage of annexin V-positive cells. Immunoblot analyses of extracts made from control untreated, MMS-treated, TTB-treated, and control DMSO-treated cells (lanes 1 to 4, respectively) using the following antibodies for detection: anti-La (F), polyclonal anti-PARP (G), and monoclonal p85-specific PARP (H). (I) Reprobing of blot B with anti-La. M, molecular size markers as indicated to the left in kilodaltons.
FIG. 3.
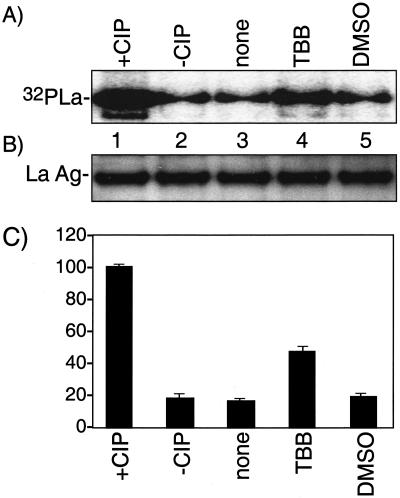
TBB decreases phosphorylation of La on serine366, a CK2 target site, in vivo. (A) Extracts were prepared from 293 cells that had been incubated with no inhibitor (lanes 1 to 3), 50 μM TBB (lane 4), or solvent alone (DMSO; lane 5) and then subjected to in vitro phosphorylation with CK2 and [γ-32P]GTP followed by IP with anti-La antibodies, sodium dodecyl sulfate-PAGE, and autoradiography. The extract in lanes 1 and 2 was treated with CIP or was mock treated, respectively, and then was effectively inhibited with phosphatase inhibitors prior to the in vitro CK2 reaction (see Materials and Methods). Exposure time was 12 h. (B) Immunoblot analyses of the immunoprecipitated La samples shown in panel A using anti-La serum and chemiluminescence for detection; exposure time was 1 min. (C) Quantitation of the 32P-La corresponding to lanes 1 to 5 of panel A represented by three separate experiments, as indicated. Results are expressed as the percentage of the radioactive counts in the +CIP lane (see the text).
As a control and to reveal the maximum amount of La potentially available for rCK2-dependent 32P phosphorylation in vitro, an extract sample was dephosphorylated by phosphatase (CIP) treatment prior to the in vitro rCK2 reaction and was compared to mock CIP treatment (Fig. 3A, lanes 1 and 2; see Materials and Methods). Quantitation (see below) indicated that CIP led to an approximately sixfold increase in the substrate activity of La in this assay, suggesting that about 85% of the La present in HeLa cells is phosphorylated, in agreement with estimates from IEF.
Extracts made from untreated, solvent-only (DMSO), and TBB-treated 293 cells were examined by the rCK2-mediated, [γ-32P]GTP-dependent phosphorylation assay. Because phosphorylation by rCK2 is specific for S366, Fig. 3A indicates that this assay monitors S366 phosphorylation specifically (Fig. 1A) (15). Further specificity was revealed by the absence of 32P-La signal when preimmune serum was used rather than anti-La for IP (data not shown). Chemiluminescence immunoblotting showed that the amount of La protein in each sample after IP was very similar, as expected (Fig. 3B). The data support the conclusion that the signal shown in Fig. 3A, lane 4, reflects increased npLa levels after TBB treatment relative to the control samples.
The results of three experiments were quantitated and are summarized in Fig. 3C. Comparison of the CIP and non-CIP samples indicates that 16% ± 3% of La is nonphosphorylated on S366, in agreement with the IEF data. The fraction of npLa increased about 2.5-fold, from ∼16% to ∼43%, after exposure of the cells to TBB. From these results we can conclude that the majority of human La protein is phosphorylated on S366, a direct CK2 target site, and that TBB leads to hypophosphorylation of this site in vivo.
Antisense-mediated knockdown of CK2β also leads to hypophosphorylation of La S366.
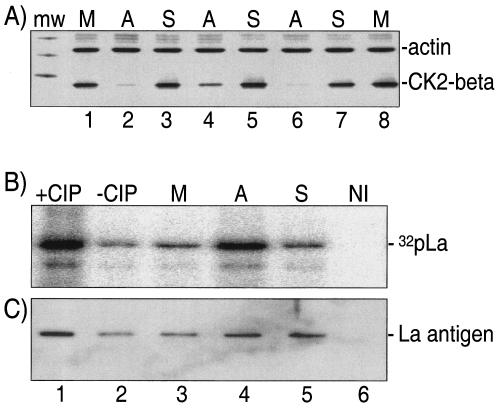
Multiple laboratories have used antisense technology to knock down CK2 in cells (10, 26, 39). 293 cells treated with sense and antisense oligonucleotides directed to CK2β under various conditions were examined for effects on CK2β and beta-actin levels, the latter of which served as an internal control (Fig. 4A). Cells treated with antisense (Fig. 4A, lanes 2, 4, and 6) but not sense (lanes 3, 5, and 7) or mock-treated cells (lanes 1 and 8) showed a decrease in CK2β but not actin. Although multiple conditions led to a specific decrease in CK2β, we chose to continue our analyses with the cells treated with 200 μg of oligonucleotides/ml for 20 h (see Fig. 4) because they maintained the highest viability (90%; data not shown).
FIG. 4.
Antisense oligonucleotide directed to CK2β decreases phosphorylation of La on S366. (A) Immunoblot to detect beta-actin and CK2β from extracts treated with oligonucleotide at 200 μg/ml for 20 h (lanes 2 to 3), 300 μg/ml for 20 h (lanes 4 to 5), or 200 μg/ml for 40 h (lanes 6 to 7); the mock treatment samples in lanes 1 and 8 were for 20 and 40 h, respectively. (B) The [γ-32P]GTP-mediated rCK2 assay was the same as that described in the legend to Fig. 3 and used extracts from the mock (M)-, sense (S)-, and antisense (A)-treated cells. The samples were transferred to nitrocellulose membranes and were visualized by autoradiography. The experiment depicted in lane 6 used nonimmune (NI) serum for IP. (C) The blot shown in panel B was incubated with anti-La and was developed for chemiluminescence to reveal the La antigen on the blot.
We performed [γ-32P]GTP-dependent in vitro phosphorylation to reveal the level of phosphorylation of La S366 in the extracts from the oligonucleotide-treated (200 μg/ml, 20 h) cells. After electrophoresis, the gel contents were electroblotted onto nitrocellulose and were visualized by autoradiography (Fig. 4B). Lane 1 of Fig. 4B reflects the [γ-32P]GTP-dependent phosphorylation achieved after dephosphorylation of endogenous La with CIP compared to that of the control reaction (Fig. 4B, −CIP, lanes 1 and 2). The relative levels of npLa in the mock treatment, antisense oligonucleotide treatment, and sense oligonucleotide treatment are shown in lanes 3, 4, and 5, respectively. Substantially more npLa was detected in the antisense-treated cells compared to that of the sense-treated cells (Fig. 4B, compare lanes 4 and 5). The nitrocellulose membrane was then assayed for total La antigen by using Go serum and chemiluminescence for detection (Fig. 4C). This assay revealed that La levels were similar in the antisense and sense samples (lanes 4 and 5). This analysis allows the conclusion that La isolated from antisense-treated cells was hypophosphorylated relative to sense-treated cells, supporting the conclusion that CK2 is responsible for phosphorylation of La S366 in vivo.
TBB does not significantly alter total levels or induce proteolytic cleavage of La.
As discussed in the Introduction, several aspects of La phosphorylation status and the pathways that may affect it in vivo remain unclear. Previous reports indicated a relationship between apoptosis and hypophosphorylation-associated proteolysis of La (4, 44). Treatment of Jurkat cells with TBB leads to proteolytic cleavage of PARP, release of cytochrome c from mitochondria, and DNA laddering, indicative of apoptosis (45). Although apoptosis-associated La hypophosphorylation may be due to phosphatase activity rather than paucity of phosphorylation (44), the present study nonetheless provided an opportunity to examine the relationship between apoptosis, hypophosphorylation, and proteolysis of La in this experimental system. This is an important issue, because cleavage of La is associated with redistribution of La to the cytoplasm (4), which could have significant effects on mRNA binding.
We examined TBB-treated 293 cells for apoptosis, employing a standard procedure that uses flow cytometry and annexin V staining to detect phosphatidylserine translocated to the outer cell membrane soon after cells initiate apoptosis (29, 49, 54). The DNA-damaging agent methyl methanesulfonate (MMS) was used as a positive control for apoptosis (31), and solvent alone was used as a negative control. The areas designated M1 in Fig. 5A to D represent unaffected cells, whereas the areas designated M2 represent annexin V-stained cells and identify the fraction of apoptotic cells. As expected, only a very small fraction of cells were in the M2 window after no treatment or mock (solvent) treatment (Fig. 5A and B). MMS treatment led to a substantial fraction of M2 cells, indicative of apoptosis (Fig. 5D). Treatment with 50 μM TBB led to an increase in annexin V-positive (M2) cells, suggestive of apoptosis (Fig. 5C).
As can be seen from the immunoblot in Fig. 5F, a single major band of approximately 47 kDa, corresponding to full-length La, was observed in untreated as well as in MMS- and TBB-treated cells, with no evidence of an apoptosis-specific cleavage product. As a control we examined PARP (∼116 kDa) and the ∼85-kDa proteolytic fragment of PARP, the latter of which is generated by caspase-mediated cleavage of full-length PARP during early apoptosis (45). As can be seen with polyclonal anti-PARP antibody (Fig. 5G), full-length PARP was readily detected in control untreated cells (lane 1) as well as in control DMSO-treated cells (lane 4). Full-length PARP was not detected in the TBB- and MMS-treated cells, which instead revealed a shorter fragment (Fig. 5G, lanes 2 and 3). We also used a monoclonal antibody that is specific for the p85 PARP fragment (i.e., does not detect full-length PARP). The p85 kDa fragment was detected in extracts from TBB-treated and MMS-treated cells (Fig. 5H, lanes 2 and 3), whereas the p85 kDa fragment was undetectable in the control cell extracts (lanes 1 and 4). As an additional quality control, we probed the immunoblot in Fig. 5G, which contained residual PARP signal, with anti-La to create Fig. 5I. This demonstrated that in extracts in which PARP cleavage was extensive, no significant differences in La levels or La cleavage products were detected. We conclude that although cleavage of PARP, which is a sensitive indicator of early apoptosis, was readily detected, cleavage of La was undetectable in TBB- or MMS-treated cells undergoing apoptosis. Thus, TBB-mediated hypophosphorylation of La was not associated with significant changes in total levels or cleavage of La.
Hypophosphorylation of La S366 is associated with increased binding to the 5′TOP-containing mRNA encoding rpL37.
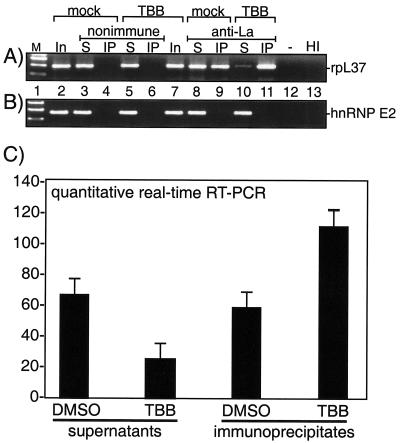
Xenopus La protein has been reported to associate with 5′TOP mRNAs (9, 11, 38). It was recently shown for human La that a significant fraction of each of the three 5′TOP mRNAs examined by RT-PCR, including rpL37 mRNA, was preferentially associated with npLa rather than pLa in HeLa cells, while two non-TOP mRNAs were not associated with either isoform of La (22). It is important to note for this study that only a fraction of rpL37 mRNA was associated with La, suggesting that npLa is limiting for L37 mRNA binding in HeLa cells (22). Here we examined if rpL37 mRNA was immunoprecipitated by anti-La or nonimmune serum in mock- and TBB-treated 293 cells (Fig. 6A, lanes 4, 6, 9, and 11) and also examined the supernatants of the IPs (lanes 3, 5, 8, and 10). The mRNAs present in an aliquot of the input material from mock- and TBB-treated cells were also examined (lanes 2 and 7). As expected, rpL37 mRNA was not immunoprecipitated from mock- or TBB-treated cells by nonimmune serum (lanes 4 and 6). Anti-La immunoprecipitated a significant amount of rpL37 mRNA from the mock-treated cells as expected (lane 9), and it immunoprecipitated approximately twice as much from the TBB-treated cells (lane 11). The greater association in TBB-treated cells versus that in control cells was also reflected by the smaller amount of rpL37 mRNA remaining in the supernatant of the TBB-treated cells compared to that in the mock-treated cells (compare lanes 8 and 10). Although hnRNP E2 mRNA, which does not contain 5′TOP, was readily detected in the input and supernatant samples, it was not immunoprecipitated by La from mock- or TBB-treated cells (Fig. 6B) as expected (22).
FIG. 6.
TBB leads to increased association of rpL37 5′TOP mRNA with La. RT-PCR analyses of RNA prepared from native WCE made from mock- and TBB-treated cells directly (lanes 2 and 7) or RNA prepared after IP with nonimmune (lanes 3 to 6) or anti-La (lanes 8 to 11) serum. Both the immunoprecipitates (lanes 4, 6, 9, and 11) and the supernatant remaining after immunoprecipitation (lanes 3, 5, 8, and 10) were examined. Lane 12 is a negative control that used water instead of RNA. The reaction shown in lane 13 was heat inactivated (HI) before the reverse transcriptase step of the reaction, using input RNA corresponding to lane 2. (A) RT-PCR detection of 5′TOP-containing mRNA encoding rpL37. (B) RT-PCR analysis of hnRNP E2 mRNA. (C) Summarized data from triplicate quantitative real-time RT-PCR analyses of rpL37 mRNA. The y axis shows units of rpL37 mRNA.
We also developed a real-time quantitative RT-PCR assay for L37 mRNA, using serial dilutions of total RNA to obtain a standard curve (data not shown). Side-by-side comparisons then demonstrated significantly more rpL37 mRNA associated with La from TBB-treated cells than that associated with DMSO control cells (Fig. 6C). Reciprocal differences in the supernatants from the TBB and control DMSO samples (Fig. 6C) further corroborated the results obtained by standard RT-PCR. These analyses indicated that TBB led to an increase in the amount of rpL37 mRNA that was associated with La. Because it had been shown that rpL37 mRNA was preferentially associated with npLa rather than pLa (22), these data suggest that increasing npLa levels leads to increased association between La and rpL37 5′TOP mRNA.
Nonphosphorylatable La A366 better competes for rpL37 mRNA than does La S366.
Although TBB increases the amount of npLa and the fraction of rpL37 mRNA associated with La, other factors are also affected by TBB and could conceivably influence rpL37 mRNA. Therefore, we asked whether a direct increase in the amount of npLa, independent of manipulating CK2 activity, would be sufficient to increase the fraction of rpL37 mRNA associated with La. For this we transfected 293 cells with a vector encoding GFP-La S366, GFP-La A366, or GFP alone and performed RT-PCR analysis for rpL37 mRNA after IP with anti-GFP.
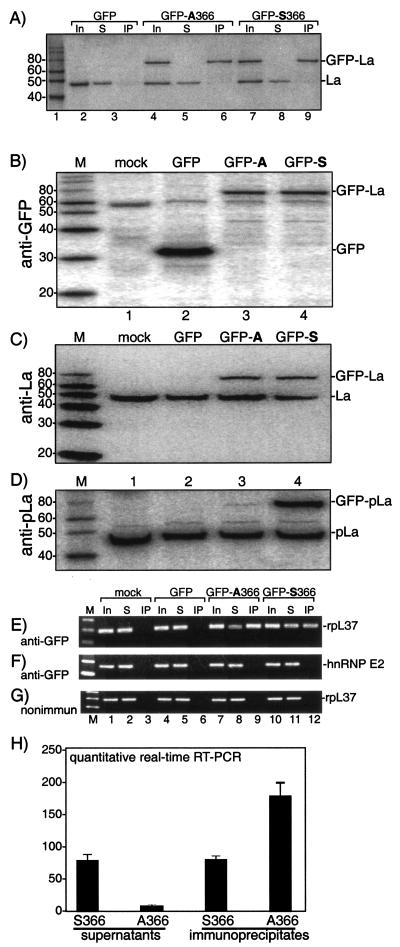
Figure 7A is an immunoblot that used anti-La for detection to examine the specificity of the anti-GFP-immunoprecipitation. Anti-GFP was specific for the fusion proteins because both the GFP-La and endogenous La proteins were detected in the inputs (lanes 1, 4, and 7), but only the GFP-La proteins were immunoprecipitated by the anti-GFP serum (lanes 6 and 9). Endogenous La remained in the supernatants, while the GFP-La proteins were quantitatively depleted (lanes 5 and 8). This revealed similar expression levels for GFP-La S366 and GFP-La A366 (Fig. 7A, lanes 4 and 7) and similar amounts of GFP-La S366 and GFP-La A366 in the IPs (lanes 6 and 9).
FIG.7.
More rpL37 mRNA associates with nonphosphorylatable La A366 than with La S366. 293 cells were mock transfected or were transfected with GFP alone or with the fusion constructs GFP-La A366 or GFP-La S366. (A) WCEs were prepared and used to examine the specificity of IP with anti-GFP; the inputs, supernatants, and IPs were analyzed by immunoblotting using anti-La for detection. (B to D) Immunoblot analyses of transfected cells used for RNA analysis below, using anti-GFP (B), anti-La (C), and anti-pLa (D) (22) for detection. RNA was prepared and used for direct RT-PCR (lanes 1, 4, 7, and 10) or RNA was prepared after IP with anti-GFP (E and F) or nonimmune (G) serum, as indicated to the left of panels B to D. (H) Summarized data from quantitative triplicate real-time RT-PCR of an anti-GFP IP experiment comparable to that of panel A above. The y axis shows units of rpL37 mRNA.
We next characterized the extracts for RNA analysis. First, immunoblotting with anti-GFP confirmed expression of the GFP-La proteins (Fig. 7B, lanes 3 and 4) and demonstrated expression of GFP in the GFP-only sample (lane 2). Immunoblotting with polyclonal anti-La (Fig. 7C) showed the expression levels of GFP-La S366 and GFP-La A366 relative to endogenous La in the extract samples used for RT-PCR (described below). Immunoblotting using AbP previously characterized to recognize phosphoserine366 La but not npLa (22) detected GFP-La S366 as being efficiently phosphorylated on S366 (Fig. 7D, lane 4) while GFP-La A366 was not recognized (lane 3), as expected. These data demonstrated S366-specific phosphorylation of GFP-La S366 and that GFP-La S366 and GFP-La A366 expressed in vivo indeed differ in their phosphorylation status.
RT-PCR analysis of the immunoprecipitates revealed more rpL37 mRNA associated with GFP-La A366 than with GFP-La S366 (Fig. 7E, compare lanes 9 and 12). This was corroborated by the data that showed less rpL37 mRNA remaining in the supernatant from the IPs of cells transfected with GFP-La A366 than with GFP-LaS366 (Fig. 7E, compare lanes 8 and 11). The same RNA samples were also examined for hnRNP E2 mRNA, which does not contain a 5′TOP motif and was not found in association with either of the GFP-La proteins, as expected (Fig. 7F). Also as expected, nonimmune serum did not IP L37 mRNA from any sample (Fig. 7G). These results demonstrated that the greater association of L37 mRNA with GFP-La A366 was specific.
The quantitative real-time RT-PCR assay convincingly demonstrated more rpL37 mRNA associated with GFP-La A366 than with GFP-La S366 in the IPs, with the expected reciprocal differences in the supernatants (Fig. 7H).
Fluorescence of GFP-La A366 and GFP-La S366 revealed indistinguishable patterns of nuclear cytoplasmic distribution in multiple cell types, as reported previously and as observed in our analysis (7 and data not shown). Thus, by all examinations thus far, GFP-La A366 and GFP-La S366 appear to differ only in their phosphorylation state and in the degree to which rpL37 mRNA is associated with them. To the extent that similar subcellular distributions reflect similar concentrations of GFP-La A366 and GFP-La S366, the cumulative data suggest that the nonphosphorylatable GFP-La A366 better competes for rpL37 mRNA than does its wild-type S366-phosphorylated counterpart, GFP-La S366. This interpretation is consistent with data that documented that npLa exhibits greater affinity than pLa for RNA in vitro and more stable association with RNA in vivo (14, 23, 37, 40).
Nonphosphorylated La alters the polysome distribution of a fraction of L37 mRNA.
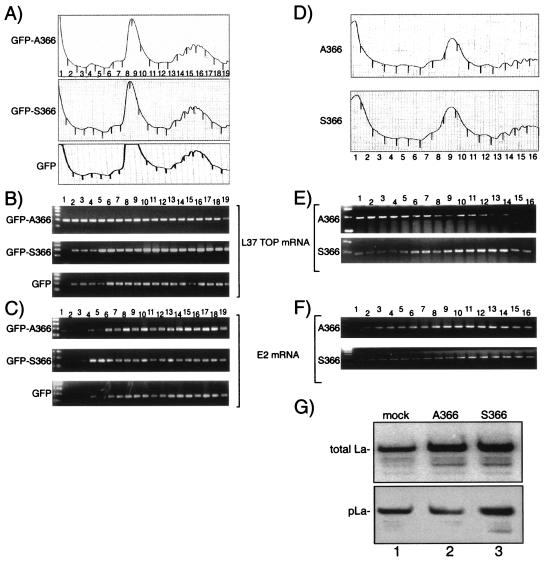
GFP, GFP-La A366, and GFP-La S366 were also examined for their effects on the polysome distribution of L37 mRNA (Fig. 8A to C). After transfection, extracts were prepared, subjected to sucrose gradient sedimentation, and examined by chromatography (Fig. 8A). The fractions collected were analyzed for L37 mRNA content by RT-PCR (Fig. 8B). Because the preponderance of published data indicate a positive effect of La on translation, whereas fewer reports indicate a negative effect, we expected to see that GFP-La A366 leads to an increase in the polysome-associated L37 mRNA. Surprisingly, we found that GFP-La A366 led to an increase in the amount of L37 mRNA that was in the slow-sedimenting material, in fractions 1 to 4, compared to that of GFP-La S366 and GFP (Fig. 8B). By contrast, the pattern obtained for hnRNP E2 mRNA did not show this effect but instead revealed a slight shift toward the slow-sedimenting material in the GFP-La S366 extract. Repeated controlled experiments corroborated these results (data not shown).
FIG. 8.
npLa alters the polysome distribution of L37 mRNA. (A to C) Data obtained after transient transfection with expression vectors encoding GFP-hLaA366, GFP-hLaS366, or GFP alone, as indicated. (D to G) Data obtained after transient transfection with expression vectors encoding native (untagged) hLaA366 or native (untagged) hLaS366 as indicated. (A and D) Sucrose gradient sedimentation profiles. (B and E) RT-PCR analyses of rpL37 mRNA. (C and F) RT-PCR analyses of hnRNP E2 mRNA. (G) Immunoblot after detection of total La antigen using polyclonal anti-La (Go) serum (upper panel) and the phosphoserine366-La fraction using phosphospecific antibodies.
We considered the possibility that while the GFP portion of the fusion protein may support specific mRNA binding by the La portion, it might confer a negative effect on polysome distribution. Therefore, we examined the effects of ectopic expression of native (untagged) La S366 and La A366 (Fig. 8D to G). Extracts from the native La S366- and La A366-transfected cell proteins yielded similar profiles (Fig. 8D) but differed in the distribution of L37 mRNA (Fig. 8E). Again, a fraction of L37 mRNA was shifted toward the slow-sedimenting fractions in the La A366 extract compared to that in the La S366 extract (Fig. 8E), and this distribution was significantly different from that of hnRNP E2 mRNA (Fig. 8F).
Immunoblotting revealed that La expressed in these cells was differentially phosphorylated, as expected (Fig. 8G). The blot was first incubated with polyclonal anti-La antibody and was developed by using 125I-protein A to reveal the total La in the extracts (Fig. 8G, upper panel). The blot was then stripped of radioactivity, incubated with the phosphoserine366-specific AbP, and developed using chemiluminescence (Fig. 8G, lower panel). While La S366- and La A366-transfected cells revealed similar amounts of total La (upper panel), the La A366 cells contained relatively less pLa (lower panel), consistent with a higher level of npLa in the La A366 cells, as expected.
DISCUSSION
La is an efficient substrate of protein kinase CK2.
A major conclusion that can be drawn from this work is that protein kinase CK2 phosphorylates hLa on S366 in vivo. This conclusion is based on the use of a highly specific CK2 inhibitor, TBB, and on established antisense-mediated knockdown of CK2β. The specificity of TBB reflects the fact that CK2 is unusual in its use of GTP and ATP as phosphoryl donors and the unique properties of its nucleotide binding site (13, 16, 18, 41, 42). Even the most sensitive of 33 other kinases tested was more than an order of magnitude less sensitive to TBB than to CK2, revealing TBB as one of the most selective inhibitors of protein kinases ever reported (5, 46). In addition, the CK2 consensus target phosphorylation site, S/TXXD/E, is unusual among protein kinases in its strong stimulation by adjacent acidic residues and the negative influence of adjacent basic residues (16, 41), a feature that closely matches the sequence context of S366, FAS366DDE. Indeed, the majority of cellular La was found phosphorylated on S366, in agreement with the noted features of CK2 that keep its catalytic activity constitutive (8, 41, 42).
It is noteworthy that although CK2 is considered constitutive, some regulatory activities have been attributed to it (1). However, turnover of 32P of cellular La (40) suggests that dephosphorylation contributes to the steady-state levels of pLa and npLa in cells and leaves open the possibility that npLa levels may be determined, at least in part, by phosphatase activity. Because the pLa fraction is large, it represents a substantial potential source of npLa that could theoretically be converted to npLa for regulatory purposes. Evidence that a PP2A-like phosphatase contributes to La dephosphorylation has been presented previously (44).
Because these and prior data suggest that the npLa is inhibitory to 5′TOP mRNA translation and pre-tRNA production, respectively, it would appear that CK2 may contribute to the ability of La to coordinately affect the production of the translational machinery under some conditions (27) and, as such, begin to elucidate a signaling pathway.
As part of this report we developed an enzymatic assay to quantitatively assess the level of S366 phosphorylation of cellular La. We note that while IEF can only reflect net charge differences between protein isoforms, the enzymatic approach assesses the phosphorylation status of S366 specifically. Thus, we believe that because of its relative ease, timeliness, increased sensitivity, and quantitatability, the enzymatic assay, if performed with appropriate calibration and other controls, is superior to IEF for determining the level of hLa S366 phosphorylation.
Uncoupling of TBB-mediated hypophosphorylation and cleavage of La.
Although apoptosis was not a central focus of this study, reports that La becomes hypophosphorylated and cleaved during apoptosis (4, 44), in addition to the demonstration that TBB leads to apoptosis (45 and this report) and La hypophosphorylation (this report), prompted us to examine if La is cleaved in TBB-treated cells and/or other cells undergoing apoptosis. This is significant because apoptosis-related proteolytic cleavage of La was associated with redistribution of La to the cytoplasm (4), which could have significant effects on mRNA binding and could provide insight into the mechanism by which TBB leads to 5′TOP mRNA binding. We were unable, despite multiple attempts, to detect any significant difference in potential cleavage products of La in TBB-treated 293 cells or in MMS-treated 293 cells, the latter of which had undergone a more substantial amount of apoptosis, even though we could readily detect substantial cleavage of PARP in TBB- and MMS-treated cells. Thus, La hypophosphorylation occurred in TBB-treated cells but was apparently uncoupled from La cleavage. We also examined La in Jurkat cells that were treated with etopside under conditions that led to apoptosis as reported previously (4), but again no evidence of cleavage was detected (data not shown). It is also noteworthy that we detected no evidence of La hypophosphorylation in MMS-treated cells undergoing apoptosis (data not shown). Thus, although we could readily detect La hypophosphorylation in the context of apoptosis in TBB-treated cells, hypophosphorylation of La was uncoupled from apoptosis in MMS-treated cells. We note that although cleavage was not detected in our study, others have also noted that PARP cleavage was more extensive (>80%) than that observed for La (35 to 40%) in the cell types examined (4). Therefore, it is possible that under our conditions, which differed from those of prior studies in the use of 293 cells (4, 44), La cleavage had not yet been initiated or was below the level of detection.
An in vivo biochemistry model of a multiclass RNA binding protein.
More rpL37 mRNA was found associated with the nonphosphorylatable protein GFP-La A366 than with GFP-La S366. GFP-La A366 and GFP-La S366 were expressed at similar levels in the transfected cells used for quantitative RT-PCR analysis of rpL37 mRNA. Moreover, GFP-La S366 was faithfully and efficiently phosphorylated in these cells, while GFP-La A366 was not. Furthermore, our fluorescence microscopy data are in agreement with a those of a report that showed that GFP-La A366 and GFP-La S366 exhibit similar nuclear cytoplasmic distribution in multiple cell types (7 and data not shown). This suggests that the isoform status of S366 is not a direct determinant of localization but may instead reflect colocalization of the appropriate kinase and phosphatase. In any case, the preferential association of rpL37 mRNA with GFP-La A366 cannot be easily explained by differential localization or concentration of these proteins but instead correlate best with their differential phosphorylation. Greater association of L37 mRNA with GFP-La A366 would appear to reflect the relatively higher affinity of npLa for RNA that has been documented previously (14, 23, 37, 40).
As a protein that interacts specifically with a distinct subset of small nuclear RNAs as well as specific subsets of cytoplasmic mRNAs, La is among a small group of RNA-binding proteins that recognizes very different classes of cellular RNAs. Although the cell biological mechanisms involved in these activities had been unclear, the finding that the differential subcellular compartmentalization of the S366 isoforms corresponds with their differential RNA ligands has provided some resolution (22).
The cumulative results suggest a model that is comprised of the following determinants: (i) differential concentrations of the La S366 isoforms in the different subcellular compartments; (ii) differential affinities of the La S366 isoforms for RNA; (iii) differential affinities of the different classes of RNAs for La; and (iv) differential concentrations of the different classes of RNA ligands in these compartments. For example, pLa is limited to the nucleoplasm, where it is far in excess of npLa and where it interacts with UUU-OH-bearing pre-tRNAs whose concentrations are also highest in this compartment. Unlike pLa, a substantial fraction of npLa is in the cytoplasm, where, in the absence of pre-tRNAs and other UUU-OH-bearing nascent pol III transcripts, it is free to associate with the pyrimidine-rich 5′TOP mRNAs, which compete more effectively for npLa than do non-TOP mRNAs. It would seem that these characteristics establish a hierarchy that determines the differential association of npLa and pLa with their respective classes of associated RNAs. Mechanisms that determine subcellular localization of the RNAs and the La proteins, as well as the kinases and putative phosphatases that control phosphorylation, are important components that remain to be characterized further.
Alteration of La S366 phosphorylation can affect 5′TOP mRNA metabolism.
Shift of rpL37 5′TOP mRNA off polysomes suggests that increased levels of npLa may have negative effects on 5′TOP mRNA translation in the system used here, although this remains to be demonstrated because we have not examined translation per se. A body of work has indicated a positive effect of Xenopus La on 5′TOP mRNA translation, including during amphibian development (9, 11, 38). Our findings are in agreement with another work that showed a negative effect of human La on 5′TOP mRNA translation (58) and further suggest that the phosphorylation status of human La S366 is a determinant of this effect. In this prior study, hLa isolated from human lymphocyte cytoplasm as well as purified human recombinant La (rLa; i.e., nonphosphorylated) specifically bound to the 5′TOP, and rLa dramatically decreased translation of the 5′TOP mRNA but not the control mRNA that had a mutated TOP element (58). Although at present we cannot explain why human La and Xenopus La exhibit different effects, continued future investigations may uncover the different mechanisms involved.
Because the preponderance of published data have indicated a positive effect of La on translation, whereas fewer reports have indicated a negative effect, we were initially surprised to find that GFP-La A366 led to a shift of L37 mRNA off of polysomes. Two aspects of this should be considered. First, human La has been found to confer negative effects on TOP mRNA translation in a TOP-specific manner (58). Second, the great majority of mRNAs whose translations have been shown to be positively affected by La contain 5′UTRs that are long and complex and are comprised of IRESs and/or upstream AUGs (3, 6, 19, 24, 28, 35, 43, 47, 52; reviewed in reference 55), whereas the 5′UTRs of regulated 5′TOP-containing mRNAs are unusually short (e.g., 30 nucleotides) (36), are devoid of upstream AUG, and initiate with an mGppp-C residue as opposed to mGppp-purine, as is the case for most non-TOP mRNAs (36). Because La exhibits preference for pyrimidines, it may recognize 7mGppp-capped TOP sequences better than other mRNAs. Previous data indicate that the SBM of npLa can interact with the 5′ triphosphate of RNA (14). Other evidence indicates that the 7mGppp cap moiety of an RNA was efficiently cross-linked to cellular La at the expense of cross-links to the CBP20 subunit of the nuclear cap binding complex (17). These observations, together with the report that the RRM of CBP20 is a close structural homolog of La RRMc (25), suggest that La may bind the 5′TOP motif in a manner that could inhibit its translation. Thus, La may have different effects on 5′TOP from its other mRNA ligands. Indeed, La has been shown to act either as a positive or negative effector, depending on the 5′UTR context and the mechanism of translation initiation (50).
In any case, we believe it is important to limit the interpretation of the present findings to the context of the experimental system used here. Because 5′TOP mRNA regulation occurs in response to nutritional stress or other signaling and because we did not attempt to induce TOP regulation, we do not know if our results would be relevant in the context of the normal physiology of TOP mRNA regulation. Notwithstanding these limitations, the present data are consistent with prior data that indicate a negative role for npLa in tRNA production and suggest a role for CK2-mediated phosphorylation of La as a means to prevent negative effects on ribosome biogenesis. Accordingly, this report provides evidence that La may coordinate certain aspects of the production of the translational machinery under some conditions (27).
Acknowledgments
We thank SMCB members for discussion and comments, L. Pinna for providing TBB, M. Avantaggiati for advice and discussion, and members of Alan Hinnebusch's laboratory at NICHD for advice and assistance with polysome profile analysis.
R.J.M. received support from the Commissioned Corps of the U.S.P.H.S.
REFERENCES
- 1.Ahmed, K., D. A. Gerber, and C. Cochet. 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol. 12:226-230. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, C., D. Sanfelice, J. Babon, G. Kelly, A. Jacks, S. Curry, and M. R. Conte. 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 14:7-12. [DOI] [PubMed] [Google Scholar]
- 3.Ali, N., G. J. Pruijn, D. J. Kenan, J. D. Keene, and A. Siddiqui. 2000. Human La antigen is required for the hepatitis C virus internal ribosome entry site (IRES)-mediated translation. J. Biol. Chem. 275:27531-27540. [DOI] [PubMed] [Google Scholar]
- 4.Ayukawa, K., S. Taniguchi, J. Masumoto, S. Hashimoto, H. Sarvotham, A. Hara, T. Aoyama, and J. Sagara. 2000. La autoantigen is cleaved in the COOH terminus and loses the nuclear localization signal during apoptosis. J. Biol. Chem. 275:34465-34470. [DOI] [PubMed] [Google Scholar]
- 5.Battistutta, R., E. De Moliner, S. Sarno, G. Zanotti, and L. A. Pinna. 2001. Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole. Protein Sci. 10:2200-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broekhuis, C. H., G. Neubauer, A. van der Heijden, M. Mann, C. G. Proud, W. J. van Venrooij, and G. J. Pruijn. 2000. Detailed analysis of the phosphorylation of human La (SS-B) autoantigen. (De)phosphorylation does not affect subcellular distribution. Biochemistry 39:3023-3033. [DOI] [PubMed] [Google Scholar]
- 8.Buchou, T., M. Vernet, O. Blond, H. H. Jensen, H. Pointu, B. B. Olsen, C. Cochet, O. G. Issinger, and B. Boldyreff. 2003. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell. Biol. 23:908-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardinali, B., C. Carissimi, P. Gravina, and P. Pierandrei-Amaldi. 2003. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J. Biol. Chem. 278:35145-35151. [DOI] [PubMed] [Google Scholar]
- 10.Chen, M., and J. A. Cooper. 1997. The beta subunit of CKII negatively regulates Xenopus oocyte maturation. Proc. Natl. Acad. Sci. USA 94:9136-9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosio, C., P. P. Boyl, F. Loreni, P. Pierandrei-Amaldi, and F. Amaldi. 2000. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 28:2927-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, G., G. Chakshusmathi, S. L. Wolin, and K. M. Reinisch. 2004. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 23:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelman, A. M., D. K. Blumenthal, and E. G. Krebs. 1987. Protein serine/threonine kinases. Annu. Rev. Biochem. 56:567-613. [DOI] [PubMed] [Google Scholar]
- 14.Fan, H., J. L. Goodier, J. Chamberlain, D. R. Engelke, and R. J. Maraia. 1998. 5′ Processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell. Biol. 18:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, H., A. L. Sakulich, J. L. Goodier, X. Zhang, J. Qin, and R. J. Maraia. 1997. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell 88:707-715. [DOI] [PubMed] [Google Scholar]
- 16.Glover, C. V., III. 1998. On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 59:95-133. [DOI] [PubMed] [Google Scholar]
- 17.Grimm, C., E. Lund, and J. E. Dahlberg. 1997. In vivo selection of RNAs that localize in the nucleus. EMBO J. 16:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra, B., and O. G. Issinger. 1999. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 20:391-408. [DOI] [PubMed] [Google Scholar]
- 19.Holcik, M., and R. G. Korneluk. 2000. Functional characterization of the X-Linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 20:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inada, M., and C. Guthrie. 2004. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl. Acad. Sci. USA 101:434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Intine, R. V., M. Dundr, T. Misteli, and R. J. Maraia. 2002. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol. Cell. 9:1113-1123. [DOI] [PubMed] [Google Scholar]
- 22.Intine, R. V., S. A. Tenenbaum, A. S. Sakulich, J. D. Keene, and R. J. Maraia. 2003. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol. Cell. 12:1301-1307. [DOI] [PubMed] [Google Scholar]
- 23.Intine, R. V. A., A. L. Sakulich, S. B. Koduru, Y. Huang, E. Pierstorrf, J. L. Goodier, L. Phan, and R. J. Maraia. 2000. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol. Cell. 6:339-348. [DOI] [PubMed] [Google Scholar]
- 24.Isoyama, T., N. Kamoshita, K. Yasui, A. Iwai, K. Shiroki, H. Toyoda, A. Yamada, Y. Takasaki, and A. Nomoto. 1999. Lower concentration of La protein required for internal ribosome entry on hepatitis C virus RNA than on poliovirus RNA. J. Gen. Virol. 80:2319-2327. [DOI] [PubMed] [Google Scholar]
- 25.Jacks, A., J. Babon, G. Kelly, I. Manolaridis, P. D. Cary, S. Curry, and M. R. Conte. 2003. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure (Cambridge) 11:833-843. [DOI] [PubMed] [Google Scholar]
- 26.Johnston, I. M., S. J. Allison, J. P. Morton, L. Schramm, P. H. Scott, and R. J. White. 2002. CK2 forms a stable complex with TFIIIB and activates RNA polymerase III transcription in human cells. Mol. Cell. Biol. 22:3757-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenan, D. J., and J. D. Keene. 2004. La gets its wings (news and views). Nat. Struct. Mol. Biol. 11:303-305. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y. K., S. H. Back, J. Rho, S. H. Lee, and S. K. Jang. 2001. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 29:5009-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koopman, G., C. P. Reutelingsperger, G. A. Kuijten, R. M. Keehnen, S. T. Pals, and M. H. van Oers. 1994. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84:1415-1420. [PubMed] [Google Scholar]
- 30.Kufel, J., C. Allmang, G. Chanfreau, E. Petfalski, D. L. Lafontaine, and D. Tollervey. 2000. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20:5415-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakin, N. D., and S. P. Jackson. 1999. Regulation of p53 in response to DNA damage. Oncogene 18:7644-7655. [DOI] [PubMed] [Google Scholar]
- 32.Madore, S. J., E. D. Wieben, and T. Pederson. 1984. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J. Biol. Chem. 259:1929-1933. [PubMed] [Google Scholar]
- 33.Maraia, R. J., and R. V. Intine. 2002. La protein and its associated small nuclear and nucleolar precursor RNAs. Gene Exp. 10:41-57. [PMC free article] [PubMed] [Google Scholar]
- 34.Maraia, R. J., and R. V. Intine. 2001. Recognition of nascent RNA by the human La antigen: conserved and diverged features of structure and function. Mol. Cell. Biol. 21:367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyuhas, O., and E. Hornstein. 2000. Translational control of TOP mRNAs, p. 671-693. In N. Sonenberg, J. W. B. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Ohndorf, U. M., C. Steegborn, R. Knijff, and P. Sondermann. 2001. Contributions of the individual domains in human La protein to its RNA 3′-end binding activity. J. Biol. Chem. 276:27188-27196. [DOI] [PubMed] [Google Scholar]
- 38.Pellizzoni, L., B. Cardinali, N. Lin-Marq, D. Mercanti, and P. Pierandrei-Amaldi. 1996. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5′ UTR of ribosomal protein mRNAs in vitro: implication of a protein factor in complex formation. J. Mol. Biol. 259:904-915. [DOI] [PubMed] [Google Scholar]
- 39.Pepperkok, R., P. Lorenz, R. Jakobi, W. Ansorge, and W. Pyerin. 1991. Cell growth stimulation by EGF: inhibition through antisense-oligodeoxynucleotides demonstrates important role of casein kinase II. Exp. Cell Res. 197:245-253. [DOI] [PubMed] [Google Scholar]
- 40.Pfeifle, J., F. A. Anderer, and M. Franke. 1987. Multiple phosphorylation of human SS-B/La autoantigen and its effect on poly(U) and autoantibody binding. Biochim. Biophys. Acta 928:217-226. [DOI] [PubMed] [Google Scholar]
- 41.Pinna, L. A. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873-3878. [DOI] [PubMed] [Google Scholar]
- 42.Pinna, L. A. 2003. The raison d'etre of constitutively active protein kinases: the lesson of CK2. Acc. Chem. Res. 36:378-384. [DOI] [PubMed] [Google Scholar]
- 43.Ray, P. S., and S. Das. 2002. La autoantigen is required for the internal ribosome entry site-mediated translation of Coxsackievirus B3 RNA. Nucleic Acids Res. 30:4500-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutjes, S. A., P. J. Utz, A. der Heijden, C. Broekhuis, W. J. van Venrooij, and G. J. Pruijn. 1999. The La (SS-B) autoantigen, a key protein in RNA biogenesis, is dephosphorylated and cleaved early during apoptosis. Cell Death Differ. 6:976-986. [DOI] [PubMed] [Google Scholar]
- 45.Ruzzene, M., D. Penzo, and L. A. Pinna. 2002. Protein kinase CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) induces apoptosis and caspase-dependent degradation of haematopoietic lineage cell-specific protein 1 (HS1) in Jurkat cells. Biochem. J. 364:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarno, S., H. Reddy, F. Meggio, M. Ruzzene, S. P. Davies, A. Donella-Deana, D. Shugar, and L. A. Pinna. 2001. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (“casein kinase-2”). FEBS Lett. 496:44-48. [DOI] [PubMed] [Google Scholar]
- 47.Shimazaki, T., M. Honda, S. Kaneko, and K. Kobayashi. 2002. Inhibition of internal ribosomal entry site-directed translation of HCV by recombinant IFN-alpha correlates with a reduced La protein. Hepatology 35:199-208. [DOI] [PubMed] [Google Scholar]
- 48.Smith, C. M., and J. A. Steitz. 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 18:6897-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sosic, D., J. A. Richardson, K. Yu, D. M. Ornitz, and E. N. Olson. 2003. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell 112:169-180. [DOI] [PubMed] [Google Scholar]
- 50.Svitkin, Y. V., L. P. Ovchinnikov, G. Dreyfuss, and N. Sonenberg. 1996. General RNA binding proteins render translation cap dependent. EMBO J. 15:7147-7155. [PMC free article] [PubMed] [Google Scholar]
- 51.Szyszka, R., N. Grankowski, K. Felczak, and D. Shugar. 1995. Halogenated benzimidazoles and benzotriazoles as selective inhibitors of protein kinases CK I and CK II from Saccharomyces cerevisiae and other sources. Biochem. Biophys. Res. Commun. 208:418-424. [DOI] [PubMed] [Google Scholar]
- 52.Trotta, R., T. Vignudelli, L. Pecorari, R. V. Intine, C. Guerzoni, G. Santilli, O. Candini, M. W. Byrom, S. Goldoni, L. P. Ford, M. A. Caligiuri, R. Maraia, et al. 2003. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell I 3:145-160. [DOI] [PubMed] [Google Scholar]
- 53.Van Horn, D. J., C. J. Yoo, D. Xue, H. Shi, and S. L. Wolin. 1997. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 3:1434-1443. [PMC free article] [PubMed] [Google Scholar]
- 54.Vermes, I., C. Haanen, H. Steffens-Nakken, and C. Reutelingsperger. 1995. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J. Immunol. Methods 184:39-51. [DOI] [PubMed] [Google Scholar]
- 55.Wolin, S. L., and T. Cedervall. 2002. The La protein. Annu. Rev. Biochem. 71:375-403. [DOI] [PubMed] [Google Scholar]
- 56.Xue, D., D. A. Rubinson, B. K. Pannone, C. J. Yoo, and S. L. Wolin. 2000. U snRNP assembly in yeast involves the La protein. EMBO J. 19:1650-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo, C. J., and S. L. Wolin. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89:393-402. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, J., A. Hayakawa, T. Kakegawa, and R. L. Kaspar. 2001. Binding of the La autoantigen to the 5′ untranslated region of a chimeric human translation elongation factor 1A reporter mRNA inhibits translation in vitro. Biochim. Biophys. Acta 1521:19-29. [DOI] [PubMed] [Google Scholar]