Abstract
Background
Atopic dermatitis (AD) is characterized by intense pruritis and is a common childhood inflammatory disease. Many factors are known to affect AD development, including the pleiotropic cytokine interleukin (IL)-4. Yet, still little is known regarding the direct effects of IL-4 on keratinocyte function. Objective and Methods: In this report, RNA-seq and functional assays were used to define the impact of the allergic environment on primary keratinocyte function and wound repair in mice.
Results
Acute or chronic stimulation by IL-4 modified expression of over 1000 genes expressed in human keratinocytes that are involved in a broad spectrum of non-overlapping functions. Among the IL-4-induced changes, repression of fibronectin critically impaired the human keratinocyte wounding response. Moreover, in mouse models of spontaneous and induced AD-like lesions there was delayed re-epithelialization. Importantly, topical treatment with fibronectin restored the epidermal repair response.
Conclusion
Keratinocyte gene expression is critically shaped by IL-4, altering cell fate decisions likely important for the clinical manifestations and pathology of allergic skin disease.
Keywords: keratinocyte, atopic dermatitis, wound healing, IL-4, RNA-seq, fibronectin
INTRODUCTION
Atopic dermatitis (AD) is one of the most common inflammatory diseases in children, and is characterized by intense pruritis (itch) and abnormal skin (reviewed in 1–3). AD is an early indicator of subsequent atopic diseases with roughly 60% of children with AD later developing airway hyperresponsiveness 4–6. The pathogenesis of AD involves impaired keratinocyte differentiation and diminished skin barrier function, resulting in dry (xerotic) skin, increased susceptibility to bacterial and viral infections, and augmented percutaneous exposure to environmental allergens 3, 7.
The Th2-type cytokine interleukin (IL)-4 has been shown to impact keratinocyte differentiation 7–9. IL-4 is a pleiotropic cytokine secreted by activated T lymphocytes and innate immune cells acting through a receptor on hematopoietic cells composed of the IL-4Rα and γc receptors (type I IL-4R) 10, 11. Keratinocytes and other non-hematopoietic cells do not express γc receptors, but can respond to IL-4 via the type II IL-4R composed of IL-4Rα and IL-13Rα112. IL-4 binding to either receptor mediates the phosphorylation and dimerization of the Signal Transducer and Activator of Transcription 6 (STAT6), regulating transcriptional activation of genes in target cells 9. Keratinocytes exposed to IL-4 exhibit reduced expression of Epidermal Differentiation Complex (EDC) genes 13, decreased expression of defensins 3, altered expression of keratins 14, and increased production of the chemokine CCL26 (eotaxin 3) 15.
IL-4 is an important target in patients, and in clinical trials of subjects with moderate-to-severe AD, disease scores improved after anti-IL-4Rα human antagonist antibody treatment 16. However, there is still little known regarding the consequences of IL-4 signaling on keratinocyte differentiation and barrier protection. The current study defines the global changes induced by IL-4 on gene expression in human keratinocytes via bioinformatics analysis of gene expression following high throughput analysis (RNA-seq). This analysis identified a wound-healing pathway, including expression of fibronectin by keratinocytes, which is repressed by IL-4 treatment and likely contributes to pathology in patients with AD. Moreover, modulating this pathway in an in vivo model improved wound repair.
MATERIALS AND METHODS
Mice
Female BALB/C and C57BL/6 mice were purchased from Harlan Bioscience (Indianapolis, Ind). The generation of Stat6VT transgenic mice was previously described 17. These mice express the human STAT6 gene with V547 and T548 mutated to alanine under transcriptional control of the CD2 locus control region. IL-4–deficient mice (Il4−/−) were purchased from The Jackson Laboratory (Bar Harbor, ME) and mated to Stat6VT transgenic mice. Stat6−/− mice were generated and backcrossed for at least 10 generations to BALB/c mice as described previously 18. Mouse ears were punched using a 2 mm punch (Kent Scientific) to determine the in vivo wound healing response. In some experiments, mice were treated for two days with ointment containing BSA (Promega) or fibronectin purified from human plasma (Sigma). To determine wounding response in an induced model of AD-like disease, WT and Stat6−/− mice were treated with the vitamin D analogue MC903 (Calcipotriol, Sigma) as previously described 19. MC903 was dissolved in 100% ethanol and topically applied on mouse ears (2 nmol in 25 μL per ear) for 5 days. Ethanol alone was used as a vehicle control. Ears were punched at day 6 after treatment. Mice were maintained in pathogen-free conditions, and all studies were approved by the Animal Care and Use Committee of the Indiana University School of Medicine.
Keratinocyte cell culture
Primary human keratinocytes were isolated from excised foreskin tissue as previously described 20, and washed with antibiotics. The tissue was minced and the individual cells were released from the tissue using trypsin digestion. Keratinocytes and fibroblasts were separated by differential resistance to treatment with EDTA. Isolated cells were grown in EpiLife Complete media (Life Technologies) with human keratinocyte (HK) growth supplement (Life Technologies) and 1000 U penicillin–streptomycin (Roche) and grown as unmanipulated cultures or immortalized by the expression of hTERT 21. To stimulate keratinocyte differentiation, HK were treated with 2 mM of CaCl2 every other day. Human keratinocytes were stimulated with 20 ng/ml or indicated concentration of recombinant human IL-4 (R&D Systems) or IL-13 (Peprotech). All HK samples were de-identified, and the institutional review board of the Indiana University School of Medicine certified these studies as exempt.
Histological and immunofluorescence analysis
Ears tissue was fixed using 10% formalin for 24 hours. Routine histological techniques were used to paraffin embed the ears, and 5-μm sections were stained with H&E or Masson’s trichrome. Ki-67 expression was analyzed by immunofluorescence using rabbit anti-human Ki67 (SP6) antibody and anti-rabbit 488nm. Quantitative digital morphometric analysis of the percentage of re-epithelialization was measured using Image J 1.43u. Percentage of re-epithelialization was calculated by dividing the total pixel length of lesion visualized at days 3 or 6 after wounding by the pixel length of lesions with incomplete re-epithelialization.
In vitro wound assay and live cell imaging
Normal human keratinocytes were cultured with a 100% confluence in plates covered or not with 1 μg/ml of BSA (Promega) or fibronectin from human plasma (Sigma-Aldrich). HK were cultured with calcium chloride alone in EpiLife Complete media and stimulated with 20 ng/ml recombinant human IL-4 as indicated. At days 2 or 5, a cell free area was created by scratching the monolayer with a 200 μl pipette tip. Time-lapse photography was performed on an automated time-lapse imaging system (Biostation IM-Q, Nikon Instruments) every 2 minutes in an environmentally controlled chamber (5% CO2, 37°C) (3.2 min) fitted with the 20× NA 0.8 lens and a DS-Qi1 CCD camera. Photoshop (Adobe) software platforms were used to export movies.
Organotypic skin equivalent
In vitro skin equivalents were constructed using normal primary fibroblasts and keratinocytes isolated from human neonatal foreskins, as previously described 22. The skin equivalents were grown at 35°C in an air incubator for 7–10 days, and fresh media was added every two days.
Cell isolation and flow cytometry
Epidermis was separated from dermis following enzymatic treatment with 400 μg/ml of liberase (Roche) for 90 minutes at 37°C. Total cells were further isolated from epidermis by using gentleMacs dissociator (Miltenyi Biotech) following manufacturer’s instruction. For detection of Stat6VT expression, splenocytes, and total epidermis and dermis cell suspension were pre-incubated with Fc-block (BD Pharmingen) for 10 min at 4°C bef ore incubation with anti-CD45, anti-CD3, anti-CD4, anti-CD8 and anti-CD19 for 30 min at 4°C. Human keratinocytes were harvested and incubated with Human TruStain FcX (Biolegend), for 10 min at 4°C. The cells were fixe d and permeabilized for intracellular staining with phospho-STAT6 Alexa 488 (BD Pharmingen) or anti-Flag APC (DYKDDDDK tag epitope) (Bioloegend) antibody before analysis by flow cytometry (Attune® Acoustic Focusing Cytometer), as described 23. The results were analyzed using FlowJo (Ashland, OR).
ELISA
Levels of CCL26 were measured in the supernatant of HK using human quantikine ELISA kit (R&D Systems). Fibronectin levels were measured in single cell suspension lysates isolated from dermis and epidermis as described above. Fibronectin levels were measured using mouse ELISA kit (Abcam) following manufacturer’s instruction.
Quantitative RT-PCR
Total RNA was isolated from cells and reverse transcribed according to manufacturer’s instructions (Invitrogen Life Technologies, Carlsbad, CA). Quantitative PCR was performed with Taqman Fast Universal PCR Master Mix and commercially available primers (Applied Biosystems). RNA was normalized to expression levels of β2-microglobulin and relative expression was calculated with the 2−ΔΔCt method.
Statistical analysis
Statistical analysis was determined using the unpaired Student’s t-test or ANOVA followed by the Tukey’s test for multiple comparisons, and a p value of less than 0.05 was considered significant.
Alignment of RNAseq reads to human genome
Total RNA was isolated from immortalized HK cultured under conditions indicated above using TRIzol reagent. RNA samples were purified using RNase-Free DNAse kit (Qiagen). RNA-Seq libraries were generated by Otogenetics using HiSeq 2000 sequencing instrument with 20 million paired-end 100 nucleotide reads per sample, with biological duplicates for each condition and results displayed as average samples. The percentage of mapped reads (50–56% of sequences) represented a 20X nucleotide coverage of the estimated transcriptome. Bioinformatics analysis was performed by the Bioinformatics Core at IUSM. Short reads (fastq files) from each sample were independently aligned to human reference genome hg19 using tophat (version 2.1.0), with default parameters and known transcriptome. Alignment results were filtered by bamutils v0.5.0 to remove reads with multiple mappings. Statistics data of the resulting alignment files were created using samtools (version 0.1.18) and bamutils (version 0.5.0). The counts of aligned reads mapping to known genes were calculated using bamutils (version 0.5.0). EdgeR (version 2.11) was used for differential expression analysis. RNA-Seq data files were submitted to GEO (accession number GSE59275). Gene ontology analysis was performed using DAVID Bioinformatics resources 24, Venn diagram analyses were generated using Venny, and heat maps were created using MultiExperiment Viewer (MEV).
RESULTS
IL-4 responses in human keratinocytes
To begin to define keratinocyte responses to IL-4, we characterized the expression of IL-4 signaling components. Immortalized human keratinocytes (HK) were cultured for five days in calcium chloride and demonstrated increased filaggrin (FLG) expression as an indicator of differentiation. Differentiated HK exhibited greater expression of STAT6, IL4R and the inhibitory IL13RA2 at day 5, contrasting largely unchanged expression of IL13RA1 during the differentiation period (Fig E1, A). Flow cytometric analysis indicated the percentage of phospho-(p)STAT6 (Tyr641)-positive keratinocytes and the mean fluorescence intensity (MFI) of pSTAT6 peaked 30 minutes after IL-4 stimulation (Fig E1, B and C). Analysis of gene expression and chemokine production demonstrated decreased expression of the EDC genes filaggrin (FLG), loricrin (LOR), and involucrin (IVL) following long term incubation with IL-4, and the induction of CCL26 expression and secretion following short and long term incubation with IL-4 (Fig E1, D–F). The parallels between these responses and those previously published for isolated keratinocytes and skin tissue validated this approach for further studies 8, 9, 25–27.
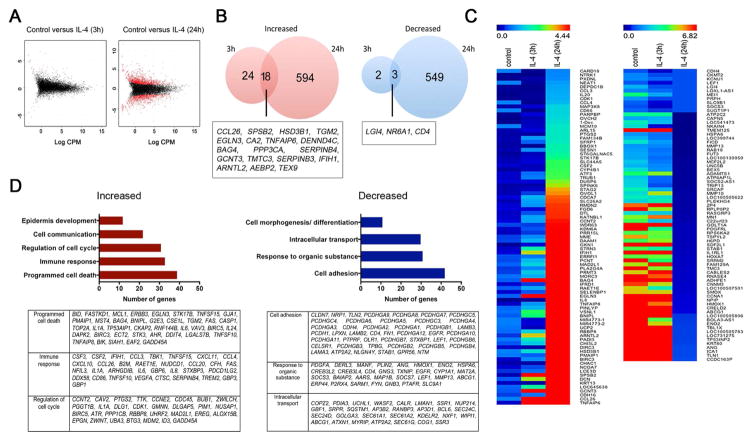
To broadly determine IL-4-stimulated transcriptional changes in HK, we performed RNA-Seq analysis. At day 2 of culture, HK were incubated in the presence or absence of IL-4 for 3 or 24 hours. After 3 hours, IL-4 stimulation resulted in at least a 2-fold change in the expression of 47 genes when compared with cells cultured with media alone (Fig 1, A and B). After 24 hours, IL-4 stimulation resulted in a differential expression of 1164 genes (Fig 1, A and B). The genes most differentially regulated at both time points are indicated by the heatmap in Fig 1, C. Among the transcripts increased at 24 hours that were classified by DAVID analysis, 39 were associated with regulation of cell death, 33 were involved with immune response and cytokine-receptor interaction, and 31 were involved with regulation of cell cycle (Fig 1, D). Transcripts for proteins involved with cell adhesion, response to organic substances, and intracellular transport and secretion were the largest functional categories of genes whose expression was decreased by IL-4.
Figure 1. IL-4 modifies human keratinocyte (HK) gene expression.
HK were differentiated for 2 days with calcium chloride and stimulated with IL-4 for 3 and 24 hours before RNA-seq analysis. (A) Graphical representation of genes that were enriched at least 2-fold in IL-4-stimulated keratinocytes (red dots). (B) Venn diagram representing the number of genes altered by IL-4 after 3 and 24 hours. (C) Heatmap comparison of gene enrichment in IL-4-stimulated HK using MeV software. (D) Gene ontology analysis using the differential expression of genes at 24 hours after IL-4. Clustering was performed using David bioinformatics database analysis. Data represent average of two biological replicates for each condition. *p<0.05 and FDR<0.1
To extend these finding, three-dimensional cultures (human skin equivalents) were treated with IL-4 for 24 hours before testing the expression of 7 IL-4-induced genes. Treatment with IL-4 resulted in a significant increase in CCL26, CA2, CISH, and SERPINB4 (Fig E2, A). Additionally, IL-4 effects on HK monolayer gene expression increased in dose dependent manner (Fig E2, B). Like IL-4, IL-13 alters gene expression in keratinocytes 12,14. To determine whether IL-4 and IL-13 have similar function in regulating HK gene expression, HK were differentiated and stimulated with IL-4 or IL-13 for 5 days before analyzing the expression of genes. IL-13 decreased FLG, IVL, LOR and FN1 similarly to IL-4 (Fig E2, C). IL-13 also resulted in a comparable increase in SERPINB3 and SERPINB4, but only a partial increase in CCL26 and CISH expression when compared to IL-4. Collectively, these data showed that acute stimulation of keratinocytes with IL-4 alters the expression of genes encoding proteins with diverse functions that could ultimately influence local inflammatory responses, alter cell proliferation and survival, and favor loss of cell adhesion.
RNA-Seq analysis of keratinocytes chronically stimulated with IL-4
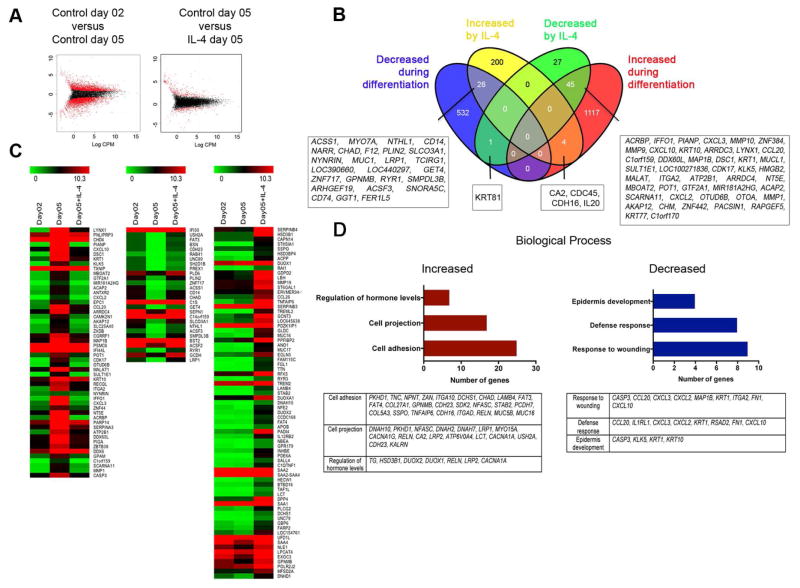
As shown in Fig. E1, D, exposure of keratinocytes to calcium chloride for 5 days resulted in altered expression of differentiation related genes. Thus, to determine whether IL-4 modifies genes expressed by keratinocytes in later phases of differentiation, we performed RNA-seq analysis of HK differentiated for five days and incubated with or without IL-4, before comparing the results to HK cultured for two days. HK differentiated for 5 days exhibited differential expression of 1724 genes when compared with HK at day 2, with IL-4 altering expression of 303 genes (Fig 2, A and B). The expression pattern for a subset of genes in IL-4-stimulated HK at day 5 of culture was similar to the expression pattern in control HK at day 2 (Fig 2, C). IL-4 also increased the expression of a set of genes compared to unstimulated HK at either time point (Fig 2, C). A Venn diagram shows the 1166 genes increased and 558 genes decreased during HK differentiation (Fig 2, B). IL-4 attenuated differentiation-induced changes in gene expression of 71 genes (45 were decreased and 26 were increased) (Fig 2, B). Moreover, IL-4 further augmented the expression of 4 genes and decreased the expression of 1 gene in calcium-differentiated HK (Fig 2, B). We also observed that IL-4 increased 200 genes and decreased 27 genes in HK that were not differentially expressed during differentiation (Fig 2, B). A gene ontology analysis of IL-4-regulated genes showed that chronic exposure to IL-4 increased transcription of 25 genes involved with cell adhesion, and 17 genes related with cell projection. IL-4 decreased transcription of 9 genes involved with wound healing and 8 genes involved in host defense (Fig 2, D).
Figure 2. Chronic IL-4 stimulation alters gene expression associated with normal differentiation in keratinocytes.
Human keratinocytes (HK) were differentiated for 5 days with calcium chloride in the presence or absence of IL-4 before RNA-seq analysis. (A) Graphical representation of genes enriched at least 2-fold in control HK at day 5 of differentiation and in IL-4-stimulated keratinocytes (red dots). (B) Venn diagram representing the number of genes differentially expressed at day 5 by keratinocytes cultured with calcium chloride alone (compared with day 2) and after IL-4. (C) Heatmap comparison using MeV software of gene enrichment in keratinocytes cultured for 2 or 5 days with calcium chloride alone or for 5 days with IL-4. (D) Gene ontology analysis performed using genes differentially expressed after 5 days of stimulation with IL-4. Clustering was performed using David bioinformatics database analysis. Data represent average of two biological replicates for each condition. p<0.05 and FDR<0.1
IL-4 impairs keratinocyte wound healing response
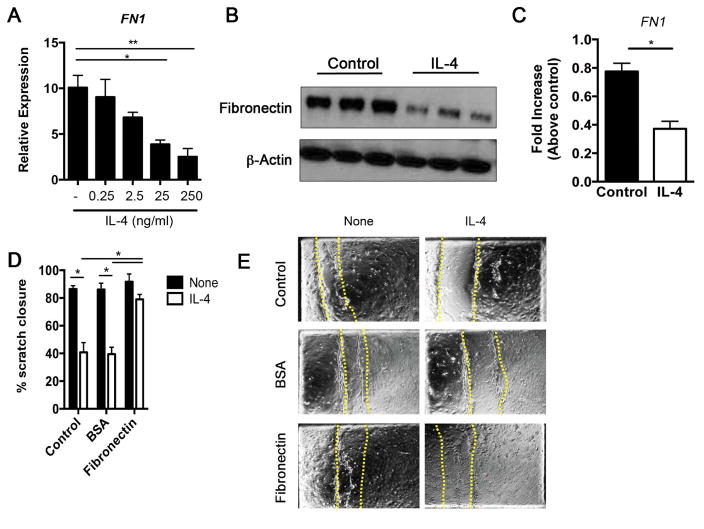
As noted above, a subset of genes regulated by IL-4 impacts the wound repair process. This observation is consistent with diminished wound repair in children that show signs of increased atopy 28, and with the diminished recovery of transepidermal water loss following injury in Stat6VT transgenic mice, a model of allergic skin inflammation 9, 27. To investigate whether IL-4-induced changes in the wound healing subset of genes alter functional activity of HK, we assayed for wound healing capacity in culture. HK were differentiated for 2 or 5 days and cultured in the presence or absence of IL-4. Scratches were made in the confluent monolayer to simulate wounding, and wound closure was monitored over 48 hours. Time-lapse analysis showed that differentiated keratinocytes migrate towards the wound area within 24 hours and partially close the wound space (Fig E3, A). Proliferative cells were observed in the area adjacent to the wound edge (Online movies) while cells at the edge guided the movement of neighboring cells. In contrast, IL-4-stimulated keratinocytes failed to migrate toward the wound edge and exhibited significantly less wound closure in both cultures when compared with the control cells (Fig E3, B). Since IL-4 decreases the potential wound healing response of HK at days 2 and 5 differentiation, we used the RNA-seq analysis to better define a target gene involved in the reduced wounding at both time points. Thus, as we observed, at days 2 and 5 of differentiation IL-4-treated HK exhibited reduced FN1 expression (Fig E3, C). Keratinocytes cultured with increasing concentrations of IL-4 exhibited reduced levels of FN1 mRNA (Fig 3, A). These cells also exhibited reduced fibronectin production (Fig 3, B), and decreased FN1 mRNA expression 3 hours after scratch of monolayers in the in vitro wound-healing assay (Fig 3, C). To further assess whether fibronectin potentiates wound-healing response in IL-4-treated HK, cells were differentiated for 5 days in control plates, BSA-coated plates or fibronectin-coated plates and cultured in the presence or absence of IL-4. A quantitative analysis showed that IL-4-treated HK cultured on a fibronectin substratum exhibited a significant increase in wound closure when compared with HK cultured on control or BSA substratum with IL-4 (Fig 3, D and E). Collectively, IL-4 potently inhibits the wound-healing capacity of keratinocytes by inhibiting fibronectin production.
Figure 3. IL-4-stimulated keratinocytes show delayed wound healing.
(A) Human keratinocytes (HK) were differentiated for 5 days with calcium chloride in the presence or absence of IL-4 before analysis of FN1 expression by qRT-PCR. (B) Cell lysates were collected and the expression of fibronectin and β-actin was determined by western blot (C) Gene expression was analyzed 3h after scratching using qRT–PCR. (D) Percentage of scratch closed after 24h of culture, and (E) microscopy of human keratinocytes differentiated with calcium chloride alone or calcium chloride plus IL-4 for 5 days in control or plates covered with a substratum of BSA or fibronectin. Dashed red lines represent original wounds. Pictures are representative of 4 fields analyzed over at least three independent experiments. Data represents the mean of three independent experiments. *p<0.05.
Impaired skin wound healing response in models of AD-like disease
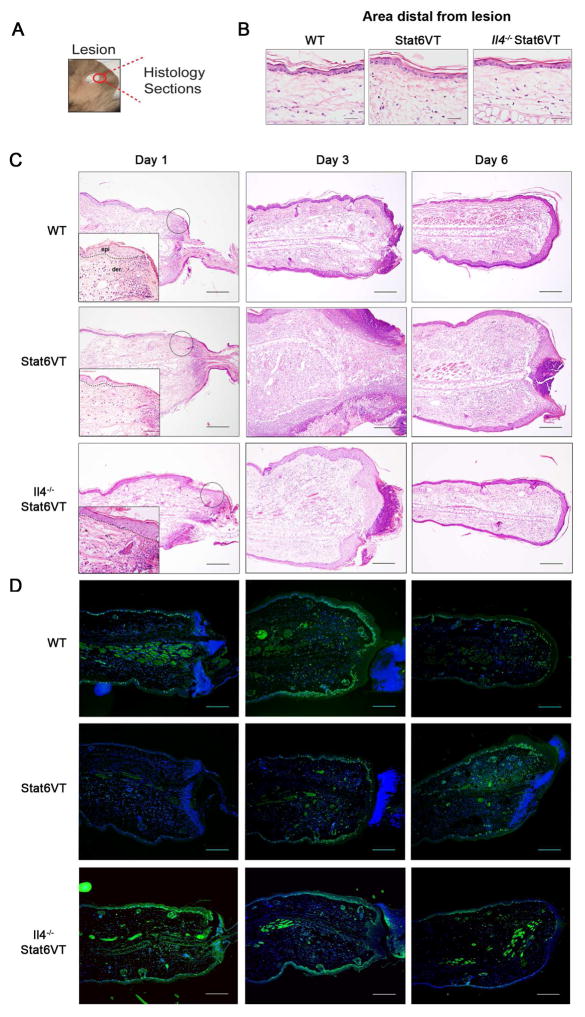
Since IL-4 decreased keratinocytes wound closure, mice with spontaneous development of AD-like lesions (Stat6VT mice) were used to test the influence of the allergic environment on in vivo wound healing. Stat6VT mice overexpress an active form of STAT6 in T cells, resulting in predominant Th2-type cytokine production and allergic skin inflammation 17, 27. Importantly for these studies, Stat6VT expression was observed in CD45+ cells, but not in CD45− cells isolated from dermis and epidermis in transgenic mice, and was predominantly observed in CD4+ cells (Fig E4). To investigate the re-epithelialization process in an atopic environment, we used an ear punch (2 mm in diameter) to wound ears of 4–6 month old WT, Stat6VT, and Il4−/− Stat6VT mice (Fig 4, A). It is important to note that there is no difference in the appearance of skin before wounding (Fig 4, B). Through histological analysis over 6 days after wounding, we monitored lesion re-epithelialization and wound closure. At day 1 following wounding, wounds in all groups of mice exhibited a neutrophilic infiltrate (Fig 4, C). At this time point, only WT and Il4−/− Stat6VT showed a denser epidermis at the border of the wounds (Fig 4, C). Further analysis in the control group (i.e. at day 3 and 6) showed reduction in cellular infiltration, and continuous re-epithelialization. In contrast, Stat6VT mice exhibited pronounced tissue inflammation associated with edema, and delayed wound closure (Fig 4, C).
Figure 4. Stat6VT mice exhibit delayed wounding healing.
Lesions were monitored by H&E in ears of WT, Stat6VT and Il4−/− Stat6VT mice at days 1, 3 and 6 after wounding (dermis (der) and epidermis (epi) separated by dashed lines). (A) Representation of ear punch. (B) Visualization of histological sections from distal areas from lesion and (C) lesion re-epithelialization and wounding closure. (D) Immunofluorescence for the proliferation marker Ki67 (green fluorescence) in the ears of WT, STAT6VT and Il4−/− Stat6VT mice in indicated days after punch. Nuclei are stained with DAPI in blue. Scale bar: 200μm. Pictures are representative of 3–4 mice from 2 independent experiments.
To determine whether differences in wound healing in WT, Stat6VT, and Il4−/− Stat6VT mice correlate with poor epithelial proliferation, ears sections were stained by immunofluorescence with the KI-67 nuclear protein expressed during cell proliferation (Fig 4, D). At all time points, all mice exhibited KI-67 positive cells in dermis and epidermis. However, WT mice and Il4−/− Stat6VT showed more intense stain in the epidermis mainly at days 1 and 3 after punch when compared to Stat6VT mice. At day 3, proliferative cells circumscribed most of the lesion in WT mice, and at day 6 the intensity of KI-67 stain was attenuated. Stat6VT mice exhibited delayed epidermal proliferation and inefficient lesion closure.
To verify whether deficient re-epithelialization also occurs in an induced model of AD-like disease, and to determine if IL-4 signaling normally regulates this process, WT and Stat6−/− mice were treated with the vitamin D analog (MC903) for 5 days prior to wounding. Wound healing was monitored at days 3 and 6 after punch and wound closure was measured by morphometric analysis. We observed a significant decrease in wound closure at day 3 in WT mice, compared to control mice treated with ethanol alone (Fig E5, A and B). Moreover, Stat6−/− mice exhibited earlier re-epithelialization when compared to WT mice treated with MC903 (Fig E5, A and B). Thus, in models of induced and spontaneous AD, an atopic environment worsens re-epithelialization and wound healing responses in the skin.
Topical fibronectin enhances skin wound healing in Stat6VT mice
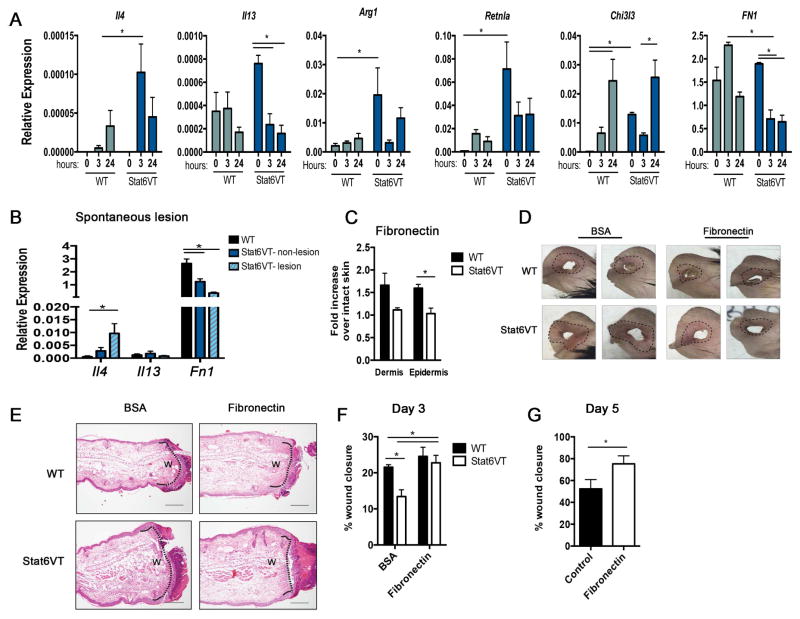
Th2-type cytokines can promote pro-fibrotic pathways involved in tissue regeneration via induction of alternatively activated (M2) macrophages differentiation 29. However, in our model, the IL-4/STAT6 axis decreases skin wound healing. To explore this apparent contradiction, we tested the expression of cytokines and M2 markers in WT and Stat6VT intact ears (0 hour), and 3 and 24 hours after punch. We observe that Arg1 (arginase), Retnla (Fizz1), and Chi3l3 (Ym1) mRNA levels were significantly increased at 0h in Stat6VT mice (Fig 5, A) when compared to WT mice. Il4, Arg1 and Retnla expression are increased at 3 and 24 hours after wounding in WT, but do not reach the levels observed in transgenic mice. Additionally we observed that Fn1 mRNA was similar at 0h in WT and Stat6VT mice (Fig 5, A). However, at 3 hours after wounding, tissue from WT mice showed an increase in fibronectin expression, and tissue from Stat6VT mice exhibited a significant reduction in Fn1 mRNA when compared to intact ears or to WT mice (Fig 5, A). Likewise, Il4 mRNA levels are increased and Fn1 mRNA levels are decreased in non-lesion and lesion from Stat6VT mice compared to intact skin from WT mice (Fig 5, B). Additionally, fibronectin levels measured by ELISA were significantly lower in epidermis of Stat6VT mice at 24 hours after wounding when compared with samples from WT mice (Fig 5, C).
Figure 5. Topical fibronectin improves wound healing potential in Stat6VT mice.
(A) Gene expression was determined by qRT-PCR in ears of WT and Stat6VT mice before (0 hour) and 3 and 24 hours after wounding, or (B) in non-lesion in WT and Stat6VT mice, and mice with spontaneous lesions. (C) Fibronectin levels were measured by ELISA in dermis and epidermis in WT and Stat6VT mice 24 hours wounding punch. Fold change was determined over intact skin (0 hours). (D) Ear representation and (E) lesions monitored by H&E in ears of WT and Stat6VT at day 3 after wounding (open wound size (w) measured by morphometry are indicated by dotted black lines). (F) Morphometric analysis of percentage of wound closure at day 3 after punch in WT and Stat6VT mice treated with ointment containing BSA or fibronectin, or (G) at day 5 after wounding in Stat6VT mice treated with ointment alone or ointment containing fibronectin. Data represents the mean of two independent experiments *p<0.05. Scale bar: 200μm. Pictures are representative of three mice from two independent experiments.
As fibronectin production was reduced in HK culture following IL-4 stimulation, and since this protein increased IL-4-stimulated HK scratch closure, we next determined whether topical application of fibronectin accelerates wound healing in allergic mice. To test the involvement of fibronectin in the Stat6VT mouse wound healing response, WT and Stat6VT mice were treated with ointment containing 10 μg of BSA or fibronectin right after (day 1) and 24 hours (day 2) after wounding. Histological analyses were performed at day 3, a period when partial re-epithelialization was observed (Fig 4, C). Macroscopic analysis revealed fibronectin treatment improved tissue recovery and erythema surrounding the lesion (Fig 5, D). In histological analysis (Fig 5, E) and morphometric analysis (Fig 5, F), topical fibronectin treatment significantly improved wound closure in Stat6VT mice, compared to control protein treated mice, but had modest effects on wild type mice. We also observed that fibronectin significantly increased wound closure in Stat6VT mice at day 5 after wounding when compared with Stat6VT mice receiving ointment alone as control (Fig 5, G). Thus, although IL-4 affects the expression of various genes directly involved in responses to wounding, the IL-4-dependent repression in fibronectin expression appears to be a major target in contributing to the reduction of wound healing potential of keratinocytes in an allergic environment.
DISCUSSION
IL-4 promotes the development of atopic diseases through affects on multiple cell types. Despite its critical role in AD, how IL-4 alters gene expression in keratinocytes has not been completely defined. In this report we use high throughput transcriptome analysis to identify IL-4-induced changes in keratinocyte gene expression at early and late stages of differentiation. Acute and chronic IL-4 stimulation resulted in extensive changes in the gene expression profile that spanned a variety of functional modules, including wound healing, a biological response impaired by IL-4. Moreover, central for IL-4-mediated effects in delayed wound healing was reduction in fibronectin production.
IL-4 alters a number of biological processes in keratinocytes, and cells in different stages of differentiation showed changes in distinct functions. Acute IL-4 stimulation (3 hours and 24 hours) resulted in alterations in genes involved in cell growth and survival, and diminished expression of genes involved in cell adhesion, which together might contribute to epidermal hyperplasia, spongiosis, and diminished barrier function, all characteristics of AD patient lesional skin 30–32. At day 5 of differentiation, IL-4 increased genes involved in cell projection and adhesion; and decreased genes participating in response to wounding and defense. In fact, a subset of genes in IL-4-stimulated differentiated HK (day 5) was similar in expression to less differentiated HK (day 2). These changes might favor a less organized epithelial structure, and contribute to an increased susceptibility to infections, also characteristics of AD lesions 33, 34. Although most of the experiments utilized immortalized human keratinocytes, and these might not entirely replicate intact skin, similar responses were observed with primary keratinocytes and skin equivalents made with primary cells. Moreover, several genes induced by IL-4, including CCL26, S100A7, S100A12, IL12RB, LCE3E, SERPINB3, SERPINB4, ADAMTS4, were also elevated in skin from patients with AD 35, 36, indicating a possible role for IL-4 in mediating many abnormalities in AD patient skin barrier.
Intense pruritis is a characteristic and often debilitating feature of AD, causing patients to scratch and consequently wound their skin (reviewed in 37). Lesions are frequently associated with areas that patients readily reach to scratch 38, 39. As our data indicates, IL-4 exposure might also contribute to AD pathogenesis by impairing appropriate wound repair. Using scratching of keratinocyte monolayers and time-lapse video microscopy, we demonstrate a significant decrease in recovery of the monolayer following IL-4 exposure. This observation could be linked to reports that children with AD demonstrating greater atopic responses had increased healing time of lesions 28. In contrast to the wound healing process observed here, recovery of barrier function following tape stripping of non-lesional skin in AD patients and healthy controls was similar 40. Koning and colleagues reported no differences in gene expression between tape-stripped non-lesional skin of AD patients versus skin from healthy controls 41, suggesting that decreased wound healing would most likely be observed in skin with a wound more severe than superficial tape stripping, or in skin with an active inflammatory response.
IL-4 has previously been shown to be a pro-healing cytokine in wound repair 29, 42–45. Our studies differ from these studies in several ways. First, we are focusing on re-epithelialization of the skin, rather than fibrosis and repair below keratinocyte layers. We observed diminished re-epithelialization in two models of AD-like skin inflammation, and impaired healing was not observed when mice had defects in IL-4 signaling, suggesting this is a consistent effect of a pro-atopic environment. Importantly, we did not observe any defects in collagen deposition in the dermis during healing (data not shown). Second, the IL-4 induced during wound repair in wild type mice is much less than observed in atopic models. We directly demonstrated that M2 gene expression, a common target of IL-4 in wound healing studies, is already higher in skin from ‘atopic’ mice. Moreover, the induction of these genes is increased in the skin of ‘atopic’ mice. Together, these observations support a model wherein a low level of IL-4, functioning particularly in the dermis, is important for a healing response. However, in an atopic environment, IL-4 is at sufficiently high concentrations to alter keratinocyte function and dermal M2 gene expression. In this respect, it is not clear that the effects of IL-4 on fibronectin production are strictly on keratinocytes. In our analyses, we did observe diminished fibronectin protein in both dermis and epidermis, suggesting that other cells might be affected by IL-4 in a similar fashion. Still, the in vivo results are consistent with the in vitro keratinocyte cultures demonstrating an important contribution of keratinocyte-produced fibronectin in re-epithelialization.
It is difficult to separate the direct effects of IL-4 in these models versus the effect of IL-4-dependent inflammation in the AD models. We observed that there was greater loss of fibronectin expression in lesional skin than in non-lesional skin of the Stat6VT mice. However, there was also greater Il4 expression in lesional skin, demonstrating how these factors are linked. Although the in vivo phenotype is consistent with the in vitro observations of keratinocyte function, we cannot exclude a contribution of additional cell types in vivo that are responding to IL-4 and mediating some of the delayed healing through secondary effects.
While IL-4 modifies many genes involved in the wound response 42, decreased fibronectin expression directly contributed to the in vitro impairment of HK monolayer wound closure. Fibronectin is a multidomain protein component of the extracellular matrix, expressed in high levels at wound sites 46, 47. It is associated with wound repair by, among other mechanisms during injury, enhancing growth factor binding stability and cell survival 48, 49. Although topical application of fibronectin in lesions of Stat6VT mice only partially restored re-epithelialization, this protein significantly improved the appearance of lesions, including decreasing erythema in the tissue. Many functions regulated by fibronectin including adhesion, migration and differentiation 50 were also functions affected in HK following IL-4 stimulation. Thus, whether fibronectin might be used to treat AD lesions or other biological defects induced by IL-4 in keratinocytes are interesting questions remaining to be investigated.
Together our findings describe a comprehensive role for IL-4 in preventing keratinocyte maturation by influencing the expression of key genes affecting skin barrier function. Induction of a subset of genes by IL-4 was confirmed in skin equivalents, and a deficient response of keratinocytes from allergic mice to wounding was also observed in vivo, further supporting a physiological role for these studies. The more detailed understanding of IL-4-regulated genetic programs in keratinocytes described in this report should lead to additional insights into the development of allergic skin disease.
Supplementary Material
Representative live imaging of wound healing in HK monolayer cultured with calcium chloride alone (Supp Movie 1 HK control) or stimulated with IL-4 (Supp Movie 1 HK IL-4) for 5 days. Time-lapse photography was performed on an automated time-lapse imaging system every 2 minutes as described in Material and Methods. Representative of 2 independent experiments.
(A) HK were differentiated with calcium chloride and gene expression was analyzed using qRT–PCR. (B) Expression of pSTAT6 Tyr641 was determined at the indicated time points after IL-4 stimulation (solid gray line-control; black line anti-pSTAT6 Tyr641). (C) pSTAT6 mean fluorescence intensity (MFI) in IL-4-stimulated HK. (D–E) Gene expression determined by qRT-PCR in HK differentiated with calcium chloride and stimulated with IL-4 for 5 days or as the indicated times. (F) Supernatants from primary keratinocyte culture were assessed for CCL26 production using ELISA. Data represent ± SEM of an average of three independent experiments. *p<0.05
(A) Gene expression was determined by qRT-PCR in organotypic skin equivalent with IL-4 for 24 hours, or (B) in HK differentiated for 5 days with indicated concentration of IL-4. (C) HK were differentiated with calcium chloride and stimulated with IL-4 or IL-13 for 2 or 5 days prior analysis of gene expression. Data represent ± SEM over two independent experiments. *p<0.05 and **p<0.01
(A) Microscopy of human keratinocytes differentiated with calcium chloride alone (control) or calcium chloride plus IL-4 for 5 days. (B) Percentage of scratch closed 24h after scratch in HK differentiated for 2 or 5 days with calcium chloride alone or calcium chloride alone plus IL-4. (C) Venn diagram representing the number of genes altered by IL-4 after 24 and 120 hours. Pictures are representative of 4 fields analyzed over at least two independent experiments. Data represents represent ± SEM over two independent experiments. *p<0.05.
(A) Epidermis was isolated from WT and Stat6VT and CD45+ and CD45− cells were gated. Expression of STAT6VT was analyzed in CD45+ and CD45− cells in epidermis (B) and in CD4+, CD8+ and CD19+ cells in spleen (C) via FLAG-tag antibody (solid gray line-WT mice; red line Stat6VT mice). FLAG-tag mean fluorescence intensity (MFI) in CD45+ and CD45+ in epidermis and dermis (D), and in lymphocytes in spleen from WT and Stat6VT mice. Data represent ± SEM over two independent experiments. *p<0.05
Ears of WT and STAT6−/− mice were treated topically for 5 days with MC903 or ethanol alone. At day 6, ears were punched and lesions were monitored by H&E at days 3 and 6 (A). Morphometric analysis of percentage of wound closure (B). Scale bar: 200μm. Pictures are representative of 3–4 mice.
Key Message.
IL-4, a critical effector cytokine in atopic dermatitis, repressed expression of genes induced during keratinocyte differentiation.
Keratinocytes exposed to IL-4 have changes in the expression of over 1000 genes.
IL-4 impaired the wound healing response of keratinocytes by decreasing fibronectin production.
Topical fibronectin increases wound healing in mice with spontaneous AD-like disease.
Acknowledgments
This work was supported by Public Health Service grants R01 AI095282 (MHK), R01 ES020866 (DS), VA CDA2 CX001019 (MJT), NIH K12 - Indiana Pediatric Scientist Award and T32 (5T32H0069047-04) (GN). Support provided by the HB Wells Center was in part from the Riley Children’s Foundation. The authors thank David H. Southern for assistance with cell culture. We thank Yunlong Liu for guidance in the bioinformatics analyses. The authors thank Drs. Janice Blum, Matthew Olson, Matthew Hufford, and Jeffrey Travers for review of this manuscript.
Abbreviations
- AD
atopic dermatitis
- BSA
bovine serum albumin
- FN
fibronectin
- HK
human keratinocyte
- STAT
signal transducer and activator of transcription
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004;113:651–7. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sehra S, Tuana FM, Holbreich M, Mousdicas N, Tepper RS, Chang CH, et al. Scratching the surface: towards understanding the pathogenesis of atopic dermatitis. Crit Rev Immunol. 2008;28:15–43. doi: 10.1615/critrevimmunol.v28.i1.20. [DOI] [PubMed] [Google Scholar]
- 3.Albanesi C, Fairchild HR, Madonna S, Scarponi C, De Pita O, Leung DY, et al. IL-4 and IL-13 negatively regulate TNF-alpha- and IFN-gamma-induced beta-defensin expression through STAT-6, suppressor of cytokine signaling (SOCS)-1, and SOCS-3. J Immunol. 2007;179:984–92. doi: 10.4049/jimmunol.179.2.984. [DOI] [PubMed] [Google Scholar]
- 4.Beltrani VS. Suggestions regarding a more appropriate understanding of atopic dermatitis. Curr Opin Allergy Clin Immunol. 2005;5:413–8. doi: 10.1097/01.all.0000182544.37724.b5. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann RL, Edenharter G, Bergmann KE, Forster J, Bauer CP, Wahn V, et al. Atopic dermatitis in early infancy predicts allergic airway disease at 5 years. Clin Exp Allergy. 1998;28:965–70. doi: 10.1046/j.1365-2222.1998.00371.x. [DOI] [PubMed] [Google Scholar]
- 6.Gustafsson D, Sjoberg O, Foucard T. Development of allergies and asthma in infants and young children with atopic dermatitis--a prospective follow-up to 7 years of age. Allergy. 2000;55:240–5. doi: 10.1034/j.1398-9995.2000.00391.x. [DOI] [PubMed] [Google Scholar]
- 7.Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–8. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Rouse BT, Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat Rev Immunol. 2010;10:514–26. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim BE, Leung DY, Boguniewicz M, Howell MD. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clin Immunol. 2008;126:332–7. doi: 10.1016/j.clim.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wills-Karp M, Finkelman FD. Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci Signal. 2008;1:pe55. doi: 10.1126/scisignal.1.51.pe55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: Signaling mechanisms and biological functions. ari. 1999;17:701–38. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 12.Kagami S, Saeki H, Komine M, Kakinuma T, Tsunemi Y, Nakamura K, et al. Interleukin-4 and interleukin-13 enhance CCL26 production in a human keratinocyte cell line, HaCaT cells. Clin Exp Immunol. 2005;141:459–66. doi: 10.1111/j.1365-2249.2005.02875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellerin L, Henry J, Hsu CY, Balica S, Jean-Decoster C, Mechin MC, et al. Defects of filaggrin-like proteins in both lesional and nonlesional atopic skin. J Allergy Clin Immunol. 2013;131:1094–102. doi: 10.1016/j.jaci.2012.12.1566. [DOI] [PubMed] [Google Scholar]
- 14.Omori-Miyake M, Yamashita M, Tsunemi Y, Kawashima M, Yagi J. In vitro assessment of IL-4- or IL-13-mediated changes in the structural components of keratinocytes in mice and humans. J Invest Dermatol. 2014;134:1342–50. doi: 10.1038/jid.2013.503. [DOI] [PubMed] [Google Scholar]
- 15.Owczarek W, Paplinska M, Targowski T, Jahnz-Rozyk K, Paluchowska E, Kucharczyk A, et al. Analysis of eotaxin 1/CCL11, eotaxin 2/CCL24 and eotaxin 3/CCL26 expression in lesional and non-lesional skin of patients with atopic dermatitis. Cytokine. 2010;50:181–5. doi: 10.1016/j.cyto.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Lewkowich IP, Lajoie S, Clark JR, Herman NS, Sproles AA, Wills-Karp M. Allergen uptake, activation, and IL-23 production by pulmonary myeloid DCs drives airway hyperresponsiveness in asthma-susceptible mice. PLoS One. 2008;3:e3879. doi: 10.1371/journal.pone.0003879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruns HA, Schindler U, Kaplan MH. Expression of a constitutively active Stat6 in vivo alters lymphocyte homeostasis with distinct effects in T and B cells. J Immunol. 2003;170:3478–87. doi: 10.4049/jimmunol.170.7.3478. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–9. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Du J, Zhu J, Yang X, Zhou B. Thymic stromal lymphopoietin signaling in CD4(+) T cells is required for TH2 memory. J Allergy Clin Immunol. 2015;135:781–91. e3. doi: 10.1016/j.jaci.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn C, Hurwitz SA, Kumar MG, Cotton J, Spandau DF. Activation of the insulin-like growth factor-1 receptor promotes the survival of human keratinocytes following ultraviolet B irradiation. Int J Cancer. 1999;80:431–8. doi: 10.1002/(sici)1097-0215(19990129)80:3<431::aid-ijc16>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–47. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sehrawat S, Reddy PB, Rajasagi N, Suryawanshi A, Hirashima M, Rouse BT. Galectin-9/TIM-3 interaction regulates virus-specific primary and memory CD8 T cell response. PLoS Pathog. 2010;6:e1000882. doi: 10.1371/journal.ppat.1000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sehrawat S. The omission of comprehensive care: an analysis of the Nursing Home Reform Act of 1987. J Gerontol Soc Work. 2010;53:64–76. doi: 10.1080/01634370802609254. [DOI] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim EJ, Lu TX, Blanchard C, Rothenberg ME. Epigenetic regulation of the IL-13-induced human eotaxin-3 gene by CREB-binding protein-mediated histone 3 acetylation. J Biol Chem. 2011;286:13193–204. doi: 10.1074/jbc.M110.210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnamurthy P, Sherrill JD, Parashette K, Goenka S, Rothenberg ME, Gupta S, et al. Correlation of increased PARP14 and CCL26 expression in biopsies from children with eosinophilic esophagitis. J Allergy Clin Immunol. 2014;133:577–80. doi: 10.1016/j.jaci.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sehra S, Yao Y, Howell MD, Nguyen ET, Kansas GS, Leung DY, et al. IL-4 regulates skin homeostasis and the predisposition toward allergic skin inflammation. J Immunol. 2010;184:3186–90. doi: 10.4049/jimmunol.0901860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci G, Patrizi A, Baldi E, Menna G, Tabanelli M, Masi M. Long-term follow-up of atopic dermatitis: retrospective analysis of related risk factors and association with concomitant allergic diseases. J Am Acad Dermatol. 2006;55:765–71. doi: 10.1016/j.jaad.2006.04.064. [DOI] [PubMed] [Google Scholar]
- 29.Knipper JA, Willenborg S, Brinckmann J, Bloch W, Maass T, Wagener R, et al. Interleukin-4 Receptor alpha Signaling in Myeloid Cells Controls Collagen Fibril Assembly in Skin Repair. Immunity. 2015;43:803–16. doi: 10.1016/j.immuni.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Benedetto A, Rafaels NM, McGirt LY, Ivanov AI, Georas SN, Cheadle C, et al. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol. 2011;127:773–86. e1–7. doi: 10.1016/j.jaci.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Benedetto A, Slifka MK, Rafaels NM, Kuo IH, Georas SN, Boguniewicz M, et al. Reductions in claudin-1 may enhance susceptibility to herpes simplex virus 1 infections in atopic dermatitis. J Allergy Clin Immunol. 2011;128:242–6. e5. doi: 10.1016/j.jaci.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malaisse J, Bourguignon V, De Vuyst E, Lambert de Rouvroit C, Nikkels AF, Flamion B, et al. Hyaluronan Metabolism in Human Keratinocytes and Atopic Dermatitis Skin Is Driven by a Balance of Hyaluronan Synthases 1 and 3. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.147. [DOI] [PubMed] [Google Scholar]
- 33.Leung DY, Gao PS, Grigoryev DN, Rafaels NM, Streib JE, Howell MD, et al. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in IFN-gamma response. J Allergy Clin Immunol. 2011;127:965–73. e1–5. doi: 10.1016/j.jaci.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, et al. Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity. 2015;42:756–66. doi: 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renard E, Poree B, Chadjichristos C, Kypriotou M, Maneix L, Bigot N, et al. Sox9/Sox6 and Sp1 are involved in the insulin-like growth factor-I-mediated upregulation of human type II collagen gene expression in articular chondrocytes. J Mol Med (Berl) 2012;90:649–66. doi: 10.1007/s00109-011-0842-3. [DOI] [PubMed] [Google Scholar]
- 36.Suarez-Farinas M, Tintle SJ, Shemer A, Chiricozzi A, Nograles K, Cardinale I, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127:954–64. e1–4. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elmariah SB, Lerner EA. The missing link between itch and inflammation in atopic dermatitis. Cell. 2013;155:267–9. doi: 10.1016/j.cell.2013.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura T, Miyazawa H. The ‘butterfly’ sign in patients with atopic dermatitis: evidence for the role of scratching in the development of skin manifestations. J Am Acad Dermatol. 1989;21:579–80. doi: 10.1016/s0190-9622(89)80235-x. [DOI] [PubMed] [Google Scholar]
- 39.Amon U, Wolff HH. Healing of chronic atopic dermatitis lesions in skin areas of paraplegia after trauma. J Dermatol. 1994;21:982–3. doi: 10.1111/j.1346-8138.1994.tb03325.x. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka M, Zhen YX, Tagami H. Normal recovery of the stratum corneum barrier function following damage induced by tape stripping in patients with atopic dermatitis. Br J Dermatol. 1997;136:966–7. [PubMed] [Google Scholar]
- 41.de Koning HD, van den Bogaard EH, Bergboer JG, Kamsteeg M, van Vlijmen-Willems IM, Hitomi K, et al. Expression profile of cornified envelope structural proteins and keratinocyte differentiation-regulating proteins during skin barrier repair. Br J Dermatol. 2012;166:1245–54. doi: 10.1111/j.1365-2133.2012.10885.x. [DOI] [PubMed] [Google Scholar]
- 42.Brauweiler AM, Goleva E, Hall CF, Leung DY. Th2 Cytokines Suppress Lipoteichoic Acid-Induced Matrix Metalloproteinase Expression and Keratinocyte Migration in Response to Wounding. J Invest Dermatol. 2015;135:2550–3. doi: 10.1038/jid.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leoni G, Neumann PA, Sumagin R, Denning TL, Nusrat A. Wound repair: role of immune-epithelial interactions. Mucosal Immunol. 2015;8:959–68. doi: 10.1038/mi.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen JE, Wynn TA. Evolution of Th2 immunity: a rapid repair response to tissue destructive pathogens. PLoS Pathog. 2011;7:e1002003. doi: 10.1371/journal.ppat.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18:1028–40. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown LF, Dubin D, Lavigne L, Logan B, Dvorak HF, Van de Water L. Macrophages and fibroblasts express embryonic fibronectins during cutaneous wound healing. Am J Pathol. 1993;142:793–801. [PMC free article] [PubMed] [Google Scholar]
- 47.Li-Korotky HS, Hebda PA, Lo CY, Dohar JE. Age-dependent differential expression of fibronectin variants in skin and airway mucosal wounds. Arch Otolaryngol Head Neck Surg. 2007;133:919–24. doi: 10.1001/archotol.133.9.919. [DOI] [PubMed] [Google Scholar]
- 48.Zhu J, Clark RA. Fibronectin at select sites binds multiple growth factors and enhances their activity: expansion of the collaborative ECM-GF paradigm. J Invest Dermatol. 2014;134:895–901. doi: 10.1038/jid.2013.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin F, Zhu J, Tonnesen MG, Taira BR, McClain SA, Singer AJ, et al. Fibronectin peptides that bind PDGF-BB enhance survival of cells and tissue under stress. J Invest Dermatol. 2014;134:1119–27. doi: 10.1038/jid.2013.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livant DL, Brabec RK, Kurachi K, Allen DL, Wu Y, Haaseth R, et al. The PHSRN sequence induces extracellular matrix invasion and accelerates wound healing in obese diabetic mice. J Clin Invest. 2000;105:1537–45. doi: 10.1172/JCI8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative live imaging of wound healing in HK monolayer cultured with calcium chloride alone (Supp Movie 1 HK control) or stimulated with IL-4 (Supp Movie 1 HK IL-4) for 5 days. Time-lapse photography was performed on an automated time-lapse imaging system every 2 minutes as described in Material and Methods. Representative of 2 independent experiments.
(A) HK were differentiated with calcium chloride and gene expression was analyzed using qRT–PCR. (B) Expression of pSTAT6 Tyr641 was determined at the indicated time points after IL-4 stimulation (solid gray line-control; black line anti-pSTAT6 Tyr641). (C) pSTAT6 mean fluorescence intensity (MFI) in IL-4-stimulated HK. (D–E) Gene expression determined by qRT-PCR in HK differentiated with calcium chloride and stimulated with IL-4 for 5 days or as the indicated times. (F) Supernatants from primary keratinocyte culture were assessed for CCL26 production using ELISA. Data represent ± SEM of an average of three independent experiments. *p<0.05
(A) Gene expression was determined by qRT-PCR in organotypic skin equivalent with IL-4 for 24 hours, or (B) in HK differentiated for 5 days with indicated concentration of IL-4. (C) HK were differentiated with calcium chloride and stimulated with IL-4 or IL-13 for 2 or 5 days prior analysis of gene expression. Data represent ± SEM over two independent experiments. *p<0.05 and **p<0.01
(A) Microscopy of human keratinocytes differentiated with calcium chloride alone (control) or calcium chloride plus IL-4 for 5 days. (B) Percentage of scratch closed 24h after scratch in HK differentiated for 2 or 5 days with calcium chloride alone or calcium chloride alone plus IL-4. (C) Venn diagram representing the number of genes altered by IL-4 after 24 and 120 hours. Pictures are representative of 4 fields analyzed over at least two independent experiments. Data represents represent ± SEM over two independent experiments. *p<0.05.
(A) Epidermis was isolated from WT and Stat6VT and CD45+ and CD45− cells were gated. Expression of STAT6VT was analyzed in CD45+ and CD45− cells in epidermis (B) and in CD4+, CD8+ and CD19+ cells in spleen (C) via FLAG-tag antibody (solid gray line-WT mice; red line Stat6VT mice). FLAG-tag mean fluorescence intensity (MFI) in CD45+ and CD45+ in epidermis and dermis (D), and in lymphocytes in spleen from WT and Stat6VT mice. Data represent ± SEM over two independent experiments. *p<0.05
Ears of WT and STAT6−/− mice were treated topically for 5 days with MC903 or ethanol alone. At day 6, ears were punched and lesions were monitored by H&E at days 3 and 6 (A). Morphometric analysis of percentage of wound closure (B). Scale bar: 200μm. Pictures are representative of 3–4 mice.