Abstract
Repair of chromosomal breaks is essential for cellular viability, but misrepair generates mutations and gross chromosomal rearrangements. We investigated the interrelationship between two homologous-repair pathways, i.e., mutagenic single-strand annealing (SSA) and precise homology-directed repair (HDR). For this, we analyzed the efficiency of repair in mammalian cells in which double-strand break (DSB) repair components were disrupted. We observed an inverse relationship between HDR and SSA when RAD51 or BRCA2 was impaired, i.e., HDR was reduced but SSA was increased. In particular, expression of an ATP-binding mutant of RAD51 led to a >90-fold shift to mutagenic SSA repair. Additionally, we found that expression of an ATP hydrolysis mutant of RAD51 resulted in more extensive gene conversion, which increases genetic loss during HDR. Disruption of two other DSB repair components affected both SSA and HDR, but in opposite directions: SSA and HDR were reduced by mutation of Brca1, which, like Brca2, predisposes to breast cancer, whereas SSA and HDR were increased by Ku70 mutation, which affects nonhomologous end joining. Disruption of the BRCA1-associated protein BARD1 had effects similar to those of mutation of BRCA1. Thus, BRCA1/BARD1 has a role in homologous repair before the branch point of HDR and SSA. Interestingly, we found that Ku70 mutation partially suppresses the homologous-repair defects of BARD1 disruption. We also examined the role of RAD52 in homologous repair. In contrast to yeast, Rad52−/− mouse cells had no detectable HDR defect, although SSA was decreased. These results imply that the proper genetic interplay of repair factors is essential to limit the mutagenic potential of DSB repair.
An increased frequency of mutations and gross chromosomal rearrangements is widely observed in a variety of tumor types and aged cells from a number of organisms (3, 9). To understand the etiology of these genetic alterations, it is critical to delineate the factors and pathways that influence the mutagenic potential of DNA damage repair. A critical type of DNA damage is a chromosomal double-strand break (DSB), which can be formed by reactive oxygen species, topoisomerase failure, radiation treatment, and DNA replication. In mammalian cells, such damage can be repaired through multiple pathways: nonhomologous end joining (NHEJ), which can result in deletions and insertions at the DSB site, or pathways that utilize homology for repair, which can also be variably mutagenic (2, 17, 26, 34).
Pathways that utilize sequence homology for repair are broadly characterized into two types based on whether homologous associations arise from strand exchange or strand-annealing activities. Homologous recombination, also called homology-directed repair (HDR), is initiated by a strand exchange protein, the prototype being RecA or its eukaryotic equivalent, RAD51 (51). Strand exchange, which involves a single strand invading a DNA duplex, results in a gene conversion, since the duplex templates repair. The mutagenic potential of HDR, which is generally considered to be quite low compared to other DSB repair pathways, is related to the choice of template, as well as to the extent or outcome of gene conversion. For example, HDR involving the identical sequence on a sister chromatid should be nonmutagenic. Because the sister chromatid is the preferred template for HDR in mammalian cells (20), HDR is presumed to be predominantly a nonmutagenic, precise type of repair. However, HDR is also well known to have the potential to result in genetic loss when the template is the allele on the homologous chromosome, given the heterozygosity between maternal and paternal chromosomes. Although apparently rare on a per allele basis, genetic loss from these repair events can arise from extensive gene conversion or crossing over (40, 50). Since this loss of heterozygosity decreases genetic variability and can uncover recessive genetic mutations, it is considered to be mutagenic.
The other DSB repair pathway involving sequence homology is single-strand annealing (SSA). SSA arises from the annealing of complementary single strands formed after resection at a DSB. Thus, when sequence repeats are present near a DSB, they can undergo SSA, resulting in a deletion of sequences between the repeats. In contrast to HDR, therefore, SSA is always mutagenic. Because ∼50% of the mammalian genome consists of repeat sequences (15), SSA is potentially an important pathway of mutagenesis. Consistent with this notion, mutagenic deletions associated with cancer and human genetic diseases often exhibit homology at the breakpoint junction, which is suggestive of SSA. For instance, both somatic and germ line rearrangements and deletions involving homologous Alu sequences are associated with human disease (8).
Although the structure-specific endonuclease ERCC1/XPF, the homologue of yeast Rad10/Rad1, has been implicated in strand-processing steps during SSA (1, 43), the protein(s) involved in the annealing step of SSA has not been as well studied in mammalian cells. The RAD52 protein is implicated in this step because Rad52 has a key role in SSA in Saccharomyces cerevisiae (52) and human RAD52 has strand-annealing activity in vitro (46, 52). In yeast, Rad52 also has a critical role in the HDR pathway (52); its role in mammalian homologous repair is uncertain, however, because Rad52 mutation in the mouse leads to only a small decrease in gene targeting (42), a pathway that utilizes other homologous-repair components.
HDR and SSA are expected to share mechanistic intermediates, such as resected single strands, and thus some protein factors, but other intermediates and factors are expected to be specific for each pathway, suggesting that the two homologous-repair pathways may be competitive. Although a weak competitive interaction of these pathways has been suggested (11, 55), a comparative analysis using mutants from each repair pathway has not been performed. Given the strikingly different mutagenic outcomes of these homologous-repair pathways, we investigated the efficiency of SSA and HDR in multiple genetic backgrounds in mammalian cells. We present evidence that RAD51 function is critical to limit the mutagenic potential of homologous repair; in the most extreme example, we found that disruption of RAD51 can lead to a >90-fold shift in pathway usage toward SSA. Additionally, we found that expression of an allele of RAD51 that is defective for ATP hydrolysis results in more extensive gene conversion, which increases the mutagenic potential of HDR. Furthermore, we present evidence that two additional protein complexes involved in DSB repair, i.e., BRCA1/BARD1 and KU70/KU80, have opposite effects on the homologous-repair pathways. Interestingly, Ku70 mutation partially suppresses the effect of BRCA1/BARD1 disruption, suggesting early and late roles for this complex. Finally, we provide evidence that mammalian RAD52 promotes SSA but not HDR, highlighting a striking difference between yeast and mammalian cells in the reliance on particular repair factors. Our results clearly demonstrate that the genetic interplay of DSB repair factors is essential to limit the mutagenic potential of homologous repair in mammalian cells.
MATERIALS AND METHODS
Plasmids and cell lines.
Green fluorescent protein gene (GFP) fragments for SA-GFP were derived from pEGFP-N1 (Clontech); hprtSAGFP was generated similarly to hprtDRGFP (36). After linearization with KpnI/SacI, the hprt vectors were electroporated into embryonic stem (ES) cells suspended in 650 μl of phosphate-buffered saline in a 0.4-cm-diameter cuvette by pulsing at 800 V and 3 μF. Puromycin was added 24 h later at 1 μg/ml for all cells except the Brca1−/− and Brca2L1/L2 cells, for which the concentration was 2 μg/ml. After 6 days of puromycin selection, 6-thioguanine was added at 10 μg/ml for all lines except Brca1−/− and Brca2L1/L2. Colonies were isolated and analyzed by Southern blotting with a GFP probe (36, 37). Mouse J1 (wild-type), Ku70−/−, Brca1−/−, and Brca2L1/L2 (also known as Brca2lex1/lex2) ES cell lines and their DR-GFP derivatives were previously described (27, 31, 32, 36, 47), as were the Ercc1−/− and Rad52−/− ES cells (33, 42).
For stable RAD51 expression in E14 ES cells containing H-DR-8mu integrated at the hprt locus (12), the P59-CAGGS-RAD51K133R and P59-CAGGS-RAD51WT cassettes (49) were targeted to the Pim1 locus. Two independently targeted clones were used in the recombination experiments. The generation of pCAGGS expression vectors for human RAD51-WT, RAD51-K133A, RAD51-K133R, KU70, BRC3, BARD1-WT, and BARD1-hB202 were previously described (36, 49, 58); for ERCC1 and RAD52, vectors were generated by PCR of cDNAs, followed by sequence confirmation.
Repair assays.
To measure the repair of an I-SceI-generated DSB, 50 μg of the I-SceI expression vector pCBASce (40) was mixed with 5 × 106 ES cells suspended in 650 μl of phosphate-buffered saline in a 0.4-cm-diameter cuvette, followed by pulsing the cells at 250 V and 950 μF, except for the ERCC1- and RAD52-deficient cell lines, which were pulsed at either 250 or 280 V. The pCAGGS expression vectors for other factors were added as follows: 30 μg for RAD51, 50 μg for BARD1, 45 μg for BRC3, and 15 μg for KU70, ERCC1, or RAD52. GFP-positive cells were quantitated by flow cytometric analysis 2 days after electroporation on a Becton Dickinson FACScan. To calculate the difference in homologous repair relative to wild-type cells (see Fig. 2 and 4d), the percentage of GFP+ cells from each individual transfection was divided by the mean value for wild-type cells transfected only with the I-SceI expression vector. The calculation for the KU-deficient cells (see Fig. 5b) is exactly analogous. To calculate the difference in homologous repair for the ERCC1- and RAD52-deficient cell lines, the percentage of GFP+ cells from individual transfections that included the complementing vector was divided by the percentage of GFP+ cells from the parallel transfection with the I-SceI expression vector alone, since transfection conditions varied between experiments. From this calculation, the relative value from the transfection of I-SceI alone in the ERCC1- and RAD52-deficient cell lines is always equal to 1, so that there is no error bar for these values. For H-DR-8mu, recombination frequencies were determined by dividing the number of G418-resistant colonies by the number of cells that survived electroporation. Homologous-repair frequencies are the mean of at least three independent transfections, and error bars represent the standard deviation from the mean. Statistical analysis of homologous-repair frequencies was performed using the unpaired t test.
FIG. 2.
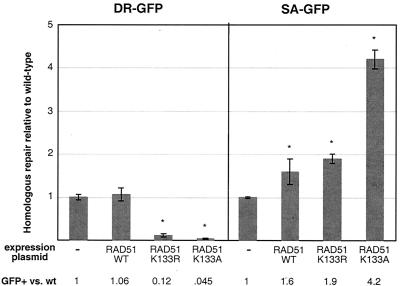
RAD51-K133R and RAD51-K133A expression reduces HDR but increases SSA. Wild-type (wt) ES cells with either the DR-GFP or SA-GFP reporter were cotransfected with the I-SceI and RAD51 expression vectors, as indicated. The efficiency of homologous repair is indicated relative to transfection with the I-SceI expression vector alone, which is set to 1. The asterisks indicate a statistically significant difference from transfection with the I-SceI expression vector alone, with P ≤ 0.0001, except for SA-GFP repair with RAD51-WT, with P = 0.02. The error bars indicate standard deviations.
FIG. 4.
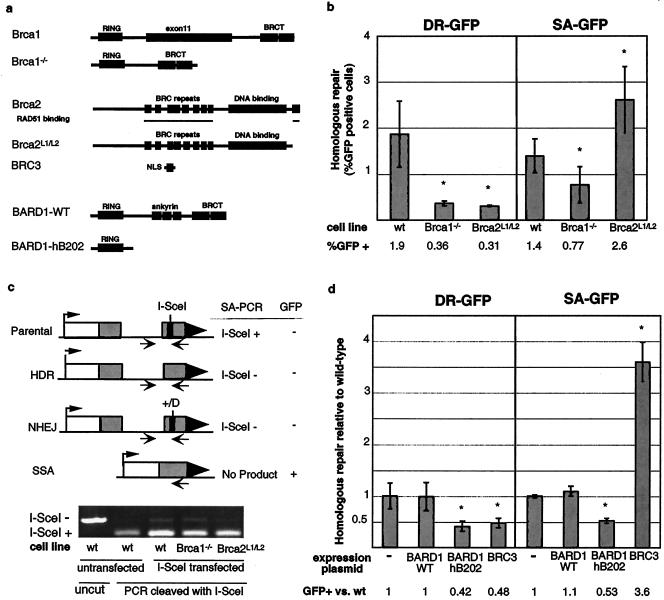
BRCA1/BARD1 promotes both HDR and SSA, whereas BRCA2, like RAD51, promotes HDR and suppresses SSA. (a) Schematic of wild-type and mutant proteins used in this analysis. Note that although the Brca1 mutant is indicated as Brca1−/− for simplicity, an exon 11-deleted peptide is expressed in this mutant (58). For BRCA2, RAD51 binding regions at the BRC repeats and C terminus are indicated. Brca2L1/L2 has the C-terminal RAD51 binding domain in exon 27 deleted. BRC3 is tagged with a nuclear localization signal (NLS). BARD1-hB202 is the N-terminal 202 amino acids of human BARD1, which interacts with BRCA1 (58). (b) Homologous repair in Brca1−/− and Brca2L1/L2 cell lines. The efficiency of homologous repair after I-SceI expression is indicated for the wild-type (wt) and mutant cell lines. SA-GFP repair frequencies are the mean (± standard deviation) of 11 and 6 independent transfections for the Brca1−/− and Brca2L1/L2 cell lines, respectively. The asterisks indicate a statistically significant difference from the wild type, with P ≤ 0.004, except for DR-GFP repair in Brca2L1/L2, with P = 0.008. (c) Analysis of I-SceI site loss in the SA-GFP reporter arising from non-SSA repair. The genomic region surrounding the I-SceI site in SceGFP3′ was PCR amplified using the primers depicted as arrows. The PCR product from untransfected wild-type cells is efficiently cleaved by I-SceI, whereas after I-SceI expression and HDR or NHEJ, a portion of the PCR product is not cleaved by I-SceI. (Note that the SSA product is not amplified because the upstream primer is lost during SSA repair.) After I-SceI expression, Brca1−/− cells have a portion of noncleaved PCR product similar to that of wild-type cells, whereas Brca2L1/L2 cells reproducibly show a smaller amount of noncleaved product, consistent with the shift toward SSA. (d) Expression of peptides predicted to interfere with BRCA1 and BRCA2 function mimics the effects of Brca1 and Brca2 mutation on homologous repair. Wild-type ES cells with either the DR-GFP or SA-GFP reporter were cotransfected with the I-SceI and BARD1-WT, BARD1-hB202, or BRC3 expression vectors, as indicated. The efficiency of homologous repair is indicated relative to transfection with the I-SceI expression vector alone, which is set to 1. The asterisks indicate a statistically significant difference from transfection with the I-SceI expression vector alone, with P ≤ 0.002, except for DR-GFP repair with BRC3, with P = 0.01.
FIG. 5.
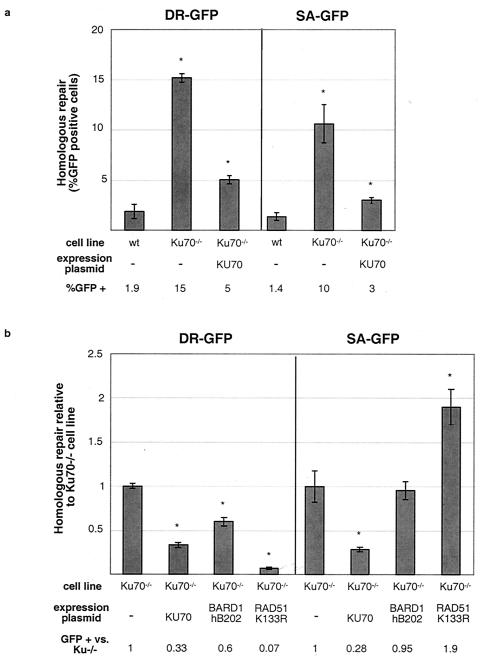
Ku70 mutation results in elevated HDR and SSA and suppresses the effect of BARD1-hB202 on homologous repair. (a) Homologous repair in Ku70−/− ES cells. The efficiencies of homologous repair after I-SceI expression are indicated for the wild-type (wt) and Ku70−/− cell lines, as well as the Ku70−/− cell line in which the I-SceI and KU70 expression vectors were cotransfected. The asterisks indicate a statistically significant difference from the wild type, with P ≤ 0.0001. The error bars indicate standard deviations. (b) Influence of BARD1-hB202 on homologous repair in Ku70−/− cells. Ku70−/− cells with either the DR-GFP or SA-GFP reporter were transfected with the I-SceI expression vector alone or together with the expression vector for KU70, BARD1-hB202, or RAD51-K133R. The efficiency of homologous repair is indicated relative to transfection with the I-SceI expression vector alone, which is set to 1. The asterisks indicate a statistically significant difference from the Ku70−/− cells transfected with the I-SceI expression vector alone, with P ≤ 0.004.
PCR analysis.
For H-DR-8mu cell lines, PCR analysis was described previously (12). Statistical analysis of the distribution of gene conversion tract lengths was performed using the unpaired t test. To determine the percent I-SceI site loss following SA-GFP repair, genomic DNA was isolated 6 days after transfection. The primer sequences for PCR were as follows: SAGFP3A, 5′-GCCCCCTGCTGTCCATTCCTTATT, and SAGFP3B, 5′-ATCGCGCTTCTCGTTGGGGTCTTT. Amplification of PCR products and determination of the fraction of I-SceI-cleaved product were performed as previously described for DR-GFP (36).
RESULTS
Reporters for DSB repair by HDR and SSA.
To investigate the genetic interrelationship of pathways of homologous DSB repair, we used chromosomal reporter substrates that allow the introduction of a DSB, i.e., DR-GFP, which assays HDR (37), and a novel reporter, SA-GFP, which assays SSA. In both reporters, the DSB is generated by the rare-cutting endonuclease I-SceI, whose 18-bp recognition sequence has been integrated within the green fluorescent protein gene (GFP) in such a way that it disrupts the gene.
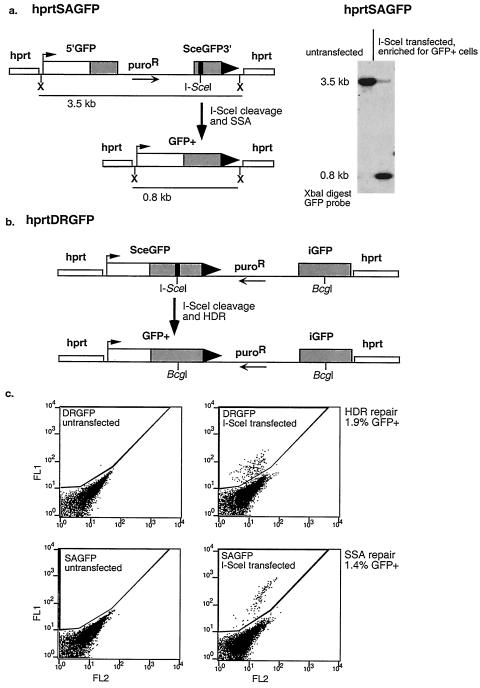
The SA-GFP reporter consists of the GFP gene fragments 5′GFP and SceGFP3′, which have 266 bp of homology (Fig. 1a). Repair of the I-SceI-generated DSB in SceGFP3′ by SSA results in a functional GFP gene when a DNA strand from SceGFP3′ is annealed to the complementary strand of 5′GFP, followed by appropriate DNA-processing steps. As a result, SSA between the homologous sequences in the GFP gene fragments produces a 2.7-kb deletion in the chromosome. The SA-GFP reporter can also be repaired by HDR, but this repair does not restore a functional GFP gene. We measured the efficiency of HDR separately using the previously described DR-GFP reporter (37). In this reporter, the I-SceI site is integrated into a full-length GFP gene (SceGFP). Repair of the I-SceI-generated DSB by HDR results in a functional GFP gene when repair is directed by the downstream internal GFP fragment, iGFP (Fig. 1b). HDR does not alter the structure of the reporter; the only change is the conversion of the I-SceI site to a BcgI site. Presumably, if SceGFP on the sister chromatid is uncleaved, it also provides a template for repair, but it is not possible to score these types of sister repair events. With both reporters, the presence of a functional GFP gene was scored in individual cells by green fluorescence using flow cytometric analysis (Fig. 1c). Furthermore, the relevant repair product was confirmed for both reporters by Southern blot analysis of genomic DNA from enriched populations of green fluorescent cells, as shown previously for DR-GFP (32, 37) and here for SA-GFP (Fig. 1a).
FIG. 1.
Reporters for measuring homologous DSB repair by SSA and HDR. (a) hprtSAGFP reporter. The structure of the reporter is shown before and after I-SceI cleavage and SSA. Note that repair by SSA results in a 2.7-kb deletion in the chromosome. Southern blot analysis is shown from an untransfected cell line with the SA-GFP reporter and from the same line after I-SceI expression and flow sorting to enrich for a pool of GFP+ cells. Large black triangles depict the 3′ end of the AFP cassette. (b) hprtDRGFP reporter. The structure of the reporter is shown before and after I-SceI cleavage and HDR. Note that repair by HDR converts the I-SceI site but otherwise maintains the structure of the reporter, as confirmed previously by Southern blot analysis (37). (c) Flow cytometric analysis of wild-type ES cells containing the hprtDRGFP and hprtSAGFP reporters targeted to the hprt locus, either untransfected or transfected with the I-SceI expression plasmid. Green fluorescence (FL1) is plotted on the y axis, with orange fluorescence (FL2) on the x axis.
To compare the efficiencies of HDR and SSA in mammalian cells with these reporters, we integrated DR-GFP and SA-GFP at the hprt locus in the mouse J1 ES cell line. The reporters were integrated separately to generate an SA-GFP-containing cell line and a DR-GFP-containing cell line. In the absence of I-SceI expression, GFP+ cells were rare or undetectable among cells carrying either reporter (≤0.01%) (Fig. 1c). Transfection of the I-SceI expression vector resulted in 1.9% GFP+ cells for the DR-GFP cell line and 1.4% GFP+ cells for the SA-GFP cell line (Fig. 1c), indicating that SSA, as measured with this reporter, is nearly as efficient as HDR in wild-type ES cells. It should be noted that the efficiency of SA-GFP repair resulting in a functional GFP gene is consistent with a mechanism involving SSA; HDR involving crossing over, which would have given an identical product, is estimated to be at least 30-fold less frequent in mammalian cells (20, 40, 50).
RAD51 promotes HDR but suppresses SSA.
We next compared the relative efficiencies of HDR and SSA in multiple genetic backgrounds, using mutant cell lines and cells expressing dominant-negative peptides. We began by examining RAD51, which forms nucleoprotein filaments on DNA, because its strand exchange activity is central to HDR. Because RAD51 has been shown to be essential for mammalian cell viability (25, 54), we investigated its function using putative dominant-negative forms. We focused on two ATPase mutants: RAD51-K133A, which is defective for ATP binding and hence is inactive for strand exchange in vitro, and RAD51-K133R, which is defective for ATP hydrolysis but competent for ATP binding and hence retains significant biological activity (28). We previously demonstrated that stable expression of RAD51-K133R in otherwise wild-type mammalian cells reduces HDR (49) so that RAD51-K133R acts as a dominant-negative peptide during HDR. The mechanism of interference is not certain; however, the mutant RAD51 peptides may form mixed nucleoprotein filaments with the endogenous wild-type RAD51 to partially disrupt its function.
To test the effects of these RAD51 alleles on both pathways of homologous repair, RAD51 expression vectors were transiently cotransfected with the I-SceI expression vector into the ES cell lines containing the integrated reporters. Under these conditions, RAD51-K133R expression reduced HDR eightfold, while additional expression of wild-type RAD51 (RAD51-WT) had little or no effect (Fig. 2), similar to previous results with cells constitutively expressing exogenous RAD51 (49). With RAD51-K133A, we found that the efficiency of HDR was reduced even more than with RAD51-K133R, i.e., 22-fold, which would be consistent with its more severely impaired biochemical activity. The magnitude of the recombination defect in this dominant-negative context suggests that complete loss of RAD51 would abrogate most or all HDR.
In contrast to the inhibition of HDR, both RAD51-K133R and RAD51-K133A enhanced the efficiency of SSA (1.9- and 4.2-fold, respectively [Fig. 2]). Expression of wild-type RAD51 also resulted in a slight (1.6-fold) increase in the efficiency of SSA, which may indicate a mild dominant-negative effect with elevated RAD51 (21, 41). Expression of yeast Rad51 in mammalian cells has been shown to reduce HDR, but SSA was not obviously increased (22); it is not clear if this is due to different experimental approaches or different allelic effects. Considering the relative efficiencies of HDR and SSA, expression of the ATPase mutants of RAD51 led to an overall shift of 16-fold (RAD51-K133R) and 93-fold (RAD51-K133A) toward mutagenic homologous repair. These results suggest that the ATP-dependent functions of RAD51 are important to promote HDR and inhibit SSA during DSB repair.
Disruption of RAD51 function shifts HDR toward more extensive gene conversion.
Although HDR is usually considered to be a precise type of repair, some outcomes of HDR, such as extensive gene conversion, have the potential to lead to genetic loss. We considered the possibility that normal RAD51 function could be important to limit the extent of gene conversion during HDR. In this regard, RAD51-K133R has been shown in vitro to promote the formation of strand invasion intermediates that are more stable than those formed by wild-type RAD51 (45); in vivo these hyperstable recombination intermediates may result in longer gene conversion tracts during HDR. Alternatively, the reduced level of HDR with RAD51-K133R may indicate impaired initiation of strand exchange, which could result in elevated strand resection prior to HDR, thereby extending the gene conversion tracts.
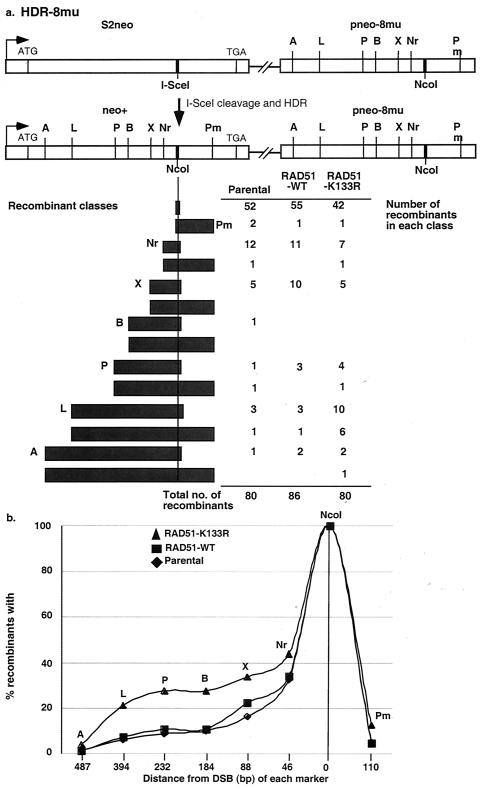
We examined the effect of RAD51-K133R on the outcome of recombination using the reporter H-DR-8mu (12) (Fig. 3a). In this reporter, which is stably integrated at the hprt locus, the I-SceI site has been introduced as a disrupting mutation into the neomycin resistance gene (S2neo). HDR of the I-SceI-induced DSB directed by the downstream internal neo fragment pneo-8mu results in the replacement of the I-SceI site with an NcoI site, which is the wild-type neo sequence at this position, giving rise to a functional neo+ gene. In addition, pneo-8mu also contains single-base-pair silent mutations, which generate restriction sites at various distances from the DSB (Fig. 3a). Incorporation of these restriction sites into the S2neo gene during recombination provides markers for the extent of gene conversion.
FIG. 3.
Expression of RAD51-K133R increases gene conversion of markers distant from the DSB during HDR. (a) Structure of the H-DR-8mu reporter and summary of gene conversion tracts in neo+ recombinants. The pneo-8mu fragment contains the wild-type NcoI site at the site of the DSB, along with 1-bp silent mutations, which create the following restriction site polymorphisms: A, ApaI; L, ApaL1; P, PstI; B, BamHI; X, XbaI; Nr, NruI; and Pm, PmlI. HDR of the I-SceI-generated DSB in S2neo, using the pneo-8mu gene as the template for repair, results in restoration of a wild-type neo+ gene, so that all recombinants have an NcoI site incorporated. HDR may be associated with incorporation of the other restriction sites. The filled bars represent the incorporation of all restriction site markers up to and including the indicated site for various recombinants derived from the parental cell line or after expression of RAD51-WT or RAD51-K133R. (b) Summary of gene conversion frequencies of each restriction site marker for the recombinants shown in panel a. The conversion frequency for each restriction site is graphed as a function of distance from the DSB.
We constitutively expressed either RAD51-K133R or, as a control, RAD51-WT in otherwise wild-type ES cells containing the H-DR-8mu reporter. By Western blot analysis, RAD51-K133R is estimated to be expressed at a level similar to that of the endogenous RAD51 (data not shown) (49). We used this constitutive-expression approach rather than transient transfection so that we could be assured that each recombinant analyzed was derived from a cell in which RAD51-K133R was expressed. We did not attempt a similar approach with RAD51-K133A, since efforts to derive cell lines with stable expression of RAD51-K133A were unsuccessful.
The RAD51-expressing H-DR-8mu cell lines were transfected with the I-SceI expression vector, and neo+ colonies were selected. As observed previously, the overall efficiency of recombination is much lower with the H-DR-8mu reporter than with the DR-GFP reporter due to the sequence heterology between the S2neo and pneo-8mu sequences (12). Expression of RAD51-K133R resulted in a further reduction in HDR of the H-DR-8mu reporter (1.6 × 10−4 compared to 5.2 × 10−4 for the parental cells). Thus, HDR between heterologous sequences is also reduced by impaired RAD51 function.
We next examined gene conversion in the residual HDR events by PCR amplification of the neo+ genes from individual recombinants and subsequent restriction digestion analysis of the PCR products (Fig. 3a). From this analysis, we found that expression of RAD51-K133R resulted in a shift toward conversion of markers more distant from the DSB (Fig. 3) (P = 0.0012). A mean gene conversion tract length of 145 bp was found in the neo+ recombinants from RAD51-K133R-expressing cells compared to only 60 and 59 bp in the recombinants from RAD51-WT-expressing cells and the parental cells, respectively (Fig. 3). Considering individual markers, the ApaLI restriction site marker, for example, which is 394 bp from the DSB, was incorporated in 23.7% of the neo+ recombinants from RAD51-K133R-expressing cells compared to 7 and 6.2% of recombinants from RAD51-WT-expressing cells and the parental cells, respectively (Fig. 3b). Thus, as well as suppressing SSA, ATP hydrolysis by RAD51 limits the extent of gene conversion during HDR. Our results indicate, therefore, that proper RAD51 function is essential to limit the mutagenic potential of homologous repair on multiple levels.
BRCA2 promotes HDR and suppresses SSA, whereas BRCA1 promotes both pathways.
We next examined the generality of the RAD51 suppression of SSA with respect to other HDR factors. Other factors with key roles in HDR are the breast cancer susceptibility genes Brca1 and Brca2, both of which have been shown to promote HDR and colocalize with RAD51 at sites of DNA damage (18). BRCA2 appears to have a direct role in modulating RAD51 activity through direct interaction; the role of BRCA1, however, is unclear, since a direct BRCA1-RAD51 interaction has not been found. As with RAD51, complete loss of BRCA1 and BRCA2 can lead to cellular lethality (18). Therefore, to test the roles of these factors in SSA, we integrated the SA-GFP reporter into mouse ES cells with hypomorphic alleles of Brca1 and Brca2 (Fig. 4a) so that the presence of an intact copy of the reporter in several independent clones was confirmed by Southern blotting (data not shown). The hypomorphic alleles in Brca1−/− cells contain a deletion in exon 11, which encodes approximately half of the protein, so that an exon 10-to-12 splice product is formed (47). The hypomorphic alleles in Brca2L1/L2 cells contain deletions of the terminal coding exon(s), so that one allele encodes a C-terminal truncation of the BRCA2 protein lacking one of the RAD51 binding motifs whereas the other appears to be essentially a null allele (10, 27).
We compared the efficiency of SSA in these mutant cells to that of wild-type cells by transfection of the I-SceI expression vector and subsequent quantification of the percentage of GFP+ cells (Fig. 4b). We found that Brca2L1/L2 cells exhibited an increased efficiency of SSA (1.9-fold increase; P = 0.002), like the RAD51-disrupted cells. In contrast, Brca1−/− cells surprisingly exhibited a decreased efficiency of SSA (1.8-fold decrease; P = 0.004). It should be noted that there is some precedent for these results (30, 55); however, the previous lack of a direct comparison of the Brca1 and Brca2 mutants, using similar repair substrates, has until now precluded a definitive conclusion about a distinct role for these proteins in DSB repair. Under the same transfection conditions, HDR, as measured with DR-GFP reporter, was reduced in both the Brca1−/− and Brca2L1/L2 cells (5.3- and 6-fold decreases, respectively), consistent with previous results (31, 32). Thus, while both hypomorphic Brca1 and Brca2 mutations impair HDR, they have opposite affects on SSA.
Since the SA-GFP reporter in each of the examined Brca1 and Brca2 mutant cell lines was randomly integrated rather than hprt targeted as in the parental cell line, we wanted to be certain that the decreased SSA in the Brca1 mutant was not due to decreased I-SceI endonuclease accessibilities at different loci. To examine this, we directly assayed loss of the I-SceI site by PCR amplification of the SceGFP3′ gene and subsequent cleavage of the PCR product by I-SceI (Fig. 4c). In this approach, DSB repair products arising from the other repair pathways, i.e., HDR and NHEJ, were specifically quantitated. The SSA product was not amplified, because the upstream primer-binding site is lost during SSA. While this assay is not a direct quantification of I-SceI endonuclease cleavage efficiency, it allows a determination of whether a change in the efficiency of SSA is specific or associated with a general change in overall repair by other pathways. Using this assay, we measured the percent I-SceI site loss for each of the transfections described in Fig. 4b. Wild-type and Brca1−/− cells had similar percentages of site loss (6 and 7%, respectively), suggesting that the SSA defect in these cells is not attributable to a decreased accessibility of the I-SceI site for cleavage; it also further suggests that decreased homologous repair is at least partially compensated for by increased NHEJ (30). By contrast, site loss is reproducibly decreased in Brca2L1/L2 cells (3%). The decrease in site loss in Brca2L1/L2 cells is consistent with the associated shift of HDR toward SSA repair (Fig. 4b).
The Brca1 and Brca2 alleles used in the above analysis maintain large portions of the BRCA1 and BRCA2 proteins. To be certain that the directions of the SSA effects we observed were not allele specific, we tested the effects of dominant-negative peptides on BRCA1 and BRCA2 function (Fig. 4a). To investigate BRCA1, we turned to its heterodimeric partner, BARD1. BRCA1 and BARD1 interact via their N-terminal RING domains, and expression of an N-terminal fragment of BARD1 (BARD1-hB202) has been shown to decrease HDR (58). We examined BRCA2 function using the BRC3 peptide, which can disrupt RAD51 function in vitro (7) and has been shown to inhibit HDR in vivo (49).
To compare the effects on both HDR and SSA, expression vectors for BARD-hB202 and BRC3 were transiently cotransfected with the I-SceI expression vector in otherwise wild-type cells containing the DR-GFP or SA-GFP reporter (Fig. 4d). Expression of BARD1-hB202 decreased the efficiencies of both HDR and SSA (2.4- and 1.9-fold decreases, respectively), similar to the Brca1−/− cell line, whereas the control full-length BARD1 had no effect. In contrast, expression of the BRC3 peptide from BRCA2 reduced HDR but also increased SSA (2-fold decrease and 3.6-fold increase, respectively), similar to the Brca2L1/L2 cell line. Together with those for the mutant cell lines, these results indicate that while BRCA1/BARD1 promotes both HDR and SSA, BRCA2 promotes HDR and suppresses SSA. These results suggest that the BRCA1/BARD1 complex functions during a step common to both homologous-repair pathways, whereas BRCA2 functions downstream with RAD51 to affect the choice between HDR and SSA.
Ku70 mutation increases both HDR and SSA and partially suppresses the repair defects of BRCA1/BARD1 disruption.
Previously, NHEJ mutants were reported to exhibit increased HDR (4, 36), with a KU mutant showing the largest increase in HDR among the tested mutants (36). The KU70/KU80 heterodimer binds DNA ends to promote their repair by NHEJ (19, 57), and this end-binding activity has been postulated to compete with HDR factors for access to DNA ends (36, 56). We were interested in determining if KU, like BRCA1/BARD1, influences steps common to both pathways of homologous repair or if its effect is limited to HDR.
To test the role of KU70 in SSA, we targeted the SA-GFP reporter to the hprt locus in Ku70−/− cells and compared the results with those for a previously described Ku70−/− cell line containing the DR-GFP reporter, also targeted to the hprt locus (36). Using each of these cell lines, the efficiency of homologous repair in cells transfected with I-SceI alone was compared to that in cells additionally transfected with a KU70 expression vector that allows partial complementation of the KU deficiency (36) (Fig. 5). We found that the Ku70−/− cells exhibited elevated levels of both SSA and HDR (eight- and sevenfold increases, respectively), each of which was partially reduced by cotransfection of the KU70 expression vector. These results suggest that KU70 acts during a step common to both homologous-repair pathways to inhibit HDR and SSA.
The above-mentioned finding that KU70 and BRCA1/BARD1 influence both HDR and SSA, albeit in opposite directions, suggested that these factors may both act during an early step of homologous repair. To investigate this further, we tested whether there is a genetic association between these factors by comparing the effect of BARD1-hB202 on homologous repair in Ku70−/− cells. In particular, we wanted to determine if the loss of KU would limit the effect of this peptide on either or both pathways. For comparison, we also tested the effect of RAD51-K133R in Ku70−/− cells, as this is expected to act at a later step. Transfection of the BARD1-hB202 expression vector into Ku70−/− cells resulted in a 1.7-fold decrease in HDR repair (Fig. 5b), a somewhat smaller effect than that seen in wild-type cells (2.4-fold) (Fig. 4d). However, transfection of the BARD1-hB202 expression vector into Ku70−/− cells had no effect on SSA, although it reduced SSA repair in wild-type cells (1.9-fold). As expected, transfection of the RAD51-K133R expression vector into Ku70−/− cells resulted in decreased HDR and increased SSA, similar to its effect on wild-type cells (14- and 1.9-fold, respectively). Thus, disruption of KU function partially suppresses the homologous-repair defects caused by BARD1-hB202 but does not suppress the homologous-repair defects caused by RAD51-K133R. These results are consistent with the notion that BRCA1/BARD1 and KU70 both act at an early step in homologous repair, whereas RAD51 acts at a step distinct from that of KU70. However, the finding that the BARD1-hB202 inhibition of HDR is not completely abolished by Ku70 mutation, indicates that BRCA1/BARD1 may also play an additional later role in HDR.
RAD52 and ERCC1 promote SSA but are not essential for HDR.
RAD52 is essential for multiple aspects of homologous repair in yeast, including SSA and HDR (34, 52), whereas mouse cells deficient in RAD52 have been shown to exhibit only a mild gene-targeting defect (42). However, the role of mammalian RAD52 in HDR and SSA has not been examined. ERCC1 is involved in nucleotide excision repair, but it has also been shown to be important for gene targeting (33), as has its yeast ortholog (34), and to promote SSA in mammalian cells (1, 43). Recently, human RAD52 has been shown to form a complex with ERCC1 and its associated factor XPF (29).
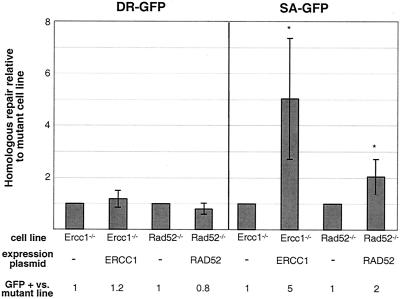
To investigate the roles of RAD52 and ERCC1 in homologous repair in mammalian cells, we integrated the DR-GFP and SA-GFP reporters into Rad52−/− and Ercc1−/− ES cells using the hprt targeting vectors. For the Rad52−/− cells, hprt-targeted clones were readily obtained; however, for the Ercc1−/− cells targeting was severely reduced, although a few targeted clones were obtained (data not shown). In the Ercc1−/− clones in which the reporter was randomly integrated, the presence of a single intact copy of each reporter was verified by Southern blotting (data not shown). The efficiency of homologous repair in cells transfected with I-SceI alone was compared to that in cells additionally transfected with the complementing expression vector (Fig. 6). We find this analysis more informative than a comparison to our wild-type line, since the Ercc1−/− and Rad52−/− cells, which are derived from a different ES cell background, appear to have a higher transfection efficiency (data not shown). In these experiments, the complemented Rad52−/− and Ercc1−/− cells had no higher efficiency of HDR than uncomplemented cells; however, SSA was increased (two- and fivefold, respectively) (Fig. 6). These results indicate that RAD52 and ERCC1 specifically promote homologous repair by SSA.
FIG. 6.
RAD52 and ERCC1 promote SSA but are not essential for HDR. The efficiency of homologous repair is indicated for the Ercc1−/− and Rad52−/− ES cells transfected with the I-SceI expression vector alone or together with the ERCC1 or RAD52 complementing expression vector, respectively. The efficiency of homologous repair in individual transfections is calculated relative to transfection with the I-SceI expression vector alone for each mutant, which is set to 1 (see Materials and Methods). SA-GFP repair frequencies are the means (± standard deviations) of seven and six independent transfections for the Rad52−/− and Ercc1−/− cell lines, respectively. The asterisks indicate a statistically significant difference from the mutant transfected with the I-SceI expression vector alone, with P ≤ 0.008.
DISCUSSION
Repair of chromosomal DSBs is essential to maintain genomic integrity, yet different repair pathways are variably mutagenic. In this report, we have examined the interrelationship of the homologous-repair pathways in mammalian cells. HDR is primarily a precise type of repair and thus much less prone than SSA to causing gross chromosomal rearrangements (39) or chromosomal deletions, as assayed in this study. However, HDR can lead to genetic loss under some circumstances, such as when conversion is extensive. Thus, the proper balance and regulation of DSB repair pathways would be predicted to be critical for the maintenance of genomic integrity. In support of this, HDR mutants exhibit chromosome aberrations and higher rates of mutagenesis (48, 53), apparently as a result of misrepair of spontaneously arising DNA lesions, including DSBs. As yet, however, a comparative analysis of distinct classes of DSB repair mutants has not been reported to provide an understanding of the interrelationship of the homologous-repair pathways in mammalian cells.
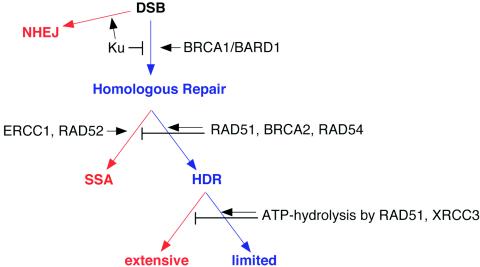
Our analysis has determined that some factors affect the competitive choice between HDR and SSA (RAD51 and BRCA2), whereas others factors promote or suppress both homologous-repair pathways (BRCA1/BARD1 and KU, respectively), and we have identified an activity that limits genetic loss from HDR (ATP hydrolysis by RAD51) (see below) (Fig. 7). Failure to specifically utilize HDR as a result of impaired RAD51 function—either directly by RAD51 mutation or indirectly by BRCA2 disruption—leads to an increased reliance on SSA. The RAD51 ATP hydrolysis mutant not only shifts homologous pathway use to SSA but also fails to limit the extent of gene conversion in the remaining HDR events, increasing the potential for genetic loss.
FIG. 7.
Genetic steps of homologous repair with distinct mutagenic consequences. Shown are different pathways of DSB repair and the roles of individual factors in pathway choice. Whereas homologous repair by HDR with limited gene conversion (blue) is a precise type of repair, SSA, NHEJ, and HDR with extensive gene conversion (red), as well as crossing over (not shown), are much more prone to being mutagenic. The arrows do not necessarily reflect a temporal order of events. Homologous-repair results were obtained using DR-GFP and SA-GFP, except for RAD54 and XRCC3 (see the text).
The finding that the proper functioning of RAD51 is essential to promote precise homologous repair indicates that control of strand exchange may be a deciding factor in suppressing mutagenic outcomes of DSB repair. BRCA2 is a particularly intriguing factor for such control, since it can interact directly with RAD51 at multiple motifs, as well as with single-stranded DNA, and thus may modulate the association of RAD51 with damaged DNA (7, 44, 59). Interestingly, we found a shift from HDR to SSA, whether BRCA2 is mutated by the loss of only the C-terminal RAD51 binding motif so that 94% of the protein coding region is left intact, including the eight BRC repeats (23, 55), or whether a single 70-amino-acid BRC repeat, which can disrupt RAD51 nucleoprotein filament formation in vitro, is overexpressed. Another Brca2 mutant, as yet uncharacterized, also shows increased deletional homologous-recombination events which may reflect SSA (23, 55). Thus, a shift from HDR to SSA appears to be a generalized outcome when RAD51 or BRCA2 function is disrupted. Consistent with this, loss of RAD54, a protein that promotes RAD51-mediated strand exchange activity in vitro (51), also results in a shift from HDR to SSA (11), although the magnitude of the shift is much smaller than that seen with RAD51 or BRCA2. Thus, this shift in homologous pathway usage appears to be a common outcome when factors that affect steps after the branch point of HDR and SSA are disrupted. Although key distinctions exist for some yeast and mammalian DSB repair factors, a similar shift to SSA has been noted in yeast for some HDR mutants (16). The shift to SSA may result from hyperresection of DSBs when RAD51 activity is impaired: whereas HDR requires minimal resection, SSA requires >2 kb of resection in our substrate. Since repetitive elements are at various distances from each other, the extent of resection is expected to be an important factor in the efficiency of SSA in the mammalian genome.
In addition to the shift to SSA, we found that genetic loss arising from more extensive gene conversion tracts also occurred when ATP hydrolysis by RAD51 was disrupted. Mutation of the RAD51 paralog XRCC3 also results in more extensive gene conversion in the residual recombinants that are obtained with this mutant, although the tracts are often discontinuous (5). As we find no evidence for discontinuous tracts with RAD51-K133R expression, RAD51 and XRCC3 appear to have different roles in the extent of conversion. Perhaps XRCC3 is essential to promote the stability of recombination intermediates and thus maintain the continuity of DNA synthesis during gene conversion, whereas ATP hydrolysis by RAD51 is important for processing recombination intermediates to limit such DNA synthesis. Alternatively, ATP hydrolysis by RAD51 may result in greater protection of DNA ends, limiting the amount of single- or double-strand resection that may occur.
In contrast to RAD51 and BRCA2, we found that BRCA1/BARD1 or KU70/KU80 affected both homologous-repair pathways in the same direction; defects in BRCA1/BARD1 decreased the efficiencies of both homologous-repair pathways, while loss of KU increased the efficiencies of both pathways. Thus, these factors appear to affect a step(s) of homologous repair that is common to both HDR and SSA, although with opposite outcomes. Furthermore, loss of KU70 partially suppresses the homologous-repair defects of BARD1 disruption, which indicates that BRCA1/BARD1 and KU70 may both be acting at an early step during homologous repair. The initial strand resection step to generate single-stranded DNA for strand exchange or annealing is one such step. Ku70-deficient yeast cells exhibit elevated rates of strand resection, indicating that Ku70 is important in limiting resection in yeast (24). Thus, while KU70 could limit strand resection, BRCA1/BARD1 could act to promote strand resection. An early role for the BRCA1/BARD1 complex in homologous repair is consistent with its rapid localization to DSBs (35). The exonuclease(s) that promotes resection has not been identified; however, BRCA1/BARD1 could affect nuclease activity or access of a nuclease to DNA ends via one of the identified BRCA1/BARD1 biochemical activities, e.g., DNA binding, ubiquitination, or phosphopeptide binding (6, 18). Unlike SSA, the inhibitory effect of BARD1 disruption on HDR is not totally abrogated by Ku mutation; thus, BRCA1/BARD1 could still have a later role in HDR, possibly through its interaction with HDR-specific factors like RAD51 or BRCA2 (18).
We also determined that mammalian RAD52 promotes SSA, although loss of RAD52 does not overtly affect HDR. That RAD52 does not play a significant role in promoting HDR in mammalian cells is surprising, given its critical role in HDR in yeast (52), its postulated role as a “gatekeeper” for HDR in mammalian cells through its biochemical activity of binding to DNA ends (56), and the fact that human RAD52 stimulates homologous pairing by human RAD51 in vitro (51). The nonessentiality of RAD52 in mammalian cells suggests this function is assumed in vivo by some other factor. BRCA2, which also binds single-stranded DNA and RAD51 and stimulates RAD51 activity (7, 44, 59), is an excellent candidate. However, a role for RAD52 in HDR may still be uncovered in other, more complex genetic contexts, as has been observed in chicken cells by combined mutation with the RAD51 paralog XRCC3 (13).
In summary, the multiple DSB repair pathways have a complex interrelationship which affects whether repair occurs faithfully. Understanding these interrelationships at the molecular level in mammalian cells is essential, given that many DSB repair mutants have markedly different phenotypes, including development defects, aging phenotypes, and tumor susceptibility (38), and in some cases even have tumors of the same tissue type yet with markedly different characteristics, i.e., as in humans for BRCA1- and BRCA2-associated tumors (14).
Acknowledgments
We thank members of the Jasin laboratory, especially Beth Elliott, Ulrica Westermark, and Mary Ellen Moynahan, for materials and discussions and Laura Niedernhofer and Roland Kanaar (The Netherlands) for the Ercc1−/− ES cell line.
This work was supported by NIH grants GM54668 and CA94060 and a P20 grant from the MSKCC Cancer and Aging Program.
REFERENCES
- 1.Adair, G. M., R. L. Rolig, D. Moore-Faver, M. Zabelshansky, J. H. Wilson, and R. S. Nairn. 2000. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19:5552-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, M. D., M. McVey, and J. J. Sekelsky. 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299:265-267. [DOI] [PubMed] [Google Scholar]
- 3.Albertson, D. G., C. Collins, F. McCormick, and J. W. Gray. 2003. Chromosome aberrations in solid tumors. Nat. Genet. 34:369-376. [DOI] [PubMed] [Google Scholar]
- 4.Allen, C., A. Kurimasa, M. A. Brenneman, D. J. Chen, and J. A. Nickoloff. 2002. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. USA 99:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenneman, M. A., B. M. Wagener, C. A. Miller, C. Allen, and J. A. Nickoloff. 2002. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell 10:387-395. [DOI] [PubMed] [Google Scholar]
- 6.Caldecott, K. W. 2003. Cell signaling. The BRCT domain: signaling with friends? Science 302:579-580. [DOI] [PubMed] [Google Scholar]
- 7.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7:273-282. [DOI] [PubMed] [Google Scholar]
- 8.Deininger, P. L., and M. A. Batzer. 1999. Alu repeats and human disease. Mol. Genet. Metab. 67:183-193. [DOI] [PubMed] [Google Scholar]
- 9.DePinho, R. A. 2000. The age of cancer. Nature 408:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Donoho, G., M. A. Brenneman, T. X. Cui, D. Donoviel, H. Vogel, E. H. Goodwin, D. J. Chen, and P. Hasty. 2003. Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer 36:317-331. [DOI] [PubMed] [Google Scholar]
- 11.Dronkert, M. L., H. B. Beverloo, R. D. Johnson, J. H. Hoeijmakers, M. Jasin, and R. Kanaar. 2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20:3147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, B., and M. Jasin. 2001. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 21:2671-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimori, A., S. Tachiiri, E. Sonoda, L. H. Thompson, P. K. Dhar, M. Hiraoka, S. Takeda, Y. Zhang, M. Reth, and M. Takata. 2001. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 20:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedenfalk, I. A. 2002. Gene expression profiling of hereditary and sporadic ovarian cancers reveals unique BRCA1 and BRCA2 signatures. J. Natl. Cancer Inst. 94:960-961. [DOI] [PubMed] [Google Scholar]
- 15.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov, E. L., N. Sugawara, J. Fishman-Lobell, and J. E. Haber. 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasin, M. 2001. Double-strand break repair and homologous recombination in mammalian cells, p. 207-235. In J. A. Nickoloff and M. F. Hoekstra (ed.), DNA damage and repair, vol. III. Humana Press, Totowa, N.J.
- 18.Jasin, M. 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21:8981-8993. [DOI] [PubMed] [Google Scholar]
- 19.Jeggo, P. A. 1998. DNA breakage and repair. Adv. Genet. 38:185-218. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, P. M., C. Allen, B. M. Wagener, Z. Shen, and J. A. Nickoloff. 2001. Overexpression of human RAD51 and RAD52 reduces double-strand break-induced homologous recombination in mammalian cells. Nucleic Acids Res. 29:4352-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, S., and B. S. Lopez. 2000. Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J. 19:3090-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larminat, F., M. Germanier, E. Papouli, and M. Defais. 2002. Deficiency in BRCA2 leads to increase in non-conservative homologous recombination. Oncogene 21:5188-5192. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 25.Lim, D.-S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16:7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, F. L., K. Sperle, and N. Sternberg. 1990. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol. Cell. Biol. 10:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morimatsu, M., G. Donoho, and P. Hasty. 1998. Cells deleted for Brca2 COOH terminus exhibit hypersensitivity to gamma-radiation and premature senescence. Cancer Res. 58:3441-3447. [PubMed] [Google Scholar]
- 28.Morrison, C., A. Shinohara, E. Sonoda, Y. Yamaguchi-Iwai, M. Takata, R. R. Weichselbaum, and S. Takeda. 1999. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol. 19:6891-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motycka, T. A., T. Bessho, S. M. Post, P. Sung, and A. E. Tomkinson. 2004. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J. Biol. Chem. 279:13634-13639. [DOI] [PubMed] [Google Scholar]
- 30.Moynahan, M. E., J. W. Chiu, B. H. Koller, and M. Jasin. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4:511-518. [DOI] [PubMed] [Google Scholar]
- 31.Moynahan, M. E., T. Y. Cui, and M. Jasin. 2001. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61:4842-4850. [PubMed] [Google Scholar]
- 32.Moynahan, M. E., A. J. Pierce, and M. Jasin. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7:263-272. [DOI] [PubMed] [Google Scholar]
- 33.Niedernhofer, L. J., J. Essers, G. Weeda, B. Beverloo, J. de Wit, M. Muijtjens, H. Odijk, J. H. Hoeijmakers, and R. Kanaar. 2001. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 20:6540-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pāques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 36.Pierce, A. J., P. Hu, M. Han, N. Ellis, and M. Jasin. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce, A. J., J. M. Stark, F. D. Araujo, M. E. Moynahan, M. Berwick, and M. Jasin. 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol. 11:S52-S59. [DOI] [PubMed] [Google Scholar]
- 39.Richardson, C., and M. Jasin. 2000. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405:697-700. [DOI] [PubMed] [Google Scholar]
- 40.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12:3831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson, C., J. M. Stark, M. Ommundsen, and M. Jasin. 2004. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene 23:546-553. [DOI] [PubMed] [Google Scholar]
- 42.Rijkers, T., J. Van Den Ouweland, B. Morolli, A. G. Rolink, W. M. Baarends, P. P. Van Sloun, P. H. Lohman, and A. Pastink. 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18:6423-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sargent, R. G., J. L. Meservy, B. D. Perkins, A. E. Kilburn, Z. Intody, G. M. Adair, R. S. Nairn, and J. H. Wilson. 2000. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 28:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin, D. S., L. Pellegrini, D. S. Daniels, B. Yelent, L. Craig, D. Bates, D. S. Yu, M. K. Shivji, C. Hitomi, A. S. Arvai, N. Volkmann, H. Tsuruta, T. L. Blundell, A. R. Venkitaraman, and J. A. Tainer. 2003. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 22:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigurdsson, S., S. Van Komen, G. Petukhova, and P. Sung. 2002. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 277:42790-42794. [DOI] [PubMed] [Google Scholar]
- 46.Singleton, M. R., L. M. Wentzell, Y. Liu, S. C. West, and D. B. Wigley. 2002. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. USA 99:13492-13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snouwaert, J. N., L. C. Gowen, A. M. Latour, A. R. Mohn, A. Xiao, L. DiBiase, and B. H. Koller. 1999. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene 18:7900-7907. [DOI] [PubMed] [Google Scholar]
- 48.Sonoda, E., M. Takata, Y. M. Yamashita, C. Morrison, and S. Takeda. 2001. Homologous DNA recombination in vertebrate cells. Proc. Natl. Acad. Sci. USA 98:8388-8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark, J. M., P. Hu, A. J. Pierce, M. E. Moynahan, N. Ellis, and M. Jasin. 2002. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 277:20185-20194. [DOI] [PubMed] [Google Scholar]
- 50.Stark, J. M., and M. Jasin. 2003. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 23:733-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung, P., L. Krejci, S. Van Komen, and M. G. Sehorn. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729-42732. [DOI] [PubMed] [Google Scholar]
- 52.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131-153. [DOI] [PubMed] [Google Scholar]
- 54.Tsuzuki, T., Y. Fujii, K. Sakumi, Y. Tominaga, K. Nakao, M. Sekiguchi, A. Matsushiro, Y. Yoshimura, and T. Morita. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93:6236-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tutt, A., D. Bertwistle, J. Valentine, A. Gabriel, S. Swift, G. Ross, C. Griffin, J. Thacker, and A. Ashworth. 2001. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20:4704-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Dyck, E., A. Z. Stasiak, A. Stasiak, and S. C. West. 1999. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398:728-731. [DOI] [PubMed] [Google Scholar]
- 57.Walker, J. R., R. A. Corpina, and J. Goldberg. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607-614. [DOI] [PubMed] [Google Scholar]
- 58.Westermark, U. K., M. Reyngold, A. B. Olshen, R. Baer, M. Jasin, and M. E. Moynahan. 2003. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 23:7926-7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, H., P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee, and N. P. Pavletich. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837-1848. [DOI] [PubMed] [Google Scholar]