Abstract
The mammalian target of rapamycin (mTOR) is a key component of a signaling pathway which integrates inputs from nutrients and growth factors to regulate cell growth. Recent studies demonstrated that mice harboring an ethylnitrosourea-induced mutation in the gene encoding mTOR die at embryonic day 12.5 (E12.5). However, others have shown that the treatment of E4.5 blastocysts with rapamycin blocks trophoblast outgrowth, suggesting that the absence of mTOR should lead to embryonic lethality at an earlier stage. To resolve this discrepancy, we set out to disrupt the mTOR gene and analyze the outcome in both heterozygous and homozygous settings. Heterozygous mTOR (mTOR+/−) mice do not display any overt phenotype, although mouse embryonic fibroblasts derived from these mice show a 50% reduction in mTOR protein levels and phosphorylation of S6 kinase 1 T389, a site whose phosphorylation is directly mediated by mTOR. However, S6 phosphorylation, raptor levels, cell size, and cell cycle transit times are not diminished in these cells. In contrast to the situation in mTOR+/− mice, embryonic development of homozygous mTOR−/− mice appears to be arrested at E5.5; such embryos are severely runted and display an aberrant developmental phenotype. The ability of these embryos to implant corresponds to a limited level of trophoblast outgrowth in vitro, reflecting a maternal mRNA contribution, which has been shown to persist during preimplantation development. Moreover, mTOR−/− embryos display a lesion in inner cell mass proliferation, consistent with the inability to establish embryonic stem cells from mTOR−/− embryos.
The mammalian target of rapamycin (mTOR) is a 300-kDa protein kinase belonging to the phosphatidylinositol 3-kinase-related family of protein kinases (23), which is emerging as a key component of an ancient nutrient and energy effector pathway (35). In metazoans, this pathway has been integrated with the phosphatidylinositide 3OH-kinase pathway to control nutrient and energy homeostasis (21, 32). Yeast TOR was initially identified as the inhibitory target of the bacterial macrolide rapamycin, produced by the soil bacterium Streptomyces hygroscopicus (15). Rapamycin forms a gain-of-function inhibitory complex with the immunophilin FKBP12, which binds to mTOR and inhibits its ability to phosphorylate downstream substrates (1, 7). Earlier studies demonstrated that two of these substrates are the 40S ribosomal protein S6 kinase 1 (S6K1) (5, 6, 20, 44) and the eukaryotic initiation factor 4E binding protein (4E-BP1) (4, 11). More recent studies showed that mTOR requires two associated proteins to signal in vivo to S6K1 and 4E-BP1, raptor (13, 25, 29) and GβL (26). Raptor appears to act as a scaffold to allow mTOR to access its substrates, whereas GβL is required to make a competent signaling complex that can respond to nutrient inputs. In parallel, it was found that the tumor suppressor complex associated with the autosomal-dominant genetic disorder tuberous sclerosis complex (36), made up of TSC1 (hamartin) and TSC2 (tuberin), acts to suppress signaling in the mTOR pathway (9, 19, 22). These studies led to the finding that the inhibitory effects of TSC1/TSC2 are elicited through the GTPase-stimulating activity of TSC2, which acts to drive the Ras homologue enriched in brain (Rheb) (50) into the inactive GDP state (10, 45, 47, 52). It is thought that Rheb in the active GTP-bound state either acts directly on mTOR or influences mTOR signaling downstream to S6K1 and 4E-BP1.
Initial studies with Saccharomyces cerevisiae suggested an essential role for TOR in cell proliferation (31). However, S. cerevisiae, unlike higher eukaryotes, has two TOR genes, TOR1 and TOR2 (31). TOR1 itself is not essential, but it shares with TOR2 a common rapamycin-sensitive essential function involved in regulating nutrient-mediated cell growth (31). In addition, TOR2 has a nonshared, independent, rapamycin-resistant essential function which is implicated in the control of actin cytoskeleton dynamics (31). Genetic studies with Caenorhabditis elegans (30) and Drosophila (41, 51) have shown that cTOR and dTOR, respectively, also play an essential role in cell growth and development in these organisms that is tightly linked to nutritional status. Recently, a search for ethylnitrosourea (ENU)-induced recessive mutations in the development of the mouse forebrain identified a mutation due to altered mTOR mRNA splicing (16). mRNA expression in mutant embryos is reduced to approximately 50% of that observed in wild-type (WT) embryos, and the corresponding protein contains a three-amino-acid insertion (16). The modified protein was shown to have severely reduced kinase activity both in vitro and in vivo, and the resulting embryos failed to develop beyond embryonic day 9.5 (E9.5) and succumbed at E12.5 (16). Hentges et al. (16) argued that this phenotype was consistent with the role of mTOR in cell growth (8). Moreover, the fact that the mouse embryo has no G1 phase until after gastrulation led them to suggest that mTOR is not required in embryogenesis until this stage (16). In support of their argument, they found that injection of WT pregnant mice with a dose of rapamycin within the human therapeutic range phenocopied the mutant.
Despite these observations, earlier findings implied that mTOR function may be critical at a much earlier stage of mouse embryogenesis. It is known that amino acids, which effect mTOR function, are permissive in vitro for the outgrowth of trophoblasts (38), the first cells to differentiate during mammalian development. Trophoblasts give rise to the outer epithelial layer of the preimplantation blastocyst, further differentiating and becoming invasive during implantation at E4.5. Studies in vivo support a role for amino acids in regulating trophoblast differentiation, as embryos which exhibit diapause or delayed implantation exhibit low rates of amino acid uptake (48). Consistent with mTOR being an amino acid effector (14, 18, 49), it has been also demonstrated that the treatment of blastocysts with rapamycin in vitro inhibits trophectoderm outgrowth (33). Recently, we established a strategy for generating a conditional allele of mTOR in mice. This approach also allowed us to delete the gene and address the role of mTOR in early mouse development.
MATERIALS AND METHODS
Targeting vector and ES cells.
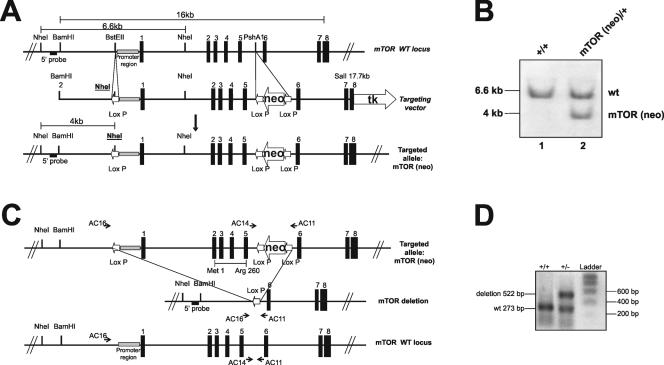
From a mouse genomic clone containing a 16.5-kb NotI insert of the 5′ mTOR sequence (129 SVJ Lambda FixII; Stratagene), a 16-kb BamHI-SalI fragment was obtained; this fragment contained the presumptive mTOR promoter region (based on the shared homology of the human and mouse genes), exons 1 to 7, and part of exon 8. Through successive subcloning steps, we introduced a loxP sequence into the BstEII site upstream of the promoter region, adding a novel NheI site used for allele genotyping. A neomycin resistance sequence (neo) driven by the phosphoglycerate kinase promoter and flanked by loxP sequences was inserted into the PshAI site of intron 6, and a thymidine kinase sequence driven by the herpes simplex virus promoter was added after the SalI site. The targeting vector was electroporated into 129SVJae embronic stem (ES) cells, and 240 G418-resistant clones were screened for homologous recombination by Southern blot analysis with a 5′ probe of 230 nucleotides from a sequence amplified from sense primer AA65 (5′-AGTAGATGAGAGCCAAGTGT-3′) and antisense primer AA17 (5′-AGTGGCTGTCCTGGCTGTGC-3′). Following NheI digestion of genomic DNA and hybridization with the 5′ probe, seven positive ES clones were obtained.
Generation of mTOR+/− mice.
Cells from an mTOR-targeted ES cell clone were injected into C57BL/6 blastocysts to produce chimeric male offspring. One chimera was backcrossed with a cytomegalovirus promoter-induced cre-transgenic mouse (46). Germ line transmission of the deleted allele was determined by PCR analysis with a sense primer located upstream of the first loxP site (primer AC16, 5′-TTCATTCCCTTGAAAGCCAGTCTCACC) and an antisense primer located in intron 5, downstream of the 3′ loxP site of the neo cassette (primer AC11, 5′-GCTCTTGAGGCAAATGCCACTATCACC). The WT allele was determined by PCR analysis with a sense primer located in the intron 5′ upstream of the 5′ loxP site in the neo cassette (primer AC14, 5′-TCATTACCTTCTCATCAGCCAGCAGTT) and primer AC11. For mouse genotyping, a multiplex PCR scheme with primers AC16, AC14, and AC11 was used. Cycling conditions were 94°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and extension at 72°C for 10 min.
Outgrowth and embryo genotyping.
Morulae and blastocysts were recovered at 2.5 and 3.5 days postcoitum, respectively, and were cultured individually in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal calf serum (FCS) on gelatinized culture plates at 37°C in 5% CO2. Embryos were recovered by scraping and genotyped by nested multiplex PCR with the following primer sets: first round—WT Forward, 5′-CCCAGCACTTGGGAATCAGACAG; mTOR− Forward, 5′-GCCCCACCCCCATAGCTTCTCTC; and WT/mTOR− Reverse, 5′-CAGGACTCAGGACACAACTAGCCC; and second round—WT Forward, 5′-GCTAGCAGTGCCCACATCATCC; mTOR− Forward, 5′-CCCGAGACAGCCTTGGCAGTTGG; and WT/mTOR− Reverse, 5′-CAGGACTCAGGACACAACTAGCCC. Single embryos transferred to PCR tubes with 0.5 to 1.5 μl of phosphate-buffered saline (PBS) were boiled for 5 min at 100°C in 12 μl of DNA-grade H2O and transferred to ice, and 3 μl of PCR master mix containing 10× buffer, primers, deoxynucleoside triphosphates, and Taq polymerase (GeneChoice, Inc., Frederick, Md.) was added. The first round of PCR was carried out at 94°C for 4 min followed by 15 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 45 s. The second round of PCR was carried out with 0.5 μl of the first-round reaction mixture as a template in a 25-μl total volume, with an initial step at 94°C followed by 27 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 45 s. Products were resolved by 1.5% agarose gel electrophoresis by standard procedures.
MEFs and ES cells.
Mouse embryonic fibroblasts (MEFs) were prepared from E13.5 embryos as previously described (3). For ES cells, embryos were flushed at the morula stage and incubated overnight in M16 medium (Sigma-Aldrich, St. Louis, Mo.). Blastocysts were transferred to DMEM containing 15% FCS, penicillin-streptomycin, glutamine, β-mercaptoethanol, gentamicin, and leukemia inhibitory factor in 6-cm dishes with 5 × 105 inactivated mouse fibroblasts and 5 × 105 inactivated 5637 cells (ATCC HTB9) to facilitate ES cell development (41). Two days later, the inner cell mass (ICM) was picked with a glass pipette, treated with trypsin for 10 min at room temperature, and further propagated on inactivated fibroblasts.
Protein extraction and Western blot analysis.
Protein extracts were prepared and Western blot analyses were carried out essentially as previously described (43). Primary antibodies were directed against phosphorylated ribosomal protein S6 (Ser240 and Ser244 [Ser240/244]; Cell Signaling), mTOR (Cell Signaling), and phosphorylated S6K1 (Thr389; Cell Signaling). Raptor antibodies were generated against an amino-terminal peptide sequence.
FACS with BrdU and 7-aminoactinomycin D labeling.
MEFs were grown for 2 days in 0.5% FCS and stimulated with 10% FCS; before being harvested at various times, MEFs were pulse-labeled with 10 mM bromodeoxyuridine (BrdU; Sigma) for 15 min, washed with PBS, and fixed in 70% cold ethanol for 30 min. DNA was denatured with 2 N HCl-0.5% Triton X-100 for 30 min followed by acid neutralization with 0.1 M Na2B4O7 · 10H2O (pH 8.5). The pelleted cells were incubated with 5 ml of anti-BrdU-fluorescein isothiocyanate antibody (Becton Dickinson) in 50 μl of 0.5% Tween 20-1% bovine serum albumin-PBS for 30 min at room temperature. After unbound antibody was washed away with PBS, the pelleted cells were resuspended in 1 ml of PBS in the presence of 5 μg of 7-aminoactinomycin D (Sigma)/ml. Samples were analyzed by fluorescence-activated cell sorting (FACS) on a FACSCalibur flow cytometer with the FL1-H and FL3-A channels.
Histological analysis.
Uteri from pregnant mice were treated for 2 h in Bouin's fixative, dehydrated, and embedded in paraffin. Sections (7 μm) were stained with hematoxylin and eosin by standard procedures.
RESULTS
Generation of a deleted mTOR allele.
To delete the mTOR gene in mice, we used a 16.0-kb DNA fragment containing the first 8 exons of the mouse gene (Fig. 1A). A targeting vector was produced by introducing a loxP site upstream of the mTOR promoter region and a neo cassette flanked by two loxP sites in the intron preceding exon 6. In this way, loxP sites flanked the putative transcription start site and the coding sequence through mTOR coding exon 5 (Fig. 1A). The construct was transfected into 129SVJae ES cells, and individual clones were screened for recombination at the homologous locus (Fig. 1B). One of the correctly targeted ES cell clones (mTORneo) was injected into C57BL/6 blastocysts to obtain chimeric mice. Subsequently, a chimeric mTORneo mouse was bred with a cytomegalovirus promoter-induced cre-transgenic mouse, which expressed Cre recombinase in the germ line (46). From this breeding, we obtained mice that had a deletion between the extreme loxP sites of the targeted mTOR allele (Fig. 1C). The mTOR+/− mice were bred with C57BL/6 mice (WT) or interbred to generate null mutants for the analysis of mTOR function during embryonic development.
FIG. 1.
Targeted disruption of the mTOR gene by the cre-loxP strategy. (A) A neomycin resistance cassette (neo) flanked by loxP sequences and a thymidine kinase expression cassette (tk) were introduced in the genomic mTOR clone for selection of ES cells. A loxP site was inserted upstream of the presumptive promoter sequence. (B) Southern blot analysis of WT and mTORneo ES cell DNAs digested with NheI and hybridized with the 5′ probe indicated in the mTOR gene map. (C) Diagram of cre-mediated full excision of the mTORneo allele. The mTOR coding sequence deleted by recombination is indicated by the first and last corresponding amino acids (Met1 and Arg260). AC16, AC14, and AC11 correspond to the oligonucleotide primers used for genotyping the WT and mTOR deletion alleles. (D) Genotyping of mouse DNA was performed by PCR with a mixture of three primers (see Materials and Methods). PCR amplification with primers AC14 and AC11 produced a 273-bp DNA fragment (lower band) for the WT mTOR allele, while PCR amplification with primers AC16 and AC11 produced a 522-bp fragment (upper band) for the excised allele.
mTOR heterozygosity does not affect cell size or proliferation.
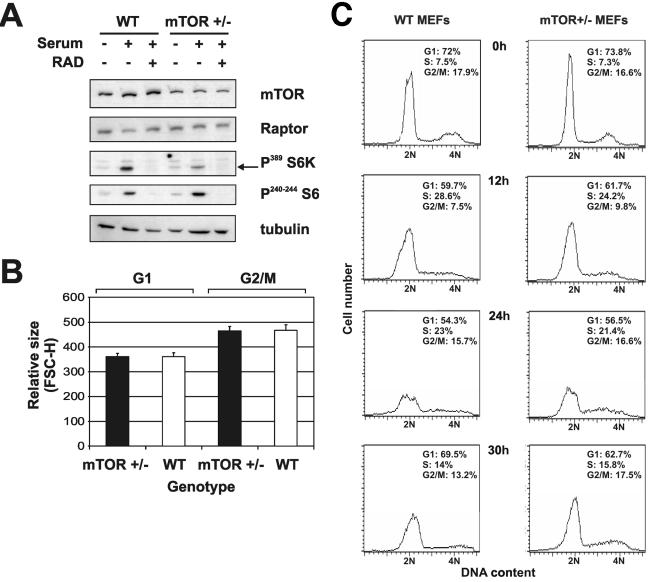
mTOR+/− mice are viable and fertile and do not present any overt phenotype. However, the ENU-induced mTOR mutation reduced mTOR expression and severely affected development (16). Moreover, small interfering RNA suppression of mTOR protein levels reduced both cell size and downstream signaling to S6K1 (25), in agreement with the finding that the absence or suppression of S6K1 also has a strong effect on cell size in Drosophila (37), mice (42), and cultured cells (8). To determine whether mTOR protein levels were decreased in mTOR+/− mice and whether any such decrease had an impact on downstream signaling or cell size, MEFs were generated from mTOR+/− and WT embryos. To assess mTOR protein levels in both cell types as well as their ability to respond to serum with respect to S6K1 and S6 phosphorylation, cells were deprived of serum for 24 h so that they were synchronously arrested in early G1 and then were stimulated to reenter the cell cycle by the exposure to serum for 90 min. Lysates prepared from both cell types were analyzed on Western blots following gel electrophoresis. As a control for mTOR function, parallel cultures were treated with the rapamycin derivative RAD001 (Everolimus; Novartis). A comparison of protein levels revealed that mTOR protein levels in mTOR+/− MEFs were approximately half those in WT MEFs, with no detectable protein of a smaller size (Fig. 2A).
FIG. 2.
Characterization of mTOR+/− MEFs. (A) Western blot analysis of protein extracts from WT and mTOR+/− MEFs was carried out with anti-mTOR, anti-raptor, anti-phosphorylated S6K (Thr389), anti-phosphorylated S6 (Ser240/244), and antitubulin antibodies. RAD, RAD001. (B) WT and mTOR+/− MEF sizes. Cells were starved for 2 days, stimulated with 10% FCS-DMEM, and harvested for analysis by flow cytometry as described in Materials and Methods. Average cell sizes for the G1 and G2/M subpopulations were inferred from the mean FSC-H value. Error bars indicate standard deviations. (C) Cell cycle histogram of WT and mTOR+/− MEFs. Percentages of cells in different phases of the cell cycle were quantified with CellQuest software.
Recently, it was reported that small interfering RNA suppression of mTOR also leads to a reciprocal decrease in the levels of its binding partner, raptor (25). However, in contrast to the amounts of mTOR, the amounts of raptor did not vary between mTOR+/− and WT MEFs (Fig. 2A). Consistent with the decrease in mTOR protein levels, there was a parallel decrease in S6K1 T389 phosphorylation, and this phosphorylation was abolished in both cell types by RAD001 (Fig. 2A). Interestingly, despite the reduced level of S6K1 T389 phosphorylation in serum-stimulated mTOR+/− MEFs, 40S ribosomal protein S6 Ser240/244 phosphorylation was unaffected (Fig. 2A).
To determine whether the reduction in mTOR protein levels and S6K1 T389 phosphorylation had an impact on cell size or cell proliferation, mTOR+/− and WT MEFs were monitored at different stages of the cell cycle. FACS analysis of gated G1 and G2/M cells by forward light scattering revealed no significant difference in cell size between the two genotypes (Fig. 2B). To analyze cell cycle progression, mTOR+/− and WT MEFs were synchronized in G1 phase of the cell cycle by serum deprivation and then induced to transit the cell cycle by the addition of fresh medium plus serum. Under conditions of serum deprivation, there was no apparent difference between the two phenotypes, as monitored by FACS analysis, with each accumulating to approximately the same extent in G1 (Fig. 2C). The readdition of fresh medium and serum induced both cell types to reenter the cell cycle, with mTOR+/− MEFs advancing through each phase at almost the same rate as their WT counterparts (Fig. 2C). Thus, despite the reduction in mTOR protein levels and S6K1 T389 phosphorylation, mTOR+/− MEFs appeared to grow and proliferate normally.
Disruption of mTOR results in early postimplantation lethality.
To analyze the effect of a complete deletion of the mTOR gene during early development, mTOR+/− mice were intercrossed. As expected, no newborn mTOR−/− mice were obtained from such intercrosses. Moreover, further analysis revealed no mTOR−/− embryos at E12.5, when ENU-induced mTOR mutants die, or E11.5, although a number of resorbed embryos could be detected (Table 1). Genotyping of E3.5 blastocysts derived from intercrossing of mTOR+/− mice gave the expected Mendelian ratio of mTOR−/−, mTOR+/−, and WT embryos (S. G. Dann and S. C. Kozma, unpublished data), consistent with the fact that in preliminary experiments, RAD001 was found to have no obvious effect on the ability of fertilized eggs to develop from the one-cell stage to early blastocysts (J.-F. Spetz, P. Svoboda, and S. C. Kozma, unpublished data). Dissection of embryos at early stages after implantation showed that E6.5 and E7.5 mTOR−/− embryos were present (Table 1) but that they were severely retarded in growth compared to either mTOR+/− or WT littermates.
TABLE 1.
Genotyping analysis of progeny of mTOR+/− intercrosses
| Embryonic stage | No. of:
|
|||
|---|---|---|---|---|
| Progeny with the following genotype:
|
Resorbed embryos | |||
| +/− | WT | −/− | ||
| E6.5 | 9 | 2 | 3a | |
| E7.5 | 8 | 3 | 4a | |
| E11.5 | 4 | 3 | 0 | 1 |
| E12.5 | 24 | 15 | 0 | 13 |
| Newborn | 68 | 27 | 0 | |
Growth was retarded.
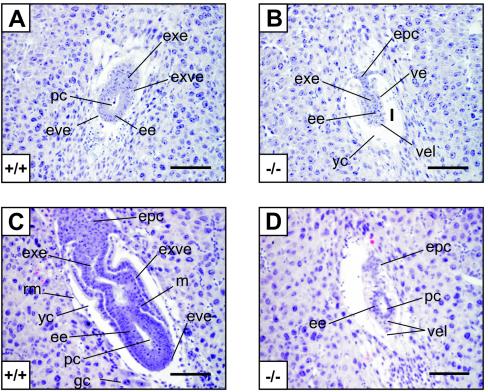
Based on these observations, thin sections of embryos from E4.5 to E7.5 were examined histologically to obtain morphological information with regard to the mTOR−/− developmental lesion (Fig. 3). Hematoxylin-eosin-stained sections of E4.5 embryos showed that all blastocysts appeared to correctly implant (data not shown). At E5.5, WT embryos displayed the characteristic elongated egg cylinder with clear extraembryonic ectoderm and embryonic ectoderm, as well as an incipient proamniotic cavity (Fig. 3A). The visceral endoderm and parietal endoderm were also well differentiated, and the yolk sac cavity was formed (Fig. 3A). At this stage, mTOR−/− embryos were severely reduced in size and did not show a clear extraembryonic or embryonic boundary, and the visceral endoderm-like cells surrounding the embryonic ectoderm were disorganized (Fig. 3B). At E6.5, WT embryos displayed a well-organized ectoderm and endoderm and an emerging mesoderm (Fig. 3C). The visceral endoderm displayed a columnar-cuboidal cell morphology in the proximal part of the egg cylinder, whereas the distal part of the epiblast displayed a squamous cell morphology (Fig. 3D). In contrast, mutant embryos exhibited a reduced embryonic region, although the proamniotic cavity was formed, and a disorganized extraembryonic ectoderm region. Moreover, the ectoplacental cone failed to elongate from E5.5 (Fig. 3D). At E7.5, mutant embryos had not progressed further; they were misoriented in the yolk sac cavity and exhibited disorganized extraembryonic and embryonic regions, with a thick layer of visceral endoderm-like cells surrounding the entire embryo (data not shown). Taken together, the data suggest that mTOR−/− embryos are arrested at E5.5, at the early egg cylinder stage.
FIG. 3.
Histological analysis of WT and mTOR−/− embryos. Hematoxylin-eosin staining of sagittal sections of normal (A and C) and presumptive mTOR−/− (B and D) E5.5 (A and B) and E6.5 (C and D) embryos within decidual tissues is shown. ee, embryonic ectoderm; epc, ectoplacental cone; eve, embryonic visceral endoderm with squamous morphology; exe, extraembryonic ectoderm; exve, extraembryonic visceral endoderm with columnar cuboidal morphology; gc, giant cell of mural trophectoderm origin; m, mesoderm; pc, proamniotic cavity; rm, Reichert's membrane; ve, visceral endoderm surrounding a reduced extraembryonic ectoderm-like region; vel, visceral endoderm-like cells with abnormal morphology; yc, yolk sac cavity. Bars, 100 μm.
mTOR is required for in vitro trophoblast development.
The particularly abnormal development of the extraembryonic ectoderm and the shortened ectoplacental cone displayed by E6.5 mTOR−/− embryos suggested a critical lesion in the development of trophoblastic cells and the establishment of the appropriate connection with the maternal circulation. In seeming agreement with this observation, Martin and Sutherland reported that rapamycin blocks amino acid-induced trophoblast outgrowth in vitro (33); however, this scenario would suggest that mTOR−/− blastocysts would have been impaired in implantation. This difference could be due to the persistence of maternally contributed mTOR transcripts and protein during early development.
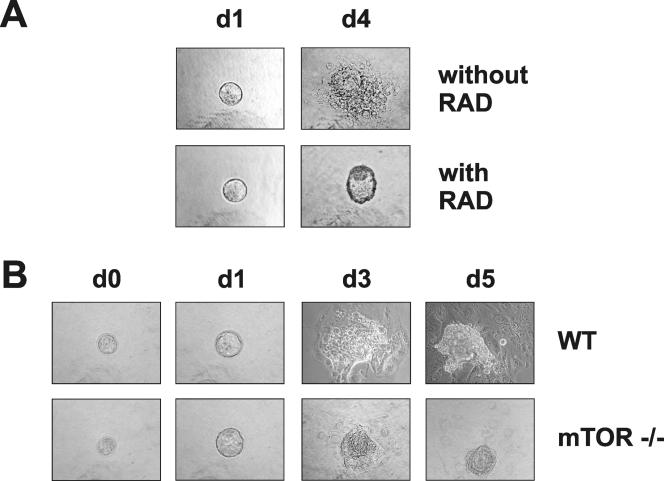
To examine this possibility, we analyzed the abilities of trophoblasts from WT blastocysts to proliferate and spread in the presence of RAD001 compared to the abilities of trophoblasts from mTOR−/−, mTOR+/−, and WT blastocysts, derived from the intercrossing of mTOR+/− mice, to proliferate and spread. On the gelatin substrate used for the outgrowth assay, morulae differentiated and began to spread and proliferate in the presence of the vehicle alone (Fig. 4A). However, in the presence of RAD001, they developed only as far as expanded blastocysts (Fig. 4A), failing to proliferate and spread (Fig. 4A), consistent with the earlier reported effects of rapamycin (33). In contrast to the findings for WT blastocysts treated with RAD001, trophoblasts from mTOR−/− blastocysts began to spread and proliferate on the gelatin surface (Fig. 4B). However, the response of mTOR−/− blastocysts was minimal compared to that of either WT or mTOR+/− blastocysts (Fig. 4B and data not shown) and failed to advance further after an additional 2 days (Fig. 4B). These data are consistent with an impairment in trophoblast outgrowth, partially compensated for by the persistence of maternally contributed mTOR transcripts during early development (see Discussion).
FIG. 4.
Phase-contrast images of embryos cultured in vitro. (A) WT embryos were explanted at E2.5 (day 0) and grown in cultures for 4 days in the absence or the presence of RAD001 (RAD). (B) WT and mTOR−/− embryos derived from mTOR+/− intercrosses were explanted at E3.5 (d1) and grown in cultures for 5 days.
mTOR is required for ICM proliferation and differentiation.
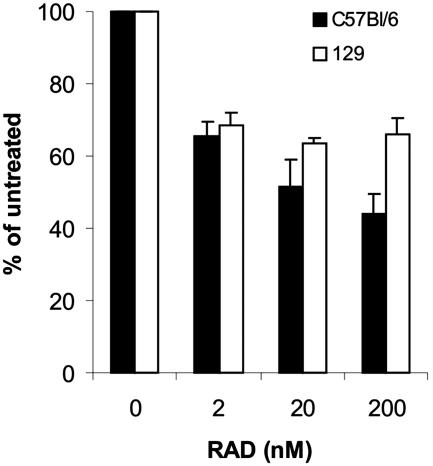
Within 4 days of in vitro embryo culturing, the ICM of mTOR+/− heterozygous and WT blastocysts differentiated into a core of ectoderm cells surrounded by a discernible visceral endoderm (Fig. 4B and data not shown). This did not appear to be the case for mTOR−/− blastocysts; in these cells, the ICM core could be detected, but the cells of the ICM failed to proliferate (Fig. 4B), suggesting an impairment in ES cell development. To examine the role of mTOR in ES cell proliferation, ES cells from C57BL/6 and 129S1 mice were treated with RAD001. Increasing concentrations of RAD001 inhibited the proliferation of both ES cell lines to approximately the same extents (Fig. 5). However, such ES cell lines are isolated from the ICM core following blastocyst outgrowth in vitro, raising the possibility that mTOR plays a more important role at an earlier stage of ICM development.
FIG. 5.
Inhibition of ES cell line proliferation by RAD001 (RAD). From each cell line, 3 × 105 cells were plated on a feeder layer in the absence or the presence of the indicated amount of RAD001. After 3 days, the cells were counted and replated under the initial conditions. Error bars indicate standard deviations.
To test this possibility, we set out to establish ES cell cultures from blastocysts obtained from intercrosses of mTOR+/− mice. In an initial analysis of 57 independent ES cell clones obtained from intercrosses of mTOR+/− mice, none was found to be mTOR −/−. In some instances, the blastocysts which were plated did not hatch from the zona pellucida and adhere to the feeder cell layer; therefore, a second set of 27 blastocysts was generated, and ES cell clones were established after the zona pellucida was removed by Tyrode treatment (39). Of the 22 clones which could be maintained in cultures, 15 grew normally, 5 developed at a slower rate, and 2 blastocysts survived without adhering to the feeder layer. Genotyping revealed that of these ES cell clones, only one of the nonadhering blastocysts was mTOR−/−. Taken together, the results suggest that the presence of mTOR is essential for the ES cells of the ICM to proliferate and differentiate.
DISCUSSION
Studies on the role of mTOR in early development are based on two observations: the ENU-induced mTOR mutant and treatment of blastocysts with rapamycin. In the first study, the authors favored a model in which mTOR did not play a major role in development until after gastrulation, when the embryo begins to increase in size (31). The data presented here show that complete deletion of the mTOR gene has a severe impact on embryonic development at a much earlier stage. Although embryos are able to implant, they appear to be strongly delayed in development and unable to advance beyond E5.5 (Fig. 3). Thus, the mTOR-null phenotype is more severe than the ENU-induced mTOR mutant phenotype and the rapamycin-induced phenotype. This difference could be explained by the facts that the ENU-induced mTOR mutant still retained 5% of mTOR mRNA and that rapamycin injection in pregnant mice was performed starting from 5.5 days postcoitum, when mTOR−/− embryos were already developmentally arrested. In contrast, in the second study, the use of rapamycin in vitro suggested that mTOR would play a much more important role in earlier development, at the stages of trophoblast differentiation and implantation of the embryo (38). In agreement with this suggestion, in vitro outgrowth studies showed that mTOR−/− blastocysts compared to either mTOR+/− or WT blastocysts are severely impaired in their ability to form trophoblasts and proliferate (Fig. 4B and data not shown). The effect is not as severe as that observed when RAD001 is added to WT blastocysts in vitro and does not prohibit in vivo implantation, most likely due to the persistence of maternal mTOR mRNA. This assumption has been substantiated in recent studies describing the pattern of mTOR gene expression in an in-depth analysis of the transcriptome during mouse preimplantation development. Hamatani et al. described nine clusters of genes which were defined by their expression profiles (12). In addition, they further grouped these nine gene clusters into three main classes, including (i) genes which were switched on during the zygotic activation, (ii) genes which were abundant in the oocyte but largely degraded during preimplantation development, and (iii) genes that followed a combination of these two patterns. The last group includes the mTOR gene, a member of gene cluster 6B, whose maternal transcript expression decreases but nevertheless remains high throughout preimplantation development. Thus, the difference in the effects of RAD001 and the loss of mTOR is most likely explained by the persistence of maternal mTOR transcripts during the early stages of embryogenesis.
Despite the effects observed in the trophoblast outgrowth assay, embryos which implanted properly appeared to have problems in further development. Pende et al. recently showed that ES cells treated with rapamycin are severely affected in their ability to upregulate the translation of 5′ terminal oligopyrimidine mRNAs (43). Likewise, we found here that ES cells derived from two distinct mouse lines are strongly inhibited in their ability to proliferate. However, the problem in ES cell development may precede the proliferative stage, based on the observation that ES cells derived from the ICM of mTOR−/− blastocysts failed to proliferate. The failure did not appear to be due to the breakdown of the zona pellucida, as removal of this membrane did not facilitate the derivation of these cells. This finding is consistent with the role of mTOR as a nutrient effector, especially of amino acids, and earlier studies showing that amino acids become important during the early blastocyst stage and continue to be important throughout hatching of the zona pellucida (38).
The importance of amino acids during preimplantation development has been supported by a number of epidemiological studies with humans, which have indicated that protein malnutrition during this stage of development may result in intrauterine retardation of growth, leading to hypertension, cardiovascular disease, or diabetes in the adult (2). Consistent with these findings, studies with rodents have shown that a low-protein diet during preimplantation development has significant effects on both the ICM and the trophectoderm (28). Female rats placed on a low-protein diet immediately following fertilization showed no differences in the numbers of blastocysts collected 4 days later. However, differences in the ICM and trophectoderm cell numbers were observed in 4.25-day-old embryos, representing middle- to late-stage blastocysts. At the morula stage, no differences were observed in either the ICM or trophectoderm cells, but early blastocysts did exhibit a reduction in the ICM cell number. Thus, consistent with the findings described here, amino acids appear to have a strong impact on embryonic development at E4-4.5. The requirement for amino acids at the early blastocyst stage may reflect the different roles that amino acids play during this time, including the dramatic increase in the rate of protein synthesis, the accumulation of protein, and the first noticeable increase in the size of the embryo (46). From early studies, it is also clear that both nonessential and essential amino acids are required for blastocyst development, with the primary beneficial effects being attributed to nonessential amino acids (34). This dependency on amino acids is coupled to the specific upregulation of a sodium-dependent amino acid transport system, which has a high affinity for leucine and tryptophan (48). Thus, the selective increase in amino acid uptake by this sodium-dependent system coincides with the time of preimplantation, when the absence of mTOR has a profound effect on blastocyst development.
The mechanism by which amino acids signal to mTOR has yet to be resolved. It is clear that the absence of amino acids blocks mTOR signaling, as judged by its ability to phosphorylate S6K1 or 4E-BP1 (14, 49). Initial studies by Iiboshi et al. demonstrated that both essential and nonessential amino acid alcohols, which competitively inhibit the aminoacylation of tRNA, block mTOR signaling (18). These findings led to the suggestion that deacylated tRNA may negatively regulate this pathway, similar to GCN2 kinase activation in S. cerevisiae by deacylated histidyl-tRNA and inhibition of global translation through the phosphorylation of initiation factor eEIF-2α (17). However, Dennis et al. previously showed that the removal of amino acids from cells has no immediate effect on aminoacylated tRNA levels (6). Instead, Dennis et al. found that amino acid deprivation led to a rapid decrease in intracellular amino acid levels, particularly those of the branched-chain amino acids, indicating that it is the intracellular pools which are critical (6). Branched-chain amino acids may be critical because their levels fall most rapidly, such that signals derived from the reduction of any amino acid could converge on a common upstream element. Kim and Sabatini recently proposed such a model (24).
Notwithstanding the absence of knowledge about the identity of the upstream signaling component, Kim et al. (25) showed that amino acids drive the mTOR-raptor complex into a competent signaling conformation, which is measured by the weaker binding of raptor to mTOR and the ability of the complex to signal downstream to S6K1. Under these conditions, mTOR can phosphorylate key effectors of cell growth, such as S6K1 and 4E-BP1, which in turn have been implicated in the upregulation of genes involved in blastocyst outgrowth, including genes for insulin-like growth factor II (40) and ornithine decarboxylase (27). Thus, the requirement for mTOR in early embryonic development most likely reflects its role as an amino acid effector facilitating the expression of gene targets involved in primordial cell lineage differentiation.
ADDENDUM
During submission of the manuscript, Murakami et al. reported the disruption of the mTOR gene by targeting of the kinase domain of the molecule (37a).
Acknowledgments
We thank M. Fitzgerald and V. Zimmerman for skillful assistance, P. Kopp for generating mutant ES cells, T. Doll and M. Lemaistre for producing ES cell lines, and M. Joaquin for help with the FACS analysis. We are also indebted to F. Frigerio, A. Di Cara, and S. Fumagalli for support with the mice. We are also grateful to P. Dennis and M. Joaquin for critical reading of the manuscript and to Sara Oakeley for preparing figures.
This work was supported by an EMBO long-term fellowship to Y.-G. G. and by the Novartis Institutes for Biomedical Research.
REFERENCES
- 1.Abraham, R. T. 2002. Identification of TOR signaling complexes: more TORC for the cell growth engine. Cell 111:9-12. [DOI] [PubMed] [Google Scholar]
- 2.Barker, D. J. 1994. Maternal and fetal origins of coronary heart disease. J. R. Coll. Physicians Lond. 28:544-551. [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 4.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 5.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 7.Dennis, P. B., and G. Thomas. 2002. Quick guide: target of rapamycin. Curr. Biol. 12:R269. [DOI] [PubMed] [Google Scholar]
- 8.Fingar, D. C., C. J. Richardson, A. R. Tee, L. Cheatham, C. Tsou, and J. Blenis. 2004. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24:200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, X., Y. Zhang, P. Arrazola, O. Hino, T. Kobayashi, R. S. Yeung, B. Ru, and D. Pan. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4:699-704. [DOI] [PubMed] [Google Scholar]
- 10.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11:1457-1466. [DOI] [PubMed] [Google Scholar]
- 11.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamatani, T., M. G. Carter, A. A. Sharov, and M. S. Ko. 2004. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6:117-131. [DOI] [PubMed] [Google Scholar]
- 13.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 14.Hara, K., K. Yonezawa, Q. P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 15.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosupressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 16.Hentges, K. E., B. Sirry, A. C. Gingeras, D. Sarbassov, N. Sonenberg, D. Sabatini, and A. S. Peterson. 2001. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc. Natl. Acad. Sci. USA 98:13796-13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iiboshi, Y., P. J. Papst, H. Kawasome, H. Hosoi, R. T. Abraham, P. J. Houghton, and N. Terada. 1999. Amino acid-dependent control of p70(s6k). Involvement of tRNA aminoacylation in the regulation. J. Biol. Chem. 274:1092-1099. [DOI] [PubMed] [Google Scholar]
- 19.Inoki, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4:648-657. [DOI] [PubMed] [Google Scholar]
- 20.Isotani, S., K. Hara, C. Tokunaga, H. Inoue, J. Avruch, and K. Yonezawa. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274:34493-34498. [DOI] [PubMed] [Google Scholar]
- 21.Jaeschke, A., P. B. Dennis, and G. Thomas. 2004. mTOR: a mediator of intracellular homeostasis. Curr. Top. Microbiol. Immunol. 279:283-298. [DOI] [PubMed] [Google Scholar]
- 22.Jaeschke, A., J. Hartkamp, M. Saitoh, W. Roworth, T. Nobukuni, A. Hodges, J. Sampson, G. Thomas, and R. Lamb. 2002. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 159:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keith, C. T., and S. L. Schreiber. 1995. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270:50-51. [DOI] [PubMed] [Google Scholar]
- 24.Kim, D. H., and D. M. Sabatini. 2004. Raptor and mTOR: subunits of a nutrient-sensitive complex. Curr. Top. Microbiol. Immunol. 279:259-270. [DOI] [PubMed] [Google Scholar]
- 25.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 26.Kim, D. H., D. dos Sarbassov, S. M. Ali, R. R. Latek, K. V. Guntur, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11:895-904. [DOI] [PubMed] [Google Scholar]
- 27.Kimball, S. R., L. M. Shantz, R. L. Horetsky, and L. S. Jefferson. 1999. Leucine regulates translation of specific mRNAs in L6 myoblasts through mTOR-mediated changes in availability of eIF4E and phosphorylation of ribosomal protein S6. J. Biol. Chem. 274:11647-11652. [DOI] [PubMed] [Google Scholar]
- 28.Kwong, W. Y., A. E. Wild, P. Roberts, A. C. Willis, and T. P. Fleming. 2000. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development 127:4195-4202. [DOI] [PubMed] [Google Scholar]
- 29.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 30.Long, X., C. Spycher, Z. S. Han, A. M. Rose, F. Muller, and J. Avruch. 2002. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 12:1448-1461. [DOI] [PubMed] [Google Scholar]
- 31.Lorberg, A., and M. N. Hall. 2004. TOR: the first 10 years. Curr. Top. Microbiol. Immunol. 279:1-18. [DOI] [PubMed] [Google Scholar]
- 32.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28:573-576. [DOI] [PubMed] [Google Scholar]
- 33.Martin, P. M., and A. E. Sutherland. 2001. Exogenous amino acids regulate trophectoderm differentiation in the mouse blastocyst through an mTOR-dependent pathway. Dev. Biol. 240:182-193. [DOI] [PubMed] [Google Scholar]
- 34.Martin, P. M., A. E. Sutherland, and L. J. Van Winkle. 2003. Amino acid transport regulates blastocyst implantation. Biol. Reprod. 69:1101-1108. [DOI] [PubMed] [Google Scholar]
- 35.Matsuo, T., Y. Kubo, Y. Watanabe, and M. Yamamoto. 2003. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22:3073-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montagne, J., T. Radimerski, and G. Thomas. 2001. Insulin signaling: lessons from the drosophila tuberous sclerosis complex, a tumor suppressor. Sci. STKE 2001:E36. [DOI] [PubMed] [Google Scholar]
- 37.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 37a.Murakami, M., T. Ichisaka, M. Maeda, N. Oshiro, K. Hara, F. Edenhofer, H. Kiyama, K. Yonezawa, and S. Yamanaka. 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24:6710-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naeslund, G. 1979. The effect of glucose-, arginine- and leucine-deprivation on mouse blastocyst outgrowth in vitro. Upsala J. Med. Sci. 84:9-20. [DOI] [PubMed] [Google Scholar]
- 39.Nicolson, G. L., R. Yanagimachi, and H. Yanagimachi. 1975. Ultrastructural localization of lectin-binding sites on the zonae pellucidae and plasma membranes of mammalian eggs. J. Cell Biol. 66:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen, F. C., L. Ostergaard, J. Nielsen, and J. Christiansen. 1995. Growth-dependent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature 377:358-362. [DOI] [PubMed] [Google Scholar]
- 41.Oldham, S., J. Montagne, T. Radimerski, G. Thomas, and E. Hafen. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14:2689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pende, M., S. C. Kozma, M. Jaquet, V. Oorschot, R. Burcelin, Y. Le Marchand-Brustel, J. Klumperman, B. Thorens, and G. Thomas. 2000. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature 408:994-997. [DOI] [PubMed] [Google Scholar]
- 43.Pende, M., S. H. Um, V. Mieulet, M. Sticker, V. L. Goss, J. Mestan, M. Mueller, S. Fumagalli, S. C. Kozma, and G. Thomas. 2004. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitoh, M., N. Pullen, P. Brennan, D. Cantrell, P. B. Dennis, and G. Thomas. 2002. Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J. Biol. Chem. 277:20104-20112. [DOI] [PubMed] [Google Scholar]
- 45.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566-571. [DOI] [PubMed] [Google Scholar]
- 46.Schwenk, F., U. Baron, and K. Rajewsky. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stocker, H., T. Radimerski, B. Schindelholz, F. Wittwer, P. Belawat, P. Daram, S. Breuer, G. Thomas, and E. Hafen. 2003. The small GTPase Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5:559-565. [DOI] [PubMed] [Google Scholar]
- 48.Van Winkle, L. J. 2001. Amino acid transport regulation and early embryo development. Biol. Reprod. 64:1-12. [DOI] [PubMed] [Google Scholar]
- 49.Xu, G., G. Kwon, C. A. Marshall, T. A. Lin, J. C. Lawrence, Jr., and M. L. McDaniel. 1998. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic beta-cells. A possible role in protein translation and mitogenic signaling. J. Biol. Chem. 273:28178-28184. [DOI] [PubMed] [Google Scholar]
- 50.Yamagata, K., L. K. Sanders, W. E. Kaufmann, W. Yee, C. A. Barnes, D. Nathans, and P. F. Worley. 1994. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J. Biol. Chem. 269:16333-16339. [PubMed] [Google Scholar]
- 51.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5:578-581. [DOI] [PubMed] [Google Scholar]