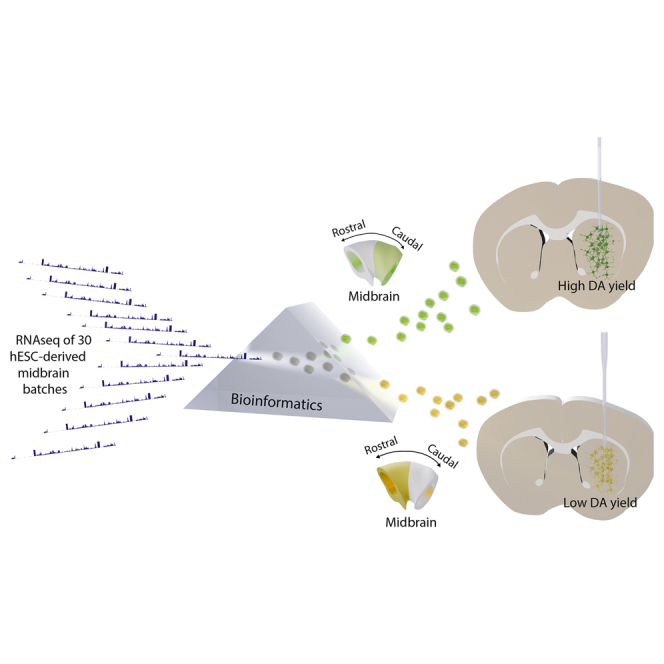
Summary
Stem cell treatments for neurodegenerative diseases are expected to reach clinical trials soon. Most of the approaches currently under development involve transplantation of immature progenitors that subsequently undergo phenotypic and functional maturation in vivo, and predicting the long-term graft outcome already at the progenitor stage remains a challenge. Here, we took an unbiased approach to identify predictive markers expressed in dopamine neuron progenitors that correlate with graft outcome in an animal model of Parkinson’s disease through gene expression analysis of >30 batches of grafted human embryonic stem cell (hESC)-derived progenitors. We found that many of the commonly used markers did not accurately predict in vivo subtype-specific maturation. Instead, we identified a specific set of markers associated with the caudal midbrain that correlate with high dopaminergic yield after transplantation in vivo. Using these markers, we developed a good manufacturing practice (GMP) differentiation protocol for highly efficient and reproducible production of transplantable dopamine progenitors from hESCs.
Keywords: Parkinson’s disease, human embryonic stem cells, hESCs, dopaminergic neurons, diencephalon, transplantation, subthalamic nucleus, STN, ventral midbrain, VM, midbrain-hindbrain boundary, MHB, FGF8, good manufacturing practice, GMP
Graphical Abstract
Highlights
-
•
Common markers of hESC-derived DA progenitors correlate poorly with graft outcome
-
•
RNA-seq reveals correlation of caudal midbrain markers with DA neuron yield in vivo
-
•
Staged application of FGF8b patterns hESC-derived progenitors to a caudal VM fate
-
•
GMP adaptation of differentiation using this approach improves transplant outcome
Kirkeby et al. show that identification and application of a set of predictive markers can help refine differentiation protocols and improve transplant outcome in a preclinical model for hESC-based treatment of Parkinson’s disease.
Introduction
Stem cell-based therapies for neurological disorders are rapidly moving toward clinical trials (Steinbeck and Studer, 2015). One major task en route to the clinic is to generate standardized GMP-grade human pluripotent stem cell (hPSC)-derived neural progenitors that will mature and function in the adult brain after transplantation. Developing meaningful quality control assays for the functionality of such clinical grade human neural stem cells poses a major hurdle since the cells are transplanted as immature progenitors and functional maturation and integration of the neurons require months in vivo. Therefore, the transplanted cells cannot be assessed for proper functionality at the progenitor stage prior to transplantation. As a surrogate for markers predicting functional properties, the field has relied on assessing the transplanted progenitors for expression of genes and proteins developmentally linked to the generation of specific neuronal subtypes. However, it is uncertain whether these markers are predictive of terminal differentiation and functional maturation, and the progenitors must therefore be taken through lengthy in vivo functional assessment to control for batch-to-batch variation and therapeutic efficacy. Identification of novel markers that can accurately predict the in vivo performance of stem cell products already at the progenitor stage in vitro would greatly reduce the number of in vivo studies to be performed during the cell development process, and it would also open up the production of standardized cell products from patient-specific cells in personalized medicine.
Parkinson’s disease (PD) is a particularly interesting target for stem cell-based therapies due to the relatively focal degeneration of a specific type of mesencephalic dopamine (mesDA) neuron, and fetal cell transplantation for PD has already been investigated in several clinical trials (Barker et al., 2015). Recent developments have resulted in a better understanding of the developmental and cellular ontogeny of mesDA neurons derived from the floor plate of the ventral mesencephalon (VM) (reviewed in Arenas et al., 2015), and this knowledge has materialized into differentiation protocols giving rise to VM progenitors from hPSCs that mature and function after transplantation (Doi et al., 2014, Kirkeby et al., 2012, Kriks et al., 2011, Chen et al., 2016). In current VM differentiation protocols, mesencephalic floor plate markers LMX1A, FOXA2, and OTX2 are commonly used to confirm the mesDA identity of progenitors in vitro prior to grafting (Doi et al., 2014, Jaeger et al., 2011, Kirkeby et al., 2012, Kriks et al., 2011). However, a new study has revealed that these and several other commonly used VM markers are also co-expressed in diencephalic progenitors of the subthalamic nucleus (STN) and therefore do not exclusively identify the mesDA domain (Kee et al., 2016).
In this study we took an unbiased approach to identify in vitro markers that can predict successful graft outcome in vivo of VM progenitors by sequencing more than 30 batches of human embryonic stem cell (hESC)-derived VM patterned cells with known graft outcomes. We found that expression of LMX1A and FOXA2 is necessary for generation of dopamine (DA) neurons after transplantation, but the levels of LMX1A, FOXA2, or CORIN did not correlate positively with the number of DA neurons in the grafts. Instead, successful in vivo outcome correlated with the expression of a set of markers enriched in the caudal part of the VM, highlighting the need for finer control of rostro-caudal patterning within VM differentiation protocols. To achieve this, we used timed delivery of FGF8b, which resulted in more precise control of the rostro-caudal patterning of VM progenitors and enabled control of differentiation toward either the rostral STN or caudal mesDA progenitor domains. Furthermore, we applied the new predictive markers to develop a good manufacturing practice (GMP) differentiation protocol for highly efficient and reproducible production of mesDA progenitors from hESCs. These cells gave rise to DA-rich grafts with extensive host brain innervation and provided functional recovery in a rat model of PD.
Results
Common In Vitro mesDA Markers Correlate Poorly with Dopaminergic Maturation In Vivo
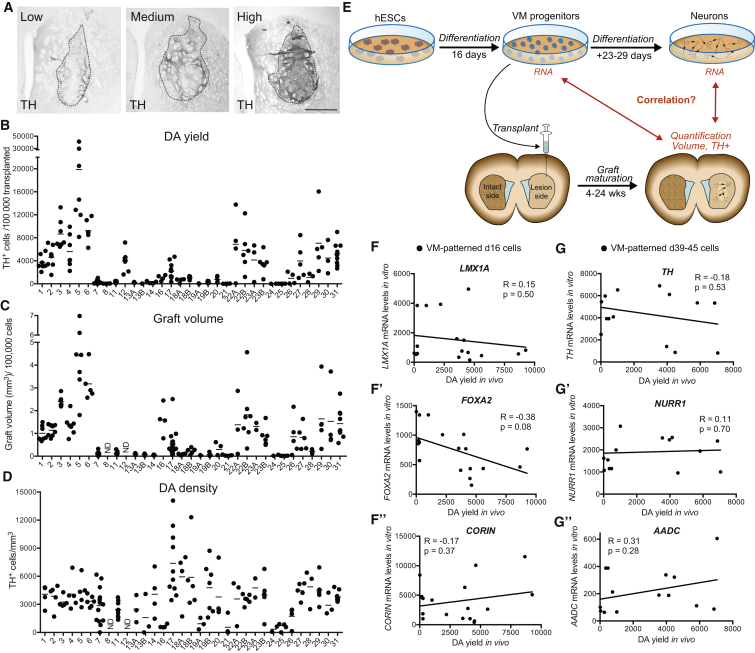
We have over the past 6 years transplanted >500 rats intracerebrally with >30 different batches of hESC-derived mesDA progenitors (Table S1). In all experiments, cells were differentiated for 16 days (d16) in vitro and transplanted into unilateral 6-OHDA lesioned rats. Although all VM-patterned cell batches were routinely assessed for high co-expression of the VM markers LMX1A, FOXA2, and OTX2 prior to grafting (≥80%), we found the in vivo outcome in terms of graft size and number of DA neurons to vary considerably between experiments (Figure 1A). To determine the level of batch-to-batch variability in in vivo DA neuron differentiation in these experiments, we quantified the total number of tyrosine hydroxylase-positive (TH+) neurons per 100,000 cells grafted (DA yield), graft volume (mm3), and DA density (TH+/mm3) for all transplants. This revealed a considerable interexperimental variability (Figures 1B–1D), highlighting a need for new markers that better predict in vivo DA differentiation and yield after grafting.
Figure 1.
VM-Patterned Batches of hESCs Result in Variable Transplantation Outcome that Is Not Correlated with Common mesDA Markers
(A) We observed a variability in graft size and number of DA neurons after in vivo maturation. Scale bar, 500 μm.
(B–D) Quantifications of (B) DA yield in individual animals from each experiment (TH+ cells per 100,000 grafted cells), (C) graft volume in individual animals based on immunostaining for human nuclei normalized to 100,000 grafted cells, and (D) DA density in grafts from individual animals (TH+ cells/mm3). See Table S1 for details on experiments.
(E) Schematic summary of study outline.
(F–F″) Mean DA yield in each graft experiment plotted versus gene expression of FOXA2, LMX1A, and CORIN in the transplanted cell population on day of transplantation (d16).
(G–G″) Mean DA yield in each graft experiment plotted versus gene expression of TH, NR4A2 (NURR1), and DDC (AADC) in the transplanted cell population after 39–45 days of in vitro maturation.
Results from Spearman correlation analysis are given as R and p values in each graph and tendencies of correlation are shown by linear regression lines. All mRNA values are shown as fold change relative to undifferentiated hESCs. Bars in (B)–(D) = mean.
We next investigated to what degree commonly used mesDA progenitor markers in vitro predicted DA yield in grafts after maturation in vivo by analyzing RNA samples collected from each individual cell batch, all containing ≥80% FOXA2/LMX1A co-expressing cells on the day of transplantation, and also from the same cells analyzed after maturation in vitro (Figure 1E). We found that FOXA2, LMX1A, and CORIN gene expression levels at the time of transplantation did not correlate significantly with DA yield in the grafts (Figure 1F). This suggests that within the FOXA2/LMX1A co-expressing progenitor cells, additional markers are needed to predict in vivo outcome.
Since terminal differentiation and assessment of postmitotic DA markers are often used to assess DAergic potential of stem cells in vitro, we investigated whether extended in vitro maturation of the progenitors into neurons for 39–45 days reflected their corresponding in vivo maturation posttransplantation. In experiments where the cells used for grafting had also been subjected to parallel terminal differentiation in vitro, we found that expression levels of DA markers TH, NR4A2 (NURR1), and DDC (AADC) did not show any statistically significant correlation to the DA yield after transplantation (Figure 1G).
RNA Sequencing Analysis Reveals that Markers of the Caudal VM Are Associated with Higher DA Yield in Vivo
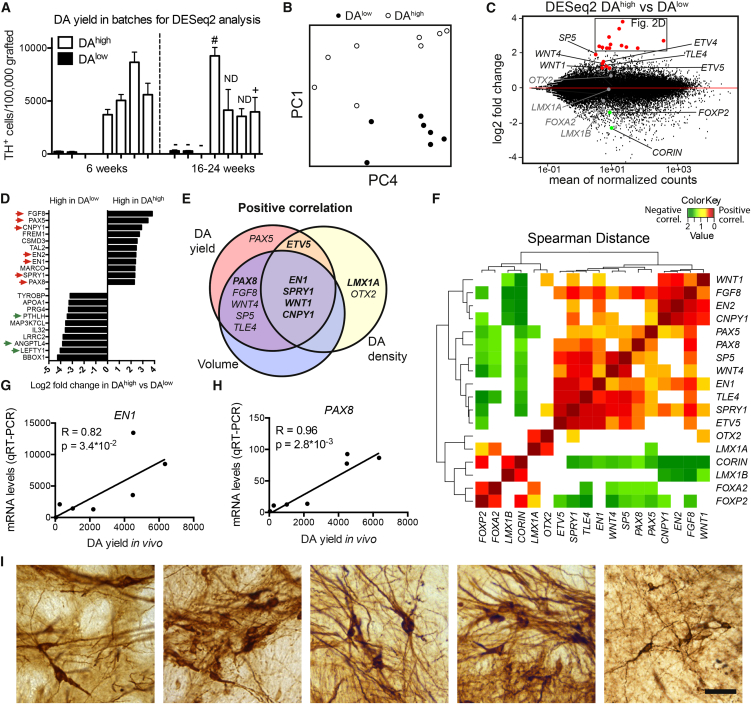
To enable an unbiased search for potential markers that positively correlate with DA yield after transplantation, we performed global gene expression profiling of cell samples collected at the day of transplantation, using RNA sequencing (RNA-seq). Experiments with d16 RNA samples were classified as giving rise to either high or low DA yield after transplantation (DAhigh with >3000 and DAlow with <500 TH+ neurons per 100,000 grafted cells, Figure 2A). Unbiased principal component analysis (PCA), including all selected RNA-seq samples, was performed to determine whether the DAhigh and DAlow cell batches could be identified by distinct gene expression profiles (Figure 2B). We observed a clustering of the DAhigh samples on the positive PC1 axis, which contained genes such as FGF8, PAX5, EN2, and CNPY1, all of which have been shown to be important for midbrain–hindbrain boundary (MHB) formation in the caudal VM (Figures 2B and S1A; Table S3). As there were clearly distinct expression profiles between the groups, we next used DESeq2 analysis to identify all differentially expressed genes between the DAhigh and DAlow samples. Interestingly, among the top ranked genes in this analysis, we again found a high representation of genes expressed in the caudal VM and MHB region to be positively associated with DA yield in vivo, i.e., PAX5, FGF8, SPRY1, EN1/2, SP5, ETV4/5, CNPY1, TLE4, and WNT1 (Figures 2C, 2D, and S1B; Tables S2 and S3). In contrast, markers of different stages of neuronal maturation were not differentially expressed between the DAhigh and DAlow group, indicating that the distinct graft outcomes between the two groups were not caused by differences in cell maturity at the time of transplantation (Figure S1D).
Figure 2.
RNA-Seq Analysis of Transplanted VM-Patterned Progenitors Reveals a Positive Correlation between DA Yield and Markers of the Caudal VM
(A) For unbiased gene expression analysis, graft experiments were divided into DAhigh and DAlow groups based on the total number of TH+ cells in the grafts. DAergic function of the grafts was assessed in grafts with long-term maturation (>16 weeks) through amphetamine-induced rotation (“−”, lack of functional recovery; “+”, functional recovery; ND, not determined) or through functional positron emission tomography (PET) imaging (#). Data are presented as mean ± SEM.
(B) PCA plot of results from RNA-seq analysis of d16 cell samples from (A) revealed clustering of DAlow and DAhigh samples on the PC1 axis.
(C) Scatter plot showing fold changes of individual genes in the DESeq2 analysis. Selected genes have been color coded and labeled. Red, higher expression in DAhigh samples (adj. p < 0.01); gray, no significant difference; green, higher expression in DAlow samples (adj. p < 0.01).
(D) Graph showing the fold changes of the top and bottom differentially expressed genes from the DESeq2 analysis comparing the DAhigh versus DAlow cell batches. Genes related to the MHB and the diencephalic domains are marked by red and green arrows, respectively.
(E) Selected gene hits were validated through direct correlation analysis. RNA levels of MHB genes and common VM markers in d16 transplanted cell batches were correlated to graft outcome, and the Venn diagram shows positive correlations defined by Spearman correlation analysis with p < 0.05 toward DA yield, DA density, and graft volume. Correlations verified by qRT-PCR are shown in boldface type. See Figure S1 and Table S2 for a complete dataset.
(F) Spearman distance analysis of RNA levels in 29 VM cell batches shows co-regulation of MHB genes, whereas LMX1A, OTX2, FOXA2, and CORIN are uncoupled or negatively coupled to the MHB gene cluster. Color coding labels only statistically significant correlations (p < 0.05).
(G and H) An unrelated set of grafting experiments validated the predictive power of key genes EN1 and PAX8 through direct correlation of mRNA levels measured by qRT-PCR of EN1 (G) and PAX8 (H) and DA yield in vivo. Results from Spearman correlation analysis are given as R and p values in each graph and correlations are visualized with linear regression lines.
(I) Representative images of TH+ neurons from five different cell batches with high expression of predictive markers reveal mature A9-like morphology of the grafted cells. Scale bar, 50 μm.
To assess the predictive value of these markers, we next performed direct Spearman correlation analysis between graft outcome and the RNA expression levels of selected genes from the PCA and DESeq2 analyses, comparing these to correlations of meso-diencephalic markers commonly used to monitor DA differentiation in vitro and in vivo (LMX1A, LMX1B, FOXA2, FOXP2, CORIN, OTX2) (Arenas et al., 2015, Kirkeby et al., 2012, Kriks et al., 2011). We correlated the genes toward graft size, DA yield, and DA density, and to validate the RNA-seq dataset, we also assessed a subset of gene correlations by qRT-PCR analysis in the same samples. These analyses showed that most of the caudal VM markers identified by DESeq2 (Figures 2C and 2D) indeed showed direct positive correlation with graft size and DA yield of the grafts (Figures 2E and S1C). A set of key markers, i.e., EN1, SPRY1, WNT1, ETV5, and CNPY1, also correlated positively with DA density (Figures 2E and S1C). This was in contrast to the broader and more widely expressed VM markers FOXA2, LMX1B, CORIN, FOXP1, and FOXP2, which showed negative correlation to both graft size and DA yield (Figure S1C), whereas LMX1A and OTX2 showed positive correlations only to DA density, but not to DA yield (Figures 2E and S1C). Other VM markers reported to be involved in mesDA neurogenesis, namely SOX6 and PBX1, did not show any correlations to DA yield or density of the grafts when analyzed in d16 VM-patterned progenitors (Figure S1C).
Next, we investigated whether the genes identified formed part of a co-regulatory network by performing Spearman correlation analysis of the expression values for each candidate gene toward the others in 29 batches of VM-patterned cells. Through hierarchical clustering of the Spearman distance between each gene, we found that all of the MHB genes associated with a high DA yield showed a pattern of co-regulation (Figure 2F). Interestingly, LMX1A and OTX2 showed little or no positive co-regulation with these genes, and CORIN, LMX1B, FOXA2, and FOXP2 were negatively correlated with several of the caudal genes, indicating that these commonly used markers are not specifically associated with a caudal VM phenotype in hESC-derived DA progenitors.
To investigate whether the newly identified markers reliably predicted a high yield of mature DA neurons after transplantation in vivo, we analyzed a separate set of seven additional transplantation experiments (not included in the DESeq2 analysis) and correlated the data to qRT-PCR analysis of the transplanted cell batches. In this independent dataset, we confirmed that the expression of key markers EN1 and PAX8 correlated positively with DA yield after transplantation (Figures 2G and 2H), thereby validating our findings from the DESeq2 analysis. Finally, we also verified that cell preparations with high expression of the predictive markers gave rise to grafts containing DA neurons with a mature A9 morphology (Figure 2I).
VM-Patterned Differentiations and Mature Grafts Contain Cells of the STN Lineage
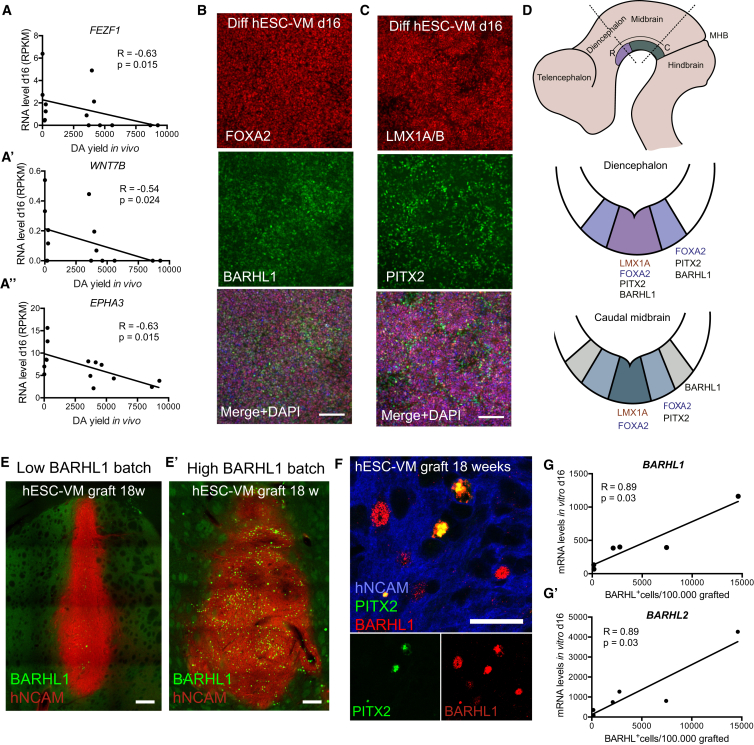
We concluded from our RNA-seq analyses that caudal VM markers correlated positively with high DA yield while expression of many classical DA neuron progenitor markers did not. Furthermore, we found that several of the most highly enriched genes in the DAlow group identified by PCA and DESeq2 analysis (LEFTY1, LEFTY2, ANGPTL4, PTHLH, and NPW) are expressed in the diencephalon and/or in structures derived from the diencephalon (Figures 2D and S1A; Table S3). A recent study (Kee et al., 2016) shows that the diencephalic STN neuron lineage develops immediately rostral to mesDA neurons and that the two are remarkably closely related, with both lineages co-expressing many genes that until now have been used as mesDA markers, including Lmx1a, Lmx1b, Foxa1, and Foxa2. Thus, we speculated that markers of the abutting STN domain might be informative of graft outcome, and RNA-seq expression values indeed revealed negative correlations between the diencephalic marker FEZF1 as well as the rostral STN markers EPHA3 and WNT7B and DA yield in grafts (Figure 3A). This led us to investigate whether some batches of VM-patterned hESCs contained STN progenitors derived from the rostral LMX1+/FOXA2+ domain, which in both mouse and human is characterized by the co-expression of BARHL1 and PITX2 (Figures 3D and S2A–S2F; Kee et al., 2016). We detected the presence of both BARHL1+ and PITX2+ cells in our differentiated cell batches on d16. The BARHL1+ cells could be either FOXA2+ or FOXA2−, and the PITX2+ cells could be LMX1A/B+ or LMX1A/B− (Figures 3B and 3C), indicating the presence of both ventral diencephalic cells and lateral mesencephalic populations in our differentiated progenitor batches (Figures 3D and S2A–S2F). We further found the presence of FOXA2+/BARHL1+/MAP2+ neurons and PITX2+/LMX1A/B+ cells in our terminally differentiated hESC cultures, indicating the differentiation of these progenitors into STN fates (Figures S2G and S2H). In animals grafted with VM-patterned cells we also detected a variable presence of BARHL1+ cells (Figures 3E and 3E′), and these cells could be either PITX2+ indicating ventral diencephalic STN fates or PITX2− indicating other lateral fates (Figure 3F). To investigate whether these markers could be used to predict the amount of non-dopaminergic lateral VM cells, as well as rostro-ventral contaminating STN cells in our grafts, we quantified the numbers of BARHL1+ cells in several of our graft experiments. Although BARHL1/2 as single markers were not predictive of graft outcome (DA yield/density or graft volume), correlation analysis showed that the density of BARHL1+ cells in the grafts indeed correlated with the expression levels of BARHL1 and BARHL2 in vitro in the differentiated cell batches at the day of transplantation (Figures 3G and 3G′). This implies that BARHL1 and BARHL2 in progenitor cultures can be used as markers to quantitatively identify several commonly occurring rostral diencephalic as well as lateral mesencephalic contaminating populations in VM-patterned hESCs both in vitro and in vivo.
Figure 3.
VM-Patterned hESC Cultures Contain Cells of Diencephalic STN Identity
(A–A″) Negative correlation between RNA levels of the diencephalic markers FEZF1, WNT7B, and EPHA3 in transplanted cells (d16) and DA yield in grafts.
(B and C) Immunostainings of VM-patterned hESC cultures (d16) reveal the presence of STN domain fates (BARHL1+/FOXA2+ and PITX2+/LMX1A/B+ cells). See Figure S2 for mature neuronal cultures.
(D) Schematic overview of expression domains of PITX2 and BARHL1 in the diencephalic STN region and in lateral midbrain domains based on data in Kee et al. (2016) and Figure S2.
(E and E′) Examples of BARHL1+ cell content in grafts derived from cell batches with low (E) or high (E′) BARHL1 RNA levels at the day of transplantation. Images are digitally stitched from multiple high-magnification images.
(F) Confocal imaging of 18-week-old grafts showing the presence of BARHL1+/PITX2+ and BARHL1+/PITX2− cells.
(G and G′) Graphs showing positive correlations between BARHL1 (G) and BARHL2 (G′) RNA levels at the time of transplantation (qRT-PCR) and the number of BARHL1+ cells in the mature grafts.
Results from Spearman correlation analysis are given as R and p values in each graph, and correlations are visualized with linear regression lines (A and G). Scale bars, 100 μm (B and C), 200 μm (E and E′), and 25 μm (F).
Patterning of VM Progenitors toward a Caudal DA Fate Can Be Controlled by Time-Dependent Exposure to FGF8b
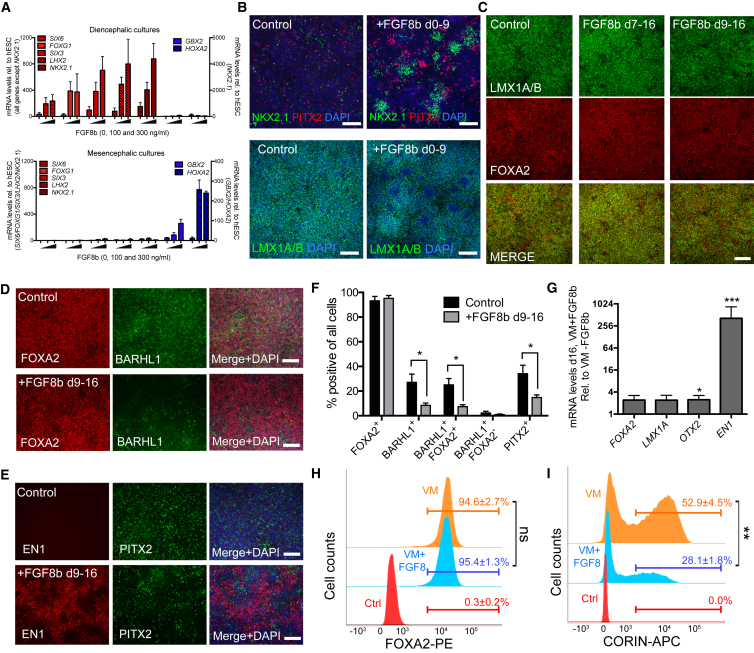
Given the variable outcome of VM-patterned hESC progenitors, we aimed to optimize and fine-tune patterning of our differentiation protocol toward the caudal DA domain of the VM, as markers of this domain correlated with high DA yield in vivo. Cells located in the caudal VM are in proximity to the MHB, and development of mesDA progenitors in this region depends on the activity of FGF8, a growth factor that is secreted from the MHB (Joyner et al., 2000). In addition, high expression of FGF8 was identified to correlate to the DAhigh group in our unbiased gene expression analyses (Figure 2D). We therefore investigated the effect of exogenous FGF8b on cultures patterned toward either ventral diencephalic (CHIR99021 [CHIR] = 0.4 μM) or ventral mesencephalic (CHIR = 0.8 μM) fates. We found that the addition of FGF8b to the differentiation medium during the early phase of neural induction and patterning (d0–d9) induced significant upregulation of forebrain markers FOXG1, SIX3, SIX6, LHX2, and NKX2-1 in diencephalic-patterned cultures, whereas hindbrain markers HOXA2 and GBX2 were upregulated in mesencephalic-patterned cultures (Figure 4A). This indicated that early exposure to FGF8b caused contamination of the cultures with several non-VM progenitor fates, and this was reflected as more heterogeneous VM cultures with clusters of NKX2.1+ and PITX2+ progenitors and patches of LMX1A/B− cells (Figure 4B). In contrast, the FOXA2+/LMX1A/B+ phenotype of the cells was maintained if FGF8b was added after the initial patterning toward VM was completed (from day 7 or 9, Figure 4C). Under these conditions, addition of 100 ng/mL FGF8b caused caudalization of the VM progenitors, which we monitored by EN1 mRNA and protein in cultures treated with FGF8b from d9 to d16 (Figures 4E and 4G). Treatment with FGF8b from d9 to d16 further caused a significant decrease in the percentage of BARHL1+/FOXA2+ and PITX2+ STN progenitors (Figures 4D–4F), indicating that late addition of FGF8 to the cultures shifted progenitor fates from anterior VM and STN fates toward caudal VM mesDA progenitor fates. This caudalization was optimal at 100 ng/mL FGF8b and increased EN1 expression was maintained >20 days after FGF8b removal (Figure S3). While the addition of FGF8b from d9 to d16 did not affect the percentage of FOXA2+ progenitors, the number of CORIN+ progenitors was reduced by 47% (Figures 4H and 4I), in accordance with the negative correlations found for CORIN in our RNA-seq analyses (Figures 2C, 2F, and S1C), indicating that CORIN expression is more strongly associated with a rostral VM and/or diencephalic fate rather than a caudal mesDA progenitor fate in hESC-derived progenitors.
Figure 4.
Timed Delivery of FGF8b Causes Fate Switch from Diencephalic to Caudal VM Progenitors
(A) qRT-PCR analysis (d10) of differentiating hESCs shows that treatment with FGF8b during VM patterning from d0 to d9 induces increased expression of forebrain markers (red) and hindbrain markers (blue) in ventral diencephalic (CHIR = 0.4 uM) and ventral mesencephalic (CHIR = 0.8 uM) cultures, respectively.
(B) Immunostainings (d16) reveal patches of PITX2+ and NKX2.1+ cells and patches of LMX1A/B− cells in VM cultures treated with FGF8b from d0 to d9.
(C) LMX1A/B+/FOXA2+ VM phenotype was maintained in cultures treated with FGF8b from d7 to d16 or d9 to d16.
(D–G) d16 cultures treated with FGF8b from d9 to d16 have decreased levels of BARHL1+ cells (D, quantified in F, n = 3) and PITX2+ cells (E, quantified in F, n = 3] and increased expression of EN1 (immunostaining in E and qRT-PCR in G).
(H and I) FACS plots of control and VM-patterned cultures with and without FGF8b treatment (d16) show unchanged percentages of FOXA2+ progenitors and (I) decreased percentages of CORIN+ progenitors. (H) and (I) show representative FACS plots with mean % ± SEM of replicate experiments (n = 3).
All scale bars, 100 μm. For all bar graphs, data are presented as mean ± SEM.
Development of a High-Yield GMP-Adapted Differentiation Protocol on Laminin-111 Matrix
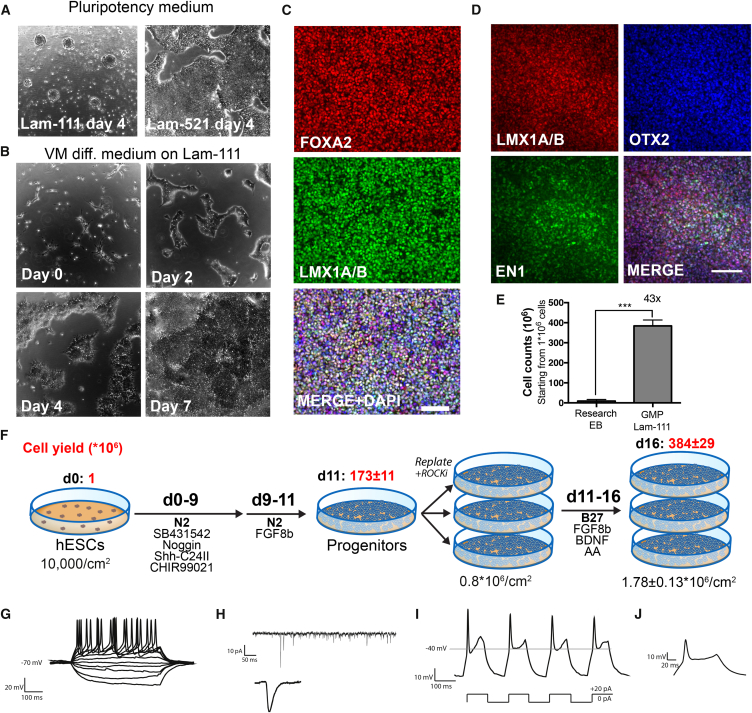
With a better understanding and control of the DA and STN differentiation, as well as new progenitor markers predicting DA yield in mature grafts, we set out to develop a GMP-compatible differentiation protocol for high and reproducible yield of caudalized mesDA VM progenitors, using the fully GMP-derived hESC line RC17. Previous research-grade VM differentiation protocols have implemented steps of embryoid body (EB) formation (Kirkeby et al., 2012) or culturing on matrigel (Kriks et al., 2011)—both of which pose problems in GMP adaptation due to difficulties in reproducibility and the content of an undefined animal-derived component, respectively. Recently various full-length recombinant laminin isoforms suitable for GMP production have been generated and successfully implemented for derivation and differentiation of hPSCs (Cameron et al., 2015, Rodin et al., 2014). We tested seven different laminin isoforms and found that four of them (Lam-111, Lam-421, Lam-511, and Lam-521) efficiently supported adherent differentiation of VM progenitors (Figures S4A and S4B). Whereas Lam-511 and Lam-521 efficiently support growth of hPSCs (Rodin et al., 2014), we found that undifferentiated hESCs detached from Lam-111-coated culture dishes when kept in pluripotency medium (Figure 5A). Conversely, plating onto Lam-111 in neural differentiation medium resulted in confluent cultures after 7 days of differentiation, accompanied by a rapid downregulation of pluripotency markers (Figures 5B and S4C). This indicated that Lam-111 selectively supported the growth of neural cells but not pluripotent stem cells.
Figure 5.
Differentiation of hESCs on a GMP-Compatible Lam-111 Matrix Produces High Yield of VM Progenitors
(A) Culturing of hESCs on Lam-111 in pluripotency medium resulted in detachment and formation of spheres, whereas pluripotent cells efficiently attached to the Lam-521 matrix.
(B) Seeding of low-density hESCs on Lam-111 matrix in neural differentiation medium resulted in confluent neuralized cultures after 7 days of differentiation.
(C and D) Cells differentiated according to the GMP protocol showed a very high co-expression of LMX1A/B and FOXA2 (C) and of OTX2, LMX1A/B, and EN1 (D) by immunostaining. Scale bars, 100 μm.
(E) Comparison of cell yield to research-grade EB-based protocol showed a 43-fold increase in differentiated cell yield from the GMP-adapted Lam-111 protocol (n = 12–20).
(F) Schematic overview of GMP differentiation protocol with average cell yields (mean ± SEM) shown in red above when starting from 1 × 106 cells plated at a density of 10,000 cells/cm2 on day 0 (n = 12).
(G) Representative trace of action potentials induced with depolarizing current injections from VM-patterned neurons generated using the GMP protocol maturated for 45 days demonstrates physiologically active neurons (n = 11/11).
(H) Spontaneous post-synaptic currents indicative of synaptic activity and input (n = 4/6).
(I) An example of rebound depolarization after brief membrane depolarization characteristic of dopaminergic phenotype (n = 4/5).
(J) Inset showing respective trace from (I) on an expanded scale.
Data are presented as mean ± SEM. See Figure S4E for statistics on electrophysiology.
We further found that differentiation in a basal medium of mixed NeuroBasal + DMEM/F12 with N2 supplement, but without B27 supplement, from day 0 to day 11 produced the highest yield of cells on day 11 (Figure S4D), and for full GMP compatibility, we switched all growth factors and chemicals in the protocol to clinical grade standard or to the highest grade available (Table S4). The protocol gave rise to cultures with a high purity of FOXA2+/LMX1A/B+ progenitors: 90.4% ± 0.9% (n = 5; Figure 5C), thereby fulfilling our standard criteria for transplantation (>80% FOXA+/LMX1A+). To ensure accurate caudalization of the VM progenitors, 100 ng/mL of FGF8b was included in the protocol from d9 to d16 of differentiation, and we verified that the cells expressed EN1 while maintaining an OTX2+/LMX1A+ phenotype (Figure 5D).
The final yield of differentiated progenitors on d16 of this protocol was >40 times higher than the yield obtained with the original EB-based research-grade differentiation protocol (Figure 5E). From this, we extrapolated that we could produce >3.8 × 108 transplantable progenitor cells in 16 days when starting from just 1 × 106 undifferentiated hESCs (Figure 5F). When terminally differentiating these progenitors in vitro, we verified that the resulting neurons showed functional properties such as evoked action potentials (n = 11/11) and received synaptic input (n = 4/5) when assessed by patch-clamp electrophysiology. In the mature fraction of neurons (resting membrane potential < −40 mV) four of seven cells further showed rebound action potentials after brief depolarization, which is a characteristic of midbrain dopamine neurons in vitro (Grace and Onn, 1989) (Figures 5G–5J and S4E).
Implementation of Novel GMP Protocol Results in Reproducible Caudal VM Differentiation with High Expression of Predictive Markers and Functional Outcome in Vivo
To monitor critical batch-to-batch variations predictive of in vivo outcome, we designed a qRT-PCR panel with commonly used mesDA markers, in which we also included the new key predictive markers (EN1, SPRY1, PAX8, CNPY1, and ETV5) to assess for correct rostro-caudal VM patterning, as well as markers to monitor the presence of any contaminating forebrain (FOXG1), hindbrain (HOXA2), or lateral (PAX6) cell populations. The specificity of all primers used was verified in samples of sub-dissected human fetal tissue (Figure S5).
To assess the reproducibility and accuracy of the new GMP protocol, we analyzed VM-patterned batches from the research-grade protocol (n = 31) and GMP protocol (n = 31) with this qRT-PCR panel. We found that while all batches of cells had very robust expression of OTX1, OTX2, LMX1A, LMX1B, and FOXA2, there was considerable variation in the expression of caudal VM markers PAX8, EN1, SPRY1, and ETV5 between batches generated with the research-grade protocol (Figure 6A). The research-grade cell batches generally fell into two categories: (1) cell batches with high levels of EN1, PAX8, ETV5, SPRY1, and HOXA2 or (2) cell batches with low levels of EN1, PAX8, ETV5, SPRY1, and HOXA2, whereas only very few batches contained high levels of caudal VM markers in the absence of HOXA2 expression (Figure 6A). These expression patterns indicated that presence of HOXA2+ hindbrain cells was necessary for induction of a full caudal VM identity in the cultures generated by the research-grade protocol. In contrast, implementation of the new GMP protocol yielded batches of cells that were more homogenous and did not contain high levels of HOXA2 contamination. Addition of FGF8b to the GMP protocol robustly induced high-level expression of the new predictive markers (EN1, PAX8, ETV5, and SPRY1) (Figure 6A).
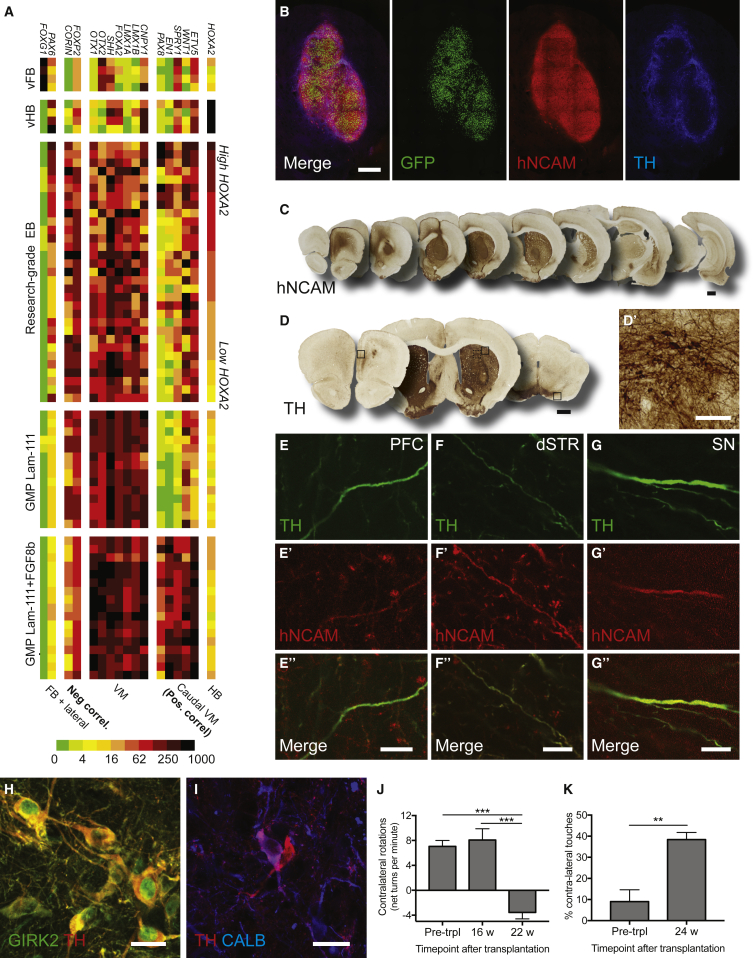
Figure 6.
Differentiation of hESCs from GMP-Compatible Protocol Produces VM Progenitors with Reproducibly High Expression of Predictive Markers that Give Rise to Functional DA Neurons In Vivo
(A) hESCs differentiated according to either research-grade EB protocol or GMP-grade Lam-111 protocol were assessed for RNA expression by qRT-PCR analysis on d16 of differentiation. Differentiations toward ventral forebrain (vFB) and ventral hindbrain (vHB) were included as controls. Addition of FGF8b to the GMP protocol (d9–d16) induced robust and reproducible induction of caudal VM markers, which we have shown to be predictive of TH-rich grafts. Values are color coded and normalized to the sample with highest expression for each gene ( = 1,000). See Figure S5 for validation of primers.
(B) Graft overview of RC17 grafts at 25 weeks post-transplantation to the 6-OHDA-lesioned rat striatum. Transplanted cells are visualized by nuclear GFP expression and are rich in hNCAM and TH.
(C, D, and D′) Overview of graft-derived innervation to the host brain (C) and graft-derived TH innervation (D and D′).
(E–G″) Double labeling of hNCAM and TH shows graft-derived TH+ fibers in the prefrontal cortex (PFC), dorso-lateral striatum (dSTR), and substantia nigra (SN).
(H and I) Many of the graft-derived TH+ neurons co-expressed GIRK2 (H) and few co-expressed CALB (I).
(J and K) Grafted animals showed complete reversal of amphetamine-induced rotations at 22 weeks after grafting, n = 8 (J), and reduction of asymmetrical paw use in the cylinder test, n = 8 (K). Data are presented as mean ± SEM.
Scale bars, 500 μm (B), 1,000 μm (C and D), 100 μm (D′), 10 μm (E″–G″), and 25 μm (H and I). Images in (B)–(D) are digitally stitched from multiple high-magnification images.
Transplantation of cells from the GMP cell line (RC17) differentiated according to the full GMP protocol resulted in neuron-rich grafts with a high number of TH+ neurons, clustering predominantly at the periphery of the graft (Figures 6B and 6D), similar to grafts of fetal VM-derived DA neurons (Li et al., 2016, Thompson et al., 2005). Quantifications showed that the DA yield (3,716 ± 1,026 TH+ per 100,000 cells grafted) was similar to that of the most successful grafts derived from our research-grade protocol, while the DA density was higher (9,100 ± 1,805 TH+/mm3) and graft size (0.37 ± 0.07 mm3/100,000 cells grafted) was lower (compared to DAhigh group of Table S1). In line with their caudal VM patterning, only few cells of the STN phenotype could be detected (Figure S6A).
Staining for hNCAM and TH showed extensive innervation of graft-derived TH+ fibers to the dorsolateral striatum and prefrontal cortex (Figures 6C–6F″), and similar to reports of fetal grafts, a minor fraction of the graft-derived fibers extended to the midbrain (Figures 6C and 6G–6G″) (Isacson and Deacon, 1996, Kirkeby et al., 2012, Thompson et al., 2009). Many of the graft-derived TH+ neurons co-expressed GIRK2, and few expressed CALB (Figures 6H and 6I). Importantly, these grafts also provided complete functional recovery in the unilateral 6-OHDA rat model of PD as assessed by recovery of amphetamine-induced rotations and reduction in asymmetric paw use in the cylinder test (Figures 6J and 6K). We further replicated the results of the GMP protocol in a second cell line (H9), where we could confirm that progenitors high in predictive markers gave rise to DA-rich grafts expressing GIRK2 and CALB and with the ability to promote functional recovery (Figures S6B–S6G).
In summary, we were able to optimize our research-grade differentiation protocol into a reproducible high-yield GMP-compatible protocol with decreased hindbrain contamination and increased expression of the caudal VM markers, which we have identified to be predictive of a good graft outcome.
Discussion
As protocols to form authentic and functional cellular derivatives from hPSCs become increasingly refined, stem cell-based therapies are rapidly moving closer to clinical trials (Chakradhar, 2016). In this development, the pre-clinical validation of cells and their in vivo performance is imperative. In some treatment paradigms, like the newly initiated pluripotent stem cell-based clinical trials for macular degeneration, fully mature and functional cells are produced in vitro and can therefore be directly assessed prior to use (Kamao et al., 2014, Schwartz et al., 2015). However, for disorders of the CNS, transplantation is performed with immature progenitors that undergo terminal differentiation and maturation after transplantation in vivo. From animal models of PD, Huntington’s disease, and stroke we know that transplanted progenitors mature and become functional only after several months in vivo (Kirkeby et al., 2012, Ma et al., 2012, Tornero et al., 2013), and from clinical trials using fetal cells in patients with PD, functional maturation of the graft has been observed to continue for several years after transplantation (Kefalopoulou et al., 2014). This complicates the assessment of the therapeutic potential of the cells prior to grafting, as the ability of stem cell-derived mesDA progenitors to terminally differentiate, mature, and significantly restore function in animal models of PD can only be determined in long-term animal studies (Grealish et al., 2014, Kriks et al., 2011).
A number of studies show that TH is aberrantly expressed by many cell types in vitro (Daadi and Weiss, 1999, Sonntag et al., 2004) and reports from older protocols show that cultures with good TH expression in vitro give rise to surprisingly few DA neurons in vivo (Barker et al., 2015, Kriks et al., 2011). We also observe aberrant TH expression in non-VM cultures and a high number of TH+/FOXA2− cells in vitro (not shown), and in line with this we found that in vitro differentiation into TH+ neurons does not correlate with the formation of TH+ neurons in vivo. We also found that although FOXA2, LMX1A, and CORIN are commonly used to identify mesDA progenitors during development (Arenas et al., 2015) and in stem cell cultures (Doi et al., 2014, Kirkeby et al., 2012, Kriks et al., 2011), they are not sufficient to predict yield or functionality of the cells in vivo. This highlights the need to validate markers that can predict functional maturation of hPSCs in vivo, rather than relying solely on markers expressed in differentiating progenitors of a specific cell lineage.
When applying an unbiased approach to identifying predictive markers of successful graft outcome, we found that markers expressed by midbrain cells close to the MHB (i.e., EN1, ETV5, CNPY1, PAX8, and SPRY1) correlated with a successful graft outcome whereas markers expressed in the diencephalic domain, such as EPHA3, FEZF1, and WNT7B, were negatively correlated with graft outcome. These findings are in line with a recent study in the mouse showing a remarkably close relationship between mesDA and STN neuronal lineages (Kee et al., 2016), where many key transcription factors, including Lmx1a, Lmx1b, Foxa1, Foxa2, Foxp1, Foxp2, Msx1, Nurr1, and Pbx1, are shared by both lineages. However, only the mesDA lineage is identified by expression of En1 and Cnpy1, which are restricted to the caudal part of the VM (Veenvliet et al., 2013).
Based on analyzing these markers we confirmed that our VM-patterned cultures contained both STN and mesDA progenitors and that their relative proportion in each batch likely contributed to the observed, but previously unexplained, variation in in vivo outcome when progenitors defined only by high co-expression of LMX1A, FOXA2, and OTX2 were transplanted. This prompted us to fine-tune the patterning to enrich for DA progenitors, which was achieved via timed, exogenous delivery of FGF8b at the progenitor stage of differentiation—a caudalizing agent that has also previously been applied in vitro (Perrier et al., 2004, Xi et al., 2012, Chen et al., 2016). This, as well as a number of additional adjustments, allowed us to generate a full GMP differentiation protocol for the production of mesDA progenitors. Key to this GMP protocol is the use of Lam-111, a physiologically relevant extracellular matrix component that is normally expressed in the developing brain and that we found to support attachment of differentiating neural progenitors but not pluripotent stem cells.
Postmortem brain analyses of patients with PD transplanted with human fetal VM tissue indicate that grafts containing approximately 50–100,000 TH+ neurons with substantial innervation of the host putamen can give long-term symptomatic relief in patients (Hauser et al., 1999, Li et al., 2016, Mendez et al., 2005). Using the laminin-based GMP protocol, the number of mesDA progenitors obtained per hESC at start of differentiation was greatly increased compared to our research-grade protocol, and the cells resulted in graft outcomes matching our best differentiations from the research-grade protocol in terms of yield, innervation, and function (Grealish et al., 2014, Grealish et al., 2015, Kirkeby et al., 2012). Given that we get an average yield of approximately 5,000 mature DA neurons per 100,000 transplanted progenitors from cell batches high in the predictive markers, manual production of cells for several hundred patients will be achievable even in small GMP laboratories without the need for automated culture systems. The protocol presented here therefore omits many of the large-scale production issues associated with other stem cell therapies going into the clinic (Chong et al., 2014). We observed in our study that a caudalized VM phenotype correlated not only with increased DA yield, but also with increased graft volume and DA density per 100,000 grafted cells. The appropriate cell dose to use in patients will be guided by these outcome parameters when evaluating the clinical cell product in pre-clinical transplantation studies.
In summary, global gene profiling of a large number of cell batches that have been transplanted into a rat model of PD, along with a refined understanding of the close relationship between DA and STN lineages during development (Kee et al., 2016), allowed us to establish a panel of markers to more precisely predict a successful graft outcome already at the progenitor stage in vitro. Although stringent in vivo assessment of safety and potency of stem cell-derived neural progenitors is still essential before use in patients, the ability to better predict graft outcome will accelerate the progression of stem cells toward clinical use as it allows for better in vitro screenings during cell production. Additionally, it can be used for batch-to-batch comparability and also to compare the cells grafted in different clinical trials. In the long term, a better prediction of in vivo maturation and functional properties of the cells already at the progenitor level will facilitate the use of autologous or individually matched cells for transplantation.
Experimental Procedures
hESC Culturing and Differentiation
H9 (WiCell) and RC17 (Roslin Cells, hPSC reg #RCe021-A) were used in this study. All grafts in Table S1 (i.e., Figures 1, 2, and 3) were derived from H9 cells differentiated toward VM progenitors according to Kirkeby et al. (2012) with minor modifications only. All grafted cell batches contained >80% FOXA2+/OTX2+/LMX1A/B+ progenitors as assessed by immunocytochemistry. Grafts in Table S1 were from H9 cells. In vitro studies in Figures 3, 4, and 6 include data from both H9 and RC17 cell lines, whereas GMP adaptation (Figures 5 and 6) was performed on RC17 cells. Pluripotent cells were maintained either on mouse embryonic feeder cells in 20% KSR or on Lam-521 in iPS Brew and passaged with EDTA once weekly. For details on research-grade EB protocol, see Kirkeby et al. (2012). For details on GMP Lam-111-based protocol, see Figure 5 and Supplemental Experimental Procedures.
Animals, Surgical Procedures, and Behavioral Testing
Adult female Sprague Dawley, Lister-hooded (Charles River) or athymic Hsd:RH-Foxn1rnu rats (Harlan Laboratories) were used as graft recipients. Animals were maintained on a 14:10-hr or 12:12-hr light:dark cycle with ad libitum access to food and water. All rats received a 6-hydroxydopamine lesion of the medial forebrain bundle and at 3–4 weeks post-lesion underwent amphetamine-induced rotations to assess extent of the lesion. Differentiated hESCs were dissociated and transplanted as previously described (Kirkeby et al., 2012); for details see Table S1. For surgical procedures, see Supplemental Experimental Procedures. Behavioral tests were performed as in Kirkeby et al. (2012) (for details, see Supplemental Experimental Procedures). All procedures were conducted in accordance with the European Union Directive (2010/63/EU) and were approved by the ethical and national committees for the use of laboratory animals at each site (Lund University, Sweden; Cardiff University, UK; and MIRCen, France).
RNA-Seq Analysis and Bioinformatics
RNA concentration and integrity were assessed on an Agilent RNA 6000 Pico chip, using the Agilent 2100 BioAnalyzer (Agilent Technologies). RNA (1 ng) was used to produce the cDNA library with Smart-Seq2 protocol (Picelli et al., 2013). The cDNA libraries were analyzed on high-sensitivity DNA chips with the Agilent 2100 BioAnalyzer. The cDNA (500 pg) was tagmented with the Nextera XT DNA sample preparation kit (Illumina) and sequenced on an Illumina HiSeq 2000 system. Bioinformatics analysis of the raw sequencing data included alignment of sequence reads to the reference human genome (hg19) and post-processing using alignment tools STAR and SAMtools. PCA was used to analyze how individual samples relate to each other based on overall gene expression profiles. The DESeq2 package without Cook’s cutoff and independent filtering (Love et al., 2014) determined the differentially expressed genes between sets of samples (Table S2).
qRT-PCR
qRT-PCR was performed using Sybr Green detection with the LightCycler 480 instrument (Roche). See Supplemental Experimental Procedures for primer sequences and details.
Immunochemical Analysis and Graft Quantifications
Cells and brains were fixed in 4% paraformaldehyde prior to staining. A complete list of antibodies and dilutions is provided in Supplemental Experimental Procedures. The number of DAB-stained TH+ neurons was counted manually at a bright-field microscope under 20× magnification for every section. Final counts were corrected for series number (1:8 or 1:12) to get an estimate of the total number of TH+ cells per animal. DA yield is presented as number of TH+ cells per 100,000 transplanted cells. Graft volumes were quantified based on DAB-stained HuNu+ cell areas in grafted sections. See Supplemental Experimental Procedures for further details.
Electrophysiology
Patch-clamp electrophysiology was performed on hESCs differentiated to a VM fate at day 45 post-differentiation. See Supplemental Experimental Procedures for detailed methods.
Statistical Analysis
All statistical methods are explained in the figure legends, with a significance level of alpha = 0.05. Unless stated otherwise, values are shown as mean ± SEM and asterisks in figures denote significance from Student’s t test between two groups. For rotational and cylinder behavioral analysis, one-way ANOVA with Bonferroni correction and paired-sampled Student’s t test were used, respectively. For all figures, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = non-significant.
Author Contributions
Conceptualization, A.K. and M.P.; Methodology, S.B.D., S.G., A.K., M.J.L., M.P., and T.P.; Investigation and Formal Analysis, T.C., S.G., A.H., A.K., N.K., M.J.L., S.N., D.R.O., and P.R. K.T.; Writing – Original Draft, A.K. and M.P.; Writing – Review and Editing, S.G., A.K., M.P., and T.P; Visualization, A.H., A.K., N.K., S.N, M.P., and K.T.; Funding Acquisition, S.B.D., M.P., and T.P.; Supervision, S.B.D., A.K., M.P., and T.P.
Acknowledgments
The authors would like to thank Michael Sparrenius, Sol da Rocha Baez, Ingar Nilsson, Ulla Jarl, Bengt Mattsson, and Marie Persson Vejgården for excellent technical assistance and Karolina Pircs for help with confocal images. This study was supported by the European Community’s 7th Framework Programme through NeuroStemcellRepair (no. 602278 to M.P. and S.B.D.), the Strong Research Environment at Lund University Multipark (Multidisciplinary Research in Parkinson’s Disease to M.P.), the Swedish Research Council (70862601/Bagadilico and K2012-99X-22324-01-5, K2014-61X-20391-08-4, and 2015-03444_3, all to M.P.), the Swedish Parkinson Foundation (Parkinsonfonden to M.P.), Knut and Alice Wallenberg’s Foundation (T.P.), and the Swedish Strategic Research Foundation (to T.P.). A.K. was supported by a postdoctoral fellowship from the Lundbeck Foundation (R44-A3856), K.T. was supported by a fellowship from the Swedish Society for Medical Research (SSMF), and S.G. was supported by a postdoctoral stipend from the Swedish Brain Foundation (Hjärnfonden). The research leading to these results has received funding from the European Research Council under the European Union’s 7th Framework Programme (FP/2007-2013)/ERC Grant Agreement no. 309712 (to M.P.).
Published: October 27, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2016.09.004.
Contributor Information
Agnete Kirkeby, Email: agnete.kirkeby@med.lu.se.
Malin Parmar, Email: malin.parmar@med.lu.se.
Accession Numbers
The accession number for the RNA-seq dataset described in this study is GEO: GSE86654.
Supplemental Information
References
- Arenas E., Denham M., Villaescusa J.C. How to make a midbrain dopaminergic neuron. Development. 2015;142:1918–1936. doi: 10.1242/dev.097394. [DOI] [PubMed] [Google Scholar]
- Barker R.A., Drouin-Ouellet J., Parmar M. Cell-based therapies for Parkinson disease—past insights and future potential. Nat. Rev. Neurol. 2015;11:492–503. doi: 10.1038/nrneurol.2015.123. [DOI] [PubMed] [Google Scholar]
- Cameron K., Tan R., Schmidt-Heck W., Campos G., Lyall M.J., Wang Y., Lucendo-Villarin B., Szkolnicka D., Bates N., Kimber S.J. Recombinant laminins drive the differentiation and self-organization of hESC-derived hepatocytes. Stem Cell Reports. 2015;5:1250–1262. doi: 10.1016/j.stemcr.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakradhar S. An eye to the future: researchers debate best path for stem cell-derived therapies. Nat. Med. 2016;22:116–119. doi: 10.1038/nm0216-116. [DOI] [PubMed] [Google Scholar]
- Chen Y., Xiong M., Dong Y., Haberman A., Cao J., Liu H., Zhou W., Zhang S.C. Chemical control of grafted human PSC-derived neurons in a mouse model of Parkinson’s disease. Cell Stem Cell. 2016;18:817–826. doi: 10.1016/j.stem.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J., Mahoney W.M., Van Biber B., Cook S.M., Palpant N.J. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daadi M.M., Weiss S. Generation of tyrosine hydroxylase-producing neurons from precursors of the embryonic and adult forebrain. J. Neurosci. 1999;19:4484–4497. doi: 10.1523/JNEUROSCI.19-11-04484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi D., Samata B., Katsukawa M., Kikuchi T., Morizane A., Ono Y., Sekiguchi K., Nakagawa M., Parmar M., Takahashi J. Isolation of human induced pluripotent stem cell-derived dopaminergic progenitors by cell sorting for successful transplantation. Stem Cell Reports. 2014;2:337–350. doi: 10.1016/j.stemcr.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace A.A., Onn S.P. Morphology and electrophysiological properties of immunocytochemically identified rat dopamine neurons recorded in vitro. J. Neurosci. 1989;9:3463–3481. doi: 10.1523/JNEUROSCI.09-10-03463.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grealish S., Diguet E., Kirkeby A., Mattsson B., Heuer A., Bramoulle Y., Van Camp N., Perrier A.L., Hantraye P., Björklund A., Parmar M. Human ESC-derived dopamine neurons show similar preclinical efficacy and potency to fetal neurons when grafted in a rat model of Parkinson’s disease. Cell Stem Cell. 2014;15:653–665. doi: 10.1016/j.stem.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grealish S., Heuer A., Cardoso T., Kirkeby A., Jönsson M., Johansson J., Björklund A., Jakobsson J., Parmar M. Monosynaptic tracing using modified rabies virus reveals early and extensive circuit integration of human embryonic stem cell-derived neurons. Stem Cell Reports. 2015;4:975–983. doi: 10.1016/j.stemcr.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R.A., Freeman T.B., Snow B.J., Nauert M., Gauger L., Kordower J.H., Olanow C.W. Long-term evaluation of bilateral fetal nigral transplantation in Parkinson disease. Arch. Neurol. 1999;56:179–187. doi: 10.1001/archneur.56.2.179. [DOI] [PubMed] [Google Scholar]
- Isacson O., Deacon T.W. Specific axon guidance factors persist in the adult brain as demonstrated by pig neuroblasts transplanted to the rat. Neuroscience. 1996;75:827–837. doi: 10.1016/0306-4522(96)00305-3. [DOI] [PubMed] [Google Scholar]
- Jaeger I., Arber C., Risner-Janiczek J.R., Kuechler J., Pritzsche D., Chen I.C., Naveenan T., Ungless M.A., Li M. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development. 2011;138:4363–4374. doi: 10.1242/dev.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner A.L., Liu A., Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr. Opin. Cell Biol. 2000;12:736–741. doi: 10.1016/s0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- Kamao H., Mandai M., Okamoto S., Sakai N., Suga A., Sugita S., Kiryu J., Takahashi M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee N., Volakakis N., Kirkeby A., Dahl L., Storvall H., Nolbrant S., Lahti L., Björklund A.K., Gillberg L., Joodmardi E. Single-cell Analysis Reveals a Close Relationship between Differentiating Dopamine and Subthalamic Nucleus Neuronal Lineages. Cell Stem Cell. 2016;20:29–40. doi: 10.1016/j.stem.2016.10.003. this issue. [DOI] [PubMed] [Google Scholar]
- Kefalopoulou Z., Politis M., Piccini P., Mencacci N., Bhatia K., Jahanshahi M., Widner H., Rehncrona S., Brundin P., Björklund A. Long-term clinical outcome of fetal cell transplantation for Parkinson disease: two case reports. JAMA Neurol. 2014;71:83–87. doi: 10.1001/jamaneurol.2013.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkeby A., Grealish S., Wolf D.A., Nelander J., Wood J., Lundblad M., Lindvall O., Parmar M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1:703–714. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Kriks S., Shim J.W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Englund E., Widner H., Mattsson B., van Westen D., Lätt J., Rehncrona S., Brundin P., Björklund A., Lindvall O., Li J.Y. Extensive graft-derived dopaminergic innervation is maintained 24 years after transplantation in the degenerating parkinsonian brain. Proc. Natl. Acad. Sci. USA. 2016;113:6544–6549. doi: 10.1073/pnas.1605245113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Hu B., Liu Y., Vermilyea S.C., Liu H., Gao L., Sun Y., Zhang X., Zhang S.C. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez I., Sanchez-Pernaute R., Cooper O., Viñuela A., Ferrari D., Björklund L., Dagher A., Isacson O. Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson’s disease. Brain. 2005;128:1498–1510. doi: 10.1093/brain/awh510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier A.L., Tabar V., Barberi T., Rubio M.E., Bruses J., Topf N., Harrison N.L., Studer L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Björklund A.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Rodin S., Antonsson L., Niaudet C., Simonson O.E., Salmela E., Hansson E.M., Domogatskaya A., Xiao Z., Damdimopoulou P., Sheikhi M. Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat. Commun. 2014;5:3195. doi: 10.1038/ncomms4195. [DOI] [PubMed] [Google Scholar]
- Schwartz S.D., Regillo C.D., Lam B.L., Eliott D., Rosenfeld P.J., Gregori N.Z., Hubschman J.P., Davis J.L., Heilwell G., Spirn M. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet. 2015;385:509–516. doi: 10.1016/S0140-6736(14)61376-3. [DOI] [PubMed] [Google Scholar]
- Sonntag K.C., Simantov R., Kim K.S., Isacson O. Temporally induced Nurr1 can induce a non-neuronal dopaminergic cell type in embryonic stem cell differentiation. Eur. J. Neurosci. 2004;19:1141–1152. doi: 10.1111/j.1460-9568.2004.03204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck J.A., Studer L. Moving stem cells to the clinic: potential and limitations for brain repair. Neuron. 2015;86:187–206. doi: 10.1016/j.neuron.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L., Barraud P., Andersson E., Kirik D., Björklund A. Identification of dopaminergic neurons of nigral and ventral tegmental area subtypes in grafts of fetal ventral mesencephalon based on cell morphology, protein expression, and efferent projections. J. Neurosci. 2005;25:6467–6477. doi: 10.1523/JNEUROSCI.1676-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.H., Grealish S., Kirik D., Björklund A. Reconstruction of the nigrostriatal dopamine pathway in the adult mouse brain. Eur. J. Neurosci. 2009;30:625–638. doi: 10.1111/j.1460-9568.2009.06878.x. [DOI] [PubMed] [Google Scholar]
- Tornero D., Wattananit S., Grønning Madsen M., Koch P., Wood J., Tatarishvili J., Mine Y., Ge R., Monni E., Devaraju K. Human induced pluripotent stem cell-derived cortical neurons integrate in stroke-injured cortex and improve functional recovery. Brain. 2013;136:3561–3577. doi: 10.1093/brain/awt278. [DOI] [PubMed] [Google Scholar]
- Veenvliet J.V., Dos Santos M.T., Kouwenhoven W.M., von Oerthel L., Lim J.L., van der Linden A.J., Koerkamp M.J., Holstege F.C., Smidt M.P. Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development. 2013;140:3373–3384. doi: 10.1242/dev.094565. [DOI] [PubMed] [Google Scholar]
- Xi J., Liu Y., Liu H., Chen H., Emborg M.E., Zhang S.C. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells. 2012;30:1655–1663. doi: 10.1002/stem.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.