Abstract
For a particular functional family of basic helix-loop-helix (bHLH) transcription factors, there is ample evidence that different factors regulate different target genes but little idea of how these different target genes are distinguished. We investigated the contribution of DNA binding site differences to the specificities of two functionally related proneural bHLH transcription factors required for the genesis of Drosophila sense organ precursors (Atonal and Scute). We show that the proneural target gene, Bearded, is regulated by both Scute and Atonal via distinct E-box consensus binding sites. By comparing with other Ato-dependent enhancer sequences, we define an Ato-specific binding consensus that differs from the previously defined Scute-specific E-box consensus, thereby defining distinct EAto and ESc sites. These E-box variants are crucial for function. First, tandem repeats of 20-bp sequences containing EAto and ESc sites are sufficient to confer Atonal- and Scute-specific expression patterns, respectively, on a reporter gene in vivo. Second, interchanging EAto and ESc sites within enhancers almost abolishes enhancer activity. While the latter finding shows that enhancer context is also important in defining how proneural proteins interact with these sites, it is clear that differential utilization of DNA binding sites underlies proneural protein specificity.
Neurogenesis in diverse organisms requires basic helix-loop-helix (bHLH) transcriptional activators that are related to the Drosophila factors encoded by the proneural genes atonal (ato) and scute (sc). A variety of functions have been assigned to different family members through genetic analyses, but not much is known of the basis for protein specificity. The proneural genes of Drosophila are essential for the specification of sense organ precursors (SOPs) of the peripheral nervous system. They are initially expressed in characteristic groups of ectodermal cells (proneural clusters) before becoming refined to the SOPs themselves. Different proneural genes specify different SOPs, so, for example, sc and other members of the ac-sc complex (AS-C) specify the precursors of the external sense organs (10, 19), while ato specifies the precursors of the chordotonal organs (stretch receptors) and the R8 photoreceptors (25). Misexpression analyses show that these different functions are to a large extent due to intrinsic differences between proteins (20, 24). However, these same analyses show that intrinsic differences are very contingent on the developmental context. For example, the same proneural protein can promote the development of different classes of peripheral nervous system (PNS) sensory neurons in different locations. Nevertheless, the conclusion is that different proneural proteins must activate both common neural target genes and distinct neuronal subtype-specific target genes in order to achieve their different developmental functions. It is not known how different neural bHLH proteins control different target genes.
Most bHLH proteins bind to a DNA sequence containing a 6-bp E box (CANNTG). Proneural proteins are class A bHLH factors (32), all of which share, at least in vitro, a preference for a DNA sequence containing the E-box variant CAGSTG (where S is C or G). Among nonneural bHLH proteins, there is evidence for differences in DNA binding site specificities between proteins of different families, corresponding to preferences for certain bases central to and flanking the CANNTG (7, 15-18, 22, 26, 27, 42). The Drosophila proteins bind DNA as heterodimers with the ubiquitously expressed bHLH protein Daughterless (Da). Sc/Da binding sites have been located in a number of proneural target gene enhancers (9, 29, 43, 45), and from these studies a proneural consensus binding site that includes preferred bases flanking the 6-bp E box core (GCAGSTGK [where K is G or T and invariant nucleotides are underlined]) has been deduced. However, studies concerning Sc target genes have not examined the issue of specificity between proneural proteins. Regulation of target genes by Ato/Da is rarely addressed or is implicitly assumed to follow the same rules as for Sc/Da. For example, some well-characterized Sc target genes are expressed widely in early neurogenesis and should be Ato targets too (i.e., common proneural target genes), but it is not known how Ato regulates them. Additionally, possible Ato-specific target genes have not been well investigated. The only Ato target for which there is information is the tachykinin receptor-related gene, TAKR86C (formerly NKD), which has an essential Ato/Da binding site in its enhancer (40). However, that report did not address the issue of selective regulation by Ato versus Sc.
Although the area is little explored, there is little indication of DNA binding differences between proneural proteins from in vitro studies (25). Consistent with this, predicted DNA-contacting residues are completely conserved between the bHLH domains of Sc and Ato (12). This suggested that Sc and Ato may not have distinct binding sequences. On this and other evidence, it has been suggested convincingly that the major determinant of proneural specificity is differential interaction with “specificity cofactors” (5, 12, 24).
Thus, it is important that proneural target genes be analyzed to understand how they are selectively regulated by Sc or Ato. Here, we show that a common proneural target gene, Bearded (Brd), is regulated by Sc/Da and Ato/Da via distinct E-box sites. We have established a preliminary Ato consensus binding sequence based on essential E boxes identified in three Ato-regulated target genes. This consensus (EAto) is different from that established for Sc (ESc), particularly in the flanking bases. Thus, despite their similar DNA binding properties in vitro, Ato and Sc utilize different binding sites in vivo even for common target gene regulation. This conclusion is substantiated by observations that these proneural binding sites are alone sufficient to confer highly specific patterns of expression on a green fluorescent protein (GFP) reporter gene. We demonstrate that differences in DNA binding sequence underlie proneural protein functional specificity in vivo.
MATERIALS AND METHODS
Fly stocks.
Brd-1.5-lacZ, Brd-1.5 E1 M-lacZ, Brd-184-lacZ, and Brd-184 E1 M-lacZ transgenic fly lines are as described previously (43) and were kindly donated by J. Posakony. Other stocks used were UAS-ato (25), UAS-sc (12), and 109-68-Gal4 (24).
Reporter plasmid construction.
For the Brd enhancer constructs, primers were designed to amplify a 1.3-kb fragment (43) (5′-GTGCTACAAACGCGTGAATTCTC-3′ and 5′-CATGGTACCTCTGCAGGTAGTTCGGG-3′) (restriction sites are underlined). The resultant fragment was cloned in pBluescript following digestion with EcoRI and Asp718I. Three further PCRs subdivided the 1.3-kb enhancer: Brd-A (531 bp) (5′-AGAGAATTCGGACAGTGAAACCTGCCA-3′ and 5′-CATGGTACCTGTCGGCAAACGAGAAAT-3′), Brd-B (493 bp) (5′-GTCGAATTCGGATTGGAATACGAATG-3′ and 5′-GTTGGTACCTGTCGGCAAACGAGAAAT-3′), and Brd-C (371 bp) (5′-CCGGAATTCGTCATATTACAACACTCG-3′ and 5′-TTAGGTACCGTTCGGGCTCTCCAAGA-3′). These were cloned in pCRIITOPO (Invitrogen). All four fragments were transferred to pHStinger (2) to give Brd-1.3-GFP, Brd-A-GFP, Brd-B-GFP, and Brd-C-GFP for germ line transformation. For sc-SMC reporter constructs, the SMC enhancer (13) was amplified and cloned into pHStinger.
Site-directed mutagenesis.
E boxes E2 and E3 in pCRIITOPO-Brd-A were mutated by using the Stratagene QuikChange mutagenesis kit. The Brd-E3 E-box sequence was changed from CATGTG to AATGTT, and the Brd-E2 sequence was changed from CACGTG to AACGTT. The appropriate fragments were then cloned in pHStinger for germ line transformation. Mutation of the sequences flanking ato-E1 and sc-E1 and E2 was carried out in a similar way with either ato-FCO-E-GFP or sc-SMC-E-GFP constructs as templates.
Drosophila germ line transformation.
Transformation plasmids were injected into w; Δ2-. flies. Transformants were selected and then outcrossed to w1118 to remove the Δ2-. element. At least two independent insertions were analyzed for each construct.
Protein purification.
pRSET-Ato, pRSET-Da, and pRSET-Sc plasmids were used to transform BL21-pLysS cells. Cultures were grown and induced with IPTG (isopropyl-β-d-thiogalactopyranoside) by standard techniques. Cell pellets were stored at −80°C, and Ni2+-nitrilotriacetic acid columns were used to purify the His-tagged proteins in 8 M urea before refolding by stepwise dialysis.
Gel retardation probes.
The following synthetic complementary 36-bp oligonucleotides containing a central 6-bp E box were used in gel retardation experiments: BrdE1GR, 5′-TGAGAGACCGAGAAACACCTGCGCGCTAGGACTCGT 3′; BrdE3GR, 5′ CTCGTTTGCCGACAACATGTGTTTAACGAGGGTCTG 3′; AtoE1GR, 5′ TGGTAGTAACCATAACAGGTGGCACGGCAGCCGCAC 3′; TAKR86CE2GR, 5′ ATGTAGTTGGGGTATCAGGTGTGCTGAACAAGGGGT 3′; and ScE1GR, 5′ CATGGCGACGCGTGGCAGGTGTATTTAGTCGAACGA 3′. E boxes are shown in boldface. In AtoE1GR, a 3′ C substitution (underlined) disrupts a Pointed binding site adjacent to the E box (47). Brd-E1,3 mutant probes have the CANNTG changed to AANNTT. For both the titration and competition experiments, the top-strand oligonucleotide was labeled prior to hybridization to the complementary oligonucleotide, which was included at a slight molar excess. The labeling reaction used [γ-33P]ATP [Amersham or Perkin-Elmer Life Sciences (UK) Ltd.] and T4 polynucleotide kinase (New England Biolabs). Unincorporated ATP was removed by using ProbeQuant G-50 microcolumns (Amersham Biosciences).
Gel retardation.
γ-33P-labeled Brd-E1 and Brd-E3 36-bp duplexes were prepared as described above and used at 0.1 nM in 20-μl binding reaction mixtures in binding buffer (10 mM Tris HCl, 1 mM dithiothreitol, and 1 mM EDTA, with 40 mM [low salt] or 100 mM [high salt] NaCl). Ato/Da or Sc/Da heterodimers were prepared by incubating the proteins at 200 nM for 20 min on ice in binding buffer, and then 0 to 100 nM Ato/Da or Sc/Da was incubated with Brd-E1 or Brd-E3 probes for 20 min on ice. An 18-μl portion of each binding reaction mixture was then electrophoresed on a 6% polyacrylamide gel with 0.5× Tris-borate-EDTA at 40 mA for 1 h at room temperature. The gels were dried before phosphorimager analysis (Molecular Dynamics). The percent bound DNA was determined in ImageQuant by quantifying the depletion of free DNA, as this gives a better estimate of complex concentration (39). The percent DNA bound versus protein concentration was plotted, and apparent Kds were determined by fitting the data to a one-site binding equation by using Prism 4 (GraphPad). For competition experiments, 5 to 100 nM unlabeled DNA was mixed with 0.1 nM γ-33P-labeled Brd-E1 before addition of Ato/Da or Sc/Da heterodimer to 200 nM.
Multimer constructs.
Synthetic oligonucleotides were designed to give, when hybridized, two repeats of 20-bp sequences containing the 6-bp E box. These were flanked by BglII and BamHI sites. For ato-E1, the nucleotides were as follows: GATCT ACCATAACAGGTGGCACGGC ACCATAACAGGTGGCACGGC GA TGGTATTGTCCACCGTGCCG TGGTATTGTCCACCGTGCCG CCTAG (E boxes are underlined).
In this case, a mutation was introduced (boldface) to ensure the disruption of an adjacent Pointed protein binding site (zur Lage et al., submitted). Similar oligonucleotides, with different 20-bp repeats, were designed for TAKR86C-E2 (GGGGGTATCAGGTGTGCTGAA) and sc-E1 (CGCGTGGCAGGTGTATTTAG). Complementary oligonucleotides were hybridized at 50 pmol/μl and phosphorylated with T4 polynucleotide kinase (New England Biolabs) before ligation (Roche). The ligated DNA was digested with BglII and BamHI and separated on a 3.5% MetaPhor agarose gel (Cambrex). Multimerized bands were excised and cloned in pBluescript (Stratagene). Clones were sequenced before transfer to pHStinger.
Immunohistochemistry.
For immunohistochemical staining, imaginal disks were dissected from wandering third-instar larvae and fixed in 3.7% formaldehyde (10 min at room temperature). Incubations with primary and secondary antibodies were carried out according to standard procedures. Primary antibodies were affinity-purified rabbit anti-Ato (1:2,000), guinea-pig anti-Sens (1:5,000; provided by H. Bellen) (37), mouse anti-β-galactosidase (1:200), mouse and rabbit anti-GFP (1:500), and mouse anti-Ac (1:50; provided by the Developmental Biology Hybridoma Bank, Iowa City, Iowa). Secondary antibodies (1:1,000) were obtained from Molecular Probes. Confocal microscopy analysis was on a Leica TCS SP microscope.
RNA in situ hybridization.
Primers were designed to amplify the GFP open reading frame. The top-strand primer was 5′-CCATGGTGAGCAAGGGCG-3′. The bottom-strand primer was 5′-GTAATACGACTCACTATAGGGCCTTGTACAGCTCGTCCATG-3′, with a T7 RNA polymerase promoter incorporated (boldface). After amplification from pHStinger DNA, the PCR product was then in vitro transcribed (Roche). The antisense RNA probe was hybridized to third-instar larval imaginal disks and detected by a tyramide labeling reaction.
RESULTS
Regulation by Ato and Sc of a common target gene, Bearded, via distinct enhancers.
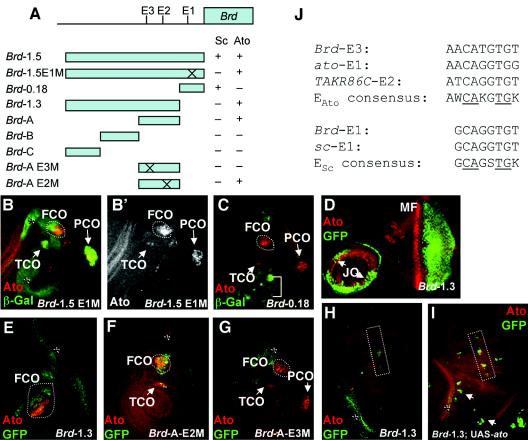
We investigated how a common proneural target gene, Brd, is regulated by Ato and Sc. A frequent assumption is that such target genes will be regulated by different proneural proteins in a common manner (i.e., via shared E boxes). Brd is expressed in all proneural clusters, where it apparently modulates lateral inhibition (30). Singson et al. (43) identified Brd enhancer sequences within a 1.5-kb proximal promoter region of the Brd gene (Fig. 1A). The proximal 184 bp of this region contains an Sc-dependent enhancer that supports reporter gene expression in Sc proneural clusters (43). This regulation is achieved via a functional E box, referred to here as Brd-E1 (GCAGGTGT, conforming to the established proneural consensus; here and below, the conserved core nucleotides are underlined). We found that the 1.5-kb promoter region also supports reporter gene expression in known Ato proneural clusters but that such expression does not depend on the Brd-E1 site. Thus, the 1.5-kb promoter region with a mutated E1 site supports β-galactosidase expression in Ato-expressing regions that give rise to chordotonal SOPs in the leg and antenna (those of the femoral, tibial, prothoracic, and Johnston's chordotonal organs) and is also expressed broadly in the eye (Fig. 1B and B′ and data not shown). Consistent with this, the activity does not reside in Brd-0.18, a 184-bp fragment that contains the Brd-E1 site (Fig. 1A and C). This suggests that Ato regulates Brd via sequences that are in the distal 1.3 kb of the promoter region. Indeed, a new GFP reporter construct containing this 1.3-kb sequence is expressed in most Ato-dependent chordotonal proneural clusters in the leg and antenna (Fig. 1D and E), as well as in the eye disk posterior to the morphogenetic furrow (Fig. 1D). Misexpression of Ato by using a proneural cluster-specific Gal4 driver line (109-68Gal4) (24) causes ectopic Brd-1.3-GFP expression, most notably in the wing disk along the developing third wing vein and in the scutellar region (Fig. 1H and I), which are the regions most susceptible to ectopic chordotonal organ production upon Ato misexpression (24). These results suggest that Brd has an Ato-dependent enhancer that is distinct from its Sc-dependent enhancer.
FIG. 1.
Separable Sc and Ato-dependent regulation of Brd. (A) Schematic diagram of Brd reporter gene constructs, indicating whether they drive Ato and/or Sc-specific expression patterns. The Brd-E1, -E2, and -E3 E boxes mentioned in the text are marked. (B to I) Brd reporter expression in Ato-dependent cells. Green: β-galactosidase in B,C and GFP in D-I. Red: Ato. (B) Brd-1.5E1 M-lacZ prothoracic leg disk, showing reporter expression in regions of the FCO tibial chordotonal organ (TCO), and prothoracic chordotonal organ (PCO). Ato is expressed in all of these areas (B′), although it does not always overlap because of dynamic Ato expression and perdurable GFP expression. The Brd fragments also support some nonproneural ectodermal expression (asterisk). (C) Brd-0.18-lacZ leg disk. Reporter expression (in Sc-dependent cells, bracket) does not coincide with Ato expression during FCO SOP formation. (D and E) Brd-1.3-GFP. (D) Eye-antennal disk, showing expression in Johnston's organ precursors (JO) and posterior to the morphogenetic furrow (MF). (E) Leg disk, showing expression in FCO as well as nonproneural ectodermal expression (asterisk). (F) Brd-A-GFP-E2 M leg disk, showing no effect of mutating E2 on Ato-dependent expression (FCO and TCO [this is a mesothoracic disk and does not have PCO]). (G) Brd-A-GFP-E3 M leg disk. Mutating E3 causes loss of FCO, TCO, and PCO expression. (H and I) Effect of Ato misexpression on Brd-1.3-GFP in wing disk. (H) In the wild type, GFP is expressed only in nonproneural ectodermal cells (asterisk). (I) When Ato is misexpressed (109-68Gal4/UAS-ato), ectopic GFP expression occurs in proneural clusters in the thorax (arrows) and third wing vein (box). (J) E-box sequences studied in this report, compared with the consensus sequences (core nucleotides are underlined).
Ato and Sc regulate Brd via different E-box sites.
When the 1.3-kb fragment was subdivided (Fig. 1A), all of the chordotonal expression of the larger construct could be located in a proximal 531-bp region immediately upstream from the Sc-dependent enhancer (Brd-A) (data not shown). This sequence contains two CANNTG sequences, E2 and E3, and further reporter constructs were made in which one or the other of these sites was destroyed by site-directed mutagenesis (Brd-A E3 M and Brd-A E2 M). While mutating E2 had no discernible effect on chordotonal GFP expression, mutation of E3 resulted in loss of GFP expression specifically in the leg and antennal chordotonal SOPs (Fig. 1F and G and data not shown). Unlike chordotonal expression, GFP expression in the eye could not be located to a smaller enhancer fragment: eye expression supported by Brd-A is substantially less than that supported by Brd-1.3, but no other eye enhancer activity was observed for Brd-B or Brd-C (data not shown).
Distinct Ato/Da and Sc/Da consensus binding sites.
Hence, direct regulation of Brd by Ato in the leg and antennal disks may be mediated by Ato/Da binding to the Brd-E3 site. This site is conserved in Drosophila pseudoobscura. The sequence of the Brd-E3 site differs from the established Sc/Da binding consensus sequence in a core base and also in the 5′ base flanking the E-box core (ACATGTGT versus GCAGSTGK) (Fig. 1J). Interestingly, the 5′ flanking base also differs in the two other confirmed Ato/Da binding sites. An ato autoregulatory enhancer (known as the ato recruitment enhancer, ato-RE [zur Lage et al., submitted]) contains an Ato/Da binding site (ato-E1) in which the 5′ base is also A, although the core sequence conforms to the Sc/Da consensus (ACAGGTGG) (Fig. 1J). Similarly, Ato/Da regulates the TAKR86C gene via an upstream E box (TAKR86C-E2) (40), in which, however, the 5′ G is replaced by a T (TCAGGTGT). These deviations from the previously deduced proneural binding consensus sequence suggest that Ato/Da regulates at least some specific downstream targets by interacting with a distinct consensus sequence. Most notably, Ato/Da binding sites differ from Sc/Da binding sites in the 5′ flanking base, where an invariant G is replaced by an A or T. Comparison of all three identified Ato/Da binding sites yields a provisional Ato/Da binding consensus sequence of AWCAKGTGK (where W is A or T and K is G or T), compared with the Sc/Da binding consensus of GCAGSTGK (Fig. 1J). We refer to these sequences as variant E boxes, with matches to the former referred to as EAto sites and matches to the former to the latter referred to as ESc sites.
No selective recognition of proneural E boxes by Sc/Da and Ato/Da in vitro.
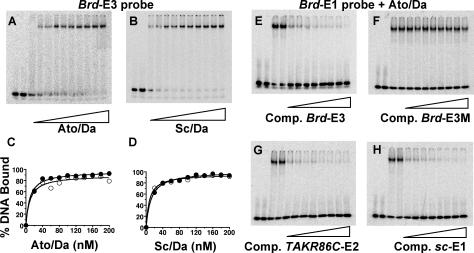
Differences in E-box binding sites between major structural and functional families of bHLH protein (e.g., Twist and Myc) have been characterized. In these cases, differential affinity to E-box variants can be observed in binding assays in vitro (8, 44), pointing to gross structural differences in DNA-protein recognition across families. No detailed comparative analysis of in vitro binding site affinities has been carried out for proteins within the proneural family. To compare the affinities of Ato/Da and Sc/Da to ESc and EAto sites (Brd-E1 and Brd-E3), gel retardation experiments were performed under conditions of limiting DNA target probe to distinguish small differences in affinity (11). In these experiments both protein dimers reproducibly bound to each site with indistinguishable affinities (apparent Kd of ca. 10 nM for each) (Fig. 2A to D). High-salt conditions to minimize nonspecific binding (31) produced results identical to those under low-salt conditions.
FIG. 2.
In vitro DNA binding analysis by gel retardation. (A to D) Relative DNA binding affinities for Brd-E1 and Brd-E3 were assessed by titration. (A) Representative titration gel for Brd-E3 with Ato/Da. (B) Representative titration gel for Brd-E3 with Sc/Da. (C) Binding curves of 0.1 nM Brd-E1 DNA probe (open circles) and 0.1 nM Brd-E3 probe (closed circles) with 0 to 200 nM Ato/Da (in 20 nM increments) in high-salt buffer. (D) Similar plot for Sc/Da. (E to H) Competition gel retardation assays with Ato/Da binding to radiolabeled 0.1 nM Brd-E1 (in high-salt buffer) competed with 0, 5, 10, 15, 20, 30, 40, 50, and 100 nM cold competitor DNA (50 to 1,000-fold competition). In each case, lanes 1 and 2 include no protein, lanes 3 and 4 include 150 nM Ato/Da and no competitor, and lanes 5 to 12 include 150 nM Ato/Da and increasing amounts of competitor DNA. Competitors used: (E) Brd-E3, (F) Brd-E3 M, (G) TAKR86C-E2, and (H) sc-E1.
Competition gel retardation experiments were used to compare binding by 36-bp duplexes containing Brd-E1, Brd-E3, ato-E1, and TAKR86C-E2 sites. We also used a well-defined ESc site from the sc autoregulatory enhancer sc-SMC-E (sc-E1, GCAGGTGT) (13). Ato/Da heterodimer binding to labeled Brd-E1 oligonucleotide was competed equally well by addition of cold competitor duplex for Brd-E1, Brd-E3, ato-E1, TAKR86C-E2, and sc-E1 (Fig. 2E, G, and H and data not shown). Duplexes with mutated E boxes (Brd-E3 M and Brd-E1 M) did not compete significantly even at a 1,000-fold excess (Fig. 2F and data not shown). Similar results were observed for competition experiments with Sc/Da heterodimer (data not shown). Thus, we could detect no difference in the in vitro binding affinities of Ato/Da and Sc/Da.
Strong selective discrimination of proneural E boxes in vivo.
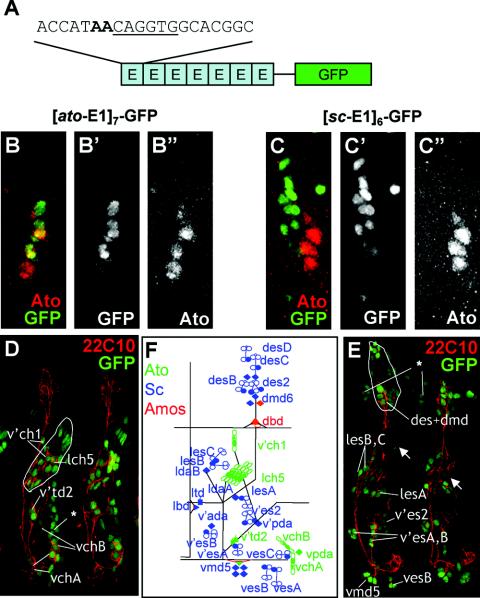
In the absence of in vitro indications for the importance of E-box sequence differences, we carried out an in vivo functional analysis. To test the contributions that the E-box variants make to enhancer specificity, we asked whether these sites could confer specific patterns of expression (and thus be differentially recognized) out of the context of their enhancers, thereby removing the contribution of other DNA binding factors. Inspired by previous findings (13), we made constructs with the GFP gene driven by artificial enhancers consisting of six to seven tandem repeats of a 20-bp sequence that includes a proneural binding site (6-bp E-box core and 7 bp of 5′ and 3′ flanking sequence) (Fig. 3A). Such artificial enhancers were constructed for an ESc site (sc-E1) and EAto sites (ato-E1 and TAKR86C-E2) to give [sc-E1]6-GFP, [ato-E1]7-GFP, and [TAKR86C-E2]6-GFP. Alignment of the 20-bp sequences used in these constructs revealed no shared motifs apart from the E boxes and their immediate flanks. Remarkably, these constructs support specific and different GFP patterns in transformed flies (Table 1).
FIG. 3.
[sc-E1]6-GFP and [ato-E1]7-GFP artificial enhancers support Sc- and Ato-dependent expression, respectively, in embryos. (A) Schematic of E-box multimer constructs (ato-E1 in this example). (B and C) Comparison of abdominal segments from stage 11 embryos (GFP, green; Ato, red). (B) [ato-E1]7-GFP, showing clear GFP/Ato overlap. (C) [sc-E1]6-GFP, showing no GFP overlap with Ato. (D and E) High-power view of two embryonic stage 16 abdominal segments (GFP, green; differentiated sensory neurons [22C10], red). (D) [ato-E1]7-GFP, showing GFP perduring in Ato-dependent sensory neurons (labeled). (E) [sc-E1]6-GFP late, showing GFP perduring in Sc-dependent sensory neurons (labeled). “Ectopic” ectodermal expression is also indicated (asterisk). (F) Schematic of abdominal sensory neurons with Sc-, Ato-, and Amos-dependent neurons in blue, green, and red, respectively.
TABLE 1.
Summary of expression patterns of E-box multimer constructs
| E box | Expression. in:
|
||||
|---|---|---|---|---|---|
| Wing | Leg | Eye | Antenna | Embryo | |
| sc-E1 | +++ (Sc) | +++ (Sc) | − | +++ (Sc) | +++ (Sc) |
| ato-E1 | + (Ato) | ++ (Ato) | + (Ato) | ++ (Ato) | +++ (Ato) |
| TAKR86C-E2. | − | − | +++ (Ato) | − | ++ (Ato; Bolwig's organ only) |
Sc and Ato, expression is broadly observed in Sc- and Ato-dependent cells, respectively.
Also expressed in embryonic muscle attachment sites.
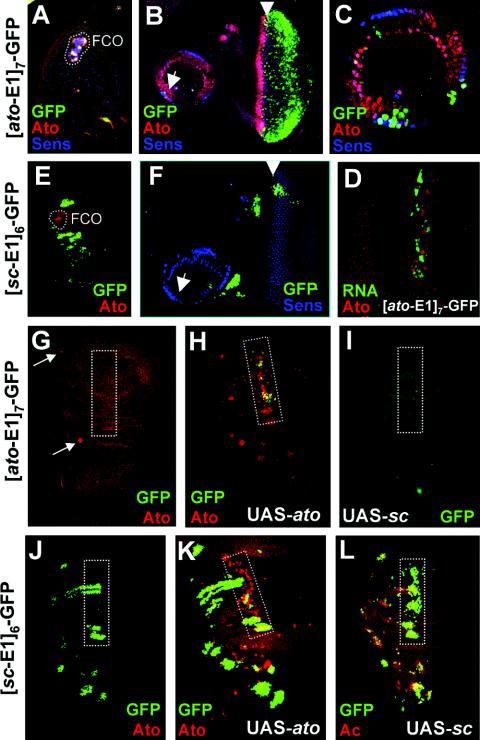
In embryos, [ato-E1]7-GFP is a very specific Ato-responsive reporter. The embryonic expression of [ato-E1]7-GFP closely followed that of Ato itself (Fig. 3B). GFP perdurability allowed these cells to be identified in late embryos by double labeling with 22C10, an antibody that detects all differentiated sensory neurons. Thus, these GFP-expressing cells differentiate as known Ato-dependent sensory neurons (25), including all chordotonal neurons, tracheal dendritic neurons, and cells of Bolwig's organ (Fig. 3D and F). Interestingly, GFP expression is generally observed in Ato-dependent SOPs but not proneural cluster cells, suggesting that there is some limitation on regulation by Ato within the latter cells. Expression was also Ato specific in imaginal disks, but generally much weaker. GFP was specifically observed in Ato-expressing femoral chordotonal organ (FCO) and Johnston's organ areas of the leg and antennal disks, although this was weak and variable (Fig. 4A to C). Moreover, expression was absent from Ato-expressing cells of the wing (Fig. 4G). Strong expression was observed in the eye (Fig. 4B). GFP protein expression in the eye does not overlap with Ato, but we could detect a close association between Ato protein and GFP mRNA in the morphogenetic furrow (Fig. 4D). This is consistent with other evidence showing that GFP fluorescence is delayed in the eye (28), and it suggests that Ato might regulate [ato-E1]7-GFP in the eye in addition to the leg and antennal disks. Consistent with this, the [ato-E1]7-GFP construct responded to UAS-ato misexpression but not to UAS-sc (Fig. 4G to I). In the wing, ectopic GFP was seen in response to the Ato misexpression, although it is apparent that GFP is not expressed in all of the sites where Ato is misexpressed (Fig. 4H). As before, GFP expression is most apparent in SOPs along the third wing vein, which is the area most responsive to Ato misexpression (24). In summary, the ato-E1 site supports expression in a pattern that suggests that it is utilized in vivo by Ato but not by Sc. Remarkably, it is not utilized by the Ato-related transcription factor Amos or Cato either, since GFP is not expressed in the patterns of these factors (20, 21).
FIG. 4.
[ato-E1]7-GFP and [sc-E1]6-GFP multimers support Ato- and Sc-dependent expression, respectively, in imaginal disks. Disks stained for GFP (green), Ato (red), Ac (red in G), and Sens (blue) as indicated. (A to D) [ato-E1]7-GFP supports weak and patchy expression in Ato-dependent areas. (A) Leg disk, showing some expression in FCO. (B) Eye-antennal disk, showing expression behind the morphogenetic furrow and in some Johnston's organ precursors (arrows). (C) Higher magnification of expression in Johnston's organ precursors. (D) High-magnification view of morphogenetic furrow, showing cytoplasmic GFP mRNA (green) in close association with nuclear Ato (red). (E and F) [sc-E1]6-GFP. (E) Leg disk, showing expression in bristle-producing areas (Sc dependent) but not in the FCO (Ato, red). (F) Eye-antennal disk, showing expression in bristle proneural clusters but generally not behind the morphogenetic furrow or in Johnston's organ. (G to I) Misexpression of Ato but not Sc causes ectopic expression of [ato-E1]7-GFP. The third wing vein is boxed. (G) Wild-type wing disk. GFP is not normally expressed, even in the two locations of wild-type Ato expression (arrows). (H) 109-68-Gal4/UAS-ato. Misexpression of Ato (red) causes GFP misexpression, but this is particularly confined to the third wing vein. (I) 109-68-Gal4/UAS-sc wing disk, showing no GFP misexpression in response to Sc misexpression. (J to L) Misexpression of Sc but not Ato causes ectopic expression of [sc-E1]6-GFP. (J) Wild-type wing disk, showing GFP in Sc-dependent proneural clusters. The third wing vein is boxed. (K) 109-68-Gal4,UAS-ato, showing little extra GFP expression in the third wing vein. (L) 109-68-Gal4,UAS-sc, showing GFP misexpression particularly along the third wing vein.
[sc-E1]6-GFP exhibits an expression pattern dramatically different from that of [ato-E1]7-GFP (Fig. 3 and 4). Indeed, there was no indication of overlap in the expression of [sc-E1]6-GFP and [ato-E1]7-GFP in any tissue. In the embryo, GFP expression was confined to a subset of SOPs and their progeny (Fig. 3C and data not shown). These SOPs do not express Ato (Fig. 3C). Since the majority of embryonic SOPs require and express either Sc or Ato (25) (Fig. 3F), this GFP pattern can be deduced to correspond to SOPs that express and require Sc function. Moreover, GFP perdurability allowed these cells to be identified in late embryos (Fig. 3E). This revealed that [sc-E1]6-GFP was activated predominantly in known Sc-dependent sensory neurons (14). Little expression was apparent in Ato- and Amos-dependent neurons, and any expression present appeared late in embryogenesis, suggesting that it was not a response to these proneural proteins. [sc-E1]6-GFP is also strongly Sc specific in imaginal disks. GFP expression that corresponds to the pattern of Sc expression was seen (Fig. 4E, F, and J). Interestingly, unlike in the embryo, expression is present in proneural cluster cells as well as SOPs (see also reference 13). However, no GFP expression was observed in ato-dependent chordotonal SOPs in the leg and antennal disks or during ato-dependent R8 photoreceptor commitment in the eye disk (Fig. 4E and F). Misexpression of Sc causes ectopic activation of [sc-E1]6-GFP (Fig. 4J and L), but misexpression of Ato does not (Fig. 4K). This provides strong evidence that sc-E1 is specifically recognized by Sc/Da during PNS neurogenesis in vivo. In summary, the E-box expression patterns strongly suggest high in vivo specificity of these ESc and EAto sites for Sc/Da and Ato/Da, respectively.
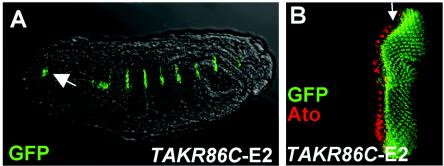
Surprisingly, the EAto site TAKR86E-E2 differed from the ato-E1 site in its ability to drive GFP expression. Strong [TAKR86E-E2]6-GFP expression was observed in the eye disk posterior to the morphogenetic furrow and in Bolwig's organ in the embryo (Fig. 5). No expression was observed in other neural cells, including chordotonal precursors, although nonproneural expression was seen in late embryogenesis in ectodermal stripes. The neural pattern supported by this E box is therefore only a subset of that supported by ato-E1. To some degree ectodermal stripes were also seen for the three other constructs (Fig. 3). This expression is reminiscent of the expression of the Ato superfamily member Delilah in muscle attachment cells and so may represent regulation by that bHLH protein. In summary, there are in vivo differences not only between Sc and Ato binding sites but also between different Ato sites.
FIG. 5.
[TAKR86C-E2]6-GFP multimer supports limited GFP expression (green). (A) Late embryo, showing GFP in epidermal stripes and in Bolwig's organ (arrow). (B) Eye disk, showing expression posterior to the morphogenetic furrow.
Altering the bases flanking the proneural E boxes disrupts enhancer function.
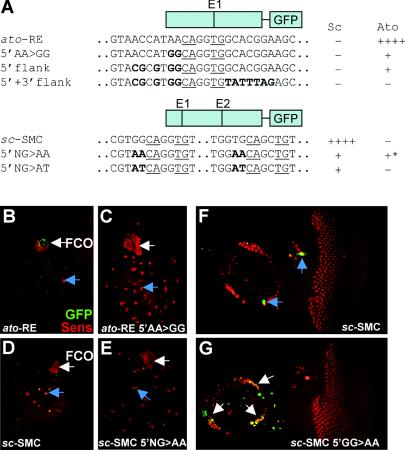
To explore further the importance of the Sc- and Ato-specific sequences, we attempted to exchange proneural binding site specificities within the context of the sc and ato autoregulatory enhancers (sc-SMC and ato-RE enhancers) (13; zur Lage et al., submitted)). The 367-bp ato-RE enhancer supports expression in the chordotonal SOPs of the leg disk (Fig. 6B), which depends on Ato/Da binding to the ato-E1 E box (zur Lage et al., submitted). Within the ato-RE enhancer, the flank of the ato-E1 E box was mutated from AACAGGTG to GGCAGGTG, thereby creating a site resembling sc-E1 (Fig. 6A, 5′AA>GG). This small change resulted in a strong reduction in Ato-dependent GFP expression (Fig. 6B and C), suggesting that the flanking AA is important for regulation by Ato. However, there was no gain of expression in an Sc-dependent pattern, and so changing the site to resemble sc-E1 did not result in regulation by Sc. A more extensive conversion of the ato-E1 5′ flanking sequence gave the same result (Fig. 6A, 5′ flank). Conversion of the entire 20 bp surrounding ato-E1 to match sc-E1 resulted in a complete loss of enhancer activity (Fig. 6A, 5′+3′flank, and data not shown). Part of the reason for this is likely to be loss of an essential ETS protein binding site adjacent to the ato-E1 site (zur Lage et al., submitted). It is striking that there is no gain of Sc-dependent expression even though the converted E-box region is now identical to the 20-bp sequence that is sufficient for Sc-dependent regulation when multimerized. Thus, this ESc site cannot function in the context of the ato-RE.
FIG. 6.
Alteration of E-box flanking nucleotides disrupts enhancer activity. (A) Schematic of the ato-RE and sc-SMC enhancer constructs, with E boxes indicated, the regions of the sequence alterations shown, and an indication of expression pattern supported. (B to I) GFP expression (green) relative to SOPs (Sens, red), in Sc-dependent cells (examples indicated with blue arrows) or Ato-dependent cells (white arrows). (B to E) Leg disks scanned using identical low laser power settings under nonsaturating conditions to show reduction of expression for the mutated enhancer relative to the wild-type enhancer. (E and F) Scanned at low laser power to show loss of expression in mutated version. (B) ato-RE-GFP in FCO precursors of leg disk. (C) ato-RE 5′AA>GG in leg disks, showing strong reduction of FCO expression. (D) sc-SMC in Sc-dependent leg bristle SOPs. (E) 5′NG>AA, showing strong reduction of bristle SOP expression. (E and F) Eye-antennal expression for sc-SMC and sc-SMC 5′NG>AA, respectively. (E) The wild type is expressed in some bristle SOPs. (F) Expression is lost from bristle SOPs in the mutated version but is gained in Ato-dependent Johnston's organ SOPs.
The 356-bp sc-SMC enhancer supports expression in Sc-dependent SOPs, which requires the function of the Sc/Da binding E-box, sc-E1, and to a lesser extent sc-E2 (13). Mutation of 2 bp flanking both sc-E1 and sc-E2 to convert them to match the flank of ato-E1 (Fig. 6A, 5′NG>AA) resulted in a substantial loss of expression in Sc-dependent SOPs, indicating a lowered ability of Sc/Da to recognize the altered sites (Fig. 6D and E and data not shown). There was no concomitant appearance of Ato-dependent expression in the leg, wing, and eye disks. In the antennal disk, however, this alteration promoted ectopic sc-SMC-GFP expression in Ato-dependent chordotonal SOPs of Johnston's organ (Fig. 6F and G). This suggests that in the context of the antenna, Ato is now able to recognize the altered E-box sites in the sc-SMC autoregulatory enhancer. At least in one context, therefore, alteration of just 2 bp flanking the E-box core is sufficient to change specificity. Interestingly, a different result is obtained when the same flanking residues are changed to match the TAKR86C-E2 site (Fig. 6A, 5′NG>AT). In this case, there was loss of Sc-dependent expression as before but no gain of Ato-dependent expression anywhere (data not shown). Overall these results suggest that the correct proneural binding site is necessary but usually not sufficient for proneural regulation of target genes. It seems that proneural proteins usually must also interact with subtype-specific DNA binding factors for correct enhancer function.
DISCUSSION
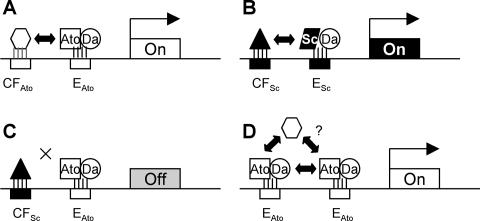
We have shown that Ato and Sc must interact with distinct DNA binding sites in vivo. In the target genes analyzed here, residues immediately flanking the 6-bp core E box allow us to define distinct EAto and ESc consensus binding sites for Ato/Da and Sc/Da, respectively. It can be deduced that these variant E boxes consist of half sites, with Sc, Ato, and Da contacting GCAG, AWCAK, and STGK, respectively. Striking affirmation of binding site differences is provided by the common target gene Brd, which is regulated by Ato and Sc in a modular fashion via distinct E boxes in different enhancers. These E-box variations are crucial for function. They are sufficient to confer proneural protein-specific expression patterns on a GFP reporter gene in isolation from other enhancer sequences. Moreover, interchanging ESc and EAto sites within proneural enhancers almost abolishes enhancer activity and so is almost equivalent to destroying the E box. This shows that the correct proneural regulation of target genes requires the presence of a specific E-box binding site in combination with the selective ability to interact with factors bound to other sites within these enhancers (Fig. 7A to C).
FIG. 7.
Schematic view of the importance of E-box variants and cofactor interactions for proneural target gene specificity. (A-C) Proneural target gene regulation requires Ato/Da or Sc/Da binding to an appropriate E-box variant (EAto or ESc) in conjunction with appropriate protein-protein interaction with cofactors (hypothetically CFAto or CFSc). (D) As suggested in the text, the need for adjacent cofactor binding sites may be overcome by tandem repeats of the E boxes. Interaction between proneural proteins may stabilize DNA binding and allow the recruitment of cofactors by protein interaction.
We show that in vivo DNA binding site differences underlie proneural specificity. Yet, paradoxically, evidence from misexpression and protein structure-function studies has strongly suggested that the target gene specificities of Ato and Sc (and their vertebrate homologues) result from specific interactions with protein cofactors and not from intrinsic differences in how proneural proteins contact DNA (12, 35). For instance, DNA-contacting residues are shared between the proteins (12), and consequently, as we show here, Ato/Da and Sc/Da have identical DNA binding properties in vitro. One way to reconcile these observations is that protein-protein interactions with cofactors may induce DNA binding specificity in proneural proteins by modifying the conformation of the DNA-contacting residues of the bHLH domain. It is also possible that DNA binding itself is not selective in vivo but that occupancy of different DNA binding sites triggers productive or unproductive conformational changes in the proneural proteins that influence their interaction with cofactor proteins. Our favored view is that specific cofactor interactions will induce distinctive DNA binding affinities; the conformational changes induced by each may be subtle individually but may be interdependent and mutually reinforcing.
Significantly, ato can rescue mutations of its mouse orthologue, Math1 (46), and vice versa (3), suggesting that DNA site preferences will be conserved among vertebrate orthologues. A number of functional E boxes have been characterized in vertebrate neural-specific genes, and in two cases the interacting bHLH protein is likely to be an Ato orthologue: an autoregulatory site in the Math1 promoter (TCAGCTGG) (23) and a proposed Xath5 site in the β3 nAChr promoter (ACAGCTGG) (41). Thus, in these cases the E boxes match EAto in the 5′ flanking base.
For correct enhancer function, proneural proteins must interact differentially with other DNA binding factors.
Although E-box consensus differences underlie specificity, enhancer context is usually also crucial for this specificity to be manifest. In swapping EAto and ESc sequences between the sc and ato autoregulatory enhancers, in only one case did we observe a corresponding “swap” in enhancer specificity: Ato could be made to regulate the sc-SMC-E enhancer via an EAto site. Otherwise, alteration of E-box flanking bases resulted in a severe loss of enhancer activity. This suggests that recruiting a different proneural protein cannot alone change the function of an enhancer. Correct proneural target enhancer function requires a combination of the correct E-box sequence and the ability to interact with other factors bound to the enhancer. This is reminiscent of the cooperative interaction between MyoD and MEF2 in myogenesis (34) and of interaction between Sc/Da and Pannier/Chip to activate ac in a specific part of the thorax (38). For the Ato enhancer we have recently shown the requirement for cooperative interaction between Ato/Da and the ETS protein Pointed, bound to a site adjacent to the EAto site (zur Lage et al., submitted). Similarly, neurogenin 2 interacts with LIM factors during the activation of subtype-specific target genes (33). Our finding that EAto and ESc sites encode much specificity in artificial enhancers suggests that tandem E boxes remove the requirement for interaction with factors bound to other DNA sites, perhaps because cooperative binding between proneural proteins themselves is then sufficient (4) and may even allow the recruitment of cofactors by protein interactions alone (Fig. 7D). Interestingly, the converse situation may also occur: for both the ato and sc enhancers, there is a low level of expression remaining after swapping of E boxes. This suggests that the original bHLH protein can be recruited to the “wrong” E-box sequence, inefficiently, by interaction with cofactors. A basis for this can be found with MyoD, where interaction with Sp1 allows MyoD to bind to a nonideal site in the human cardiac α-actin promoter (6).
Another indication of the importance of enhancer context is that parent enhancers support patterns different from those of the isolated E boxes, at least in the case of Ato. [ato-E1]7-GFP is widely expressed in Ato-specific regions in the embryo, whereas the parent ato-FCO-E enhancer is limited to a small subset of chordotonal SOPs (zur Lage et al., submitted). In disks, ato-E1 drives expression relatively poorly in FCO precursors compared with ato-FCO-E. TAKR86C is even more extreme: the TAKR86C enhancer is normally active only in a single embryonic chordotonal precursor (the P cell) (40), but the TAKR86C-E2 site drives Ato-dependent expression in the larval and adult eye and not in the P cell. Clearly, the parent enhancers must have other regulatory inputs that restrict expression.
Proneural selective E-box binding sites.
Despite the importance of enhancer context and interaction with other factors, the ESc and EAto sequences support strikingly specific expression patterns when taken out of their enhancers. First, all tandem repeat E-box constructs tested are activated almost exclusively during PNS neurogenesis, despite the presence of some 24 class A factors in Drosophila (32). None are activated during CNS neurogenesis, myogenesis, or mesoderm formation, even though AS-C proteins function during the former two processes. In the case of two sites, sc-E1 and ato-E1, expression is remarkably consistent, with regulation solely by Ato or Sc, respectively, in PNS neurogenesis; the sites alone must contain all of the information necessary for specific recognition. It is remarkable that ato-E1 does not respond in vivo to the Ato-related protein Cato or Amos, even though the latter has a basic region almost identical to that of Ato and might be expected to have the same DNA binding properties (20). The main exception to this specificity is the presence of expression in embryonic ectodermal stripes. These resemble muscle attachment sites, suggesting recognition by the Ato superfamily member Delilah (1, 32). The conclusion is that tandem duplications can overcome the need for DNA binding sites for other factors. Cooperative binding of proneural proteins may negate the need for cofactor interactions, or, as suggested above, cooperative binding may allow the recruitment of cofactors directly (Fig. 7).
There are dramatic differences between the two Ato sites tested. Unlike ato-E1, the TAKR86C-E2 site drives expression in only a subset of Ato locations; it appears to be photoreceptor specific despite containing a good class A core E-box match (CAGGTG). This opens up the possibility that there may be different subtypes of Ato binding sites. The spatially restricted recognition of TAKR86C-E2 also implies that cellular context is important in how different sites are recognized. One may speculate, for instance, that eye-specific DNA binding properties of Ato may be conferred by interaction with PAX6 proteins (36). Interestingly, diversity of E-box expression patterns correlates with variability in the consensus sequences. The ESc consensus sequence (based on some 23 sites) is less variable than the Ato/Da consensus, even though the latter is based on only three sites. We suggest that regulatory fine-tuning by E-box variation is more important for Ato target genes than for Sc target genes.
In summary, the E-box sequences and their flanking bases contain impressively sufficient information for regulation by specific proneural proteins. However, there is further complexity: at least the two Ato sites tested support different patterns and have a different relationship with their parent enhancers. Subtle variations in regulation by proneural proteins may therefore contribute to variations in target gene expression; indeed, there may be no such thing as a typical target site or target gene. This may also be true for common target genes: despite the modular regulation of Brd, we do not rule out the possibility that within the spectrum of proneural E boxes there are some sites that are jointly recognized by Sc and Ato in vivo and that this would be another mechanism for regulating common target genes.
Acknowledgments
We are indebted to James Posakony (University of California at San Diego) for providing the original Brd-lacZ stocks and the pHStinger vector and to Bassem Hassan for unpublished information about the ato upstream region. We thank Tom Carter for his excellent technical help with the multimer experiments.
A.P.J. is a Senior Fellow of The Wellcome Trust (grant 042182).
REFERENCES
- 1.Armand, P., A. C. Knapp, A. J. Hirsch, E. F. Wieschaus, and M. D. Cole. 1994. A novel basic helix-loop-helix protein is expressed in muscle attachment sites of the Drosophila epidermis. Mol. Cell. Biol. 14:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barolo, S., L. A. Carver, and J. W. Posakony. 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29:726-732. [DOI] [PubMed] [Google Scholar]
- 3.Ben Arie, N., B. A. Hassan, N. A. Bermingham, D. M. Malicki, D. Armstrong, M. Matzuk, H. J. Bellen, and H. Y. Zoghbi. 2000. Functional conservation of atonal and Math1 in the CNS and PNS. Development 127:1039-1048. [DOI] [PubMed] [Google Scholar]
- 4.Bengal, E., O. Flores, P. N. Rangarajan, A. Chen, H. Weintraub, and I. M. Verma. 1994. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proc. Natl. Acad. Sci. USA 91:6221-6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand, N., D. S. Castro, and F. Guillemot. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurol. 3:517-530. [DOI] [PubMed] [Google Scholar]
- 6.Biesiada, E., Y. Hamamori, L. Kedes, and V. Sartorelli. 1999. Myogenic basic helix-loop-helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac alpha-actin promoter. Mol. Cell. Biol. 19:2577-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwell, T. K., L. Kretzner, E. M. Blackwood, R. N. Eisenman, and H. Weintraub. 1990. Sequence-specific DNA binding by the c-Myc protein. Science 250:1149-1151. [DOI] [PubMed] [Google Scholar]
- 8.Blackwell, T. K., and H. Weintraub. 1990. Differences and similarities in DNA-binding preferences of myoD and E2A protein complexes revealed by binding site selection. Science 250:1104-1110. [DOI] [PubMed] [Google Scholar]
- 9.Cabrera, C. V., and M. C. Alonso. 1991. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products in Drosophila. EMBO J. 10:965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campuzano, S., and J. Modolell. 1992. Patterning of the Drosophila nervous system: the achaete-scute gene complex. Trends Genet. 8:202-208. [DOI] [PubMed] [Google Scholar]
- 11.Carey, J. 1991. Gel retardation. Methods Enzymol. 208:103-117. [DOI] [PubMed] [Google Scholar]
- 12.Chien, C.-T., C.-D. Hsiao, L. Y. Jan, and Y. N. Jan. 1996. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc. Natl. Acad. Sci. USA 93:13239-13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culí, J., and J. Modolell. 1998. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signalling. Genes Dev. 12:2036-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dambly-Chaudière, C., and A. Ghysen. 1987. Independent subpatterns of sense organs require independent genes of the achaete-scute complex in Drosophila larvae. Genes Dev. 1:297-306. [Google Scholar]
- 15.Ellenberger, T., D. Fass, M. Arnaud, and S. C. Harrison. 1994. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8:970-980. [DOI] [PubMed] [Google Scholar]
- 16.Ferre D'Amare, A. R., G. C. Prendergast, E. B. Ziff, and S. K. Burley. 1993. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363:38-45. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, F., D. H. Crouch, P. Jayaraman, W. Clark, D. A. F. Gillespie, and C. R. Goding. 1993. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 12:5075-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher, F., and C. R. Goding. 1992. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the CANNTG motif. EMBO J. 11:4103-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghysen, A., C. Dambly-Chaudière, L. Y. Jan, and Y. N. Jan. 1993. Cell interactions and gene interactions in peripheral neurogenesis. Genes Dev. 7:723-733. [DOI] [PubMed] [Google Scholar]
- 20.Goulding, S. E., P. zur Lage, and A. P. Jarman. 2000. amos, a proneural gene required for olfactory sense organs that is regulated by lozenge. Neuron 25:69-78. [DOI] [PubMed] [Google Scholar]
- 21.Goulding, S. E., N. M. White, and A. P. Jarman. 2000. cato encodes a basic-helix-loop-helix transcription factor implicated in the correct differentiation of Drosophila sense organs. Dev. Biol. 221:120-131. [DOI] [PubMed] [Google Scholar]
- 22.Halazonetis, T. D., and A. N. Kandil. 1991. Determination of the c-MYC DNA-binding site. Proc. Natl. Acad. Sci. USA 88:6162-6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helms, A. W., A. L. Abney, N. Ben Arie, H. Y. Zoghbi, and J. E. Johnson. 2000. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127:1185-1196. [DOI] [PubMed] [Google Scholar]
- 24.Jarman, A. P., and I. Ahmed. 1998. The specificity of proneural genes in determining Drosophila sense organ identity. Mech. Dev. 76:117-125. [DOI] [PubMed] [Google Scholar]
- 25.Jarman, A. P., Y. Grau, L. Y. Jan, and Y. N. Jan. 1993. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73:1307-1321. [DOI] [PubMed] [Google Scholar]
- 26.Jennings, B. H., D. M. Tyler, and S. J. Bray. 1999. Target specificities of Drosophila Enhancer of split basic helix-loop-helix proteins. Mol. Cell. Biol. 19:4600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kophengnavong, T., J. E. Michnowicz, and T. K. Blackwell. 2000. Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations. Mol. Cell. Biol. 20:261-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar, J. P., F. Hsiung, M. A. Powers, and K. Moses. 2003. Nuclear translocation of activated MAP kinase is developmentally regulated in the developing Drosophila eye. Development 130:3703-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunisch, M., M. Haenlin, and J. A. Campos Ortega. 1994. Lateral inhibition mediated by the Drosophila neurogenic gene Delta is enhanced by proneural proteins. Proc. Natl. Acad. Sci. USA 91:10139-10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai, E. C., R. Bodner, J. Kavaler, G. Freschi, and J. W. Posakony. 2000. Antagonism of notch signaling activity by members of a novel protein family encoded by the bearded and enhancer of split gene complexes. Development 127:291-306. [DOI] [PubMed] [Google Scholar]
- 31.Lane, D., P. Prentkl, and M. Chandler. 1992. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol. Rev. 56:509-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ledent, V., and M. Vervoort. 2001. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11:754-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, S.-K., and S. L. Pfaff. 2003. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38:731-745. [DOI] [PubMed] [Google Scholar]
- 34.Molkentin, J. D., and E. N. Olson. 1996. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 93:9366-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakada, Y., T. L. Hunasker, M. Hencke, and J. E. Johnson. 2004. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development 131:1319-1330. [DOI] [PubMed] [Google Scholar]
- 36.Niwa, N., Y. Hiromi, and M. Okabe. 2004. A conserved developmental program for sensory organ formation in Drosophila melanogaster. Nat. Genet. 36:293-297. [DOI] [PubMed] [Google Scholar]
- 37.Nolo, R., L. A. Abbott, and H. J. Bellen. 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102:349-362. [DOI] [PubMed] [Google Scholar]
- 38.Ramain, P., R. Khechumian, K. Khechumian, N. Arbogast, C. Ackermann, and P. Heitzler. 2000. Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol. Cell 6:781-790. [DOI] [PubMed] [Google Scholar]
- 39.Revzin, A. 1989. Gel electrophoresis assays for DNA-protein interactions. BioTechniques 7:346-355. [PubMed] [Google Scholar]
- 40.Rosay, P., J. F. Colas, and L. Maroteaux. 1995. Dual organisation of the Drosophila neuropeptide receptor NKD gene promoter. Mech. Dev. 51:329-339. [DOI] [PubMed] [Google Scholar]
- 41.Roztocil, T., L. Matter-Sadzinski, M. Gomez, M. Ballivet, and J. M. Matter. 1998. Functional properties of the neuronal nicotinic acetylcholine receptor beta3 promoter in the developing central nervous system. J. Biol. Chem. 273:15131-15137. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu, T., A. Toumoto, K. Ihara, M. Shimizu, Y. Kyogoku, N. Ogawa, Y. Oshima, and T. Hakoshima. 1997. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 16:4689-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singson, A., M. W. Leviten, A. G. Bang, X. H. Hua, and J. W. Posakony. 1994. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 8:2058-2071. [DOI] [PubMed] [Google Scholar]
- 44.Van Antwerp, M. E., D. G. Chen, C. Chang, and E. V. Prochownik. 1992. A point mutation in the MyoD basic domain imparts c-Myc-like properties. Proc. Natl. Acad. Sci. USA 89:9010-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Doren, M., P. A. Powell, D. Pasternak, A. Singson, and J. W. Posakony. 1992. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaete. Genes Dev. 6:2592-2605. [DOI] [PubMed] [Google Scholar]
- 46.Wang, V. Y., B. A. Hassan, H. J. Bellen, and H. Y. Zoghbi. 2002. Drosophila atonal fully rescues the phenotype of Math1 null mice: new functions evolve in new cellular contexts. Curr. Biol. 12:1611-1616. [DOI] [PubMed] [Google Scholar]
- 47.zur Lage, P. I., L. M. Powell, D. R. A. Prentice, and A. P. Jarman. EGF receptor signalling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev. Cell, in press. [DOI] [PubMed]