Abstract
The anti-bacterial and anti-biofilm activities of cationic amphiphilic methacrylate polymers against cariogenic bacterium S. mutans were investigated. Cationic homopolymer PE0 and copolymer PE31 containing 31 mole% of ethyl methacrylate were synthesized by reversible addition-fragmentation chain transfer polymerization. These polymers displayed bactericidal activity towards S. mutans and prevented biofilm formation by killing the planktonic bacteria. At a concentration of 1000 μg/mL when incubated for 2 hours the polymers reduced more than 80% of biofilm biomass. When the polymer assay solution with the biofilm was vigorously mixed using a pipette for 30 seconds, more than 50% of biofilm mass was removed at a polymer concentration of 250 μg/mL. Chlorhexidine and a cationic surfactant failed to reduce the biofilm mass at the same concentration. PE0 was the most effective in removing biofilm and did not show any significant cytotoxicity to human gingival fibroblast and periodontal ligament stem cells when incubated for 10 minutes.
Keywords: antimicrobials, anti-biofilm, S. mutans, amphiphilic copolymers
Graphical abstract
INTRODUCTION
Dental caries, also known as cavities or tooth decay, continues to be a major oral problem in today’s society.1–2 From WHO statement, worldwide, 60–90% of school children and nearly 100% of adults have dental caries, which often leads to pain and discomfort.3 This is the most prevalent chronic disease in both children and adults: it is about five times as common as asthma and seven times as common as hay fever.1, 4 Dental caries are due to the demineralization and destruction of tooth surface by means of bacterial activity.5 Oral bacteria accumulate on and adhere to the tooth surface in order to form dental biofilms, or dental plaque, which harbor cariogenic bacteria such as Streptococcus mutans.6–7 When the cariogenic bacteria metabolize fermentable carbohydrates acid is produced as a byproduct.8 The tooth surface is mostly composed of hydroxyapatite which is eroded by the acidic environment that is created by the cariogenic bacteria. Ultimately, the erosion destroys the tooth’s enamel and the underlying dentin, and in other words dental caries have been formed. Therefore, control of cariogenic bacterial growth in the oral cavity and eradiation of dental biofilms formed on the tooth surface are essential steps to prevent tooth decay in order to maintain good oral health.
Host-defense antimicrobial peptides (AMPs) are a key component in the innate immune system that defends the host against bacterial invasion.9–10 AMPs are secreted small proteins, only a few ten thousand Daltons in size. While no common sequence has been identified, many AMPs share cationic amphiphilic structures composed of cationic and hydrophobic amino acid residues. These peptides inhibit bacterial growth by non-specifically disrupting bacterial cell membranes rather than specific ligand binding. Although AMPs are good candidates for new antimicrobial agents, their implementation as effective antimicrobials has been hampered by several drawbacks including low stability due to proteolytic degradation and high manufacturing cost. In order to overcome these drawbacks, a new approach is the development of synthetic mimics of AMPs based on small molecules or polymers.11–13 We have previously developed synthetic methacrylate random copolymers consisting cationic and hydrophobic side chains as a synthetic mimic of cationic amphiphilic AMPs.14–15 These random copolymers are designed to mimic the membrane-disruptive mechanism of AMPs, but not necessarily secondary structures of AMPs such as α-helices.15 These AMP-mimetic methacrylate polymers showed antimicrobial activity against a broad spectrum of human bacterial pathogens including E. coli and S. aureus.
In the oral environment, AMPs also play an important role in maintaining a healthy, anti-cariogenic oral flora by controlling bacterial growth and biofilm formation.16–17 Naturally occurring and synthetic AMPs have shown their ability to kill cariogenic planktonic bacteria or bacteria in oral biofilms to prevent biofilm formation or eradicate biofilms.18–20 Accordingly, we hypothesize that the AMP-mimetic antimicrobial polymer would be an effective agent in controlling cariogenic bacterial growth and associated biofilm formation. Previously small molecule AMP-mimetics have been examined for their effectiveness against oral bacteria21 but the investigation of AMP-mimetic polymers on their activity against oral bacteria and biofilms is unexplored. Owing to the chemical diversity of synthetic polymers, we believe that AMP-mimetic polymers will be a new platform to design and develop active ingredients for use in oral care products such as mouthwash and toothpaste in order to promote oral health.
In this study, we tested the hypothesis and evaluate the potential use of polymers as an active ingredient in oral care products including mouthwash and toothpaste. We examined the in vitro anti-bacterial and anti-biofilm effect of AMP-mimetic methacrylate polymers against cariogenic bacterium S. mutans. We also investigated eradication of biofilm by rinsing and swishing with polymer solution for 30 seconds as a “liquid toothbrush”. Our study indicates that the polymers can inhibit the growth of planktonic S. mutans and prevent S. mutans biofilm formation. The polymers are also capable of removing a significant amount of biomass of S. mutans biofilms.
EXPERIMENTAL SECTION
Materials
4-amino-1-butanol, di-tert-butyl dicarbonate, and triethylamine were purchased from Acros Organics. 2,2′-azobisisobutyronitrile (AIBN), chlorhexidine diacetate salt hydrate and the bee venom toxin melittin (purity > 85 %) were purchased from Sigma-Aldrich Co. LLC. Vancomycin hydrochloride was purchased from MB Biomedicals, LLC. 2-cyanoprop-2-yl-dithiobenzoate was purchased from Strem Chemicals, Inc. Trifluoroacetic acid (TFA) and solvents were purchased from Thermo Fisher Scientific, Inc. The chemicals were used without further purification, with the exception of methacryloyl chloride and ethyl methacrylate (EMA), which were purchased from Acros Organics and distilled before use. 1H NMR was performed using a Varian MR400 (400 MHz) and analyzed using VNMRJ 3.2 and MestReNova. Gel permeation chromatography (GPC) analysis was performed using a Waters 1515 HPLC instrument equipped with Waters Styragel (7.8 × 300 mm) HR 0.5, HR 1, and HR 4 columns in sequence and detected by a differential refractometer (RI). Streptococcus mutans ATCC®25175™ was used in this study to evaluate antimicrobial and anti-biofilm activity of polymers. Human red blood cells (RBCs) (leukocytes reduced adenine saline added) were obtained from the American Red Cross Blood Services Southeastern Michigan Region and used prior to the out date indicated on each unit. Human gingival fibroblasts (hGF) (ScienCell™ Research Laboratories) and periodontal ligament stem cells22 were used to evaluate cytotoxicity of polymers.
Polymer synthesis
Methacrylate homo- and random copolymers were synthesized by reversible addition fragmentation chain transfer (RAFT) polymerization (Figure 1(A)). 4-((tert-butoxycarbonyl) amino)butyl methacrylate (Boc-ABMA) was synthesized according to the previous report.23–24 Boc-ABMA and ethyl methacrylate (EMA) monomers (0 or 30 mole% of EMA relative to total amount of monomers), RAFT chain transfer agent 2-cyanoprop-2-yl-dithiobenzoate (10 mole% relative to total amount of monomers) and AIBN (1 mole% relative to total amount of monomers) were dissolved in acetonitrile. The mixture was flushed with nitrogen gas for 5 minutes, then stirred at 70°C. After 16 hours, the reaction was stopped by cooling the vial in a dry ice-acetone bath. The polymer was isolated by evaporating the acetonitrile under reduced pressure, and then the residue was dissolved in dichloromethane and precipitated in excess hexanes twice to remove unreacted monomers and impurities. The obtained Boc-protected polymer was mixed with methyl 3-mercaptopropionate (MMP), and then dissolved in trifluoracetic acid (TFA). TFA is harmful and corrosive; an exposure of TFA should be limited. After stirring for 30 min, the TFA was removed by blowing with nitrogen gas. The gas containing TFA was passed through a sodium hydroxide solution in a fume hood. The residue was dissolved in methanol, and the deprotected polymers were collected by precipitating in excess diethyl ether. Subsequently, the polymer precipitates were dissolved in distilled water and lyophilized to yield light, fluffy fibrous polymers. The Boc-protected copolymer was characterized by GPC analysis to measure the number average molecular weight (Mn) and the weight average molecular weight (Mw) calculated using a calibration curve based on 10 standard samples of poly(methyl methacrylate), MW 500–50,000 (Agilent Technologies, M-L-10, no. PL2020-0100). The Boc-protected and de-protected polymers were also characterized by 1H NMR analysis to determine the mole percentage of EMA (MPethyl), the degree of polymerization (DP), and consecutive the number average molecular weight (Mn). The MPethyl was determined by comparing integrated peaks of butylene groups of Boc-ABMA and ethylene groups of EMA in the 1H NMR spectra. The DP was calculated by comparing integrated peaks of phenyl group of chain transfer agent at the polymer ω-end and side chains in the 1H NMR spectra (Fig. S1 in Supporting Information (SI)).
Figure 1.
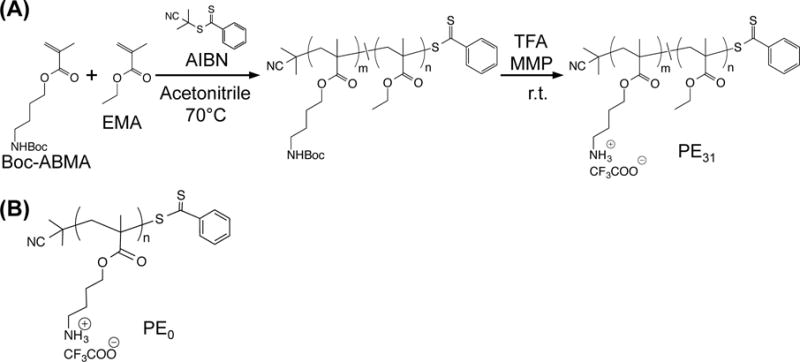
Antimicrobial peptide-mimetic methacrylate polymers. (A) Synthesis of cationic amphiphilic methacrylate random copolymer (PE31) by reversible addition fragmentation chain transfer (RAFT) polymerization. (B) Chemical structure of cationic homopolymer PE0.
1H NMR (CDCl3, 400 MHz) for homopolymer (PE0), Boc-protected, DP= 14.1, MPethyl= 0 mole%, Mn (GPC) = 2200, PDI (Đ) = 1.32: δ 7.90–7.26 (m), 5.25–4.70 (brs), 4.20–3.85 (m), 3.25–2.90 (brs), 2.15–1.77 (m), 1.70–1.32 (m), 1.38–1.20 (brs), 1.20–0.75 (m).
1H NMR (CD3OD, 400 MHz) for homopolymer (PE0), de-protected, DP= 13.9, MPethyl= 0 mole%: δ 7.90–7.38 (m), 4.20–3.83 (brs), 3.18–2.85 (m), 2.20–1.60 (m), 1.59–1.22 (m), 1.20–0.76 (m).
1H NMR (CDCl3, 400 MHz) for copolymer (PE31), Boc-protected, DP= 15.0, MPethyl= 30.3 mole%, Mn (GPC) = 2800, PDI (Đ) = 1.12: δ 7.90–7.26 (m), 5.25–4.70 (brs), 4.20–3.83 (m), 3.25–2.92 (brs), 2.15–1.77 (m), 1.70–1.45 (m), 1.42 (brs), 1.32–1.17 (m), 1.16–0.80 (m).
1H NMR (CD3OD, 400 MHz) for copolymer (PE31), de-protected, DP= 15.9, MPethyl= 30.6: δ 8.18–7.35 (m), 4.25–3.83 (m), 3.09–2.84 (m), 2.16–1.82 (m), 1.80–1.65 (m), 1.46–1.17 (m), 1.13–0.80 (m).
Preparation of solutions
AMP-mimetic polymers and vancomycin were dissolved in 0.01% acetic acid at 10 mg/mL (polymers) or 1 mg/mL (vancomycin) to create a stock solution, and serial dilutions of antimicrobials were prepared from stock solution by dilution with 0.01% acetic acid. Chlorhexidine diacetate salt hydrate and cetyltrimethylammonium bromide were dissolved in pure water at 10 mg/mL as a stock solution, and serial dilutions were prepared by dilution with water.
Antimicrobial and bactericidal assays
The minimum inhibitory concentration (MIC) against S. mutans were determined in a standard microbroth dilution assay according to the Clinical and Laboratory Standards Institute guidelines with suggested modifications by R.E.W. Hancock Laboratory (University of British Columbia, British Columbia, Canada)25 and Giacometti et al26. S. mutans cultured in Todd Hewitt broth (THB) at 37°C with 5% CO2. An overnight (approximately 18 h) culture of S. mutans was regrown to exponential phase (OD600 of 0.5–0.7) and diluted with the THB to give the bacterial suspension with approximately 6.0 × 105 cfu/mL as final concentration. After serial dilutions, antimicrobials (10 μL) were prepared on a 96-well sterile round-bottom polypropylene plate and the bacterial suspension (90 μL) was added and incubated for 18 h at 37°C with 5% CO2. 0.01 % acetic acid or pure water as a solvent control. The MIC was defined as the lowest polymer concentration to completely inhibit visible bacterial growth. Bacterial growth was detected at OD600 using WPA S800 visible spectrophotometer (Biochrom). Continuously, the minimum bactericidal concentration (MBC) was determined.21 The MBC was defined as the lowest polymer concentration to kill a particular bacterium. After the MIC assay, bacterial solution from each well was mixed thoroughly and diluted 100-fold with THB to sub-inhibitory concentrations to remove the effect of polymers or antibiotics due to carryover of these antimicrobials. Then, the diluted bacterial suspension (100 μL) was streaked onto Todd Hewitt agar plates, and incubated at 37°C with 5% CO2 overnight. Based on the dilution factor and the volume of inoculation, the formation of one colony in the plate requires 1000 viable bacterial cells per one milliliter in the original bacterial suspension before dilution. In other words, if less than 1000 colony forming unit, cfu/mL of bacteria remained after polymer or antibiotic treatment, there would be no colony in an agar plate. The MBC was determined as the lowest polymer concentration at which no colony of viable cells in an agar plate was detected, indicating that the number of viable bacterial cells was less than 1000 cfu/mL. The MIC and MBC assays were independently repeated three times using different stock solutions in duplicate on different days. It should be noted that polymers are soluble to an assay medium and did not cause any precipitation under the assay condition (Fig. S2 in SI).
Bactericidal kinetics
An overnight (approximately 18 h) culture of S. mutans was regrown to exponential phase (OD600 of 0.5–0.7) in Todd Hewitt broth (THB) at 37°C with 5% CO2 and diluted with the THB to give approximately 6.0 × 105 cfu/mL as final concentration. At the time of 0 minutes, each antimicrobial was added to the bacterial solution in sterile polystyrene microtubes at a concentration twice the MIC and incubated at 37 °C with orbital shaking (180 rpm). 0.01 % acetic acid was used as a control. Aliquots from each solution were drawn at appropriate time intervals and immediately diluted with PBS at least 100-fold to remove the effects of the antimicrobials. The solutions (100 μL) were then streaked onto Todd Hewitt agar plates and incubated at 37°C with 5% CO2 overnight to determine viable bacterial cell number. As described in the antimicrobial and bactericidal assays, the formation of one colony in the plate requires 1000 viable bacterial cells per one milliliter in the original bacterial suspension before dilution. Therefore, the actual number of bacteria is not possibly determined if less than 1000 cfu/mL of bacterial cells remained. Each assay was independently repeated three times using different stock solutions on different days.
Hemolysis assay
Human red blood cells from healthy donors (RBCs; 1 mL) were suspended in 9 mL of PBS buffer (pH 7.4) and centrifuged at 660 × g for 5 min. The supernatant was removed by pipetting and RBCs were re-suspended in PBS. This procedure was repeated two additional times. The number of RBCs in resulting suspension was counted by a counting chamber and diluted in PBS to give 3.0 × 108 cells/mL as a final concentration. After serial dilutions of polymers and chlorhexidine (10 μL) were prepared on a 96-well sterile round-bottom polypropylene plate, the RBC suspension (90 μL) was added and incubated at 37°C with orbital shaking (180 rpm). Triton X-100 (0.1% v/v in water) was used as the positive lysis control and 0.01% acetic acid and pure water were used as negative control. The bee venom toxin melittin was also tested as reference standard. After incubation for 1 h, the plate was centrifuged at 1000 × g for 5 min and supernatant (6 μL) from each well was diluted with PBS buffer (100 μL) in a 96-well sterile flat-bottom polystylene plate. The absorbance of the released hemoglobin at 415 nm was measured using Varioskan Flash microplate reader (Thermo Fisher). The percentage of hemolysis was calculated relative to the positive control Triton X-100 (100%) and negative control solvents (0%). The percentage difference between two negative control solvents, 0.01% acetic acid and pure water, was less than 1.5%. The HC50 was defined as the polymer concentration causing 50% hemolysis. The HC50 or hemolysis% at highest concentration if the hemolysis% showed below 50% was reported. Each hemolysis assay was independently repeated three times using different stock solutions in triplicate on different days.
Biofilm inhibition assay
The anti-biofilm activity of polymers against biofilm formation was measured using the microdilution method20 with small modifications. An overnight (approximately 18 h) culture of S. mutans in Todd Hewitt broth (THB) was diluted with the 1/4 Todd Hewitt Yeast Extract medium (THB + 5 mg/mL yeast extract) plus 30 mmol/L sucrose solution (1/4 THYE+S) to give OD600= 0.01. The 1/4 THYE+S broth was used to promote S. mutans biofilm formation.27 After, serial dilutions of antimicrobials (10 μL) were prepared on a 96-well sterile flat-bottom polystylene cell culture plate. The bacterial suspension (90 μL) was added and incubated for 24 h at 37°C with 5% CO2. 0.01 % acetic acid or pure water was used as a solvent control. After 24 h incubation, the OD600 was recorded using Varioskan Flash microplate reader to assess bacterial cell growth. After, Planktonic cells were carefully removed and the remaining biofilm biomass was measured by crystal violet (CV) staining. The minimum biofilm inhibitory concentration (MBIC), defined as the lowest polymer concentration to inhibit biofilm formation less than 5% relative to the control solvents (100%), was measured. Each assay was independently repeated three times using different stock solutions in triplicate on different days.
Biofilm eradication assay
Similar to biofilm inhibition assay, an overnight (approximately 18 h) culture of S. mutans in THB was diluted with the 1/4 THYE+S to give OD600= 0.01. Then 100 μL of bacterial suspension was added to 96-well sterile flat-bottom polystylene cell culture plate and incubated for 24 h at 37°C with 5% CO2 to form S. mutans biofilm on the bottom of wells. After 24 h incubation, planktonic cells were carefully removed and the biofilms were washed three times with PBS buffer (pH 7.4). After fresh PBS (90 μL) was placed in each wells, the serial dilutions of antimicrobials including CTAB (10 μL) were added and incubated for 2 h or 24 h at 37°C without shaking. 0.01 % acetic acid or pure water was used as a solvent control. It should be noted that the percentage difference between the two control solvents, 0.01% acetic acid and pure water, was less than 1% (Fig. 5B). In order to model swishing liquid in the mouth, antimicrobial solution was vigorously mixed upon repeatedly dispensing and drawing the solution using a pipette ~30 times for 30 seconds instead of static incubation. After treatment, the remaining biofilm biomass was measured by crystal violet (CV) staining. Each assay was independently repeated three times using different stock solutions in triplicate on different days.
Figure 5.
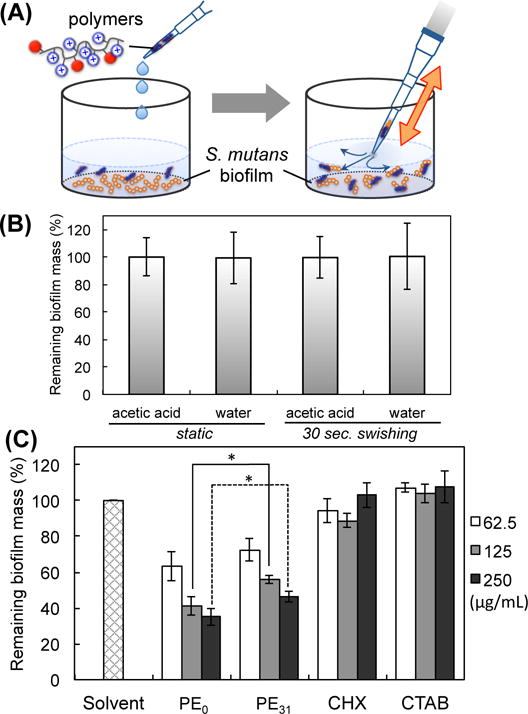
Eradiation of S. mutans biofilm by swishing treatment with antimicrobial polymers for 30 seconds. (A) Schematic presentation of swishing treatment of biofilm by vigorously pipetting for 30 seconds. (B) The effect of solvents (0.001% acetic acid and water) on biofilm eradication. (C) The biomass of remaining biofilm after 30 seconds treatment of polymers, chlorhexidine (CHX) and surfactant cetyltrimethylammonium bromide (CTAB) measured by CV staining. [Polymer, CHX, or CTAB] = 62.5, 125 and 250 μg/mL. *p< 0.05.
CV staining.20, 28–29
The plates with biofilms were washed three times with PBS buffer (pH 7.4) and then fixed with methanol for 15 min. The biofilm mass in each well was quantified by staining with 0.1 % (w/v) crystal violet for 10 min at room temperature and washed three times with water to remove extra dye. Then 200 μL of 95 % ethanol was added to each crystal violet-stained well, and the plate was kept at room temperature for 30 min to elute CV. Eluted CV/ethanol solution (100 μL) from each well was transferred to new 96-well sterile flat-bottom polystylene plate, then the OD595 was measured using Varioskan Flash microplate reader. The percentage of biofilm mass was calculated relative to the negative control solvents (100%).
Viability of bacterial cells in remaining biofilm
S. mutans biofilm was prepared on glass bottom dish (glass diameter= 10 mm) by incubation of S. mutans (OD600= 0.01) in the 1/4 THYE+S broth for 24 h at 37°C with 5% CO2. After washing with PBS buffer (pH 7.4), the biofilm was treated by adding antimicrobials at a concentration of 125 μg/mL and vigorously mixing using a pipette ~30 times for 30 seconds in order to model swishing in the mouth. 0.01 % acetic acid was used as a solvent control. The remaining biofilm was stained for 15 min in the dark using a LIVE/DEAD® BacLight™ Bacterial Viability kit (Invitrogen) following a standard protocol. The S. mutans cells in the biofilms were imaged using confocal laser scanning microscope (Nikon Eclipse Ti300 confocal instrument), live bacteria were stained with the SYTO® 9 green fluorescent nucleic acid stain and dead bacteria were stained with the propidium iodide red/orange fluorescent dye.
Cytotoxicity assay
Human gingival fibroblasts (hGF) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco® Life Technologies). Periodontal ligament stem cells (PDLSC)22 were cultured in alpha Modified Eagle’s Medium (α-MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco® Life Technologies) and then incubated at 37°C with 5% CO2. Cells were seeded at 1 × 104 cells/well in 96-well sterile flat-bottom polystylene cell culture plate and cultured for 24 h. Then cells were exposed to antimicrobials at a concentration of 62.5, 125 and 250 μg/mL for 10 min. 0.01 % acetic acid and pure water were used as solvent controls. After 10 min treatment, antimicrobial solutions were removed and cells were washed with PBS. The cells’ viability after exposure to the antimicrobials was measured using Cell Proliferation Kit II (Roche Applied Sciences) by measuring the amount of the metabolized formazan at 450 nm following a standard protocol from product. The percentage of cells viability was determined relative to the control solvents (100% cell viability). The percentage difference between two control solvents, 0.01% acetic acid and pure water, was less than 3.0%. Each assay was independently repeated two times (against hGF) or three times (against PDLSC) using different stock solutions in triplicate on different days.
RESULTS AND DISCUSSION
Polymer Design and Synthesis
To mimic the cationic amphiphilic property of AMPs, we have designed and developed methacrylate random copolymers with cationic and hydrophobic groups in the side chains.23–24 In general, traditional antimicrobial polycations are high molecular weight polymers bearing quaternary ammonium groups modified with long alky chains.30 Our polymers have primary ammonium groups in the side chains, which mimic the lysine residue rich in AMPs. The molecular weight of polymers is also low (2000–3000), which is similar to the size of AMPs9–10. The cationic groups of polymers are designed to enhance the binding of polymers to anionic bacterial membranes by electrostatic interactions. Because the bacterial membranes are rich in negatively charged lipids as compared to the human cell membranes, the cationic polymers exhibit selectivity for bacterial cells over human cells. The hydrophobic groups drive the insertion of polymer chains into the hydrophobic core of membranes. This causes membrane disruption and ultimately cell death. A recent study on direct observation of bacterial membranes by AFM suggested that amphiphilic polycations act by disrupting membranes.31
The antimicrobial methacrylate copolymers were synthesized by reversible addition-fragmentation chain transfer (RAFT) polymerization (Fig. 1).23 The monomer with boc-protected amine groups 4-((tert-butoxycarbonyl)amino)butyl methacrylate (Boc-ABMA) and hydrophobic monomer ethyl methacrylate (EMA) were copolymerized using AIBN as a radical imitator and 2-cyanoprop-2-yl-dithiobenzoate as a chain transfer agent, yielding a copolymer with 30.6 mole% of hydrophobic ethyl group (PE31) (See Table S1 in Supporting Information for characterization of Boc-protected polymers). Similarly, a cationic homopolymer (PE0) was also synthesized from only Boc-ABMA. The dispersity (Đ) of boc-poretcted polymers was 1.12 for PE31 and 1.32 for PE0, respectively. The protecting boc-group was removed by treating the polymers in TFA. 1H NMR analysis indicated that the degree of polymerization (DP) of PE31 and PE0 was 15.9 and 13.9, respectively (Table 1) (see Fig. S1 in SI for 1H NMR spectra), giving the molecular weights of 2500 and 2400 for PE31 and PE0 polymers, respectively. It should be noted that the molecular weights of polymers do not include the molecular weight of trifluoroacetate CF3COO− (113 g/mol) to compare with the molecular size of AMPs.
Table 1.
Polymer characterization, antimicrobial activity of polymers, chlorhexidine (CHX), and vancomycin (VAN) against planktonic S. mutans, and hemolytic activity.
| Polymer/antimicrobial | MPethyla (mol. %) | DPb | Mn (NMR)c | Đd | MICe (μg/mL) | MBCf (μg/mL) | HC50g (μg/mL) |
|---|---|---|---|---|---|---|---|
| 2,400 | >1000 | ||||||
| PE0 | 0 | 13.9 | 1.32 | 52.1 ± 14.7 | 62.5 ± 0.0 | ||
| (4,000) | (0.9 ± 0.1%) | ||||||
| 2,500 | >1000 | ||||||
| PE31 | 30.6 | 15.9 | 1.12 | 7.8 ± 0.0 | 10.4 ± 3.7 | ||
| (3,800) | (26.9 ± 9.6%) | ||||||
| CHXh | – | – | 626 | – | 0.3 ± 0.1 | 4.2 ± 2.5 | 133 ± 23.6 |
| VANi | – | – | 1,486 | – | 0.8 ± 0.0 | 2.3 ± 1.1 | n.d.j |
MPethyl: Mole percentage of ethyl group in a polymer chain determimed by 1H NMR.
DP: The number average degree of polymerization determimed by 1H NMR.
Mn (NMR): The number average molecular weight (Mn) without trifluoroacetate calculated based on the molecular weight of monomers, MPmethyl and DP. The molecular weight including trifluoroacetate is given in parenthesis.
Đ: The dispersity of boc-protected polymers calculated as Mw/Mn using Mw and Mn values determined by GPC.
MIC: minimum inhibitory concentration.
MBC: minimum bactericidal concentration, the lowest concentration to reduce viable bacterial cells less than 1000 cfu/mL.
HC50: concentration causing 50% lysis (hemolysis) of red blood cells (RBCs) relative to Trinton-X (positive control, 100 %) and solvents (negative control, 0.001% acetic acid for PE0, PE31 and VAN and water for CHX, 0 %). The hemolysis % at 1000 μg/mL is given in parenthesis.
Chlorhexidine diacetate.
Vancomycin hydrochloride.
Not determined.
Growth inhibition of planktonic S. mutans
We first evaluated the ability of polymers to inhibit growth of planktonic S. mutans. S. mutans is a Gram-positive, facultative anaerobic spherical bacterium (coccus) which grow both aerobically and anaerobically owing to the switchable respiratory system depending on the oxygen level. Normally, oxygen is abundant in the human mouth. However, S. mutans in the deeper layers of biofilm or dental plaque is exposed to a lower level of oxygen or the bacteria are in an anaerobic enviroment.32 Therefore, S. mutans was cultured at 37°C with 5% CO2, in our study presented here, which has been a standard culture condition found in literature.27 The minimum inhibitory concentration (MIC), defined as the lowest polymer concentration to completely inhibit visible bacterial growth, was determined in order to measure the antimicrobial activity using the turbidity-based micro-dilution assay (Table 1).25–26 The methacrylate polymers both showed antimicrobial activity against S. mutans. Specifically, PE31 showed high activity (MIC= 7.8 μg/mL), while cationic homopolymer analogous PE0 showed lower activity (MIC= 52.1 μg/mL). This suggests that the hydrophobicity of polymers enhances the activity, which parallels our previous results pertaining to the antimicrobial activity of polymer analogues against other bacteria.23–24 This is likely due to increased ability of polymers to disrupt bacterial cell membranes.33 The antiseptic antibacterial agent chlorhexidine and antibiotic vancomycin were also tested for their antimicrobial activity under the same condition for comparison purposes. Chlorhexidine is a low molecular weight compound with cationic charges, which interacts with the cell membrane and enters the cells to kill them by precipitating the cytoplasmic contents.34 It should be noted that chlorhexidine has been used as an active agent in oral rinse products (prescribed by a dentist) to treat gingivitis and periodontitis35, but not intended to prevent or treat dental caries. Chlorhexidine and vancomycin showed inhibitory effects against the growth of S. mutans, which was illustrated by an MIC of 0.3 μg/mL and 0.8 μg/mL, respectively.
Bactericidal activity against planktonic S. mutans
The bactericidal effect of polymers was also assessed by determining the minimum biocidal concentration (MBC) at which no colony formation was detected with detection limit of 1000 cfu/mL under the assay condition. The MBC of PE31 and PE0 polymers were 10.4 and 62.5 μg/mL, which are close to the MIC values (7.8 and 52.1 μg/mL). This result indicates that these polymers inhibit bacterial growth by killing. The bactericidal activity of antimicrobial agents has been reported for AMPs18–19 and other synthetic mimetics21. On the other hand, the MBC values of chlorhexidine and vancomycin were 4.2 and 2.3 μg/mL, respectively. These MBC values are substantially higher than the MIC values (0.3 μg/mL and 0.8 μg/mL), indicating that these antimicrobials are rather bacteriostatic.
Killing kinetics
To determine the rate of bacterial killing, we monitored the reduction in the number of viable bacterial cells in the presence of polymers of time (Fig. 2). The number of viable bacteria decreased by >99.8% in 120 min for PE31 and 180 min for PE0 at twice the MIC values. The higher rate of PE31 than PE0 may reflect the higher bactericidal activity (lower MBC values) of PE31 than PE0. Interestingly, the time course of PE31 is almost the same as that of PE0 for the first 30 minutes, but begins to deviate at 60 minutes. As the generation (doubling) time of S. mutans under this assay condition is about 70 minutes, we speculate that the change of PE31 in the killing rate may be related to the cell growth or cell duplication process. However, the antimicrobial mechanism of polymers is not clear at this point. On the other hand, chlorhexidine and vancomycin did not change the number of viable bacteria at twice the MIC values, indicating that these antimicrobials are bacteriostatic.
Figure 2.
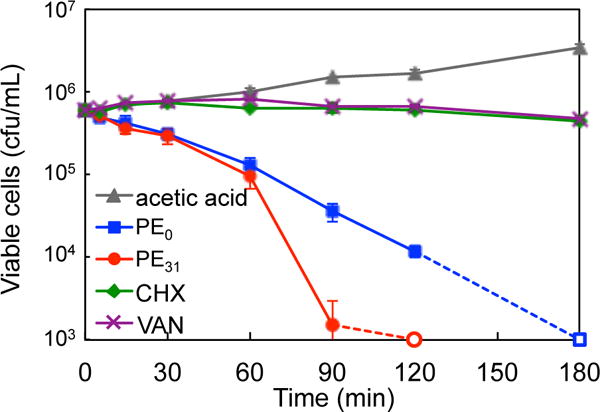
Bactericidal kinetics of polymers, chlorhexidine (CHX), and vancomycin (VAN) against S. mutans at twice MIC. The bacterial cell counts below 1000 cfu/mL are presented by open markers at 1000 cfu/mL. 0.001% acetic acid was tested as solvent control. [PE0]= 125 μg/mL, [PE31]= 15.6 μg/mL, [CHX]= 0.8 μg/mL and [VAN]= 1.6 μg/mL.
Hemolytic activity
To evaluate toxicity of polymers to human cells, the hemolytic activity of polymers was determined using human red blood cells (RBCs) (Table 1). PE0 did not show any significant lysis of RBCs even at the highest concentration (1000 μg/mL) tested in this assay while PE31 caused ~26% hemolysis at 1000 μg/mL (see Fig. S3 in SI for hemolysis curve). On the other hand, chlorhexidine caused significant hemolysis with the HC50 value of 133 μg/mL, in which chlorhexidine caused 50% hemolysis. For comparison, the HC50 value of lytic peptide melittin was 3.8 μg/mL. In summary, PE31 and PE0 showed potent antimicrobial activity against planktonic S. mutans with high selectivity over human RBCs.
Inhibition of S. mutans biofilm formation
We further evaluated the ability of polymers to prevent the formation of S. mutans biofilm. We used 1/4 THYE+S broth for this assay to promote S. mutans biofilm formation, while nutrient-rich Todd Hewitt broth (THB) was used for antimicrobial assay with planktonic S. mutans.27 The polymers were added to S. mutans bacterial solution and incubated for 24 hours. The biofilm formation was quantified by the crystal violet (CV) staining method, which reflects the total biomass of biofilms.28–29 The polymers inhibited the formation of biofilms in a concentration-dependent manner (Fig. 3). The minimum biofilm inhibitory concentration (MBIC) that inhibited biofilm formation less than 5% relative to the control solvents was 6.3 μg/mL for PE31 and 8.3 μg/mL for PE0, respectively. For comparison, the MBIC value of chlorohexidine was 1.0 μg/mL. Additionally, in order to probe into the inhibition mechanism, the relationship between bacterial growth and biofilm inhibition was examined. If the formation of a biofilm was prevented due to killing of bacteria by the polymers, the bacterial growth in solution should be correlated to the inhibition in biofilm formation. If the polymers were to interfere the biological processes of biofilm formation such as production of extracellular matrix biopolymers, the biofilm formation would be inhibited, but bacteria would be viable. To test these models, the bacterial growth was determined by measuring the turbidity (optical density) of bacterial solution in the wells after incubation with the polymers.18, 20 The result indicates that the bacteria growth curves PE31 of and PE0 polymers closely matched with the biofilm biomass curves of each of them, suggesting that the inhibition of polymer-mediated biofilm is due to the inhibition of bacteiral growth (Fig. 3).
Figure 3.
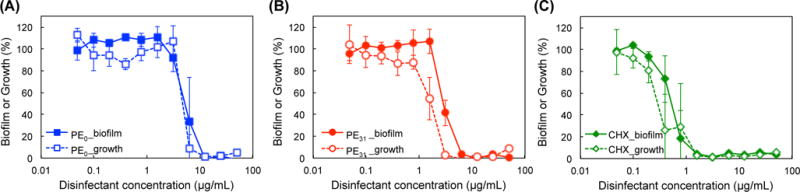
Prevention of S. mutans biofilm formation by antimicrobial polymers.
After 24 h incubation with PE0 (A), PE31 (B) or chlorhexidine (CHX) (C), the formation of biofilm was determined by CV staining (filled markers), and planktonic bacterial growth in solution was determined by OD600 (opened markers), relative to solvents as 100% (0.001% acetic acid for PE0 and PE31 and water for CHX).
Eradication of S. mutans biofilm
To evaluate the ability of polymers to eradicate biofilms, the polymers were incubated with one-day matured S. mutans biofilm for 2 hours, and the remaining biofilm biomass was determined by CV staining (Fig. 4). As the polymer concentration was increased, the biofilm biomass was reduced by up to 80 to 85%, and only 15–20% of biofilm mass remained at the polymer concentration of 1000 μg/mL. On the other hand, chlorhexidine was not effective in removing biofilms. To examine the effect of incubation time with the polymers, the biofilm reduction was also measured after 24-hour incubation. Although the biofilm biomass was further reduced, the biofilm reduction from 2-hour incubation is relatively small, indicating that most biofilm biomass was removed within the fist 2 hours. Interestingly, PE31 and PE0 did show little or no difference in biofilm eradication, although the antimicrobial activity of PE31 against planktonic S. mutans is higher than PE0. This may suggest that the cationic functionality of polymers is a dominant factor in the eradication of S. mutans biofilm rather than the hydrophobicity of polymers. The cationic properties of polymers may favor binding of polymers to negatively charged biopolymers in the extracellular biofilm matrix, which may ultimately cause physical disruption of biofilm matrix. Alternatively, the polymer might kill bacteria in biofilm, which triggered biofilm dispersion. The anti-biofilm mechanism of polymers will be examined and discussed in detail below.
Figure 4.
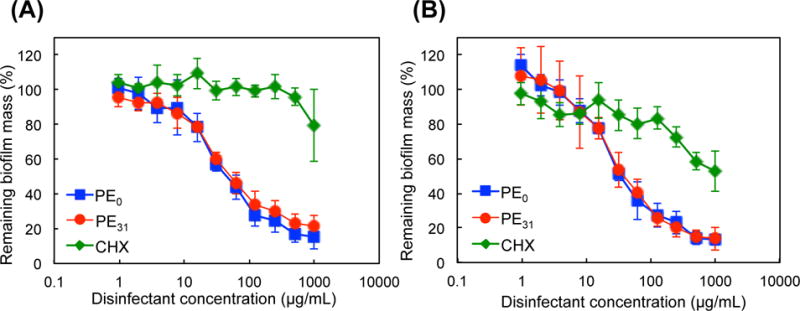
Eradication of pre-formed S. mutans biofilm by antimicrobial polymers.
One-day matured S. mutans biofilm was incubated with the polymers PE0 and PE31 or chlorhexidine (CHX), and the mass of remaining biofilm was determined by CV staining after 2- (A) or 24- (B) hour incubation, relative to solvent treatment as 100% (0.001% acetic acid for PE0 and PE31 and water for CHX).
Eradication of S. mutans biofilm by rinsing and swishing
The results described above indicated that the polymers can reduce the mass of S. mutans biofilms. We question if these polymers could be used as active ingredients in mouthwash, which could potentially work as a “liquid toothbrush” in order to remove S. mutans biofilm and thus reduce the risk of dental decay. In order to test this idea, we designed an in vitro assay to simulate rinsing and swishing of a mouth using mouthwash. S. mutans biofilm was prepared on a bottom of 96-well plates. The polymer solution was added to the well and vigorously mixed by repeatedly dispensing and drawing the solution using a pipette ~30 times for 30 seconds (Fig. 5A). In this procedure, we paid special attention to avoid direct contact of a pipet tip to the biofilm so that the biofilm was not scratched or damaged physically. We used the treatment time of 30 seconds because it is commonly suggested by commercial mouthwash products as an adequate time to rinse. The biofilm biomass was not changed by the treatment with water or 0.001% acetic acid (vehicle for the polymers) after static incubation or vigorously mixing, indicating that S. mutans biofilm is resistant to rinsing and swishing with solvents (Fig. 5B). However, PE31 and PE0 removed 40–60% of biofilm mass at 62.5, 125, and 250 μg/mL (Fig. 5C). PE0 reduced the biofilm mass more than PE31 at 125, and 250 μg/mL. On the other hand, chlorhexidine did not reduce the biofilm mass in the range of concentrations used in this study. We wondered if the biofilm eradication is due to the detergent-like property of amphiphilic polymers. If so, S. mutans biofilm may be also susceptible to a cationic surfactant. To test this hypothesis, we examined the biofilm eradication activity of conventional surfactant cetyltrimethylammonium bromide (CTAB). CTAB did not remove S mutant biofilm (Fig. 5C), suggesting that polymeric structures are required to remove biofilm mass.
Viability of bacteria in remaining S. mutans biofilms
As indicated above, the polymers can remove significant amounts of biofilm mass when swished with solution containing the polymers. However, did the polymers kill the bacteria in the biofilm? To answer this question, we examined the cell viability of S. mutans in the remaining biofilms by confocal laser scanning fluorescence microscopy.19–20 The control biofilm showed a thick layer of bacteria containing many viable cells (green-colored). The biofilm treated with PE0 and PE31 at 125 μg/mL showed a lower fluorescence intensity than the control, reflecting the biofilm eradiation by the polymers (Fig. 6). The volume of biofilm treated with PE0 is smaller than that of PE31, which is in agreement with the results presented in Figure 5 illustrated that PE0 removed more biofilm mass than PE31. However, many bacteria in the remaining biofilm treated by PE0 were viable. This result indicates that PE0 can remove the biofilm mass, but it cannot kill the bacteria embedded in the biofilm. This result also indicates that it is unlikely that PE0 eradicates biofilms by killing bacteria in the biofilm. The remaining biofilm after PE31 treatment contains more dead bacteria with fewer viable bacteria than the PE0-treated biofilm. The result indicates that PE31 has the potential to kill more bacteria in the biofilm than PE0, which reflects the higher bactericidal activity of PE31 against planktonic S. mutans. In contrast, while chlorhexidine failed to remove the biofilm, most bacteria in the biofilm were dead. Although the polymer concetration of 125 μg/mL was well above the MBC values of PE0 (62.5 μg/mL) and PE31 (10.4 μg/mL), the polymers could not kill S. mutans in the biofilms. One possibility is the low killing rate of polymers. The results on the bactericidal kinetics of polymers (Figure 2) suggested that it takes 60–90 minutes for the polymers to effectively kill S. mutans (> 90% killing) at the concentrations of twice MIC values. Therefore, the antimicrobial action of polymers may inherently require relatively longer period of time to be effective, and the treatment time of 30 seconds may be too short for the polymers to kill bacteria. Another possibility is that the biopolymer network of biofilm matrix traps the polymer chains well and prevents the diffusion of polymer chains to reach the embedded bacteria. In contrast, beause of small molecualer size, chlorhexidine could penetrate into the biofilm matrix and kill the bacteria effectively. While the mechanism is not clear at this point, the results described above suggest that the cationic polymer structures play an important role in the removal of the biofilms. We speculate that the interaction of cationic polymer chains with biofilm matrix physically makes the biofilm integrity weak, facilitating biofilm eradiation by the polymers. In this study, we prepared an S. mutnas biofilm model in a culture medium. However, interaction of S. mutans with salivary components may induce changes in gene expression that could potentially influence not only biofilm maturation, but also the composition and biological activity of biofilms.6 To better evaluate a therapeutic potential of the polymers, further investigation on the ability of polymers to eradicate oral biofilm is needed in the presence of human saliva.
Figure 6.
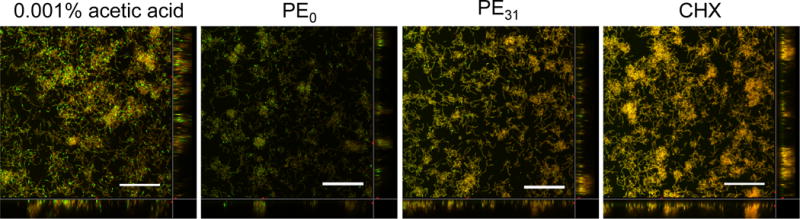
S. mutans biofilm images after treatment with antimicrobial polymers and chlorhexidine (CHX). The biofilms after antimicrobial treatment were stained by a LIVE/DEAD® staining kit and imaged by confocal scanning laser microscopy. Green color (SYTO® 9) showed live bacterial cells, and red/orange color (propidium iodide) showed dead bacterial cells. 0.001% acetic acid was tested as solvent control. [Polymer or CHX] = 125 μg/mL. Bar = 30 μm.
Cytotoxocixy of polymers against oral cells
Finally, we determined the cytotoxicity of polymers against human gingival fibroblasts (hGFs) and periodontal ligament stem cells (PDLSCs)22 (Fig. 7). These cells were incubated with polymers or chlorhexidine for 10 min according to the previous studies in literature regarding the cytotoxicity test of commercial toothpastes36 and mouthwashes37–38. PE0 did not show any significant cytotoxicity against both cell lines, while PE31 reduced the cell viability by 60% at 250 μg/mL. Chlorhexidine also showed cytotoxicity against both cell lines and reduced the cell viability by ~50% at 250 μg/mL. The cytotoxicity of PE31 is comparable to chlorhexidine commonly used in mouthwash for treatment of periodontal disease. When the PDLSCs were exposed to the polymers for 24 hours, the cell viability further decreased (Fig. S4) as comapred to 10 minute incunbation, and the half maximal lethal concentrations (LC50) of PE0 and PE31 were 80 and 45 μg/mL, respectively. For comparison, the LC50 value of chlorhexidine was 13 μg/mL under the same condition. It should be noted that the typical concentration of chlorhexidine is 0.12–0.20% (1.2–2.0 mg/mL), which is significantly higher than the concentrations used in this study. In addition, mouthwash contains other ingredients including surfactants, which likely bind to chlorhexidine and mask the antimicrobial activity and cytotoxicity of chlorhexidine as well. Therefore, the presented results of chlorhexidine and polymers may not directly reflect the cytotoxicity of these compounds when used in mouthwash formula. The investigation of the biological activity of polymers and chlorhexidine in mouthwash is beyond the scope of this study, but a subject of future studies.
Figure 7.
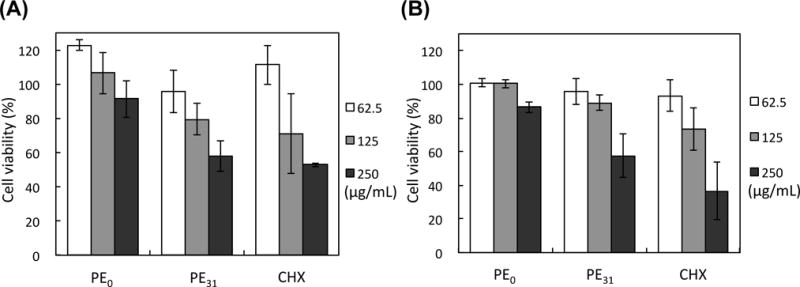
Cytotoxicity of antimicrobial polymers and chlorhexidine (CHX) against human gingival fibroblasts (hGFs) (A) and periodontal ligament stem cells (PDLSCs) (B). Cell viability was determined against hGFs and PDLSCs after incubation with the polymers or chlorhexidine for 10 min, relative to solvents as 100% cell viability (0.001% acetic acid for PE0 and PE31 and water for CHX).
CONCLUSIONS
In summary, the anti-bacterial and anti-biofilm activities of AMP-mimetic methacrylate polymers against cariogenic bacterium S. mutans were investigated. The polymers inhibited the growth of planktonic S. mutans as well as prevented the formation of S. mutans biofilm. The polymers also removed significant biomass of one-day matured S. mutans biofilms. The homopolymer PE0 did not show any significant cytotoxicity to human gingival fibroblast and periodontal ligament stem cells for 10 minutes exposure. Amphiphilic copolymer PE31 reduced the cell viability of these cells under the same condition, and the cytotoxicity level of PE31 was comparable to chlorhexidine. The results support our hypothesis on controlling S. mutans growth and biofilm formation by AMP-mimetic antimicrobial polymers. However, the activity of polymers in an environment close to oral cavity needs to be thoroughly studied to evaluate the potential of antimicrobial polymers as active ingredients of oral care products.
Supplementary Material
Acknowledgments
The authors acknowledge the National Science Foundation for NSF CAREER Award (DMR-0845592), NIH U01DE023771, and Department of Biologic and Materials Sciences, School of Dentistry, University of Michigan. This work was also supported by JSPS Postdoctoral Fellowships for Research Abroad (No. 26-774 to H.T.). We thank Dr. Hamid Mortazavian at the University of Michigan School of Dentistry for help on GPC measurement. We also thank Dr. Christopher Fenno at University of Michigan School of Dentistry for providing S. mutans, Dr. Robertson Davenport at University of Michigan Hospital for providing RBCs, Dr. William Giannobile at University of Michigan School of Dentistry for providing human gingival fibroblast, and Dr. Brian Clarkson and Dr. Jun Liu at University of Michigan School of Dentistry for providing periodontal ligament stem cells.
Footnotes
Supporting Information
Supporting information (SI) includes characterization of boc-protected polymers, 1H NMR spectra, polymer solubility in medium, dose-response hemolysis curve, and results of cytotoxicity evaluation after 24 hour incubation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dye BA, Tan S, Smith V, Lewis BG, Barker LK, Thornton-Evans G, Eke PI, Beltrán-Aguilar ED, Horowitz AM, Li C-H. Vital Health Stat. 2007;11:1–92. [PubMed] [Google Scholar]
- 2.Dye BA, Arevalo O, Vargas CM. Int J Paediatr Dent. 2010;20:132–143. doi: 10.1111/j.1365-263X.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- 3.World Helth Organization. http://www.who.int/mediacentre/factsheets/fs318/en/
- 4.Benjamin RM. Public Health Rep. 2010;125:158–159. doi: 10.1177/003335491012500202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selwitz RH, Ismail AI, Pitts NB. Lancet. 2007;369:51–59. doi: 10.1016/S0140-6736(07)60031-2. [DOI] [PubMed] [Google Scholar]
- 6.Krzyściak W, Jurczak A, Kościelniak D, Bystrowska B, Skalniak A. Eur J Clin Microbiol Infect Dis. 2014;33:499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sbordone L, Bortolaia C. Clin Oral Investig. 2003;7:181–188. doi: 10.1007/s00784-003-0236-1. [DOI] [PubMed] [Google Scholar]
- 8.Robinson C, Shore RC, Brookes SJ, Strafford S, Wood SR, Kirkham J. Crit Rev Oral Biol Med. 2000;11:481–495. doi: 10.1177/10454411000110040601. [DOI] [PubMed] [Google Scholar]
- 9.Zasloff M. Nature. 2002;415:389–95. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 10.Brogden KA. Nature Reviews Microbiology. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 11.Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. J Am Chem Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 12.Tew GN, Scott RW, Klein ML, Degrado WF. Acc Chem Res. 2009;43:30–39. doi: 10.1021/ar900036b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Punia A, He E, Lee K, Banerjee P, Yang N-L. Chem Commun. 2014;50:7071–7074. doi: 10.1039/c4cc01583e. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda K, Caputo GA. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2013;5:49–66. doi: 10.1002/wnan.1199. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi H, Palermo EF, Yasuhara K, Caputo GA, Kuroda K. Macromol Biosci. 2013;13:1285–1299. doi: 10.1002/mabi.201300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batoni G, Maisetta G, Brancatisano FL, Esin S, Campa M. Curr Med Chem. 2011;18:256–279. doi: 10.2174/092986711794088399. [DOI] [PubMed] [Google Scholar]
- 17.da Silva BR, de Freitas VAA, Nascimento-Neto LG, Carneiro VA, Arruda FVS, de Aguiarc ASW, Cavada BS, Teixeira EH. Peptides. 2012;36:315–321. doi: 10.1016/j.peptides.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Wei GX, Campagna AN, Bobek LA. J Antimicrob Chemother. 2006;57:1100–1109. doi: 10.1093/jac/dkl120. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Tao R, Tong ZC, Ding YL, Kuang R, Zhai SF, Liu J, Ni LX. Peptides. 2012;33:212–219. doi: 10.1016/j.peptides.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Shang DJ, Liang H, Wei S, Yan X, Yang QZ, Sun Y. Appl Microbiol Biotechnol. 2014;98:8685–8695. doi: 10.1007/s00253-014-5927-9. [DOI] [PubMed] [Google Scholar]
- 21.Beckloff N, Laube D, Castro T, Furgang D, Park S, Perlin D, Clements D, Tang H, Scott RW, Tew GN, Diamond G. Antimicrob Agents Chemother. 2007;51:4125–4132. doi: 10.1128/AAC.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi ST. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 23.Nadres ET, Takahashi H, Kuroda K. J Polym Sci, Part A: Polym Chem. 2017;55:304–312. [Google Scholar]
- 24.Palermo EF, Vemparala S, Kuroda K. Biomacromolecules. 2012;13:1632–1641. doi: 10.1021/bm300342u. [DOI] [PubMed] [Google Scholar]
- 25.Hancock REW. Hancock Laboratory Methods. Vol. 2014. Depertment of Microbiology and Immunology, University of British Columbia; British Columbia, Canada: 2001. p http://cmdr.ubc.ca/bobh/methods.htm. [Google Scholar]
- 26.Giacometti A, Cirioni O, Barchiesi F, Del Prete MS, Fortuna M, Caselli F, Scalise G. Antimicrob Agents Chemother. 2000;44:1694–1696. doi: 10.1128/aac.44.6.1694-1696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraigsley AM, Tang K, Lippa KA, Howarter JA, Lin-Gibson S, Lin NJ. Macromol Biosci. 2012;12:1706–1713. doi: 10.1002/mabi.201200214. [DOI] [PubMed] [Google Scholar]
- 28.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. J Microbiol Methods. 2000;40:175–179. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 29.Peeters E, Nelis HJ, Coenye T. J Microbiol Methods. 2008;72:157–165. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Munoz-Bonilla A, Fernandez-Garcia M. Prog Polym Sci. 2012;37:281–339. [Google Scholar]
- 31.Wang CZ, Zolotarskaya OY, Nair SS, Ehrhardt CJ, Ohman DE, Wynne KJ, Yadavalli VK. Langmuir. 2016;32:2975–2984. doi: 10.1021/acs.langmuir.5b04247. [DOI] [PubMed] [Google Scholar]
- 32.Ahn SJ, Burne RA. J Bacteriol. 2007;189:6293–6302. doi: 10.1128/JB.00546-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda K, Caputo GA, DeGrado WF. Chem Eur J. 2009;15:1123–1133. doi: 10.1002/chem.200801523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammadi Z, Abbott PV. Int Endod J. 2009;42:288–302. doi: 10.1111/j.1365-2591.2008.01540.x. [DOI] [PubMed] [Google Scholar]
- 35.Strydonck DAC, Slot DE, Van der Velden U, Van der Weijden F. J Clin Periodontol. 2012;39:1042–1055. doi: 10.1111/j.1600-051X.2012.01883.x. [DOI] [PubMed] [Google Scholar]
- 36.Ghapanchi J, Kamali F, Moattari A, Poorshahidi S, Shahin E, Rezazadeh F, Khorshidi H, Jamshidi S. J Int Oral Health. 2015;7:9–43. [PMC free article] [PubMed] [Google Scholar]
- 37.Ghabanchi J, Moattari A, Tadbir AA, Mardanı M, Shakib M. Aust J Basic Appl Sci. 2012;6:318–320. [Google Scholar]
- 38.Schmidt J, Zyba V, Jung K, Rinke S, Haak R, Mausberg RF, Ziebolz D. Drug Chem Toxicol. 2016;39:322–330. doi: 10.3109/01480545.2015.1121274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


