Abstract
Background
Primary immunodeficiency diseases (PIDDs) are clinically and genetically heterogeneous disorders thus far associated with mutations in more than 300 genes. The clinical phenotypes derived from distinct genotypes may overlap. Genetic etiology can be a prognostic indicator of disease severity and can influence treatment decisions.
Objective
To investigate the ability of whole-exome screening methods to detect disease-causing variants in individuals with PIDDs.
Methods
Individuals with PIDDs from 278 families from 22 countries were investigated using whole-exome sequencing (WES). Computational CNV prediction pipelines and an exome-tiling chromosomal microarray were also applied to identify intragenic copy number variants (CNVs). Analytic approaches initially focused on 475 known or candidate PIDD genes, but were non-exclusive and were further tailored based upon clinical data, family history and immunophenotyping.
Results
A likely molecular diagnosis was achieved in 110 (40%) unrelated probands. Clinical diagnosis was revised in about half (60/110) and management was directly altered in nearly a quarter (26/110) of families based on the molecular findings. Twelve PIDD-causing CNVs were detected, including seven smaller than 30 Kb that would not have been detected with conventional diagnostic CNV arrays.
Conclusion
This high-throughput genomic approach enabled detection of disease-related variants in unexpected genes, permitted detection of low-grade constitutional, somatic and revertant mosaicism, and provided evidence of a mutational burden in mixed PIDD immunophenotypes.
Keywords: Primary immunodeficiency disease, whole-exome sequencing, copy number variants
Introduction
Primary immunodeficiency diseases (PIDDs) are clinically and genetically heterogeneous. The severe end of the clinical spectrum includes acutely life-threatening conditions. Milder disorders may present with frequent, severe, and/or unusual infections, autoimmune, autoinflammatory or lymphoproliferative phenomena, and with or without dysmorphic features.1 Underdiagnosis and diagnostic delay can contribute to morbidity and mortality.2-9 Identification of the molecular cause and disease mechanism(s) may enable early protective interventions and potentially targeted or curative therapy.10-17 Additionally, in individuals with radiosensitive PIDDs, interventions such as radiographic imaging and the use of DNA-damaging radiomimetic drugs in pre-treatment conditioning must be addressed.
Mutations in more than 300 genes are thus far known to cause PIDDs. Diagnostic molecular testing is further complicated by overlapping clinical phenotypes.1 Genetic testing for PIDD-associated gene variants using iterative Sanger sequencing of single genes can be time-consuming and costly. Furthermore, analysis of very rare disease-related genes is not always available in diagnostic laboratories. While the utility of targeted capture of a panel of known PIDD genes has been demonstrated, this method has limitations, for example the inability to detect new disease genes.18-20 Whole-exome sequencing (WES) has the potential to provide rapid molecular diagnoses and improve diagnostic yield,21, 22 which is particularly useful in clinically and genetically heterogeneous disorders such as PIDD.
In an unbiased approach to disease gene discovery and molecular diagnosis in a large cohort (n=278) with a broad range of PIDD phenotypes, we assayed all known relevant genes using WES. Study participants had been evaluated previously with conventional tools, including clinically available immunological assays and genetic tests, but lacked a molecular diagnosis at the time of enrollment. WES and Sanger sequencing detected single nucleotide variants (SNVs) and small indels. Multiplex ligation-dependent probe amplification (MLPA),23 chromosomal microarray and tailored bioinformatic analysis of WES data enabled detection of both intragenic and larger copy number variants (CNVs). Gene variant evaluation was performed according to recommended genetic guidelines, which include information concerning variant type, its relevance to phenotype, segregation, predicted impact, and the biological evidence for a definitive or potential functional effect.
Methods
Clinical samples
Study centers included the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) at Baylor College of Medicine (BCM), and the Center for Human Immunobiology, Division of Immunology Allergy and Rheumatology at Texas Children's Hospital (TCH) in Houston USA, and Oslo University Hospital (OUS), Oslo, Norway. A total of 278 PIDD families from 22 countries consecutively recruited from 2010 to 2015 participated. The 278 probands ranged in age from seven weeks to 71 years with a mean of 14.7 years. The 78 probands (47 males, 31 females) tested in Oslo ranged in age from four months to 71 years with a mean of 18.4 years. At BHCMG, the 200 probands (114 males, 86 females) ranged in age from seven weeks to 60 years with a mean of 13.2 years.
Only clinically well-characterized affected individuals for whom appropriate conventional diagnostic investigations, including immunological and clinically available genetic laboratory testing, had been previously performed were included. The diagnostic assessment was performed according to the ESID Registry diagnosis criteria (http://esid.org/Working-Parties/Registry/Diagnosis-criteria),24 and/or the joint AAAAI and ACAAI Practice parameter for the diagnosis and management of primary immunodeficiency.25, 26 Disease manifestations ranged from mild to severe. Based on the clinical and immunologic presentation at time of inclusion, probands were assigned to one of ten PIDD subgroups (Table 1): i) antibody deficiency (humoral deficiency that does not fulfill the diagnostic criteria for CVID), ii) autoimmune disease, iii) autoinflammatory disorder, iv) severe combined immunodeficiency (SCID), v) combined immunodeficiency (not SCID) and selective T cell deficiency, vi) common variable immunodeficiency (CVID), vii) defect in innate immunity including mucocutaneous candidiasis, hyper IgE syndrome, Mendelian susceptibility to mycobacterial disease, and complement deficiency, viii) lymphoproliferative disease, hemophagocytic lymphohistiocytosis (HLH) and NK cell deficiency, ix) neutrophil defect or congenital condition with bone marrow failure such as dyskeratosis congenita and Fanconi-like phenotype, anemia and thrombocytopenia x) immuno-osseous dysplasia, chromosomal disorder or other syndromal PIDD. Due to lack of a molecular diagnosis at enrollment, rather than using the molecular based IUIS classification system 27, these subgroups above were employed, as they were constructed based upon the presenting phenotype and not ultimate genetic diagnosis.
Table 1.
Total number of individuals and families WES tested at the two different study centers
| Clinical phenotypesa per family | BHCMGb Houston, USA | Oslo, Norway | Total | |
|---|---|---|---|---|
| i | Antibody deficiency | 14 | 2 | 16 |
| ii | Autoimmune disease | 27 | 5 | 33 |
| iii | Autoinflammatory disorder | 7 | 8 | 15 |
| iv | SCID | 6 | 4 | 10 |
| v | Combined or selective T cell defect | 40 | 16 | 56 |
| vi | CVID | 13 | 7 | 20 |
| vii | Defect in Innate immunity | 14 | 7 | 21 |
| viii | Lymphoproliferative or NK cell defect | 53 | 9 | 62 |
| ix | Bone marrow failure or neutrophil defect | 12 | 17 | 29 |
| x | Syndromal PIDD | 14 | 3 | 17 |
| Sum ready WES analyzed families | 200 | 78 | 278 | |
| Total number of individuals WES analyzed | 400 | 83 | 483 |
The PIDD subgroups are based on the proband's clinical presentation pre-WES.
Baylor-Hopkins Center for Mendelian Genomics (BHCMG) at Baylor College of Medicine (BCM)
Whole exome sequencing
For affected individuals, WES was performed using genomic DNA extracted from whole-blood prior to hematopoietic stem cell transplantation (HSCT). WES of 78 probands (Table 1), four unaffected family members, and one additional affected family member was performed at the Department of Medical Genetics, OUS in Oslo. WES of 196 unrelated probands, 177 unaffected family members, and 27 additional affected family members was performed in Houston at the BHCMG.28 In four of the 200 families tested at the BHCMG (Table 1), obligate carriers but not probands underwent WES. Nine of the samples were tested by WES at both places but were assigned to the center where the first analysis was performed, and confirmed concordance of sequencing standards.
At BCM-HGSC, exome capture was performed using the in-house developed BCM-HGSC Core design (52 Mb, Roche NimbleGen, Inc), as has been previously thoroughly described 21, 29, 30 In Oslo, exome capture was performed using the 50 Mb SureSelect Human All Exon kit v.5 (Agilent technologies, CA, USA). For 13 affected individuals from whom blood samples yielded a low DNA concentration (<10 ng/uL) exome capture was performed using the 45 Mb Nextera Rapid Capture Exome (Illumina, CA, USA). The three different capture methods provide similar output.31, 32 Sample preparation was performed according to the manufacturer's recommendations. For both locations, the captured exome was sequenced on the Illumina HiSeq 2500 platform (Illumina, CA, USA), with 100 or 125 bp paired-end reads resulting in an average coverage of 100x, and >90% of bases with >20x coverage. Details for data processing of the exome sequence is given in the Supplementary Methods. For both centers the bioinformatics pipelines were applied based on the GATK best practices and ANNOVAR.33
Identification of potential PIDD-causing variants
Initially, rare variants were selected based on ESP, 1000 Genomes (October 2013), Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org), and in-house databases that include WES data from phenotypically well-characterized individuals. Details for variant evaluation are given in the Supplementary Methods. All affected individuals were initially screened for variants in a list of 475 known or candidate PIDD genes, designated PIDD(475), which was collated from gene lists available at Resource of Asian Primary Immunodeficiency Disease (http://rapid.rcai.riken.jp/RAPID) and International Union of Immunological Societies (http://www.iuisonline.org/iuis/),1 and supplemented with genes known to affect telomere length or cause Fanconi anemia. If no PIDD-causing variants were identified from the PIDD(475) list, complete exome data were investigated for potential PIDD-causing variants. In the individuals who received a primary molecular diagnosis based on the PIDD(475) list, the complete exome data were also reviewed for occurrence of additional and modifying gene variants. Referring clinician(s) and molecular geneticists collaborated to assess the relevance of detected variants to the phenotype. In the exome-wide investigations, variants of interest were selected based on rarity, previously published cases with the same gene variants, and evaluation of possible genotype-phenotype correlation based on gene function, pathway, expression pattern and results from model organisms. Additional support for variants of potential impact was sought via computational prediction tools (PhyloP,34 GERP,35 SIFT,36 PolyPhen-2,37 LRT,38 and MutationTaster,39).
For potential disease-causing variants, the proband and available affected and unaffected family members underwent Sanger sequencing to confirm co-segregation of detected variants with disease phenotypes (see Supplementary Methods for details). More than 1,000 individual DNA samples were collected from the 278 PIDD families during the study. Altogether 400 samples underwent WES (Table 1). The remaining were collected and stored for Sanger segregation testing, and in total 367 samples were used for Sanger segregation testing from family members of the 110 probands with a likely or potential molecular diagnosis presented in this paper.
The functional impact of the detected variants was evaluated based on criteria (Table E1) adapted from the guidelines recommended by the American College of Medical Genetics and Genomics (ACMG),40 see Supplementary Methods. The reported potential disease-causing variants were of class 5 (definitive pathological), class 4 (likely pathological) and class 3, but class 3 variants were only reported when the affected gene was consistent with the phenotype or parts of the phenotype in the affected individual. When possible, further supporting data were generated through functional studies, such as cDNA sequencing to confirm aberrant splicing using RNA extracted from PaxGene blood samples. The reported potential disease-causing variants were not present in homozygous state, and allele frequency was less than 0.0001 in heterozygous/hemizygous state (for dominant/X-linked inheritance respectively) in the ExAC database (as of March 2016), with the exception of modifying variants with known functional effect. Some candidate genes belonged to an established potential PIDD gene list.41 Additional lines of evidence included biological validation consistent with known clinically acceptable measures of a disease gene's function or at least in part with guidance proposed for unique discoveries in single cases,42 presence of PIDD gene protein homologues with relevant expression patterns belonging to the PIDD gene interactome, and segregation with identical immunophenotype in several individuals within the same family and/or other families. Given the residual uncertainty, we refer to the potentially causal linkage to disease as “likely”. For otherwise previously unreported genes and pending complete biological validation,42,41 these genes are referred to in this paper as “potentially novel” in order to allow for the consideration of overall diagnostic yield appropriate fulfillment of established criteria.
Copy number variants (CNVs)
Two methods were used to identify CNVs: computational CNV prediction and microarray CNV detection. CNV prediction was initially performed for the probands. Microarray CNV detection was applied in selected cases where no disease-causing variants were detected after exome sequencing or when CNV prediction data indicated the presence of a relevant CNV (see Supplementary Methods for details). One third of probands had been evaluated via a diagnostic CNV microarray prior to inclusion via 180K aCGH (Agilent technologies, CA, USA) or SNP Array 6.0 (Affymetrix, CA, USA), which did not provide a definitive diagnosis by itself.
Results
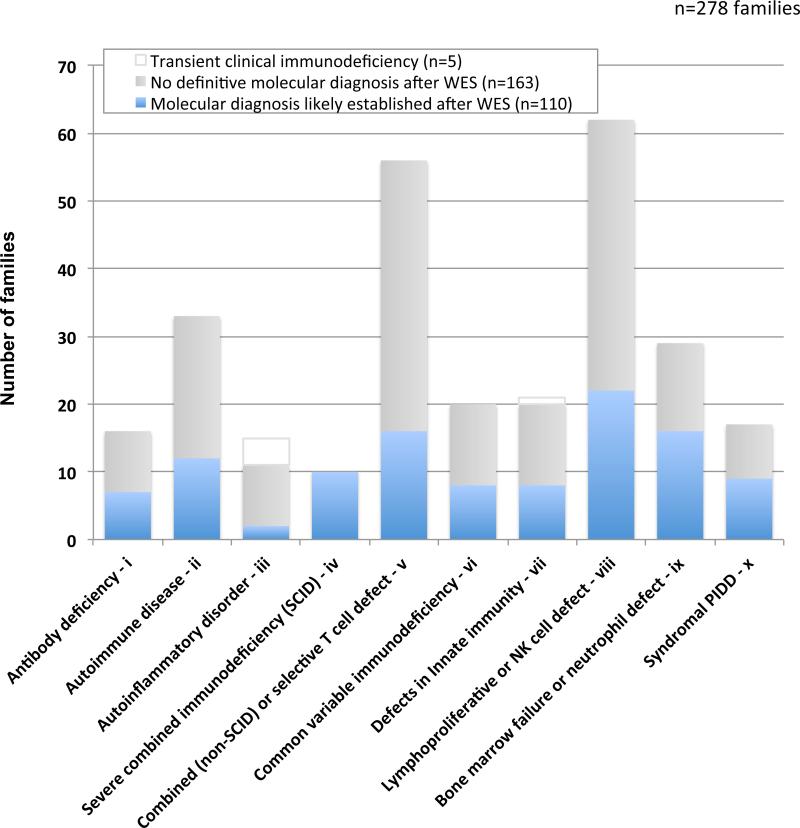
In 278 PIDD families who underwent whole exome sequencing, a likely molecular diagnosis that explains all or part of the phenotype (PIDD and non-PIDD) was established in 110 (40%) (Figure 1;Table 2). Diagnostic yield varied by PIDD subgroup (Figure 2). The highest diagnostic rate was in severe combined immunodeficiency (100%), followed by bone marrow failure (55%) and syndromal PIDD (53%). The lowest diagnostic rate was in the autoinflammatory subgroup (13%), even when transient disease was excluded (18%).
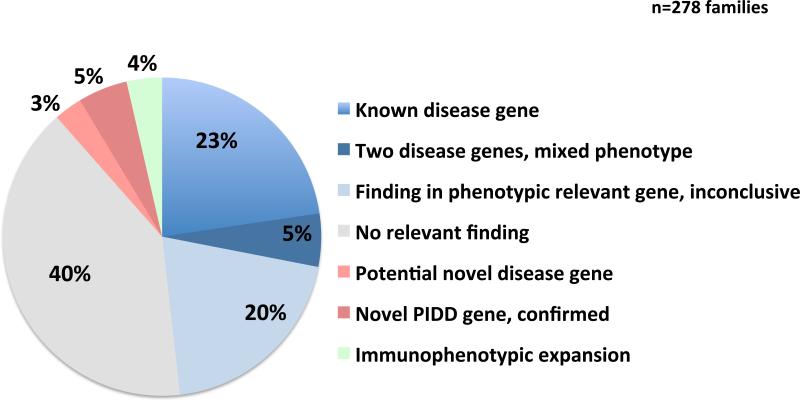
Figure 1.
Status of molecular diagnosis after WES of 278 PIDD families. Findings are grouped in categories based on whether the detected disease causing variant(s) a) affects known disease genes, b) are located in more than one gene contributing to the assumed mixed phenotype (PIDD or non-PIDD) in the individual, c) affects a confirmed novel disease gene d) affects a potentially novel PIDD gene, e) leads to immunophenotypic expansion: a modified immune phenotype observed in the affected individual, modified from that which is historically characteristic of the particular gene, f) affects a phenotypic relevant gene, that were not presumably the major disease etiology, or only one deleterious variant in a disease relevant AR gene (in total 57 cases), or g) no identifiable molecular explanation was identified using the technologies applied here (111 cases).
Table 2.
Number of WES tested family members and detection rate
| Family members WES tested | Total number of families | Molecular diagnosis established | Total detection rate |
|---|---|---|---|
| One individual onlya | 192 | 70 (36%) | 36% |
| Proband + 1 healthy parent or other unaffected relativesb | 25 | 7 (28%) | 43% (17/40) |
| Proband + affected relatives | 15 | 10 (67%) | |
| Trio: Proband + both parents | 39 | 17 (44%) | 42% (23/55) |
| Trio + affected relatives | 6 | 2 (33%) | |
| Trio + unaffected relatives | 10 | 4 (40%) | |
| Sum all family WES studies | 287 | 110 | 40% |
Figure 2.
Diagnostic yield in the PIDD cohort (n=278 families) by subgroup distribution based on the proband's diagnosis prior to WES+CNV testing.
Genetic spectrum of disease-associated variants
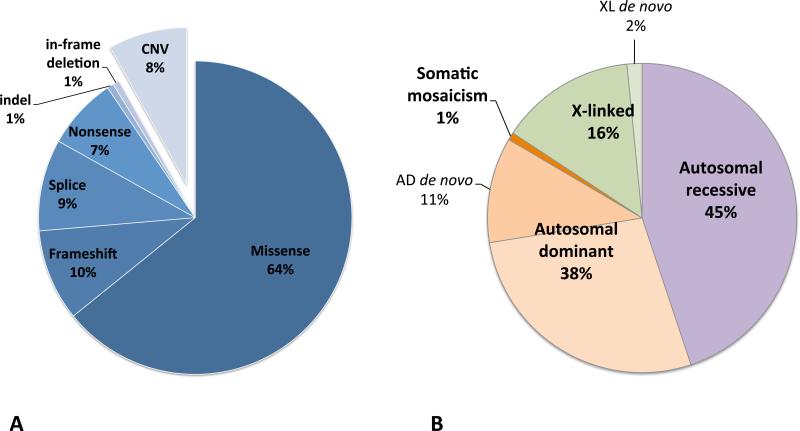
In the 110 probands, 148 different disease-associated, potentially causative or contributing variants were identified in 88 genes (Figure 3 and Table E1). Forty-six variants had previously been reported as disease-causing in HGMD, and another six were located in the same nucleotide position or induced a change in the same codon as a previously reported HGMD variant (Table E1). In addition, 36 other variants met criteria for ACMG class 5 (pathogenic), and 10 for class 4 (likely pathogenic)., and the rest were classified as variants of unknown significance (VUS), class 3. Altogether 98 variants (66%) were either reported before and/or likely or certain disease-causing. The 148 detected variants included 136 single nucleotide variants (SNVs): 95 missense, 13 frameshift, 11 nonsense, 14 splice variants, two indels and one in-frame deletion (Figure 4). Many of these variants likely explain the PIDD immunophenotype and some may also explain non-PIDD features of the clinical phenotype.
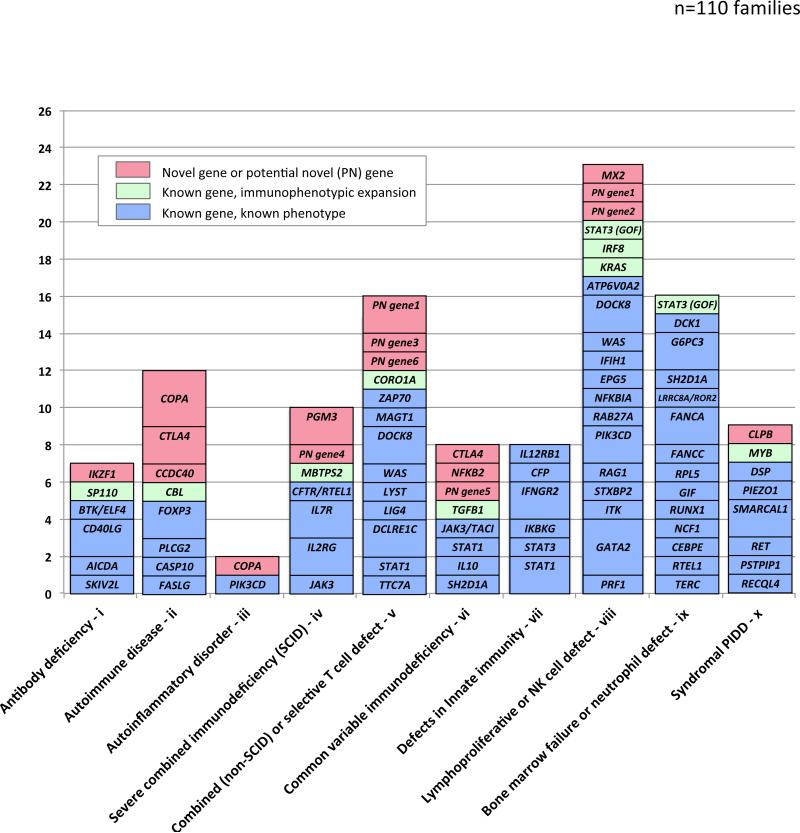
Figure 3.
Distribution of main likely disease genes in 111 families with established molecular diagnosis after WES+CNV testing by PIDD subgroup prior to testing.
Figure 4.
Disease-associated variants and inheritance patterns observed among the 110 families in the PIDD cohort where a likely molecular diagnosis was established. (A) Spectrum of disease-associated variants detected in the PIDD cohort. In total 148 different variants were identified among the 110 families where a likely molecular diagnosis was established. The SNVs are classified based on assumed effect on protein. The indel led to a frameshift and stop codon and the in-frame deletion is previously reported to be disease causing by altered protein function. With exception of the DKC1 duplication, all 12 CNVs are deletions. All CNVs are assumed to result in loss of function. (B) Inheritance patterns observed among the 110 families in the PIDD cohort where a likely molecular diagnosis was established. The mosaic variant in KRAS is only compatible with life in somatic mosaic state, thus a Mendelian inheritance pattern is not applicable.
PIDD-causing CNVs were detected in 12 of 110 families (11%). The CNVs constituted 12 (8%) of the 148 likely disease-causing variants (Figure 4) and involved 11 genes: DKC1 (MIM: 305000), DOCK8 (twice) (MIM: 243700), FANCA (MIM: 227650), IKZF1 (MIM: 603023), IL7R (MIM: 608971), MAGT1 (MIM: 300853), MYB (MIM: 189990), NCF1 (MIM: 233700), PGM3 (MIM: 615816), SMARCAL1 (MIM: 242900) and TERC (MIM: 127550) (Table 3). Homozygosity and hemizygosity for CNVs as well as compound heterozygosity for a CNV and a SNV were observed in 12 cases (Table 3), three of which have been published previously.43-45 Disease-causing CNVs ranged in size from 1.6 Kb to 3.4 Mb, averaging 0.45 Mb with a median of 19.7 Kb. Seven CNVs (58%) were less than 30 Kb in size and would not have been detected by standard diagnostic microarrays (Table 3). The WES data were screened for CNVs by computational prediction.43
Table 3.
Disease causing copy number variants detected in the PIDD cohort
| Gender | Age | Diagnosis pre-WES (PIDD subgroup) | PIDD gene involved | Inheritance pattern for disease | Mutation type | Size of CNV | State | Affected family members |
|---|---|---|---|---|---|---|---|---|
| Male | 13y | Hoyeraal Hreidarssons syndrome, X linked (ix) | DCK1 | XL | Duplication | 14 kb | Hemi | 3 |
| Male | NA | Combined immunodeficiency (v) | DOCK8 | AR | Deletion | 355 kb | Hom | 2 |
| Female | 5y | HLH and NK cell defect (viii) | DOCK8 | AR | Deletion | 84 kb | Hom | 1 |
| Female | 34y | Fanconi anemia, mild (ix) | FANCA | AR | Deletion | 22-24.6 kb | Het | 2 |
| Male | 30y | Agammaglobulinemia (i) | IKZF1 | AD | Deletion | 15 kb | Het | 1 |
| Female | 2y | SCID, later debut (iv) | IL7R | AR | Deletion | 224 kb | Het | 1 |
| Male | 16y | Immunodeficiency, X linked, extensive warts (v) | MAGT1 | XL | Deletion | 16 kb | Hemi | 2 |
| Female | 8y | Immunodeficiency, progressive bone marrow failure. Short stature, dysmorphic facial features. (x) | MYB | AD | Deletion | 3.4 Mb | Het | 1 |
| Female | 71y | Chronic Granulomatous Disease (ix) | NCF1 | AR | Deletion | 15 kb | Hom | 1 |
| Male | 6y | T and B cell deficiency and neutropenia (iv) | PGM3 | AR | Deletion | 1.24 Mb | Het | 3 |
| Female | 4y | Immunosseous dysplasia (x) | SMARCAL1 | AR | Deletion | 1.6 kb | Hom | 1 |
| Male | 49y | Dyskeratosis Congenita, progressive bone marrow failure, Short telomere lengths (ix) | TERC | AD | Deletion | 3 kb | Het | 1 |
CNVs predicted as potentially disease-associated were confirmed either by an exon-tiling CNV microarray, MLPA or PCR-based breakpoint analysis. Disease-causing CNVs in PGM3,45 FANCA, SMARCAL1, NCF1, TERC, MAGT1, and IKZF1 46 were detected by computational prediction (Figures E1-E3) and in IL7R, MYB and DKC1 by exon-wise CMA (Figure E1). The two homozygous DOCK8 deletions were first noted on a SNP array used for WES quality control and subsequently visualized by HMZDelFinder (Figure E4). The CNVs in NCF1 47 and DKC1 were confirmed by MLPA.
The DKC1 duplication was detected by CMA in samples from the family reported in 1970 by Hoyerall et al,48 the first report of dyskeratosis congenita, subsequently designated Hoyeraal-Hreidarsson syndrome (MIM: 305000). Our bioinformatic CNV tools were unable to detect this duplication of this X-linked gene, even in retrospect. PCR located the duplication within the gene. Although it is unclear how the duplication affects gene function, the detected CNV is the likely cause of the disease in the family because DKC1 is the only X-linked gene known to cause the specific phenotype. All three female carriers had a 100% skewed X-chromosome inactivation pattern. cDNA sequencing of one carrier failed to identify an aberrant DKC1 product, in keeping with inactivation of the mutant allele.
Mendelian patterns in PIDD
Among the hereditability of variants observed (n = 126), autosomal recessive (AR) inheritance was most frequent (n = 57, 45%), followed by autosomal dominant (AD)(n = 49, 38%, excluding one case of somatic mosaicism,) and X-linked inheritance (n = 20, 16%; Figure 4, Table E1).1 Where parental samples were available, the likely disease-causing variants with dominant inheritance were de novo in fourteen probands (Table E1). This corresponds to a de novo mutation rate for autosomal dominant mutations of at least 29%. Reduced penetrance or parental mosaicism was present in nine families. Two variants in X-linked genes occurred de novo in affected males (Table E1). In four families obligate carriers were sequenced because affected probands were deceased or had undergone HSCT (families 90, 93, 95, and 111, Table E1). A molecular diagnosis was established in three of these families, with variants in DKC1, FANCC (MIM: 227645) or IL2RG (MIM: 300400). In a single family, two individuals suspected to have identical disease in fact had different PIDDs: one caused by a homozygous SNV in RAG1 (individual 81.5)(MIM: 601457), and the other by a homozygous CNV in DOCK8 (proband 81.1). In another family two affected cousins with neutropenia and a congenital heart defect were suspected to have identical mutations but in fact did not (family 91, Table E1 and Figure E5).
A potential mutational burden
More than one gene may contribute to the PIDD phenotype in a given family. The presence of a dual molecular diagnosis is considered likely when diagnostic criteria for both genes can be fulfilled by the single patient. In our cohort, this was observed in 12 (11%) of families with a likely molecular diagnosis. For example, in one family three deceased males (proband 93.1, Table E1, Figure E5) had immunodeficiency, leukemia, lymphoma, anemia, and solid tumors. The affected individuals shared a SH2D1A variant c.80G>A, p.Gly27Asp (NM_002351) causing X-linked lymphoproliferative syndrome (MIM: 308240). An additional variant (c.989T>C, p.Ile330Thr (NM_001018113) in FANCB (MIM: 300515), the gene for X-linked Fanconi anemia that maps 2.5 Mb from SH2D1A, co-segregated and may have contributed to the complex phenotype (Figure E5). As another example, in another family, an adult female (proband 27.1, Table E1) presented with combined immunodeficiency, low absolute lymphocyte counts, very low levels of CD8+ T cells, poor phytohemagglutinin (PHA) mitogen proliferative response, disseminated HPV infection, recurrent respiratory infections, vaccine-induced varicella, and cryptococcal meningitis. WES identified compound heterozygous disease-causing variants in ZAP70 (p.Gly245Arg in the SH2 domain and p.Pro502Leu in the kinase domain). Hypomorphic variants in ZAP70 have been reported in individuals with a later onset immunodeficiency,49 and may explain this proband's clinical presentation, low CD8+ T cell counts, and combined immunodeficiency. In addition, she harbored a homozygous missense variant in RNF168, the gene associated with another PIDD, RIDDLE syndrome (MIM: 611943). Radiation sensitivity analyses performed by colony survival assay (CSA)50 of the proband's EBV transformed lymphoblasts demonstrated a reduced survival fraction (8%) and exhibited increased radiosensitivity, a hallmark of RIDDLE syndrome and to our knowledge not observed in ZAP70-deficient individuals. Of interest, the patient also developed HPV-associated extensive scalp squamous cell cancer. All variants co-segregated with the disease in the family (Figure E5). Impaired function of both proteins may underlie the blended phenotype.21, 22 In 12 of the 110 families in which the proband's disorder was attributed to deleterious variants in a disease gene, variants in additional genes were considered having a potentially modifying effect (Figure 1, Figure E5, and Table E1 – marked with footnote fp).
Clan genomics and founder mutations
In families with known consanguinity or from areas where consanguineous marriages are common, disease-causing homozygous variants were observed as expected (Table E1; 5.1, 30.1, 31.1, 40.1, 64.1, 81.1 and 81.5, 85.1, 91.4, 98.1, 101.1, 108.1, 110.1, 114.1, 122.1, 123.1 and 124.1). As an unexpected finding, we were also able to use WES to detect genomic regions with region of absence of heterozygosity (AOH) in individuals from presumed outbred populations, potentially reflecting distant parental kinship (Table E1; 33.1, 33.4, 39.1, 56.1, 87.1, 100.1 and 105.1), consistent with the Clan Genomics hypothesis.51
Variable expressivity
Reduced penetrance or variable expressivity was observed in families for which probands (Table E1; 10.1, 11.1, 14.1, 15.1, 20.1, 47.1, 79.1) had a likely autosomal dominant disease-causing variant in CTLA4 (MIM: 123890), PIK3CD (MIM: 602839), NFKB2 (MIM: 164012), FASLG (MIM: 134638), and COPA (MIM: 601924). Some heterozygous mutation carriers in these families appeared clinically unaffected.52 For example, proband 20.1 presented with neutropenia, enteropathy, and joint disease, and harbored a missense mutation in FASLG, causing autoimmune lymphoproliferative syndrome (ALPS). Sanger sequencing confirmed that the variant was inherited from the father, who had only minor symptoms and laboratory signs of disease, suggesting reduced expressivity or lack of environmental triggering. Splicing variants were also identified that may be incomplete and cause milder phenotypes (Figure E6). Milder disease could as well be due to low-grade mosaicism, such as in the PIK3CD family (proband 67.1) with the ratios of variant to total reads 37%, 54%, and 42% in the three affected siblings but only 15% in their mildly affected father. (Table E1; 67.1).
Mendelian exceptions - somatic and revertant mosaicism
In three families, affected individuals displayed a milder phenotype and had findings suggesting somatic revertant mosaicism. Their less severe phenotypes could be related to a reduced number of cells harboring mutant alleles (Figure E7). In the first family, a six month old boy (proband 59.1) presented with eczema, diarrhea, BCG vaccination associated infection, recurrent severe staphylococcal infections and eosinophilia but normal T, B and NK cell numbers. A previously reported X-linked disease-causing IKBKG mutation (c.1167dupC, p.Glu390Argfs*5(NM_001099857)) was identified; his unaffected mother was a heterozygous carrier (69/284 reads by WES). Interestingly, the IKBKG mutant allele was observed in only 35% (55/159) of reads in the boy's DNA extracted from blood (Table E1). This finding argues for revertant mosaicism, which could explain his relatively mild phenotype compared to classic IKBKG associated immunodeficiency (MIM: 300291) in males. Revertant mosaicism and milder phenotypes were also detected in two other families with disease-causing variants in FANCA (Figure E7) and IL7R44.
Female subject 65.1 (Table E1) serves as an example of somatic mosaicism with severe clinical consequences. She presented with recurrent infections in early childhood and subsequently developed lymphadenopathy, hepatosplenomegaly, and ALPS. She died at 4 years of age with a clinical phenotype reminiscent of HLH. WES identified a de novo variant in exon 2 of KRAS in 15/110 reads (13.6%) (c.37G>T, p.Gly13Cys (NM_004985)). Mosaicism in blood was confirmed by Sanger sequencing (Figure E7). The child had no dysmorphic features and a normal head circumference/stature. The variant was likely confined to her hematopoietic cells. Mosaicism for the identical variant was previously reported in a child with ALPS-like disease.53 The phenotype is now designated RAS-associated autoimmune leukoproliferative disorder (RALD) (MIM: 614470).
Unexpected genomic and correlated clinical findings
We included affected individuals from all subgroups of PIDD through an unbiased genomic approach. In addition to using the PIDD(475) gene list we further examined other disease-related OMIM genes and all detected variants in other genes in an effort to identify new PIDD genes. For 90 out of 278 probands, variants in disease-causing genes with known, new or expanded phenotypes were observed (Table E1).30, 54, 55 Expansion of the immunophenotypic spectrum was associated with nine OMIM genes (SP110 (MIM: 235550), CBL (MIM: 613563), MBTPS2 (MIM: 308205), CORO1A (MIM: 615401), STAT3 (MIM: 147060, 615952), IRF8 (MIM: 614894), KRAS, MYB, and TGFB1 (MIM: 131300)) (Figure 3). Unexpected rare congenital syndromes were also detected in our cohort. In six families, variants were detected in OMIM genes which have not previously been reported in association with immunodeficiency or bone marrow failure. These variants may or may not explain the immunological defect, or perhaps only parts of the affected individual's phenotype. This could be the case for example in proband (113.1) with pathological CFTR variants and recurrent pulmonary infections. Some cases may represent multi-disease burden, others true phenotypic expansions. Biological studies of relevant gene products in the immune system are in progress in such families. The insights derived from immunophenotypic expansion can be substantial and may relate to the specifics of individual mutations or broadened clinical presentations.
For example, proband 115.1 (Table E1) presented with a SCID-like immunophenotype shortly after birth, as well as microcephaly, severe scaling erythrodermatitis, conjunctivitis, keratitis, and oligoclonal T cells – all features of Omenn syndrome – but with a normal IgE level and eosinophil count. He was ultimately found to have ichthyosis follicularis, alopecia and photophobia (IFAP) syndrome, due to alteration of MBTPS2, which has been previously associated with immunodeficiency, albeit not of this severity (Figure E8).56, 57 Other unexpected variants, which explained all or parts of the probands’ phenotypes, were identified in RET (MIM: 142623), PLXNA1 (MIM: 601055), ATP6V0A2 (MIM: 611716), FBN1 (MIM: 154700), MX2 (MIM: 147890), CFTR (MIM: 219700), CCDC40 (MIM: 613808), and CLPB (MIM: 16271). None of these variants would have been identified if the samples were analyzed with a gene panel restricted to targeted capture of known PIDD genes only.
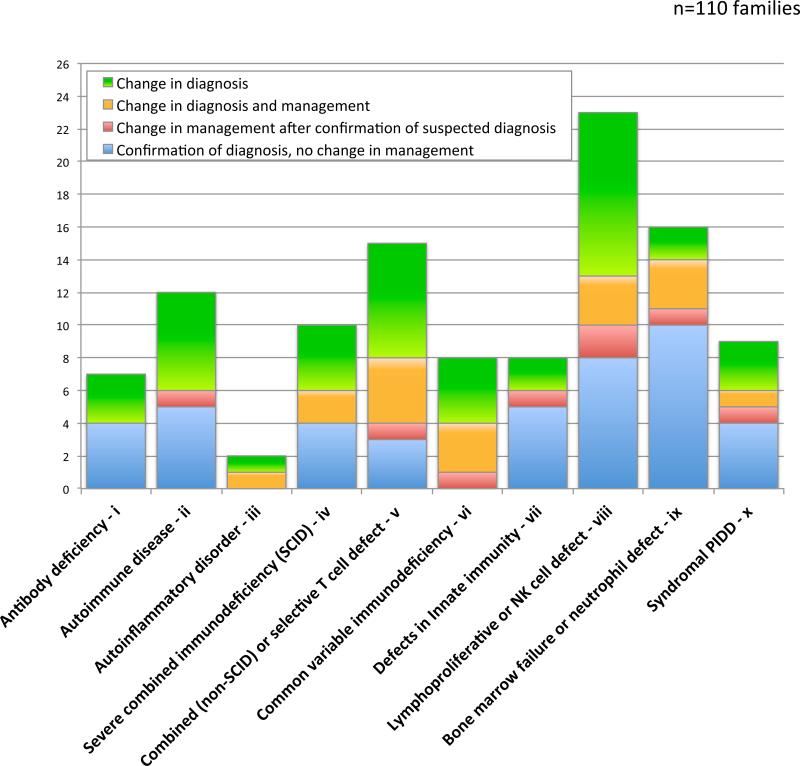
Change in diagnosis and management after WES
In 55% (60/110) of the families in whom a likely molecular diagnosis was identified by WES, the prior PIDD diagnosis based on clinical features and immunophenotyping was altered (Figure 5). Post-WES revision of diagnosis was observed across all PIDD subgroups. A likely etiological diagnosis was established in approximately half (range 18% to 100%) of the affected individuals in each PIDD subgroup prior to WES. In 25% of cases (26 families, 32 affected individuals), WES findings directed a significant change in management (Figure 5). Ten individuals underwent hematopoietic stem cell transplantation (HSCT) primarily guided by WES findings. In four additional individuals, WES results supported a previous decision to treat with HSCT. Six probands had already undergone HSCT. For one of them, proband 115.1 with a molecular diagnosis of MBTPS2-IFAP, HSCT might not have been performed if the molecular diagnosis had been known.
Figure 5.
Alteration of specific diagnosis and change in management due to molecular findings by WES+CNV testing. The distribution is shown by PIDD subgroup which was based on clinical presentation at time of inclusion.
Recent and new disease genes
Twenty-five percent of molecular findings were in genes reported as disease-causing during the last five years; five in 2011, four in 2012, seven in 2013, three in 2014 and three in 2015 (Table E1). Eight were new disease genes (CCDC40,58 CLPB,59, 60 COPA,52 CTLA4,61, 62 IKZF1,46 MX2,63, 64 NFKB2,65-67 PGM3,45, 68, 69 at the time of detection and were either confirmed biologically by us or others. Six likely candidate PIDD genes were also identified in this study (listed as potentially novel; Figure 3). These are rare, damaging, conserved variants that fit biologically with the immunophenotype, but full biological confirmation to comply with proposed criteria 42 remains presently incomplete. This group of patients is included so as not to underestimate the potential impact of WES as applied to PIDD. One example of a biologically confirmed new disease gene identified by WES comes from two families in our cohort with autosomal recessive variants in PGM3. Functional studies detected reduced specific phosphoglucomutase enzymatic activity.45 Additional PIDD affected individuals with PGM3 mutations were reported simultaneously,68, 69 and more than 10 families have been published since.19, 45, 68-70 New and potentially novel PIDD genes were found in 21 families in our cohort (19%), and biological investigations to confirm these associations according to published standards 42 have been performed or are in progress.
In general, and as would be expected, every exome tested contained several rare, exonic variants (MAF < 0.001 in 1000 Genomes data, in-house databases, ESP and ExAC) in PIDD genes and HGMD-reported variants with MAF <0.05. With the 148 likely causative variants excluded, the majority of the other rare variants were definitively not disease-causing, but some represented single variants in AR trait genes where disease genes were compatible with the affected individual's phenotype. These are included in Figure 1 among the 20% families with findings in a phenotypically relevant gene, but findings are considered inconclusive and molecular diagnoses classified as unsolved.
Discussion
Using genomic approaches, we identified the potential molecular basis for disease in 40% of 278 probands with various PIDD phenotypes of unknown etiology who had previously been investigated with conventional methods (albeit with some variation across the 24 different hospital centers contributing patients to this study). We found evidence for a mutational burden effect,71 which might contribute to disease variability and underlie blended immunophenotypes, as well as somatic and revertant mosaicism. With no preset bioinformatic cut-off level for variant reads, we were able to detect mosaicism below 15% (KRAS, Figure E7). Several disorders with significant nonimmunological features, such as primarily skin or intestinal diseases, were diagnosed. Some cases might represent the phenotypic expansion of a known disorder to include immunodeficiency. For autosomal dominant (AD) traits, we noted reduced penetrance associated with variant alleles. For autosomal recessive (AR) traits, we identified founder variants in outbred populations. Our approach resulted in the expansion of the phenotypic spectrum associated with nine previously known disease genes (Figure 3), and the identification of eight new PIDD genes previously reported by us (PGM3,45 COPA,52 IKZF1,46) or others (CTLA4,61, 62 NFKB2,65-67 CLPB,59, 60), new non-PIDD genes (CCDC40,58 MX2,63, 64)reported by others and detected in our cohort as well, and six potentially novel candidate genes. All of these new disease genes and potential novel candidate genes affected 21 probands (Figure 2, Table E1). We found PIDD relevant variants in 88 different genes, 25% of which were first reported during the last five years (Table E1). Twelve probands had presumably phenotypically relevant sequence alterations in more than one gene, compatible with a blended phenotype in 11%. As this has not previously been reported for major PIDD cohort studies, variations in PIDD phenotypes may need to be revisited. Importantly, molecular findings resulted in a revised diagnosis in 60 families and altered management in 32 cases (Figure 5).
In contrast to a previous report,22 we found an abundance of disease-causing CNVs. Disease-causing CNVs were detected in 11% of the families (Table 3), half of which would not have been identified by conventional chromosomal arrays. A broad spectrum of likely disease-related SNV types was observed, but the paucity of indels in our cohort deviates from the data reported for selected PIDD genes in HGMD. This finding may reflect characteristics of some immunodeficiency related genes or perhaps challenges in algorithm detection of indels from WES data. However, the distribution of SNV types we report is similar to the results demonstrated using WES in other heterogeneous Mendelian cohorts.22
X-linked disorders contribute to the male excess in population-based PIDD studies.2 In our cohort with 58% molecularly undiagnosed males, many individuals had already undergone testing for X-linked PIDD genes such as BTK, WAS, IL2RG, IPEX, DKC1, TAZ and CD40L. However, 16% of the potential causal variants in our cohort were located in these and other X-linked genes, including variants in BTK (c.141+11C>T) (MIM: 300755) and IL2RG (c.924+5G>A) not detected by diagnostic Sanger sequencing. The same was true for the variant in JAK3 (c.1695C>A) (MIM: 600802) undetected by Sanger sequencing prior to WES. The indels in CTLA4 and CD40LG (MIM: 308230) were inadequately annotated on the initial WES data but visualized in IGV and correctly classified using Sanger sequencing. This emphasizes the ongoing value and importance of validation. The mosaic variants in FANCA, IL7R and KRAS had not been detected without WES, but were visualized by Sanger sequencing as small peaks on electropherograms, which could easily have been misinterpreted as artifact.
Of the 364 genes in our PIDD(475) list with known inheritance patterns, 23 (6%) are associated with X-linked, 76 (21%) with AD, 247 (68%) with AR, and with 18 (5%) AD or AR inheritance. The distribution of inheritance patterns we observed mirrors the inheritance patterns documented for known PIDD genes with slight skewing toward variants in AR genes and an increased trend towards variant discovery in AD genes. In our families with documented AD or AR inheritance, we identified disease-related variants in 38% and 45% of families, respectively. In cohorts with molecularly confirmed Mendelian disorders, including neurological and neurodevelopmental disorders associated with reduced reproductive fitness, the trend is reversed, with a 3:2 ratio of AD and AR disorders and more than 70% de novo variants in the AD genes.22, 72, 73 In contrast, fewer than 30% of variants were proven to be de novo amongst the AD PIDD genes identified in this study. Even after including potential de novo variants in AD PIDD genes for whom parental samples were missing (GATA2 in proband 88.1, IKZF1 in 1.1, RPL5 (MIM: 612561) in 99.1, TERC in 63.1, STAT3 in 66.1, STAT1 (MIM: 614162) in 50.1, 60.1 and 37.1, Table E1) the percentage (45%) was substantially lower than seen in neurodevelopmental disorders.
Immunologically important genes are numerous and spread throughout the genome. Caution is required in attributing causality to variants identified by WES because even rare inherited or de novo variants may not cause disease. Widespread use of WES in diagnostics and research reveals that even variants listed as deleterious in HGMD can be misclassified.74 A case in point is the intronic BTK variant c.141+11C>T (NM_000061.2) (Table E1) detected in individuals without antibody deficiencies and reported frequently (160 hemizygotes) in healthy males in the ExAC database. Another example is the affected male in our cohort with an X-linked, maternally inherited missense variant in WAS (exon 10 c.995T>C, NM_000377) (MIM: 301000). This p.Val334Ala variant has previously been reported to be disease-related (CM072118; HGMD), but has a minor allele frequency (MAF) of 0.005. With 115 hemizygotes reported in ExAC, it seems unlikely that such a large number of affected males with classic Wiskott Aldrich syndrome or X-linked thrombocytopenia are included among the individuals in the ExAC database. Nonetheless, while the data argue that the variant may not be a primary cause of disease, we cannot exclude the possibility that the variant may have a milder or disease-modifying pathogenic effect. We did not identify any other disease-causing variants in this individual, and he is not included in the 110 families with a likely established molecular diagnosis. Every exome contained several rare, exonic variants in PIDD genes and HGMD-reported variants that were definitively not disease-causing or only one variant in an AR trait gene which could theoretically contribute to the observed clinical phenotype. This general observation underscores the importance of evaluating a genetic variant in relationship to the clinical phenotype, especially when limited biological or functional evidence for causality exists.75 Optimal utilization of WES data in PIDD therefore requires a dynamic iterative collaboration between clinicians, laboratory immunologists, molecular geneticists and bioinformaticians. The standards for biological consideration of novel single gene discoveries, as exemplified by PIDD, have been proposed and represent an important threshold to clear when evaluating candidate genes.42 We have elected not to include the names of ‘potentially novel’ otherwise unreported PIDD-candidate genes in the present work in deference to these published criteria and their current status as not yet biologically confirmed. Investigations and/or collaborative evaluations of these candidates are currently in progress in order to provide the necessary mutational impact testing and biological validation.
A few factors were considered that could potentially affect discovery rates. The ages of the probands among the families who received a likely or potential molecular diagnosis from our study were similar to those who did not. Thus, clear Mendelian traits were observed across all age groups and not confined to the youngest children with PIDD (Table E1). In Houston, the overall discovery rate was 37.5% (75/200). Among the 200 families, 204 additional family members underwent WES, which may have improved the discovery rate (Table 2). Mainly singleton cases were WES tested in Oslo; however the overall discovery rate was still 44.9% (35/78). In general, across all subgroups WES testing of more than one affected individual in the same family increased the diagnostic rate, as demonstrated in Table 2 and Table E1, and a larger proportion of potentially novel genes were discovered among those families. In 21 families, more than one affected family member underwent WES as part of our study. Among all 110 families that received a likely or potential molecular diagnosis, 12 had more than one affected family member WES tested, and 5 of these had disease related variants in novel PIDD genes. Formal WES trios (proband plus both parents) may have increased the discovery rate, but not to the extent as observed in other cohorts.76-78 Since the numbers are small (Table 2), however, definitive conclusions cannot be made. Finally, the fact that all patients were evaluated according to diagnostic criteria and conventional methods prior to inclusion, many of the more straight forward molecular diagnoses had been already made, and those patients did not contribute to this study. Thus, if all presenting PIDD patients were included, discovery rates in a PIDD cohort would have likely been higher.
Primary immunodeficiency diseases with severe, opportunistic infections represent distinct phenotypes with strong genetic components and simple Mendelian inheritance patterns, even in PIDD subgroups historically considered as complex polygenic disorders, such as CVID. 79, 80 Conversely, due to heterogeneity, known occurrence of genetic modifiers and susceptibility factors (e.g. MBL deficiency, (MBL2, MIM:614372)), reduced penetrance and variable expressivity between mutation carriers even within families, and strong influence by environmental factors (bacteria, viruses, fungi, nutrition status, age), the delineation of genetic causality in each individual can be challenging. Our 40% gene variant identification rate is high compared to other WES studies of various non-PIDD disease cohorts.21, 22 However, the rate is lower than for WES trios in cohorts with severe intellectual disability 76, 81 and brain abnormalities.82 Those conditions are characterized by highly penetrant gene variants, with skewing towards de novo deleterious variants in AD genes, and perhaps less influenced by environmental factors such as infection-triggered immunodysregulation. Further biological verification of causality will be needed for the variants classified as VUS, and despite our effort taking the genome-wide approach, additional disease-causing variants may be discovered even among the 40% with an assumed molecular diagnosis.
For the 60% of families in this cohort who still lack a molecular diagnosis (Figure 1), further genomic studies are indicated in an effort to identify variants in non-captured regions, including regulatory regions, low-grade mosaicism, small CNVs and indels. Some of these families may also have polygenic PIDD, which remains an active area of investigation. A stepwise approach for PIDD diagnostics with targeted next generation sequencing (NGS),20 followed as needed by WES and/or whole-genome sequencing (WGS) has been previously proposed.19 Our findings support such an approach, although use of WES and/or WGS may ultimately become preferred to targeted multi-gene panel testing as technologies advance to provide results in a comparable timeframe. The need for rapid analysis is emphasized by the fact that acquisition of the proper diagnosis directly altered management of 25% of probands in our cohort. Targeted gene panel testing currently has advantages in terms of high coverage of genes of interest (including deep intronic regions), ease of data interpretation, and rapid turnaround time. Targeted capture of a limited number of PIDD genes can often deliver molecular diagnostic results within weeks or days, at a lower cost, while gene hunting with inadequate coverage of regions of interest may highlight pitfalls associated with currently available WES and WGS.18-20, 83-86 WES, however, allows for detection of variants in potentially novel gene candidates. It can uncover mutations in other genes, such as CFTR, that mimic PIDD. Our PIDD(475) gene list was an efficient means for detecting causative variants in known genes in 77 probands. We have also demonstrated the utility of WES combined with tools for intragenic CNV detection to identify small CNVs. It remains unknown whether targeted gene panels will prove more cost-effective than WES, as various laboratory-based, insurance-related, and nationally-implemented policies factor greatly influence cost analyses. WGS is not capture based, unlike targeted gene panel testing and WES, allowing for direct discovery of structural abnormalities, such as CNVs and inversions, and SNVs within nonexonic genomic regions, albeit currently at a substantially higher cost.87
Based on our data and a review of the literature, we suggest a two-tiered approach: 1) Initial testing with a limited capture kit targeting a) common PIDD-causing genes, b) actionable gene variants, and c) genes causing severe phenotypes and requiring a rapid molecular diagnosis, such as disorders characterized by low TRECs on newborn screening for SCID 88 and lymphoproliferative disorders.89 Optimal probe design of a targeted capture kit to ensure sufficient coverage of the targeted genes may enable replacement of Sanger sequencing by rapid testing on a bench-top NGS platform.20 Given substantial read depth-of-coverage, targeted gene test data may allow for detection of intragenic CNVs and low-grade mosaicism. 2) If no PIDD-causing variants are identified by the above testing, we suggest proceeding to a genome-wide approach using either WES or WGS with a large, frequently updated PIDD gene candidate list such as PIDD(475) that includes other extended immunologically PIDD-relevant genes as a bioinformatic filter in the initial variant screening.41 Genome-wide data can be used to identify AOH regions that can help guide the search for disease-causing variants in consanguineous families and in individuals from outbred, but homogeneous populations.
Supplementary Material
Clinical implications.
With a rate of likely molecular diagnosis in 40%, WES in tandem with CNV screening tools is effective for detecting a broad spectrum of disease-causing variants in individuals with PIDD.
Capsule summary.
Whole-exome sequencing and CNV screening underlines the genetic component of PIDDs. Revised clinical diagnosis in half and altered management in a quarter of the families emphasizes the relevance of a molecular diagnosis.
Acknowledgements
A special thanks to the patients and their families for participating in the study. Thanks to the patient organizations, Norwegian Association for Immunodeficiencies (http://www.immunsvikt.no/) and Jeffrey Modell Foundation (JMF). The Baylor-Hopkins Center for Mendelian Genomics is supported by the National Human Genome Research Institute and the National Heart Lung and Blood (NHLBI) (U54HG006542). Funding for the work performed in Oslo was supported by the South-Eastern Norway Health Authority, and A.S-P received research scholarship from the American Women's club of Oslo.
Abbrevations
- aCGH
array comparative genomic hybridization
- AD
Autosomal dominant
- ALPS
Autoimmune lymphoproliferative syndrome
- AOH
Absence of heterozygosity
- AR
Autosomal recessive
- CMA
Chromosomal microarray analysis
- CNV
Copy number variant
- CVID
Common variable immunodeficiency
- HLH
Hemophagocytic lymphohistiocytosis
- HPV
Human papillomavirus
- HSCT
Hematopoietic stem cell transplantation
- MAF
Minor allele frequency
- MPLA
Multiplex ligation-dependent probe amplification
- NGS
Next generation sequencing
- PCR
Polymerase chain reaction
- PHA
Phytohemagglutinin
- PIDD
Primary immunodeficiency disease
- SCID
Severe combined immunodeficiency
- SCN
Severe congenital neutropenia
- SNP
Single nucleotide polymorphism
- SNV
Single nucleotide variant
- WES
Whole-exome sequencing
- WGS
Whole-genome sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc., is a member of the Scientific Advisory Board of Baylor Miraca Genetics Laboratories, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Baylor Miraca Genetics Laboratory (BMGL:http://www.bmgl.com/BMGL/Default.aspx).
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Browser, http://browser.1000genomes.org/index.html
Arterosclerosis Risk In Communities (ARIC), http://www2.cscc.unc.edu/aric/
Baylor-Hopkins Center for Mendelian Genomics, https://mendeliangenomics.org
Centers for Mendelian Genomics, http://www.mendelian.org
Baylor Miraca Genetics Laboratories, formerly known as the Medical Genetics
Laboratories, Baylor College of Medicine, http://www.bcm.edu/geneticlabs/
dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
HUGO Gene Nomenclature Committee (HGNC) http://www.genenames.org
Integrative Genomics Viewer (IGV), http://www.broadinstitute.org/igv/
NHLBI Exome Sequencing Project (ESP), http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu/
Clinical diagnosis criteria for the ESID Registry http://esid.org/Working-Parties/Registry/Diagnosis-criteria
Supplemental data – Online Repository
The Online Repository include one table (Table E1), further method descriptions, and eight extra figures (Figures E1-E8).
References
- 1.Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, et al. Primary Immunodeficiency Diseases: an Update on the Classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. Journal of clinical immunology. 2015 Oct 19; doi: 10.1007/s10875-015-0201-1. PubMed PMID: 26482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stray-Pedersen A, Abrahamsen TG, Froland SS. Primary immunodeficiency diseases in Norway. Journal of clinical immunology. 2000 Nov;20(6):477–85. doi: 10.1023/a:1026416017763. PubMed PMID: 11202238. [DOI] [PubMed] [Google Scholar]
- 3.Al-Herz W, Zainal ME, Alenezi HM, Husain K, Alshemmari SH. Performance status and deaths among children registered in Kuwait National Primary Immunodeficiency Disorders Registry. Asian Pac J Allergy. 2010 Jun-Sep;28(2-3):141–6. PubMed PMID: WOS:000283405200007. English. [PubMed] [Google Scholar]
- 4.Gathmann B, Mahlaoui N, Ceredih, Gerard L, Oksenhendler E, Warnatz K, et al. Clinical picture and treatment of 2212 patients with common variable immunodeficiency. The Journal of allergy and clinical immunology. 2014 Jul;134(1):116–26. doi: 10.1016/j.jaci.2013.12.1077. PubMed PMID: 24582312. [DOI] [PubMed] [Google Scholar]
- 5.de Pagter AP, Bredius RG, Kuijpers TW, Tramper J, van der Burg M, van Montfrans J, et al. Overview of 15-year severe combined immunodeficiency in the Netherlands: towards newborn blood spot screening. European journal of pediatrics. 2015 Sep;174(9):1183–8. doi: 10.1007/s00431-015-2518-4. PubMed PMID: 25875249. Pubmed Central PMCID: 4539359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Matteo G, Giordani L, Finocchi A, Ventura A, Chiriaco M, Blancato J, et al. Molecular characterization of a large cohort of patients with Chronic Granulomatous Disease and identification of novel CYBB mutations: an Italian multicenter study. Molecular immunology. 2009 Jun;46(10):1935–41. doi: 10.1016/j.molimm.2009.03.016. PubMed PMID: 19410294. [DOI] [PubMed] [Google Scholar]
- 7.Koker MY, Camcioglu Y, van Leeuwen K, Kilic SS, Barlan I, Yilmaz M, et al. Clinical, functional, and genetic characterization of chronic granulomatous disease in 89 Turkish patients. The Journal of allergy and clinical immunology. 2013 Nov;132(5):1156–63. e5. doi: 10.1016/j.jaci.2013.05.039. PubMed PMID: 23910690. [DOI] [PubMed] [Google Scholar]
- 8.Fattahi F, Badalzadeh M, Sedighipour L, Movahedi M, Fazlollahi MR, Mansouri SD, et al. Inheritance pattern and clinical aspects of 93 Iranian patients with chronic granulomatous disease. Journal of clinical immunology. 2011 Oct;31(5):792–801. doi: 10.1007/s10875-011-9567-x. PubMed PMID: 21789723. [DOI] [PubMed] [Google Scholar]
- 9.Chan A, Scalchunes C, Boyle M, Puck JM. Early vs. delayed diagnosis of severe combined immunodeficiency: a family perspective survey. Clinical immunology. 2011 Jan;138(1):3–8. doi: 10.1016/j.clim.2010.09.010. PubMed PMID: 21035402. Pubmed Central PMCID: 3022101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S, Thrasher AJ, Gaspar HB. Gene therapy for monogenic disorders of the bone marrow. British journal of haematology. 2015 Jun 5; doi: 10.1111/bjh.13520. PubMed PMID: 26044877. [DOI] [PubMed] [Google Scholar]
- 11.Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014 Jun 12;510(7504):235–40. doi: 10.1038/nature13420. PubMed PMID: 24870228. Pubmed Central PMCID: 4082311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacein-Bey Abina S, Gaspar HB, Blondeau J, Caccavelli L, Charrier S, Buckland K, et al. Outcomes following gene therapy in patients with severe Wiskott-Aldrich syndrome. JAMA. 2015 Apr 21;313(15):1550–63. doi: 10.1001/jama.2015.3253. PubMed PMID: 25898053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pai SY, Logan BR, Griffith LM, Buckley RH, Parrott RE, Dvorak CC, et al. Transplantation outcomes for severe combined immunodeficiency, 2000-2009. The New England journal of medicine. 2014 Jul 31;371(5):434–46. doi: 10.1056/NEJMoa1401177. PubMed PMID: 25075835. Pubmed Central PMCID: 4183064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montiel-Equihua CA, Thrasher AJ, Gaspar HB. Gene therapy for severe combined immunodeficiency due to adenosine deaminase deficiency. Current gene therapy. 2012 Feb 1;12(1):57–65. doi: 10.2174/156652312799789253. PubMed PMID: 22348551. [DOI] [PubMed] [Google Scholar]
- 15.Henderson C, Goldbach-Mansky R. Monogenic autoinflammatory diseases: new insights into clinical aspects and pathogenesis. Current opinion in rheumatology. 2010 Sep;22(5):567–78. doi: 10.1097/BOR.0b013e32833ceff4. PubMed PMID: 20671522. Pubmed Central PMCID: 3020910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milman N, Andersen CB, Hansen A, van Overeem Hansen T, Nielsen FC, Fledelius H, et al. Favourable effect of TNF-alpha inhibitor (infliximab) on Blau syndrome in monozygotic twins with a de novo CARD15 mutation. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2006 Dec;114(12):912–9. doi: 10.1111/j.1600-0463.2006.apm_522.x. PubMed PMID: 17207093. [DOI] [PubMed] [Google Scholar]
- 17.Aksentijevich I, Masters SL, Ferguson PJ, Dancey P, Frenkel J, van Royen- Kerkhoff A, et al. An autoinflammatory disease with deficiency of the interleukin-1-receptor antagonist. The New England journal of medicine. 2009 Jun 4;360(23):2426–37. doi: 10.1056/NEJMoa0807865. PubMed PMID: 19494218. Pubmed Central PMCID: 2876877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijman IJ, van Montfrans JM, Hoogstraat M, Boes ML, van de Corput L, Renner ED, et al. Targeted next-generation sequencing: a novel diagnostic tool for primary immunodeficiencies. The Journal of allergy and clinical immunology. 2014 Feb;133(2):529–34. doi: 10.1016/j.jaci.2013.08.032. PubMed PMID: 24139496. [DOI] [PubMed] [Google Scholar]
- 19.Moens LN, Falk-Sorqvist E, Asplund AC, Bernatowska E, Smith CI, Nilsson M. Diagnostics of primary immunodeficiency diseases: a sequencing capture approach. PloS one. 2014;9(12):e114901. doi: 10.1371/journal.pone.0114901. PubMed PMID: 25502423. Pubmed Central PMCID: 4263707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Mousa H, Abouelhoda M, Monies DM, Al-Tassan N, Al-Ghonaium A, Al- Saud B, et al. Unbiased targeted next-generation sequencing molecular approach for primary immunodeficiency diseases. The Journal of allergy and clinical immunology. 2016 Feb 23; doi: 10.1016/j.jaci.2015.12.1310. PubMed PMID: 26915675. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England journal of medicine. 2013 Oct 17;369(16):1502–11. doi: 10.1056/NEJMoa1306555. PubMed PMID: 24088041. Pubmed Central PMCID: 4211433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014 Nov 12;312(18):1870–9. doi: 10.1001/jama.2014.14601. PubMed PMID: 25326635. Pubmed Central PMCID: 4326249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation- dependent probe amplification. Nucleic Acids Res. 2002 Jun 15;30(12):e57. doi: 10.1093/nar/gnf056. PubMed PMID: 12060695. Pubmed Central PMCID: 117299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies). Clinical immunology. 1999 Dec;93(3):190–7. doi: 10.1006/clim.1999.4799. PubMed PMID: 10600329. [DOI] [PubMed] [Google Scholar]
- 25.Bonilla FA, Khan DA, Ballas ZK, Chinen J, Frank MM, Hsu JT, et al. Practice parameter for the diagnosis and management of primary immunodeficiency. The Journal of allergy and clinical immunology. 2015 Nov;136(5):1186–205. e1–78. doi: 10.1016/j.jaci.2015.04.049. PubMed PMID: 26371839. [DOI] [PubMed] [Google Scholar]
- 26.Bonilla FA, Bernstein IL, Khan DA, Ballas ZK, Chinen J, Frank MM, et al. Practice parameter for the diagnosis and management of primary immunodeficiency. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2005 May;94(5 Suppl 1):S1–63. doi: 10.1016/s1081-1206(10)61142-8. PubMed PMID: 15945566. [DOI] [PubMed] [Google Scholar]
- 27.Bousfiha A, Jeddane L, Al-Herz W, Ailal F, Casanova JL, Chatila T, et al. The 2015 IUIS Phenotypic Classification for Primary Immunodeficiencies. Journal of clinical immunology. 2015 Nov;35(8):727–38. doi: 10.1007/s10875-015-0198-5. PubMed PMID: 26445875. Pubmed Central PMCID: 4659854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews Genetics. 2011 Nov;12(11):745–55. doi: 10.1038/nrg3031. PubMed PMID: 21946919. [DOI] [PubMed] [Google Scholar]
- 29.Lupski JR, Gonzaga-Jauregui C, Yang Y, Bainbridge MN, Jhangiani S, Buhay CJ, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome medicine. 2013;5(6):57. doi: 10.1186/gm461. PubMed PMID: 23806086. Pubmed Central PMCID: 3706849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stray-Pedersen A, Jouanguy E, Crequer A, Bertuch AA, Brown BS, Jhangiani SN, et al. Compound heterozygous CORO1A mutations in siblings with a mucocutaneous-immunodeficiency syndrome of epidermodysplasia verruciformis-HPV, molluscum contagiosum and granulomatous tuberculoid leprosy. Journal of clinical immunology. 2014 Oct;34(7):871–90. doi: 10.1007/s10875-014-0074-8. PubMed PMID: 25073507. Pubmed Central PMCID: 4386834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulonen AM, Ellonen P, Almusa H, Lepisto M, Eldfors S, Hannula S, et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome biology. 2011;12(9):R94. doi: 10.1186/gb-2011-12-9-r94. PubMed PMID: 21955854. Pubmed Central PMCID: 3308057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shigemizu D, Momozawa Y, Abe T, Morizono T, Boroevich KA, Takata S, et al. Performance comparison of four commercial human whole-exome capture platforms. Scientific reports. 2015;5:12742. doi: 10.1038/srep12742. PubMed PMID: 26235669. Pubmed Central PMCID: 4522667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. PubMed PMID: 24475911. Pubmed Central PMCID: 3922167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome research. 2010 Jan;20(1):110–21. doi: 10.1101/gr.097857.109. PubMed PMID: 19858363. Pubmed Central PMCID: 2798823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome research. 2005 Jul;15(7):901–13. doi: 10.1101/gr.3577405. PubMed PMID: WOS:000230424000001. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003 Jul 1;31(13):3812–4. doi: 10.1093/nar/gkg509. PubMed PMID: 12824425. Pubmed Central PMCID: 168916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010 Apr;7(4):248–9. doi: 10.1038/nmeth0410-248. PubMed PMID: 20354512. Pubmed Central PMCID: 2855889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome research. 2009 Sep;19(9):1553–61. doi: 10.1101/gr.092619.109. PubMed PMID: 19602639. Pubmed Central PMCID: 2752137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014 Apr;11(4):361–2. doi: 10.1038/nmeth.2890. PubMed PMID: 24681721. [DOI] [PubMed] [Google Scholar]
- 40.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine. 2015 May;17(5):405–24. doi: 10.1038/gim.2015.30. PubMed PMID: WOS:000354096900013. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itan Y, Casanova JL. Novel primary immunodeficiency candidate genes predicted by the human gene connectome. Frontiers in immunology. 2015 Apr 1;6 doi: 10.3389/fimmu.2015.00142. PubMed PMID: WOS:000354902900001. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. Journal of Experimental Medicine. 2014 Oct 20;211(11):2137–49. doi: 10.1084/jem.20140520. PubMed PMID: WOS:000345268700002. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samarakoon PS, Sorte HS, Kristiansen BE, Skodje T, Sheng Y, Tjonnfjord GE, et al. Identification of copy number variants from exome sequence data. BMC genomics. 2014;15:661. doi: 10.1186/1471-2164-15-661. PubMed PMID: 25102989. Pubmed Central PMCID: 4132917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bayer DK, Martinez CA, Sorte HS, Forbes LR, Demmler-Harrison GJ, Hanson IC, et al. Vaccine-associated varicella and rubella infections in severe combined immunodeficiency with isolated CD4 lymphocytopenia and mutations in IL7R detected by tandem whole exome sequencing and chromosomal microarray. Clinical and experimental immunology. 2014 Dec;178(3):459–69. doi: 10.1111/cei.12421. PubMed PMID: 25046553. Pubmed Central PMCID: 4238873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stray-Pedersen A, Backe PH, Sorte HS, Morkrid L, Chokshi NY, Erichsen HC, et al. PGM3 mutations cause a congenital disorder of glycosylation with severe immunodeficiency and skeletal dysplasia. American journal of human genetics. 2014 Jul 3;95(1):96–107. doi: 10.1016/j.ajhg.2014.05.007. PubMed PMID: 24931394. Pubmed Central PMCID: 4085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, et al. Loss of B Cells in Patients with Heterozygous Mutations in IKAROS. The New England journal of medicine. 2016 Mar 17;374(11):1032–43. doi: 10.1056/NEJMoa1512234. PubMed PMID: 26981933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayrapetyan A, Dencher PC, van Leeuwen K, de Boer M, Roos D. Different unequal cross-over events between NCF1 and its pseudogenes in autosomal p47(phox)-deficient chronic granulomatous disease. Biochimica et biophysica acta. 2013 Oct;1832(10):1662–72. doi: 10.1016/j.bbadis.2013.05.001. PubMed PMID: 23688784. [DOI] [PubMed] [Google Scholar]
- 48.Hoyeraal HM, Lamvik J, Moe PJ. Congenital hypoplastic thrombocytopenia and cerebral malformations in two brothers. Acta paediatrica Scandinavica. 1970 Mar;59(2):185–91. doi: 10.1111/j.1651-2227.1970.tb08986.x. PubMed PMID: 5442429. [DOI] [PubMed] [Google Scholar]
- 49.Picard C, Dogniaux S, Chemin K, Maciorowski Z, Lim A, Mazerolles F, et al. Hypomorphic mutation of ZAP70 in human results in a late onset immunodeficiency and no autoimmunity. European journal of immunology. 2009 Jul;39(7):1966–76. doi: 10.1002/eji.200939385. PubMed PMID: 19548248. [DOI] [PubMed] [Google Scholar]
- 50.Huo YK, Wang Z, Hong JH, Chessa L, McBride WH, Perlman SL, et al. Radiosensitivity of ataxia-telangiectasia, X-linked agammaglobulinemia, and related syndromes using a modified colony survival assay. Cancer research. 1994 May 15;54(10):2544–7. PubMed PMID: 8168076. [PubMed] [Google Scholar]
- 51.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011 Sep 30;147(1):32–43. doi: 10.1016/j.cell.2011.09.008. PubMed PMID: 21962505. Pubmed Central PMCID: 3656718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watkin LB, Jessen B, Wiszniewski W, Vece TJ, Jan M, Sha Y, et al. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune- mediated lung disease and arthritis. Nat Genet. 2015 Jun;47(6):654–60. doi: 10.1038/ng.3279. PubMed PMID: 25894502. Pubmed Central PMCID: 4513663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niemela JE, Lu L, Fleisher TA, Davis J, Caminha I, Natter M, et al. Somatic KRAS mutations associated with a human nonmalignant syndrome of autoimmunity and abnormal leukocyte homeostasis. Blood. 2011 Mar 10;117(10):2883–6. doi: 10.1182/blood-2010-07-295501. PubMed PMID: 21079152. Pubmed Central PMCID: 3062298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015 Jan 22;125(4):591–9. doi: 10.1182/blood-2014-09-602763. PubMed PMID: 25359994. Pubmed Central PMCID: 4304103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegel AM, Stone KD, Cruse G, Lawrence MG, Olivera A, Jung MY, et al. Diminished allergic disease in patients with STAT3 mutations reveals a role for STAT3 signaling in mast cell degranulation. The Journal of allergy and clinical immunology. 2013 Dec;132(6):1388–96. doi: 10.1016/j.jaci.2013.08.045. PubMed PMID: 24184145. Pubmed Central PMCID: 3881191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bornholdt D, Atkinson TP, Bouadjar B, Catteau B, Cox H, De Silva D, et al. Genotype-phenotype correlations emerging from the identification of missense mutations in MBTPS2. Human mutation. 2013 Apr;34(4):587–94. doi: 10.1002/humu.22275. PubMed PMID: 23316014. [DOI] [PubMed] [Google Scholar]
- 57.Corujeira S, Agueda S, Monteiro G, Canelhas A, Sampaio M, Rocha R, et al. Expanding the phenotype of IFAP/BRESECK syndrome: a new case with severe hypogammaglobulinemia. European journal of medical genetics. 2013 Nov;56(11):603–5. doi: 10.1016/j.ejmg.2013.09.005. PubMed PMID: 24090718. [DOI] [PubMed] [Google Scholar]
- 58.Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat Genet. 2011 Jan;43(1):79–84. doi: 10.1038/ng.727. PubMed PMID: 21131974. Pubmed Central PMCID: 3132183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders C, Smith L, Wibrand F, Ravn K, Bross P, Thiffault I, et al. CLPB variants associated with autosomal-recessive mitochondrial disorder with cataract, neutropenia, epilepsy, and methylglutaconic aciduria. American journal of human genetics. 2015 Feb 5;96(2):258–65. doi: 10.1016/j.ajhg.2014.12.020. PubMed PMID: 25597511. Pubmed Central PMCID: 4320254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wortmann SB, Zietkiewicz S, Kousi M, Szklarczyk R, Haack TB, Gersting SW, et al. CLPB mutations cause 3-methylglutaconic aciduria, progressive brain atrophy, intellectual disability, congenital neutropenia, cataracts, movement disorder. American journal of human genetics. 2015 Feb 5;96(2):245–57. doi: 10.1016/j.ajhg.2014.12.013. PubMed PMID: 25597510. Pubmed Central PMCID: 4320260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schubert D, Bode C, Kenefeck R, Hou TZ, Wing JB, Kennedy A, et al. Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nature medicine. 2014 Dec;20(12):1410–6. doi: 10.1038/nm.3746. PubMed PMID: 25329329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuehn HS, Ouyang W, Lo B, Deenick EK, Niemela JE, Avery DT, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014 Sep 26;345(6204):1623–7. doi: 10.1126/science.1255904. PubMed PMID: 25213377. Pubmed Central PMCID: 4371526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibbs DC, Orlow I, Kanetsky PA, Luo L, Kricker A, Armstrong BK, et al. Inherited genetic variants associated with occurrence of multiple primary melanoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2015 Jun;24(6):992–7. doi: 10.1158/1055-9965.EPI-14-1426. PubMed PMID: 25837821. Pubmed Central PMCID: 4452425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011 Nov;43(11):1108–13. doi: 10.1038/ng.959. PubMed PMID: 21983787. Pubmed Central PMCID: 3251256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen K, Coonrod EM, Kumanovics A, Franks ZF, Durtschi JD, Margraf RL, et al. Germline mutations in NFKB2 implicate the noncanonical NF-kappaB pathway in the pathogenesis of common variable immunodeficiency. American journal of human genetics. 2013 Nov 7;93(5):812–24. doi: 10.1016/j.ajhg.2013.09.009. PubMed PMID: 24140114. Pubmed Central PMCID: 3824125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Hanson S, Gurugama P, Jones A, Clark B, Ibrahim MA. Novel NFKB2 mutation in early-onset CVID. Journal of clinical immunology. 2014 Aug;34(6):686–90. doi: 10.1007/s10875-014-0064-x. PubMed PMID: 24888602. [DOI] [PubMed] [Google Scholar]
- 67.Lougaris V, Tabellini G, Vitali M, Baronio M, Patrizi O, Tampella G, et al. Defective natural killer-cell cytotoxic activity in NFKB2-mutated CVID-like disease. The Journal of allergy and clinical immunology. 2015 Jun;135(6):1641–3. doi: 10.1016/j.jaci.2014.11.038. PubMed PMID: 25605273. [DOI] [PubMed] [Google Scholar]
- 68.Sassi A, Lazaroski S, Wu G, Haslam SM, Fliegauf M, Mellouli F, et al. Hypomorphic homozygous mutations in phosphoglucomutase 3 (PGM3) impair immunity and increase serum IgE levels. The Journal of allergy and clinical immunology. 2014 May;133(5):1410–9. 9, e1–13. doi: 10.1016/j.jaci.2014.02.025. PubMed PMID: 24698316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Yu X, Ichikawa M, Lyons JJ, Datta S, Lamborn IT, et al. Autosomal recessive phosphoglucomutase 3 (PGM3) mutations link glycosylation defects to atopy, immune deficiency, autoimmunity, and neurocognitive impairment. The Journal of allergy and clinical immunology. 2014 May;133(5):1400–9. 9, e1–5. doi: 10.1016/j.jaci.2014.02.013. PubMed PMID: 24589341. Pubmed Central PMCID: 4016982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernth-Jensen JM, Holm M, Christiansen M. Neonatal-onset T(−)B(−)NK(+) severe combined immunodeficiency and neutropenia caused by mutated phosphoglucomutase 3. The Journal of allergy and clinical immunology. 2016 Jan;137(1):321–4. doi: 10.1016/j.jaci.2015.07.047. PubMed PMID: 26409661. [DOI] [PubMed] [Google Scholar]
- 71.Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell reports. 2015 Aug 18;12(7):1169–83. doi: 10.1016/j.celrep.2015.07.023. PubMed PMID: 26257172. Pubmed Central PMCID: 4545408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, et al. A de novo paradigm for mental retardation. Nat Genet. 2010 Dec;42(12):1109–12. doi: 10.1038/ng.712. PubMed PMID: 21076407. [DOI] [PubMed] [Google Scholar]
- 73.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014 Jul 17;511(7509):344–7. doi: 10.1038/nature13394. PubMed PMID: 24896178. [DOI] [PubMed] [Google Scholar]
- 74.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. The New England journal of medicine. 2010 Apr 1;362(13):1181–91. doi: 10.1056/NEJMoa0908094. PubMed PMID: 20220177. Pubmed Central PMCID: 4036802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duzkale H, Shen J, McLaughlin H, Alfares A, Kelly MA, Pugh TJ, et al. A systematic approach to assessing the clinical significance of genetic variants. Clinical genetics. 2013 Nov;84(5):453–63. doi: 10.1111/cge.12257. PubMed PMID: 24033266. Pubmed Central PMCID: 3995020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. The New England journal of medicine. 2012 Nov 15;367(20):1921–9. doi: 10.1056/NEJMoa1206524. PubMed PMID: 23033978. [DOI] [PubMed] [Google Scholar]
- 77.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012 Nov 10;380(9854):1674–82. doi: 10.1016/S0140-6736(12)61480-9. PubMed PMID: 23020937. [DOI] [PubMed] [Google Scholar]
- 78.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015 Mar 12;519(7542):223–8. doi: 10.1038/nature14135. PubMed PMID: 25533962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. The Journal of allergy and clinical immunology. 2011 Jun;127(6):1360–7. e6. doi: 10.1016/j.jaci.2011.02.039. PubMed PMID: 21497890. Pubmed Central PMCID: 3646656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Schouwenburg PA, Davenport EE, Kienzler AK, Marwah I, Wright B, Lucas M, et al. Application of whole genome and RNA sequencing to investigate the genomic landscape of common variable immunodeficiency disorders. Clinical immunology. 2015 Oct;160(2):301–14. doi: 10.1016/j.clim.2015.05.020. PubMed PMID: 26122175. Pubmed Central PMCID: 4601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012 Nov 10;380(9854):1674–82. doi: 10.1016/S0140-6736(12)61480-9. PubMed PMID: WOS:000310951400029. English. [DOI] [PubMed] [Google Scholar]
- 82.Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Akdemir ZC, et al. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron. 2015;88(3):499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stoddard JL, Niemela JE, Fleisher TA, Rosenzweig SD. Targeted NGS: A Cost-Effective Approach to Molecular Diagnosis of PIDs. Frontiers in immunology. 2014;5:531. doi: 10.3389/fimmu.2014.00531. PubMed PMID: 25404929. Pubmed Central PMCID: 4217515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raje N, Soden S, Swanson D, Ciaccio CE, Kingsmore SF, Dinwiddie DL. Utility of next generation sequencing in clinical primary immunodeficiencies. Current allergy and asthma reports. 2014 Oct;14(10):468. doi: 10.1007/s11882-014-0468-y. PubMed PMID: 25149170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Picard C, Fischer A. Contribution of high-throughput DNA sequencing to the study of primary immunodeficiencies. European journal of immunology. 2014 Oct;44(10):2854–61. doi: 10.1002/eji.201444669. PubMed PMID: 25154746. [DOI] [PubMed] [Google Scholar]
- 86.Platt C, Geha RS, Chou J. Gene hunting in the genomic era: Approaches to diagnostic dilemmas in patients with primary immunodeficiencies. The Journal of allergy and clinical immunology. 2014 Aug;134(2):262–8. doi: 10.1016/j.jaci.2013.08.021. PubMed PMID: 24100122. Pubmed Central PMCID: 3976463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC bioinformatics. 2014;15:247. doi: 10.1186/1471-2105-15-247. PubMed PMID: 25038816. Pubmed Central PMCID: 4122774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kwan A, Abraham RS, Currier R, Brower A, Andruszewski K, Abbott JK, et al. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. JAMA. 2014 Aug 20;312(7):729–38. doi: 10.1001/jama.2014.9132. PubMed PMID: 25138334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu H, et al. Comprehensive Clinical Evaluation of Primary Immunodeficiency by Next Generation Sequencing. 2015 Submitted. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.