Abstract
Background
The causes of intellectual disability (ID) are diverse and de novo mutations are increasingly recognised to account for a significant proportion of ID.
Methods and results
In this study, we performed whole exome sequencing on a large cohort of patients with ID or neurodevelopmental delay and identified four novel de novo predicted deleterious missense variants in HECW2 in six probands with ID/developmental delay and hypotonia. Other common features include seizures, strabismus, nystagmus, cortical visual impairment and dysmorphic facial features. HECW2 is an ubiquitin ligase that stabilises p73, a crucial mediator of neurodevelopment and neurogenesis.
Conclusion
This study implicates pathogenic genetic variants in HECW2 as potential causes of neurodevelopmental disorders in humans.
INTRODUCTION
Intellectual disability (ID) is a common and aetiologically heterogeneous disorder, affecting 1% of the general population.1 A significant fraction of profound ID is caused by genomic alterations, with cytogenetically detectable anomalies found in up to 15% of cases and copy number variants in up to 15%–20% of individuals.2,3 Recent progress in whole exome sequencing (WES) have rapidly advanced our identification of genetic causes of ID, establishing de novo mutations as an important and common cause of ID4–6 in 16%–55% of cases.7–10 Many novel candidates genes have been identified through WES and have advanced our understanding of potential molecular mechanisms implicated in neurodevelopmental processes.11–13
In this study, we identified novel de novo missense variants in HECW2 in six independent probands of 3309 total patients with neurodevelopmental delays or ID referred for clinical WES. HECW2, also known as NEDL2, is one of the nine members of the Nedd4 family of HECT domain E3 ubiquitin ligases.14 E3 ligases control specificity of the ubiquitin modification of proteins targeted for degradation. HECW2 acts on a diverse group of proteins including p73, a member of the p53 family with specific neurodevelopmental expression.15 We identified multiple and recurrent de novo, novel variants in HECW2 in six unrelated probands with neurodevelopmental disorders, supporting its role as a potential cause of developmental delay and ID.
MATERIALS AND METHODS
Informed consent was obtained from all participants’ parents included in this study. This study was approved by the Institutional Review Board of Columbia University.
Whole exome sequencing
Genomic DNA was extracted from whole blood from affected children and their parents. Exome sequencing was performed on exon targets captured using the Agilent SureSelect Human All Exon (V.4) (50 Mb) kit or the Clinical Research Exome kit (Agilent Technologies, Santa Clara, California, USA). Whole exome sequencing data for all sequenced family members were analysed using GeneDx’s XomeAnalyzer (a variant annotation, filtering and viewing interface for WES data), which includes nucleotide and amino acid annotations, population frequencies (NHLBI Exome Variant Server and 1000 Genomes databases), in silico prediction tools, amino acid conservation scores and mutation references. Variants were filtered based on inheritance patterns, variant type, population frequencies and gene lists of interest in relation to the patient’s major phenotypic features, as appropriate. The full sequencing methodology and variant interpretation protocol has been previously described.11 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/). As part of our analysis, individuals with de novo variants occurring in the same gene are examined for overlapping clinical features and were Sanger-sequenced to confirm that the variant is present in the proband and neither parent and is de novo.
RESULTS
Exome sequencing was performed in 3309 probands with neurodevelopmental disorders sequenced in a single clinical laboratory; most of the probands had previous non-diagnostic genetic testing including chromosome microarrays. WES of the six probands identified with a HECW2 de novo variant produced an average of 11.5 GB of sequence per sample. Among the 3309 probands with neurodevelopmental disorders, there were six probands with four unique de novo variants in HECW2. All variants were confirmed with Sanger sequencing in the proband and the parents. The false discovery rate corrected q-value of the observing six de novo missense variants in this population is 6.20e–07, using the Transmission and De novo Association algorithm,16 and the default parameters for missense variation in neurodevelopmental cases are γ=4.7 and β=1, with a missense mutation rate of 4.85e–05 for HECW2.17
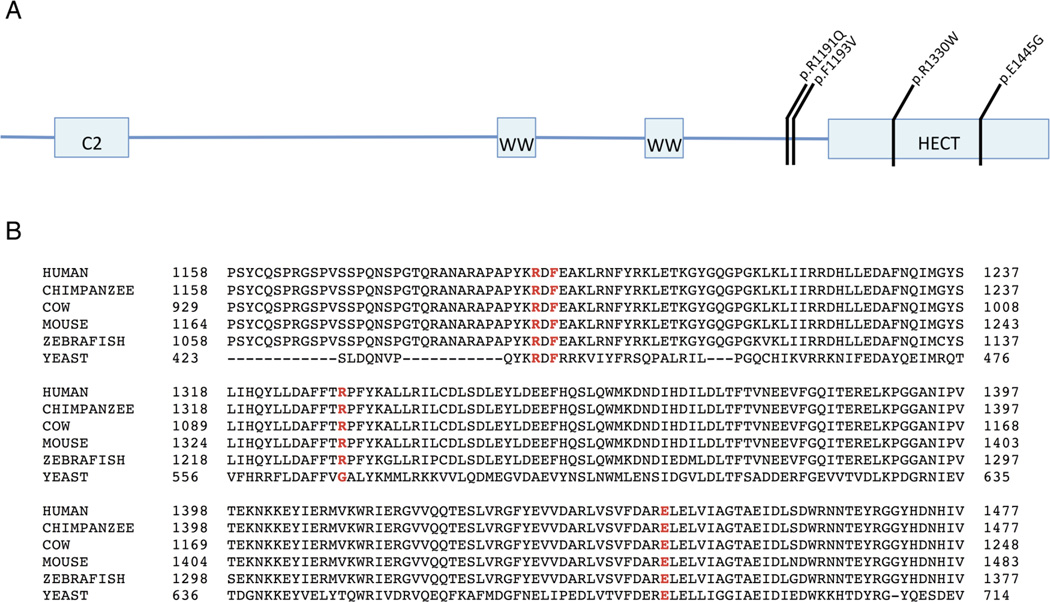
All variants are novel and none have been described in the Exome Aggregation Consortium,18 although the Exome Variant Server reports a synonymous variant at codon 1193.19 The variants were also absent in our own internal database consisting of 9651 adult unaffected controls. Residual Variation Intolerance score for this gene is −2.47 (0.98%), indicating that the gene is intolerant to variation,20 with a misZ of 3.11 (3-sigma conservation) and the pLI or probability of being intolerant to a loss-of-function allele is 1.21 Eight deletions that encompass HECW2 are reported in ClinVar, with six of those individuals demonstrating a phenotype of developmental delay and/or significant developmental morphological phenotypes. The four de novo missense variants are all predicted to be deleterious and damaging to protein function by Polyphen, Provean, SIFT and other prediction algorithms22–24 (table 1). One of the missense variants, R1330W, lies within the HECT domain of the protein (figure 1).
Table 1.
Predictions of pathogenicity for the four novel, de novo missense variants in HECW2
| Amino acid alternation |
Patients | Nucleotide change |
Protein domain |
SIFT prediction |
Polyphen prediction |
Likelihood ratio test |
Mutation taster |
Meta-SVM | MetaLR | Provean |
|---|---|---|---|---|---|---|---|---|---|---|
| p.Arg1191Gln | 1 and 2 | c.3572 G>A | Inter-domain | Deleterious | Probably damaging |
Deleterious | Disease causing |
Deleterious | Deleterious | Deleterious |
| p.Phe1193Val | 3 and 4 (twins) |
c.3577 T>G |
Inter-domain | Deleterious | Probably damaging |
Deleterious | Disease causing |
Deleterious | Deleterious | Deleterious |
| p.Arg1330Trp | 5 and 6 | c.3988 C>T |
HECT | Deleterious | Probably damaging |
Deleterious | Disease causing |
Deleterious | Deleterious | Deleterious |
| p.Glu1445Gly | 7 | c.4334 A>G |
HECT | Deleterious | Probably damaging |
Deleterious | Disease causing |
Deleterious | Deleterious | Deleterious |
LR, logistic regression; SVM, support vector machine.
Figure 1.
(A) Location of de novo variants in HECW2 and (B) conservation of these amino acids across species are shown.
Age of patients with de novo HECW2 missense variants ranges from 18 months to 11 years (table 2). All patients have developmental delay and hypotonia and are all severely neurologically impaired. Common clinical features include seizures (five probands), cortical visual impairment (three probands), autism (two probands), dysmorphic features (five probands) (figure 2) and problems with feeding requiring gastrostomy tube (three probands).
Table 2.
Phenotypic characteristics of patients with de novo HECW2 missense variants
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | |
|---|---|---|---|---|---|---|---|
| Age | 11 yo | 9 yo | 33 mos | 33 mos | 18 mos | 3 yo | 6 yo |
| Gender | Female | Male | Female | Female | Female | Male | Female |
| Mutation | c.3572G>A: p.Arg1191Gln |
c.3572G>A: p.Arg1191Gln |
c.3577T>G: p.Phe1193Val |
c.3577T>G: p.Phe1193Val |
c.3988C>T: p.Arg1330Trp |
c.3988C>T: p.Arg1330Trp |
c.4334A>G p.Glu1445Gly |
| Birth weight | 3175 g | 3941 g | 2412 g | 2155 g | 3544 g | 4196 g | 2353 g |
| Current growth parameters |
At 10 yo Wt=34.56 kg (36%ile) Ht=141 cm (35%ile) |
Wt=25.4 kg (21%ile) Ht=131 cm (32%ile) OFC=52.8 cm (50%ile) |
Wt=13.8 kg (60%ile) Ht=90 cm (23%ile) OFC=48 cm (39%ile) |
Wt=12.8 kg (34%ile) Ht=87.9 cm (10%ile) OFC=50 cm (86%ile) |
At 15 mos Wt=10.22 kg (59% ile) Ht=79.2 cm (88%ile) OFC=46 cm (65% ile) |
At 23 mos Wt=12.56 kg (69%ile) Ht=90 cm (87%ile) OFC=48 cm (48%ile) |
Wt=28.8 kg (<3%ile), Ht=104 cm (<3%ile), OFC=49.5 cm (<3%ile) |
| DD | + | + | + | + | + | + | + |
| ID | + (Moderate) | + (IQ=55) | NA | NA | NA | + | + |
| Autism | + | + | NA | NA | NA | NA | − |
| Age at sitting | 9 mos | 11–12 mos | Unable | Unable | Unable | Unable | Unable |
| Age at walking |
3 yo | 3 yo with braces | Unable | Unable | Unable | Unable | Unable |
| Age at talking | Non-verbal | 2 yo; currently has ~15 words | Non-verbal | Non-verbal | One word at 17 mos |
Non-verbal | Non-verbal |
| Hypotonia | + | + | + | + | + | + | + |
| Seizures | Generalised tonic clonic seizures at 5 yo (now seizure free) |
EEG showed excessive slowing with abnormal burst discharges |
Currently intractable tonic seizures |
Currently intractable tonic seizures |
− | Infantile spasms starting ~5 mos; has been a few months seizure free |
EEG showed multifocal and generalised epileptiform and slow spike-wave discharges with diffuse background slowing |
| Abnormal behaviours |
Hand flapping, self-injurious behaviours, aggressive behaviour, inappropriate laughter |
Self-stimulatory behaviour | Rocking and flapping hand behaviours |
Rocking and flapping hand behaviours |
Repetitive hand movements, self-injurious behaviours |
− | Self-stimulatory rocking, sucking on fingers |
| Other neurological problems |
Wide-based gait, exercise intolerance |
− | − | − | − | − | Choreiform movements |
| Visual problems |
Strabismus | None | Strabismus, nystagmus with fevers |
Strabismus, nystagmus with fevers |
CVI | CVI | Optic atrophy and CVI |
| Brain MRI | Normal | Normal | Mild ventriculomegaly and increased extra-axial fluid |
Mild ventriculomegaly and increased extra-axial fluid |
Normal | Cerebral atrophy, thin corpus callosum |
Progressive cerebral atrophy with mild cerebellar loss, atrophy of visual pathways, several small arachnoid cysts |
| GI problems | GERD, constipation | None | G-tube fed | G-tube fed | Requires burping regularly |
G-tube fed | G-tube, gastroparesis, constipation |
| Dysmorphic features |
Midface retrusion, thin vermillion upper lip |
Flat nasal bridge, mild epicanthal folds, telecanthus, thick eyebrows, synophrys, short, upturned nose with bulbous nasal tip, midface hypoplasia, full lower lip, widely spaced teeth, tongue protrusion, thick supraorbital ridge, deep set eyes, mouth is wide and down turned, prominent central incisors |
Deep set eyes, ptosis, prominent forehead |
Deep set eyes, ptosis, prominent forehead |
Slightly large ears, upturned nose with bulbous nasal tip |
Sparse eyebrows, slightly depressed nasal bridge, upturned nasal tip |
Hypotonic facies, highly arched palate |
| Other | − | Joint laxity in knees, ankles, pain insensitivity | Osteopenia, frequent waking at night |
Osteopenia, frequent waking at night |
− | Hyperglutaminaemia, scrotal hypoplasia, pectus excavatum |
Gracile bones, mild osteopenia throughout with no definite dysplasia. Cardiomyopathy started prenatally, heart block, prolonged QT interval |
| Other genetic findings |
− | − | − | − | − | 439 Mb 15q11.2 duplication; de novo p. G184E SLC7A7 variant |
Heterozygous G200V variant in EIF2B2 gene |
%ile, percentile; −, absent; +, present; CVI, cortical visual impairment; GERD, gastro-oesophageal reflux disease; GI, gastrointestinal; G-tube, gastrostomy tube; Ht, height; ID, intellectual disability; mos, months old; NA, not available; OFC, occipital frontal circumference; Wt, weight; yo, years old.
Figure 2.
Photographs showing the facial features of probands with variants in HECW2. (A) Photograph showing patient 1 with midface retrusion and thin vermillion upper lip. (B) Photograph showing patient 2 with features including flat nasal bridge, mild epicanthal folds, telecanthus, thick eyebrows, synophrys, short, upturned nose with bulbous nasal tip, widely spaced teeth and deep set eyes. (C and D) Photographs showing patients 3 and 4, respectively, with deep set eyes, ptosis and a prominent forehead. (E and F) Photographs showing patient 5 with slightly large ears and an upturned nose with bulbous nasal tip. (G and H) Photographs showing patient 6 with sparse eyebrows, slightly depressed nasal bridge and upturned nasal tip.
The two unrelated probands (patients 1 and 2) with the same de novo p.Arg1191Gln variant have autism and similar dysmorphic features with midface retrusion. Patient 2 had an abnormal EEG as an infant, with excessive slowing throughout and abnormal burst discharges. Patient 1 had absence and generalised tonic clonic seizures starting at age 4 years with an abnormal EEG (slow for age showing runs of rhythmic theta activity with intermixed spikes). She also has absent speech, a broad-based ataxic gait, extremely high pain tolerance, aggressive and self-injurious behaviour and strabismus. She was clinically felt to have a phenotype similar to Angelman syndrome and had negative Angelman molecular testing.
The p.Phe1193Val variant was identified once in a set of monozygotic twins (patients 3 and 4) with generalised hypotonia with severe kyphosis, strabismus, nystagmus and mildly dysmorphic features (highly arched palate, prominent forehead and deep set eyes). MRI showed non-specific cortical atrophy, with resultant ex-vacuo dilatation of ventricles and increased extra-axial fluid. One twin (patient 3) had intrauterine growth restriction and neonatal hypoglycaemia and the other twin (patient 4) had hypothermia after birth. Both twins were able to be breast-fed for 7 months but had feeding regression which led to all feeds being given through gastrostomy tube. At approximately 30 months of age, they both began having intractable tonic clonic seizures.
Two unrelated patients (patients 5 and 6) carry the same p. Arg1330Trp variant. Both have cortical visual impairment. Patient 5 has abnormal behaviours with repetitive movements and self-injurious behaviours. Patient 6 had a history of infantile spasms and now at 3 years of age has intractable tonic seizures as well as feeding difficulties requiring a gastrostomy tube. MRI demonstrated loss of cerebral hemispheric white matter under-operculisation and thin corpus callosum. Patient 6 also had hyperglutaminaemia and on chromosome microarray analysis had a 439 Mb 15q11.2 duplication that contains four genes (arr 15q11.2(22,770,421–23,209,654)x3) and a de novo p.G184E SLC7A7 variant. SLC17A7 encodes a vesicular glutamate transporter expressed in the brain that plays a role in synaptic transmission. To our knowledge, no mutations in SLC17A7 have been associated with a specific human disease. This variant was not observed in approximately 115 000 alleles in the ExAC dataset. We cannot exclude the possibility that this variant contributes to the phenotype in this patient.
The p.Glu1445Gly variant was also identified once in patient 7 with hypotonia, epileptic encephalopathy and cortical visual impairment. This patient also has choreiform movements. MRI showed generalised volume loss with non-specific white matter changes. The patient has gastroparesis and constipation and has a gastrostomy tube and also has cardiomyopathy. A heterozygous variant in the EIF2B2 gene was also found in this patient. Typically, EIF2B2-related disorders are inherited in an autosomal recessive manner and this patient was not found to have a second variant in the gene.
DISCUSSION
In our series of 3309 patients with neurodevelopmental delays, we identified six probands, each with one of four de novo, novel missense variants in HECW2 in highly conserved regions of the gene. Two of the variants are recurrent. The clinical characteristics shared by all the patients include severe developmental delay and hypotonia. Seizures, cortical visual impairment, autism, stereotypical and repetitive behaviours and difficulty with feeding are common associated phenotypes observed in the majority of the patients. MRIs in some patients are largely normal and showed only non-specific cortical atrophy with ex-vacuo ventriculomegaly.
HECW2 is an E3 ubiquitin ligase and contains a N-terminal C2 domain, two internal WW domains that bind to PY motifs of substrate proteins and a C-terminal HECT domain (figure 1). HECW2 is expressed largely in brain, lung and heart tissue.25
HECW2 is extremely intolerant of haploinsufficiency and the patients with microdeletions that encompass HECW2 have overlapping phenotypes, suggesting that the missense variants have a functional consequence. However, functional studies will be necessary to assess the missense alleles for loss or gain of function or dominant negative mechanism.
The amino acids identified in this study in individuals with neurodevelopmental deficits are highly conserved, with alterations predicted to be extremely deleterious (figure 1). The protein prediction algorithms consistently predict that these missense variants are harmful to protein structure and function.
The main known function of HECW2 is to stabilise and enhance transcriptional activity of p73.25 p73 is a member of the p53 family of tumour suppressors that promotes cell cycle arrest and apoptosis and also has an essential role in neurodevelopment.15 Loss of HECW2 activity could lead to decreased p73 activity.
Mouse knockout models of p73 show profound central nervous system abnormalities with hippocampal dysgenesis, cortical thinning, progressive communicating hydrocephalus and small olfactory bulbs and subventricular zones.26,27 The brain pathology is progressive, with worsening cortical loss as the mice age.28 p73 is specifically involved in embryonic neurogenesis and maintenance of adult neural stem cells (NSCs). Neurospheres derived from p73−/− cells are smaller than wild type, demonstrating decreased proliferation and survival of neural progenitors and have decreased capacity for self-renewal.29 p73-deficient animals are born with decreased pool of neural progenitors and continue to display defects in adult neurogenesis and long-term maintenance of NSCs.27,30,31 Additionally, neurons and oligodendrocytes that differentiate from p73−/− progenitors are abnormal, showing deficiencies in both quality and quantity. Neurons have defects in neurite outgrowth and synaptic connectivity.27,32
p73 is an essential regulator of embryonic neurogenesis, neuronal differentiation and organisation and maintenance of adult NSCs.33 HECW2 stabilises and enhances p73 transcriptional activity and aberrant p73 activity could at least partially explain the severe neurological phenotype we observed.
HECW2 also interacts with other regulators of cell growth and differentiation. HECW2 has recently been shown to co-precipitate with the APC/C-Cdh1 complex that mediates the transition from metaphase to anaphase. Overexpression or underexpression of HECW2 leads to early or delayed activation of the complex, respectively, and resultant cell cycle instability.34 In addition to its role in cell cycle regulation, the APC/C-Cdh1 complex is highly expressed in brain, specifically in post-mitotic neurons.35 APC/C-Cdh1 is involved in neuronal patterning and regulating axon growth in the developing and mature brain. In vivo knockdown of Cdh1 in developing animal models caused aberrant axonal growth trajectories and fibre defasciculation.36 Further work is necessary to determine if this functional role of the APC/CDH1 complex requires HECW2 interaction and regulation.
SUMMARY
Four novel de novo predicted deleterious missense variants in HECW2 were identified in six probands with severe developmental delay, hypotonia and dysmorphic features and was frequently associated with seizures, cortical visual impairment, autism, stereotypical and repetitive behaviours and difficulty with feeding. Two of the missense variants were recurrent and HECW2 is intolerant of haploinsufficiency. The phenotype of these patients overlaps with patients with contiguous gene deletions of HECW2 and additional functional data are necessary to evaluate the E3 ubiquitin ligase activity of these alleles and their effects on downstream proteins such as p73 and APC/C-Cdh1 in neurogenesis and neuronal differentiation.
Acknowledgments
The authors thank the families for their generous contribution. They would like to thank Christina Rigelsky of the Cleveland Clinic for some assistance with proband 7’s testing.
Funding This work was supported in part by a grant from the Simons Foundation.
Footnotes
Contributors Study concept and design: ERB and WKC. Recruitment of patients and collection of clinical information: MTC, CE, YS, DAS, JW, NHR, FB, SD, PM, CGB, MB, CM, LAW and CJT. Acquisition of data: ERB, MTC, CE, YS, DAS, JW, NHR, FB, SD, PM, CGB, MB, CM, LAW, CJT and WKC. Analysis and interpretation of data: ERB, MTC, YS, KR, FM and WKC. Drafting of the manuscript: ERB. Critical revision of the manuscript: ERB, MTC, CE, YS, DAS, JW, NHR, FB, SD, PM, CGB, MB, CM, LAW, CJT, YS, KR, FM and WKC. Study supervision: WKC.
Competing interests MTC, KR and FM are employees of GeneDx. WKC was previously an employee of GeneDx.
Patient consent Parental/guardian consent obtained.
Ethics approval Columbia University.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement The variants reported in this study have been deposited in ClinVar.
REFERENCES
- 1.Srivastava AK, Schwartz CE. Intellectual disability and autism spectrum disorders: causal genes and molecular mechanisms. Neurosci Biobehav Rev. 2014;46(Pt 2):161–174. doi: 10.1016/j.neubiorev.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellison JW, Rosenfeld JA, Shaffer LG. Genetic basis of intellectual disability. Annu Rev Med. 2013;64:441–450. doi: 10.1146/annurev-med-042711-140053. [DOI] [PubMed] [Google Scholar]
- 3.Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 4.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 5.Ku CS, Polychronakos C, Tan EK, Naidoo N, Pawitan Y, Roukos DH, Mort M, Cooper DN. A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol Psychiatry. 2013;18:141–153. doi: 10.1038/mp.2012.58. [DOI] [PubMed] [Google Scholar]
- 6.Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BB, Kleefstra T, Brunner HG, Vissers LE, Veltman JA. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 8.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Ropke A, Moog U, Riess A, Thiel CT, Tzschach A, Wiesener A, Wohlleber E, Zweier C, Ekici AB, Zink AM, Rump A, Meisinger C, Grallert H, Sticht H, Schenck A, Engels H, Rappold G, Schrock E, Wieacker P, Riess O, Meitinger T, Reis A, Strom TM. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 9.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, del Rosario M, Hoischen A, Scheffer H, de Vries BB, Brunner HG, Veltman JA, Vissers LE. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 10.Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, Vertino-Bell A, Smaoui N, Neidich J, Monaghan KG, McKnight D, Bai R, Suchy S, Friedman B, Tahiliani J, Pineda-Alvarez D, Richard G, Brandt T, Haverfield E, Chung WK, Bale S. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka AJ, Cho MT, Millan F, Juusola J, Retterer K, Joshi C, Niyazov D, Garnica A, Gratz E, Deardorff M, Wilkins A, Ortiz-Gonzalez X, Mathews K, Panzer K, Brilstra E, van Gassen KL, Volker-Touw CM, van Binsbergen E, Sobreira N, Hamosh A, McKnight D, Monaghan KG, Chung WK. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am J Hum Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang L, Cho MT, Retterer K, Folk L, Humberson J, Rohena L, Sidhu A, Saliganan S, Iglesias A, Vitazka P, Juusola J, O’Donnell-Luria AH, Shen Y, Chung WK. Mutations in ARID2 are associated with intellectual disabilities. Neurogenetics. 2015;16:307–314. doi: 10.1007/s10048-015-0454-0. [DOI] [PubMed] [Google Scholar]
- 13.Damseh N, Simonin A, Jalas C, Picoraro JA, Shaag A, Cho MT, Yaacov B, Neidich J, Al-Ashhab M, Juusola J, Bale S, Telegrafi A, Retterer K, Pappas JG, Moran E, Cappell J, Anyane Yeboa K, Abu-Libdeh B, Hediger MA, Chung WK, Elpeleg O, Edvardson S. Mutations in SLC1A4, encoding the brain serine transporter, are associated with developmental delay, microcephaly and hypomyelination. J Med Genet. 2015;52:541–547. doi: 10.1136/jmedgenet-2015-103104. [DOI] [PubMed] [Google Scholar]
- 14.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 15.Killick R, Niklison-Chirou M, Tomasini R, Bano D, Rufini A, Grespi F, Velletri T, Tucci P, Sayan BS, Conforti F, Gallagher E, Nicotera P, Mak TW, Melino G, Knight RA, Agostini M. p73: a multifunctional protein in neurobiology. Mol Neurobiol. 2011;43:139–146. doi: 10.1007/s12035-011-8172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, State MW, Devlin B, Roeder K. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnstrom K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH, Jr, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale BM, Daly MJ. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Exome Aggregation Consortium (ExAC) Cambridge, MA: http://exac.broadinstitute.org. [Google Scholar]
- 19.Serve EV. NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: 2015. [Google Scholar]
- 20.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T, O’Donnell-Luria A, Ware J, Hill A, Cummings B, Tukiainen T, Birnbaum D, Kosmicki J, Duncan L, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Cooper D, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki M, Levy Moonshine A, Natarajan P, Orozco L, Peloso G, Poplin R, Rivas M, Ruano-Rubio V, Ruderfer D, Shakir K, Stenson P, Stevens C, Thomas B, Tiao G, Tusie-Luna M, Weisburd B, Won H-H, Yu D, Altshuler D, Ardissino D, Boehnke M, Danesh J, Roberto E, Florez J, Gabriel S, Getz G, Hultman C, Kathiresan S, Laakso M, McCarroll S, McCarthy M, McGovern D, McPherson R, Neale B, Palotie A, Purcell S, Saleheen D, Scharf J, Sklar P, Patrick S, Tuomilehto J, Watkins H, Wilson J, Daly M, MacArthur D. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 25.Miyazaki K, Ozaki T, Kato C, Hanamoto T, Fujita T, Irino S, Watanabe K, Nakagawa T, Nakagawara A. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem Biophys Res Commun. 2003;308:106–113. doi: 10.1016/s0006-291x(03)01347-0. [DOI] [PubMed] [Google Scholar]
- 26.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 27.Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A, Bode D, Dobbelstein M, Bruck W, Moll UM. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–1829. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pozniak CD, Barnabé-Heider F, Rymar VV, Lee AF, Sadikot AF, Miller FD. p73 is required for survival and maintenance of CNS neurons. J Neurosci. 2002;22:9800–9809. doi: 10.1523/JNEUROSCI.22-22-09800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Cano L, Herreros-Villanueva M, Fernandez-Alonso R, Ayuso-Sacido A, Meyer G, Garcia-Verdugo JM, Silva A, Marques MM, Marin MC. p73 deficiency results in impaired self-renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Dis. 2010;1:e109. doi: 10.1038/cddis.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A, Mak TW, Wojtowicz MJ, Miller FD, Kaplan DR. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol. 2010;20:2058–2065. doi: 10.1016/j.cub.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 31.Fatt MP, Cancino GI, Miller FD, Kaplan DR. p63 and p73 coordinate p53 function to determine the balance between survival, cell death, and senescence in adult neural precursor cells. Cell Death Differ. 2014;21:1546–1559. doi: 10.1038/cdd.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niklison-Chirou MV, Steinert JR, Agostini M, Knight RA, Dinsdale D, Cattaneo A, Mak TW, Melino G. TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor. Proc Natl Acad Sci USA. 2013;110:18952–18957. doi: 10.1073/pnas.1221172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niklison-Chirou MV, Killick R, Knight RA, Nicotera P, Melino G, Agostini M. How does p73 cause neuronal defects? Mol Neurobiol. 2015 doi: 10.1007/s12035-015-9381-1. http://dx.doi.org/10.1007/s12035-015-9381-1. [DOI] [PubMed] [Google Scholar]
- 34.Lu L, Hu S, Wei R, Qiu X, Lu K, Fu Y, Li H, Xing G, Li D, Peng R, He F, Zhang L. The HECT type ubiquitin ligase NEDL2 is degraded by anaphase-promoting complex/cyclosome (APC/C)-Cdh1, and its tight regulation maintains the metaphase to anaphase transition. J Biol Chem. 2013;288:35637–35650. doi: 10.1074/jbc.M113.472076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters JM. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci USA. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]