Abstract
Nuclear factor Y (NF-Y) is an evolutionarily conserved trimeric transcription factor complex present in nearly all eukaryotes. The heterotrimeric NF-Y complex consists of three subunits, NF-YA, NF-YB, and NF-YC, and binds to the CCAAT box in the promoter regions of its target genes to regulate their expression. Yeast and mammal genomes generally have single genes with multiple splicing isoforms that encode each NF-Y subunit. By contrast, plant genomes generally have multi-gene families encoding each subunit and these genes are differentially expressed in various tissues or stages. Therefore, different subunit combinations can lead to a wide variety of NF-Y complexes in various tissues, stages, and growth conditions, indicating the potentially diverse functions of this complex in plants. Indeed, many recent studies have proved that the NF-Y complex plays multiple essential roles in plant growth, development, and stress responses. In this review, we highlight recent progress on NF-Y in Arabidopsis thaliana, including NF-Y protein structure, heterotrimeric complex formation, and the molecular mechanism by which NF-Y regulates downstream target gene expression. We then focus on its biological functions and underlying molecular mechanisms. Finally, possible directions for future research on NF-Y are also presented.
Keywords: nuclear factor Y, CCAAT box, Arabidopsis, flowering time, hypocotyl elongation, abiotic response
Main Text
Nuclear factor Y (NF-Y) is widespread in plants, animals, and other eukaryotes, and is also termed CCAAT Binding Factor (CBF) or Heme Activator Protein (HAP). The NF-Y complex consists of the subunits NF-YA (CBF-B/HAP2), NF-YB (CBF-A/HAP3), and NF-YC (CBF-C/HAP5), all of which are necessary for binding to the CCAAT box (Nardini et al., 2013). Individual subunits of NF-Y cannot regulate transcription independently; instead, they must function in heterodimers or heterotrimers. In yeast and mammals, each subunit of NF-Y is encoded by a single gene, but these genes have multiple splicing forms and undergo various post-translational modifications (Li et al., 1992a; Mantovani, 1999). In mammals, the physiological function and underlying molecular mechanism of the NF-Y complex has been extensively characterized in different cellular processes, such as endoplasmic reticulum stress, DNA damage, and cell cycle regulation (Oldfield et al., 2014; Benatti et al., 2016; Dolfini et al., 2016). In plants, each subunit of NF-Y is encoded by multiple members, which further form different sub-families (Petroni et al., 2012). A previous classification system described 36 NF-Y members (10 NF-YA, 13 NF-YB, and 13 NF-YC) in Arabidopsis (Siefers et al., 2009). This system considered the plant homologs of Negative Cofactors (NC2), DNA Polymerase II Subunit B3 (Dpb3), and Dpb4 as members of the NF-Y family; however, NC2, Dpb3p, and Dpb4p do not functionally overlap with NF-Y. NC2 associates with TATA-binding protein (TBP) to bind TATA boxes (Kamada et al., 2001), whereas Dpb3p and Dpb4p participate in the complex with DNA polymerase and the complex of chromatin remodeling (Ohya et al., 2000; Hartlepp et al., 2005). Therefore, the current consensus on Arabidopsis NF-Y members excludes AtNF-YC11/B12/B13 (NC2 subfamily) and AtNF-YC10/C13/B11 (Dpb3/4 subfamily), but does include AtNF-YC12. Thus, in the updated scheme, Arabidopsis has 30 members of the NF-Y family, 10 from each family (Petroni et al., 2012).
The initial reports of NF-Y genes in plants date back to the 1990s (Li et al., 1992b; Albani and Robert, 1995; Edwards et al., 1998). In the past decades, many studies have shed light on the biological functions of individual NF-Y subunits in Arabidopsis and other plant species, showing that this complex acts in gametogenesis, embryogenesis, seed development, flowering time regulation, primary root elongation, abscisic acid signaling, drought resistance, the endoplasmic reticulum stress response, hypocotyl elongation, and so on (Mantovani, 1999; Gusmaroli et al., 2001; Petroni et al., 2012 and references therein). All these studies suggest that the NF-Y gene family is a powerful and mysterious gene family and important for many aspects of plant life.
The Protein Structure of NF-Y Subunits
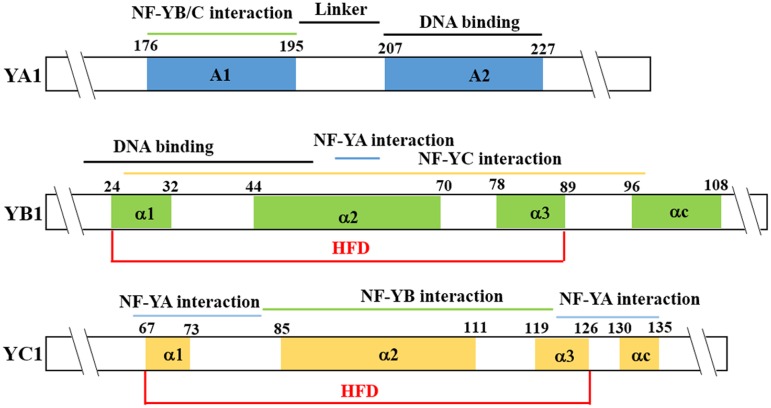
The A subunits of NF-Y generally localize to the nucleus and most NF-YA proteins can bind to the CCAAT cis-element in the promoter regions of target genes, but with different affinities (Calvenzani et al., 2012; Petroni et al., 2012; Laloum et al., 2013; Nardini et al., 2013). Protein structure analysis indicated that two conserved α-helix domains (A1 and A2) are present in the core regions of NF-YA subunits (Figure 1). The 20-amino-acid α helix A1 is in the N-terminal of the core region and functions in the interaction with NF-YB and NF-YC subunits, whereas the 21-amino-acid α helix A2 is in the C-terminal and provides sequence-specificity for recognition and binding of the CCAAT cis-element (Petroni et al., 2012; Laloum et al., 2013).
FIGURE 1.
Nuclear factor Y (NF-Y) protein structure. The NF-YA conserved region is composed of two α-helices. The A1 helix is in the N-terminal of the core region and functions in interaction with NF-YB and NF-YC. The A2 is in the C-terminal and functions in specific recognition of the CCAAT element. The Histone Fold Domain (HFD) domain of the NF-YB and NF-YC proteins contains the DNA-binding domain and protein-protein interaction domain. The diagram shows AtNF-YA1, AtNF-YB1, and AtNF-YC1 as examples.
The NF-YB and NF-YC subunits contain the highly conserved Histone Fold Domain (HFD, also termed as Histone Fold Motif, HFM) (Figure 1), which functions in protein-DNA and protein-protein interactions (Frontini et al., 2004; Kahle et al., 2005; Laloum et al., 2013). The HFD domains of NF-YB and NF-YC subunits are more closely related to the core histone H2B and H2A, respectively (Dolfini et al., 2012; Petroni et al., 2012; Laloum et al., 2013). Typically, the HFD domain of NF-YB/YC is formed by a minimum of three α-helices (α1, α2, and α3) separated by two loops, similar to the HFD domains of histones H2B and H2A, and even similar to the HFD domain of the β and α subunits of NC2 (Arents and Moudrianakis, 1995; Mantovani, 1999). In NF-YB, the α1 helices contain the putative DNA-binding domain. In NF-YC, the α1 helices contain a few conserved amino acids which supposed to be the putative DNA-binding domains (Laloum et al., 2013). Indeed, the α1 helix makes essential stabilizing contacts with the DNA backbone flanking the central CCAAT pentamer (Nardini et al., 2013). NF-YB and NF-YC form a heterodimer via their HFDs, specifically via α2 and α3 (Kim et al., 1996; Zemzoumi et al., 1999; Frontini et al., 2004). The fourth helix of the conserved domain of NF-YB and NF-YC (called αC) has been proposed to function in interactions with other protein in mammals (Romier et al., 2003; Laloum et al., 2013). Formation of NF-YB/YC heterodimers is necessary for the translocation of NF-YB from the cytoplasm to the nucleus since NF-YB family members lack a nuclear localization signal (NLS), in contrast to the NF-YA and NF-YC subunits (Liu and Howell, 2010; Hackenberg et al., 2012). The HFD domains of NF-YB and NF-YC subunits assemble on a head-to-tail fashion to form a dimer, thus producing a structural scaffold for DNA-binding and interaction with NF-YA subunits (Nardini et al., 2013).
Crystal structure analysis of the NF-Y/CCAAT complex in mammals indicated that NF-YA binds to NF-YB/YC and inserts an α-helix into the minor groove of DNA, thus providing a sequence-specific contact to the CCAAT box. By contrast, NF-YB/NF-YC subunits form heterodimers that bind to the DNA sugar-phosphate backbone, mimicking the nucleosome H2A/H2B-DNA assembly (Romier et al., 2003; Nardini et al., 2013; Nardone et al., 2016). NF-YB/NF-YC interacts with DNA through non-specific HFD-DNA contact, which is similar to those of the core histone H2A/H2B. NF-YB/NF-YC also interact with NF-YA to produce the NF-Y complex with its sequence-specific recognition properties and nucleosome-like capabilities of non-specific DNA-binding. Thus combinations of subunits lead to stable DNA-binding activity of the NF-Y complex (Oldfield et al., 2014). Recently, the first crystal structure of the Arabidopsis NF-YB6/NF-YC3 dimer has been solved, which revealed that AtNF-YB6 and AtNF-YC3 subunits interact in a head-to-tail fashion and form a classical histone-like pair (Gnesutta et al., 2016; Nardone et al., 2016). The structural information on the NF-Y/CCAAT complex has given crucial insight into the molecular mechanism responsible for the architecture of the NF-YB/NF-YC heterodimer and the NF-Y heterotrimer, and also for the capacity of recognition and binding to DNA of NF-Y complexes.
NF-Y Protein Complexes
There are 30 predicted NF-Y members in the Arabidopsis genome; in theory, this could result in about 1000 heterotrimeric combinations (Petroni et al., 2012). To define the molecular mechanism of NF-Y in plants, two fundamental questions about NF-Y complexes should be considered. First, how many heterotrimeric combinations actually exist in vivo and, second, what determines the specificity of those interactions and of DNA-binding. To define the unique NF-Y complexes, tissue- and development-specific expression patterns for all the subunits of Arabidopsis NF-Y have been investigated using stable promoter:beta-glucuronidase (GUS) fusion reporter lines (Siefers et al., 2009). The different subunits of NF-Y are specifically expressed in different tissues and organs during special developmental stages (Gusmaroli et al., 2001, 2002; Siefers et al., 2009; Cao et al., 2011; Sorin et al., 2014), or in response to environmental changes (Pant et al., 2009; Des Marais et al., 2012), suggesting that only some combinations of NF-Y subunits can be assembled and act in different developmental stages or under certain stimuli or conditions. In addition, yeast two-hybrid and three-hybrid systems have also been used to detect protein-protein interactions and formation of heterotrimers between Arabidopsis NF-Y subunits (Calvenzani et al., 2012; Hackenberg et al., 2012; Sato et al., 2014). Yeast two-hybrid assays indicated that most of the NF-Y subunits of the same type (A-A, B-B, or C-C) could not form homodimers or heterodimers, and NF-YA and NF-YB show little or no interaction. However, NF-YB and NF-YC subunits could interact in many combinations and form different heterodimers (Calvenzani et al., 2012; Hackenberg et al., 2012). These systematic studies of the protein interactions between NF-Y subunits have facilitated the discovery of complete NF-Y complexes, but tissue, development, and even time-dependent specific combinations should also be considered.
Research based on yeast three-hybrid and co-immunoprecipitation assays showed that NF-Y could function as a NF-YA-YB-YC heterotrimeric complex in vivo (Table 1) (Hou et al., 2014; Sato et al., 2014). In theory, Arabidopsis NF-Y subunits can form 1000 different heterotrimeric combinations (Petroni et al., 2012); however, only a few NF-Y complexes have been verified through molecular and biochemical studies (Table 1). One possible reason is that some or many heterotrimeric of NF-Y may be transient and highly dynamic in vivo, and thus hard to detect. Nevertheless, with the emergence of new experimental methods and in-depth studies of NF-Y family members, more and more NF-Y complexes are being identified and explored.
Table 1.
Molecular function of different nuclear factor Y (NF-Y) protein complexes.
| Protein complex | Molecular function | Reference |
|---|---|---|
| AtNF-YA1-B6-C10 AtNF-YA2-B2/5/6-C10 AtNF-YA4/7-B9-C10 | Heat and drought response | Sato et al., 2014 |
| AtNF-YA5-B9-C9 | Blue light and abscisic acid responses. | Warpeha et al., 2007 |
| AtNF-YA2-B10-C2 | Unknown | Hackenberg et al., 2012 |
| AtNF-YA4-B10-C2 | Unknown | Hackenberg et al., 2012 |
| AtNF-YA2-B2/B3-C9-CO-RGA | Regulate flowering time via regulating the expression of SOC1/FT | Hou et al., 2014; Xu F. et al., 2016 |
| AtNF-YA4-B3-C2-bZIP28 | Regulate ER stress | Liu and Howell, 2010 |
| AtNF-YA2-B3-C10-DREB2A | Regulate heat tolerance via regulating the expression of HsfA3 | Sato et al., 2014 |
| AtNF-YB2, B3-C3, C4, C9-CO | Regulate flowering time via regulating the expression of FT | Kumimoto et al., 2010 |
| AtNF-YB9-C2-bZIP67 | Regulate seed development via activating expression of CRC | Yamamoto et al., 2009 |
| AtNF-YB9-PIF4 | Regulate hypocotyl elongation via regulating the expression of IAA19 | Huang et al., 2015b |
| AtNF-YA2-RGA | Regulate flowering time | Hou et al., 2014 |
| AtNF-YC1-CO | Regulate flowering time | Wenkel et al., 2006 |
| AtNF-YC3-RGA | Regulate flowering time | Hou et al., 2014 |
| AtNF-YC9-RGL2 | Regulate seed germination | Liu et al., 2016 |
In the past few years, a growing number of studies have reported that subunits of NF-Y not only form heterodimeric and heterotrimeric complexes, but also interact with other proteins in various kinds of complexes. For example, multiple NF-YC subunits (C1, C3, C4, and C9) and NF-YB subunits (B2 and B3) can interact with CONSTANS (CO) to regulate flowering time (Wenkel et al., 2006; Kumimoto et al., 2010; Cao et al., 2014; Xu F. et al., 2016). In addition, basic region/leucine zipper motif (bZIP) type transcription factors bZIP28 and bZIP67 interact with NF-Y subunits to regulate endoplasmic reticulum (ER) stress and seed development, respectively (Yamamoto et al., 2009; Liu and Howell, 2010). Phytochrome interacting factor 4 (PIF4) interacts with LEAFY COTYLEDON1 (LEC1/NF-YB9) to regulate hypocotyl elongation (Huang et al., 2015b). REPRESSOR OF ga1-3 (RGA) and RGA-LIKE2 (RGL2) interact with NF-YA2, NF-YB2/B3, or NF-YC3/C4/C9 to regulate gibberellin (GA) and photoperiod-mediated flowering time, or abscisic acid (ABA) and GA signaling pathways during seed germination, respectively (Hou et al., 2014; Liu et al., 2016). Studies of the interaction of NF-Y with other proteins regulating target gene expression have largely improved our understanding of the physiological roles of NF-Y in many biological activities (Table 1).
NF-Y Regulates the Expression of Downstream Target Genes
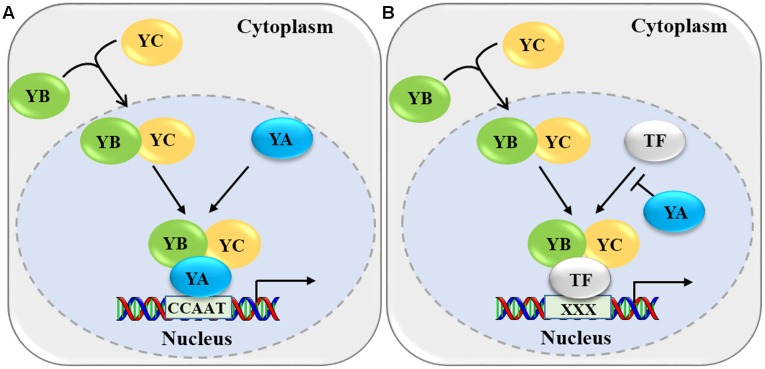
Since NF-Y regulates gene expression mainly as a protein complex, it is important to understand its specific regulatory mechanisms. Previous reports have proposed that NF-Y modulates expression of downstream target genes mainly by two mechanisms (Figure 2). In the first mechanism, the NF-YB-YC heterodimer assembles in the cytoplasm, then translocates into the nucleus, where it interacts with NF-YA to form an active heterotrimer (Figure 2A) (Hackenberg et al., 2012; Laloum et al., 2013). The NF-YA-YB-YC complex mainly binds to the CCAAT box in the promoter regions of the downstream target genes through NF-YA, and regulates the expression of target genes, which is a highly conserved transcriptional regulation mechanism for NF-Y complexes in yeast, mammals, and plants (Mantovani, 1999; Frontini et al., 2004; Liu and Howell, 2010; Dolfini et al., 2012; Petroni et al., 2012; Hou et al., 2014; Sato et al., 2014). For example, the AtNF-YA4-YC2-YB3 complex binds to the CCAAT cis-element in the promoter region of BINDING PROTEIN 3 (BiP3) through AtNF-YA4 and up-regulates the expression of ER stress-induced genes (Liu and Howell, 2010). In addition, the AtNF-YA6 subunit interacts with the AtNF-YB6/YC3 dimer and the resulting trimer can directly bind CCAAT cis-elements, even a probe containing the CCAAT box derived from the human Heat Shock Protein70 (HSP70) promoter (Nardone et al., 2016). Most NF-Y complexes bind to the CCAAT box mainly through NF-YA; however, in plants, a recent study in rice reported that OsNF-YB1 might bind to the CCAAT box of Sucrose Transporter1 (SUT1), SUT3, and SUT4 (Bai et al., 2016). However, the in vitro and in vivo data are not sufficient to prove OsNF-YB1 binding to CCAAT box directly, since some sequences flanking the CCAAT box could bind via NF-YB (Nardini et al., 2013).
FIGURE 2.
Two molecular mechanisms by which NF-Y modulates gene expression. (A) The NF-YB-YC heterodimers translocate into the nucleus and interact with NF-YA to form an active heterotrimer. This heterotrimer binds to the CCAAT box in the promoter regions of the target genes and mediates their expression. (B) The NF-YB-YC heterodimer interacts with the transcription factor (TF) to form a NF-YB-YC-TF heterotrimer and binds to specific cis-element (indicated by XXX). NF-YA can competitively inhibit the interaction of the TF with NF-YB/NF-YC.
In the second molecular mechanism, the NF-YB/NF-YC heterodimer associates with specific transcription factors to form a complex (NF-YB-YC-transcription factor), and regulates the expression of their target genes through the binding of transcription factors to specific cis-elements in the promoters of various target genes (Wenkel et al., 2006; Yamamoto et al., 2009; Kumimoto et al., 2010) (Figure 2B). In this mechanism, NF-YA probably could suppress the formation of the NF-YB-YC-transcription factor trimeric complex by preventing the combination of transcription factors with NF-YB-YC. Although this kind of transcriptional regulation has been proposed by many studies, while the direct molecular evidences are still required since the opinion of compete of YA with TF derived from supposes upon some opposite observations. For example, AtB9-C2-bZIP67 directly binds to the ABA-response elements (ABREs) through bZIP67 in the promoter regions of SUCROSE SYNTHASE 2 (SUS2) and CRUCIFERIN C (CRC), then activates the expression of SUS2 and CRC and promotes seed development (Yamamoto et al., 2009). However, NF-YA subunits strongly inhibited the expression of CRC by competing with bZIP67 to form an NF-YA-YB9-YC2 complex, which indicated different members of the NF-Y subunits play distinct roles in plant development and growth (Adrian et al., 2010). In mammals, genome-wide association with selected transcription factors indicated that the NF-Y complex directly binds to the CCAAT box and associates with other cis-elements by interacting with different transcription factors (Dolfini et al., 2016; Zambelli and Pavesi, 2016). Recently, a ChIP-sequencing approach in rice identified genome-wide downstream targets of OsNF-YB1, indicating that OsNF-YB1 directly binds to the CCAAT box and associates with the GCC box by interacting with an ERF-type transcription factor (Xu J.J. et al., 2016).
Some special transcriptional regulatory mechanisms do not fit these two major regulatory models in Arabidopsis. For instance, AtNF-YA2 can directly bind to the NFYBE cis-element (not the CCAAT box) of the SUPPRESOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) promoter to regulate the expression of SOC1 (Hou et al., 2014). In addition, a recent study indicated that NF-YC subunits (C1, C3, C4, and C9) directly interact with HISTONE DEACETYLASE 15 (HDA15), recognize the promoter of hypocotyl elongation-related genes, and repress their expression through the deacetylation of H4 in a light-dependent manner (Tang et al., 2016). A previous study also showed that NF-Y directly activates the expression of SOC1 partly through the H3K27 demethylase RELATIVE OF EARLY FLOWERING 6 (REF6; Hou et al., 2014). Therefore, the NF-Y complex may function together with various epigenetic factors to regulate the transcription of downstream target genes. Certainly, deeper research will discover more and more novel mechanisms.
The Biological Functions of NF-Y Subunits
Embryo Development and Seed Germination
NF-Y transcription factors play crucial roles in embryogenesis. The first identified and extensively studied NF-Y subunit acting in embryogenesis and seed maturation is NF-YB9, which was identified as LEAFY COTYLEDON1 (LEC1; West et al., 1994; Lotan et al., 1998; Lee et al., 2003). NF-YB9/LEC1 plays multiple essential roles in embryogenesis and post-embryonic development in Arabidopsis, where it is required to maintain the fate of embryonic cells and prevent immature seeds from germinating prematurely (Meinke et al., 1994; West et al., 1994; Lotan et al., 1998; Lee et al., 2003; Suzuki et al., 2007; Yamamoto et al., 2009; Junker et al., 2012; Mu et al., 2013). NF-YB9 and LEC1-LIKE (L1L/NF-YB6) affect embryo development through induction of genes related to embryogenesis and cellular differentiation (Lotan et al., 1998; Lee et al., 2003; Huang et al., 2015a).
Based on the tissue-specific expression patterns and mutant phenotypes, many Arabidopsis NF-Y genes participate in embryo development. For example, NF-YA1, YA2, A3, A4, A6, A7, A8, and A9 are expressed in the embryo and may affect embryo development (Siriwardana et al., 2014). Moreover, the transgenic plants overexpressing NF-YA1, A5, A6, or A9 show hypersensitivity to ABA during seed germination and promotion of the vegetative-to-embryonic transition. However, single or double mutants of the four NF-YA genes do not have detectable phenotypes (Mu et al., 2013). NF-YA3 and A8 show their highest expression in the embryo from the globular to torpedo stages. The nf-ya3 nf-ya8 double mutants are embryo lethal, but the nf-ya3 and nf-ya8 single mutants do not display an obvious phenotype, indicating that NF-YA3 and NF-YA8 function redundantly in early embryogenesis of Arabidopsis (Fornari et al., 2013). Therefore, multiple individual NF-YA subunits likely play redundant roles in embryo development and seed germination.
Recent research indicates that NF-YC subunits are also involved in seed germination through ABA responses and different NF-YC subunits have both unique and opposing functions in ABA-mediated seed germination (Kumimoto et al., 2013). For example, Arabidopsis nf-yc4 single mutants are hypersensitive to ABA during seed germination (Warpeha et al., 2007), but nf-yc3 nf-yc9 double mutants are hyposensitive to ABA (Kumimoto et al., 2013). Although extensive genetic evidence indicated that many individual NF-Y subunits function in embryonic and seed development, and seed germination, most studies focus on single subunits; therefore, how different subunits act in the NF-Y complexes to regulate these process remains elusive.
Hypocotyl Elongation under Light Signaling Pathway
NF-Y also functions in plant photomorphogenesis, especially in hypocotyl elongation (Leyva-González et al., 2012; Mu et al., 2013; Huang et al., 2015b; Myers et al., 2016). Previous genetic evidence showed that overexpression of AtNF-YA1 and NF-YA9 changed cell identity, and led to the hypocotyl becoming greener and swollen (Mu et al., 2013). In addition, overexpression of most NF-Y A subunits significantly reduced hypocotyl elongation (Leyva-González et al., 2012; Myers et al., 2016). However, the underlying molecular mechanism by which different A-type subunits of NF-Y regulate hypocotyl elongation requires further investigation.
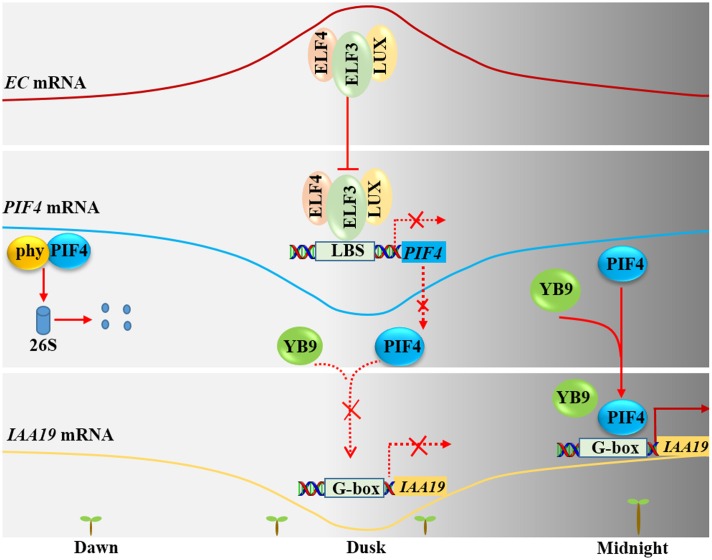
After seed imbibition, induced expression of NF-YB9/LEC1 resulted in longer hypocotyls, compared with non-induced seedlings, and mutants of NF-YB9 had short hypocotyls (Junker et al., 2012; Huang et al., 2015b). In addition, genome-wide chromatin immunoprecipitation (ChIP) microarray (ChIP-chip) analysis revealed that a number of auxin and cell elongation-related genes (such as YUC10, IAA5, and IAA19) are putative target genes of NF-YB9/LEC1 (Junker et al., 2012). Moreover, previous studies reported that the evening complex (EC, ELF4-ELF3-LUX) binds to the promoter regions of PIF4/PIF5 through the transcription factor LUX to repress the expression of PIF4 and PIF5 in the evening (Nusinow et al., 2011). Further study found that PIF4 could interact with NF-YB9 to coordinately regulate the expression of IAA19 and thus control the elongation of the hypocotyl in the dark (Figure 3) (Huang et al., 2015b). At dawn, PIF4 protein interacts with photoactivated phytochrome and then is degraded through the 26S proteasome pathway. With prolonged illumination, the ELF4-ELF3-LUX complex accumulates, and reaches a peak at dusk. The accumulated ELF4-ELF3-LUX complex can repress the transcription of PIF4, thus suppressing the binding of the PIF4-NF-YB9 complex to the promoter region of IAA19 and inhibiting its expression. Therefore, Arabidopsis hypocotyl elongation is inhibited at this stage. At midnight, the levels of the ELF4-ELF3-LUX EC decrease, allowing the restoration of PIF4 mRNA levels and accumulation of PIF4 protein. So, PIF4 can recruit NF-YB9 to form a complex and target the promoter region of IAA19, positively regulating IAA19 expression. Thus, Arabidopsis hypocotyl elongation gradually increases at this stage (Figure 3).
FIGURE 3.
Nuclear factor Y regulates hypocotyl elongation through PIF4. At dawn, Phytochrome interacting factor 4 (PIF4) is degraded by the 26S proteasome. At dusk, the transcription level of PIF4 is repressed by evening complex (ELF4-ELF3-LUX). Therefore, NF-YB9/LEC1 cannot be recruited to the promoter of IAA19. At midnight, the transcription level of PIF4 increases along with decreased abundance of the ELF4-ELF3-LUX complex. Meanwhile, NF-YB9/LEC1 is recruited to the promoter of IAA19 through its interaction with PIF4, thus increasing the transcription level of IAA19 and promotes hypocotyl elongation.
Recent reports demonstrated that five NF-YC subunits (C1, C3, C4, C6, and C9) also function in hypocotyl elongation by interacting with HDA15 (Myers et al., 2016; Tang et al., 2016). The triple mutant of nf-yc3 nf-yc4 nf-yc9 displayed longer hypocotyls under blue, and red light conditions; therefore these NF-YC subunits positively regulate photomorphogenesis (Myers et al., 2016; Tang et al., 2016). Although all three types of NF-Y subunits function in hypocotyl elongation, the underlying molecular mechanism by which NF-Y complexes regulate photomorphogenesis and hypocotyl elongation remains elusive.
Flowering Time Regulation
The timing of the transition from vegetative to reproductive development is crucial for reproductive success of flowering plants. In Arabidopsis, the major flowering pathways include photoperiod and GA pathways, which promote flowering in response to seasonal changes in day length and the endogenous content of GA, respectively. The interactions of these flowering pathways regulate the expression of two floral pathway integrators, FT and SOC1, which in turn activate the genes involved in the formation of floral meristems (Brambilla and Fornara, 2016). Overexpression of many individual NF-Y subunits (such as NF-YA1, YA4, NF-YB1, B2, B3, NF-YC1, C2, C3, C4, and C9) altered flowering time, indicating that NF-Y complexes regulate flowering time by a highly redundant and complicated mechanism. Multiple individual subunits of NF-Y can interact with CO and affect the transcript levels of FT and SOC1, two key integrators in the flowering time pathway, thus resulting in early or late flowering (Figure 4A) (Kumimoto et al., 2010; Cao et al., 2014; Hou et al., 2014; Xu F. et al., 2016).
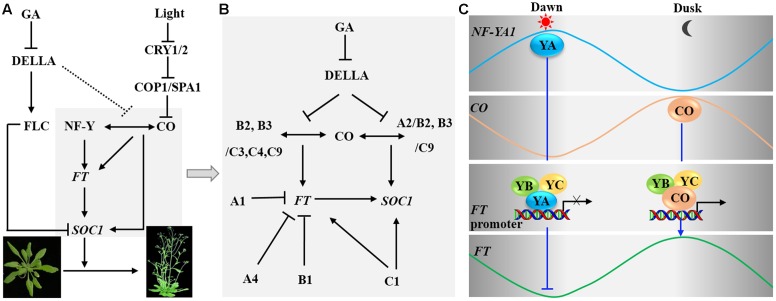
FIGURE 4.
Nuclear factor Y controls flowering time. (A) In the photoperiod pathway, the NF-Y complex can interact with CONSTANS (CO) protein, then binds the promoter region of FT and enhances its transcription. In the gibberellin (GA) signaling pathway, DELLA interacts with FLC and enhances transcriptional repression of FT and SUPPRESOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) by FLC. DELLA also represses the interaction of CO with NF-YB2, thus preventing the CO-NF-Y complex from promoting expression of FT and SOC1. (B) A diagram indicating the detailed molecular mechanism is shown in the region in gray in (A). NF-YA1, YA4, and YB1 repress the expression of FT and thus delay flowering; NF-YC1 increases the transcription of FT and SOC1, thus promoting flowering. NF-YB2/B3 and YC3/YC4/YC9 can form a complex with CO and regulate the expression of FT. Moreover, NF-YA2/YB2 and NF-YB3/YC9 interact with CO and regulate the expression of SOC1. (C) CO and NF-Y regulate FT expression. At dawn, NF-YA1-YB-YC form a heterotrimeric complex that binds to the promoter of FT thus repressing its transcription. At dusk, CO competes with NF-YA1 to form a heterotrimeric complex NF-YB-YC-CO to promote the transcription of FT.
In the photoperiod-dependent pathway, AtNF-YB2 and B3 have additive roles in the positive regulation of FT expression and flowering time (Kumimoto et al., 2008). Further studies indicate that the NF-YC subunits AtNF-YC3, C4, and C9 interact with AtNF-YB2, B3, and CO to form NF-YB-YC-CO complexes, and NF-YB and NF-YC subunits are required for the transcriptional activation of FT mediated by CO under long-day (LD) conditions (Kumimoto et al., 2010) (Figure 4B). In addition, two CO-Responsive Elements (CORE1 and 2) present in the proximal region and the CCAAT box present in the distal region of the FT promoter are crucial for regulation of FT expression (Adrian et al., 2010; Tiwari et al., 2010; Cao et al., 2014). CO directly binds to CORE2, interacts with NF-Y complexes, which can bind to the CCAAT box, resulting in chromatin loops, and positively regulates the expression of FT (Tiwari et al., 2010; Cao et al., 2014). Whether CO associates with the CORE2 in the NF-YB-YC-CO complex and how protein binding to the CORE can drive FT expression remain to be investigated (Adrian et al., 2010). Conversely, some NF-Y subunits can delay flowering by repressing the transcription of FT. For example, the overexpression of AtNF-YA1, A4, and AtNF-YB1 reduced the transcript level of FT and postponed flowering (Wenkel et al., 2006; Nelson et al., 2007). Further studies have shown that NF-YA might delay flowering by preventing the interaction between CO and NF-YB/NF-YC. For instance, overexpression of NF-YA1 and A4 prevents the interaction between CO and the NF-YB/NF-YC heterodimer, finally reducing the expression of FT and postponing flowering (Wenkel et al., 2006; Nelson et al., 2007; Leyva-González et al., 2012). Thus, different subunits of NF-Y might act in different complexes and have opposite effects on the final phenotypic output.
In the regulation of flowering time, both the NF-YB-YC-CO complexes and NF-YA-YB-YC complexes can bind to FT promoter (Adrian et al., 2010; Cao et al., 2014; Brambilla and Fornara, 2016). Indeed, both the transcript and protein levels of CO and NF-Y undergo rhythmic changes during the day; thus the complexes of NF-YA-YB-YC and the complexes of NF-YB-YC-CO likely change dynamically over time (Figure 4C). The protein level of CO decreases gradually from night to dawn (Song et al., 2015). Meanwhile, increasing NF-YA1-YB-YC forms a trimer and binds to the FT promoter to repress FT expression (Wenkel et al., 2006). In addition, CO protein accumulates gradually during the day and its expression peaks at dusk; thus CO can compete with NF-YA1 to form the trimeric complex NF-YB-YC-CO and promote the transcription of FT leading to peak expression at dusk (Wenkel et al., 2006).
NF-Y also affects flowering time by interacting with DELLA protein in the GA-dependent pathway (Figure 4). A study using ChIP showed that the NF-YA2/B2/C9 complex could interact with CO and DELLA and bind to the NFYBE cis-element in the promoter of SOC1 through NF-YA2. The NF-Y complex then recruits REF6 to demethylate H3K27me3, thus promoting SOC1 expression and leading to early flowering (Hou et al., 2014). Under LD conditions, RGA (a member of the DELLA protein family) not only interacts with NF-YB2 and CO but also represses the interaction of CO with NF-YB2. This interaction was tested through in vitro pull-down assays, but in vivo tests need further investigate (Hou et al., 2014). In addition, a previous study reported that NF-YB2 and CO form a complex with part of NF-YC family members to affect FT expression (Kumimoto et al., 2010). Therefore, the effect of RGA on CO-NF-Y binding to the promoter of SOC1 very likely represses the activity of CO in activating FT expression (Xu F. et al., 2016).
Abiotic Stress
Arabidopsis NF-Y also has an important role in the responses to abiotic stresses, such as drought, salt, cold, and heat (Table 2). Many NF-YA, NF-YB, and NF-YC subunits play key roles in the drought response (Nelson et al., 2007; Li et al., 2008; Hackenberg et al., 2012). Overexpressing NF-YA5 reduced leaf water loss and increased resistance to drought stress compared with the wild type, whereas nf-ya5 mutants were more sensitive to drought stress, which suggests that NF-YA5 positively regulates drought stress responses (Li et al., 2008). Beside NF-YA5, overexpression of other NF-YA genes such as YA2, A3, A7, and A10 also increased the tolerance to drought, which suggests that NF-Y subunits share overlapping functions in stress responses (Leyva-González et al., 2012). In addition to NF-YA, NF-YB is also involved in Arabidopsis drought stress tolerance. Overexpression of NF-YB1 enhanced plant drought resistance (Nelson et al., 2007). However, this did not affect the expression of genes involved in ABA signaling, indicating that NF-YB1 might regulate drought stress responses independent of ABA signaling (Nelson et al., 2007).
Table 2.
The functions of NF-Y genes in Arabidopsis.
| Gene name | Other name | Biological function | Reference |
|---|---|---|---|
| AtNF-YA1 | AtHAP2a | Flowering time regulation; salt stress response; seed morphology regulation; early embryogenesis. | Wenkel et al., 2006; Li et al., 2013; Mu et al., 2013 |
| AtNF-YA2 | AtHAP2b | Flowering time regulation; root growth. | Sorin et al., 2014; Hou et al., 2014 |
| AtNF-YA3 | AtHAP2c | Early embryogenesis; nitrogen nutrition. | Fornari et al., 2013 |
| AtNF-YA4 | Flowering time regulation; ER stress. | Liu and Howell, 2010; Leyva-González et al., 2012 | |
| AtNF-YA5 | Drought resistance; early embryogenesis. | Warpeha et al., 2007; Li et al., 2008; Mu et al., 2013 | |
| AtNF-YA6 | ABA response and seed germination; blue light signaling transduction; plant fertility. | Mu et al., 2013 | |
| AtNF-YA8 | Early embryogenesis. | Fornari et al., 2013 | |
| AtNF-YA9 | Embryogenesis; seed morphology regulation; seed germination. | Mu et al., 2013 | |
| AtNF-YA10 | Root growth. | Sorin et al., 2014 | |
| AtNF-YB1 | AtHAP3a | Flowering time regulation; drought resistance. | Wenkel et al., 2006; Nelson et al., 2007 |
| AtNF-YB2 | AtHAP3b | Root development; flowering time regulation. | Kumimoto et al., 2008; Ballif et al., 2011;Xu F. et al., 2016 |
| AtNF-YB3 | Flowering time regulation; ER stress; Heat stress response. | Liu and Howell, 2010; Sato et al., 2014 | |
| AtNF-YB6 | LEC1-L | Early embryogenesis. | Lee et al., 2003 |
| AtNF-YB9 | LEC1 | Early embryogenesis; hypocotyl elongation. | Warpeha et al., 2007; Junker et al., 2012; Huang et al., 2015b |
| AtNF-YC1 | AtHAP5a | Flowering time regulation; cold tolerance | Hackenberg et al., 2012; Shi et al., 2014 |
| AtNF-YC2 | AtHAP5b | Flowering time regulation; ER stress; drought resistance. | Liu and Howell, 2010; Hackenberg et al., 2012 |
| AtNF-YC3 | Flowering time regulation. | Kumimoto et al., 2010 | |
| AtNF-YC4 | Flowering time regulation; increasing protein and decreasing starch levels. | Kumimoto et al., 2010 | |
| AtNF-YC9 | AtHAP5c | Flowering time regulation; chlorophyll biosynthesize. | Warpeha et al., 2007; Kumimoto et al., 2010; Hou et al., 2014 |
| AtNF-YC10 | DPB3-1 | Heat stress response | Sato et al., 2014 |
A previous study demonstrated that overexpression of NF-YC1 improved freezing resistance, whereas nf-yc1 mutants exhibited decreased freezing resistance, which suggests that NF-YC1 positively regulates freezing responses (Shi et al., 2014). Further research found that NF-YC1 improved freezing resistance by binding to the CCAAT cis-elements in the promoter region of Xyloglucan Endotransglucosylase/Hydrolase 21 (XTH21) in Arabidopsis (Shi et al., 2014). Further, overexpression of NF-YA1 enhanced plant resistance to salt stress by increasing the expression of Abscisic Acid Insensitive3 (ABI3) and Abscisic Acid Insensitive5 (ABI5; Li et al., 2013). In addition, the NF-YA2-B3-C10 ternary complex enhanced the expression of the heat stress-inducible gene HEAT SHOCK FACTOR A3 (HsfA3) during heat stress responses in cooperation with DREB2A (Sato et al., 2014).
Other Functions of NF-Y
In addition to these above functions, NF-Y also plays roles in other plant processes. For example, NF-YA2, A3, and A5 participate in nitrogen nutrition (Zhao et al., 2011; Leyva-González et al., 2012); NF-YA2, NF-YA10, and NF-YB2 function in the control of primary root growth (Ballif et al., 2011; Sorin et al., 2014); NF-YB1 affects shoot apical meristem growth (Wenkel et al., 2006); NF-YC4 is involved in starch and protein metabolism (Li et al., 2015); and NF-YA5/B9/C9 associates with chlorophyll biosynthesis (Warpeha et al., 2007).
Perspectives
In plant, the different individual subunits of NF-Y have attracted extensive attention in recent years, but the many functions of the NF-Y complex remain only partially defined. In contrast to yeast and mammals, plants have a large NF-Y family, generally having multi-gene families encoding each subunit. This provides more combinations of NF-YA-YB-YC complexes in different developmental stages or under certain conditions, and increases functional complexity. This indicates that NF-Y is widely involved in the intricate regulatory processes in plants, compared with its more narrow roles in other organisms.
The downstream target genes and upstream regulators of NF-Y, and its functional redundancy and specificity are largely unknown. The diversity of NF-Y subunits and their many potential combinations, as well as likely redundant and divergent functions, provide a substantial challenge for work aiming to tease apart the functions of the different subunits. In mammals, NF-Y is a pioneer factor that binds to the CCAAT box in the core promoter and enhancer region (Nardini et al., 2013; Oldfield et al., 2014), but its targets remain elusive in plants. In addition, other fundamental questions about NF-Y, such its transcriptional regulation, and post-translational modification, need to be investigated. NF-Y complexes definitely function as essential regulatory hubs for many processes in plant, but its functional redundancy remains a problem for further investigation. Therefore, future studies on NF-Y will play an indispensable part in plant science.
Author Contributions
HZ and GL conceived the manuscript; HZ and DW drafted the manuscript. HZ, GL, FK, HZ, and KL edited the draft, and all authors approved the final version of manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer JPL and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank Xingliang Hou, from South China Botanical Garden, Chinese Academy of Science, for critical reading.
Footnotes
Funding. This work was supported by grants from the National Natural Science Foundation of China (31670249, 31271309) and 1000-Talents Plan for young researchers of China (to GL), by the Outstanding Young Scientist Award Fund (JQ201208) and the Taishan Scholar Program of Shandong Provincial Government (to GL).
References
- Adrian J., Farrona S., Reimer J. J., Albani M. C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22 1425–1440. 10.1105/tpc.110.074682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani D., Robert L. S. (1995). Cloning and characterization of a Brassica napus gene encoding a homologue of the B subunit of a heteromeric CCAAT-binding factor. Gene 167 209–213. 10.1016/0378-1119(95)00680-X [DOI] [PubMed] [Google Scholar]
- Arents G., Moudrianakis E. N. (1995). The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. U.S.A. 92 11170–11174. 10.1073/pnas.92.24.11170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai A. N., Lu X. D., Li D. Q., Liu J. X., Liu C. M. (2016). NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 26 384–388. 10.1038/cr.2015.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif J., Endo S., Kotani M., MacAdam J., Wu Y. (2011). Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol. Biochem. 49 579–583. 10.1016/j.plaphy.2011.01.013 [DOI] [PubMed] [Google Scholar]
- Benatti P., Chiaramonte M. L., Lorenzo M., Hartley J. A., Hochhauser D., Gnesutta N., et al. (2016). NF-Y activates genes of metabolic pathways altered in cancer cells. Oncotarget 7 1633–1650. 10.18632/oncotarget.6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V., Fornara F. (2016). Y flowering? Regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species. Biochim. Biophys. Acta 10.1016/j.bbagrm.2016.10.009 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Calvenzani V., Testoni B., Gusmaroli G., Lorenzo M., Gnesutta N., Petroni K., et al. (2012). Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS ONE 7:e42902 10.1371/journal.pone.0042902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Kumimoto R. W., Gnesutta N., Calogero A. M., Mantovani R., Holt B. F., III. (2014). A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26 1009–1017. 10.1105/tpc.113.120352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Kumimoto R. W., Siriwardana C. L., Risinger J. R., Holt BF. 3rd (2011). Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS ONE 6:e21805 10.1371/journal.pone.0021805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D. L., McKay J. K., Richards J. H., Sen S., Wayne T., Juenger T. E. (2012). Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 24 893–914. 10.1105/tpc.112.096180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini D., Gatta R., Mantovani R. (2012). NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 47 29–49. 10.3109/10409238.2011.628970 [DOI] [PubMed] [Google Scholar]
- Dolfini D., Zambelli F., Pedrazzoli M., Mantovani R., Pavesi G. (2016). A high definition look at the NF-Y regulome reveals genome-wide associations with selected transcription factors. Nucleic Acids Res. 44 4684–4702. 10.1093/nar/gkw096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D., Murray J. A., Smith A. G. (1998). Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 117 1015–1022. 10.1104/pp.117.3.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari M., Calvenzani V., Masiero S., Tonelli C., Petroni K. (2013). The Arabidopsis NF-YA3 and NF-YA8 genes are functionally redundant and are required in early embryogenesis. PLoS ONE 8:e82043 10.1371/journal.pone.0082043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontini M., Imbriano C., Manni I., Mantovani R. (2004). Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 3 217–222. 10.4161/cc.3.2.654 [DOI] [PubMed] [Google Scholar]
- Gnesutta N., Saad D., Chaves-sanjuan A., Mantovani R., Nardini M. (2016). Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the lec1 family of NF-Y subunits in plants. Mol. Plant 10.1016/j.molp.2016.11.006 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Gusmaroli G., Tonelli C., Mantovani R. (2001). Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene 264 173–185. 10.1016/S0378-1119(01)00323-7 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G., Tonelli C., Mantovani R. (2002). Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283 41–48. 10.1016/S0378-1119(01)00833-2 [DOI] [PubMed] [Google Scholar]
- Hackenberg D., Wu Y., Voigt A., Adams R., Schramm P., Grimm B. (2012). Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 5 876–888. 10.1093/mp/ssr107 [DOI] [PubMed] [Google Scholar]
- Hartlepp K. F., Fernandez-Tornero C., Eberharter A., Grune T., Muller C. W., Becker P. B. (2005). The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions. Mol. Cell. Biol. 25 9886–9896. 10.1128/MCB.25.22.9886-9896.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Zhou J., Liu C., Liu L., Shen L., Yu H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5: 4601. 10.1038/ncomms5601 [DOI] [PubMed] [Google Scholar]
- Huang M., Hu Y., Liu X., Li Y., Hou X. (2015a). Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front. Plant Sci. 6:955 10.3389/fpls.2015.00955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Hu Y., Liu X., Li Y., Hou X. (2015b). Arabidopsis LEAFY COTYLEDON1 Mediates postembryonic development via interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell 27 3099–3111. 10.1105/tpc.15.00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A., Monke G., Rutten T., Keilwagen J., Seifert M., Thi T. M., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71 427–442. 10.1111/j.1365-313X.2012.04999.x [DOI] [PubMed] [Google Scholar]
- Kahle J., Baake M., Doenecke D., Albig W. (2005). Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol. Cell. Biol. 25 5339–5354. 10.1128/MCB.25.13.5339-5354.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K., Shu F., Chen H., Malik S., Stelzer G., Roeder R. G., et al. (2001). Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 106 71–81. 10.1016/S0092-8674(01)00417-2 [DOI] [PubMed] [Google Scholar]
- Kim I. S., Sinha S., de Crombrugghe B., Maity S. N. (1996). Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 16 4003–4013. 10.1128/MCB.16.8.4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumimoto R. W., Adam L., Hymus G. J., Repetti P. P., Reuber T. L., Marion C. M., et al. (2008). The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228 709–723. 10.1007/s00425-008-0773-6 [DOI] [PubMed] [Google Scholar]
- Kumimoto R. W., Siriwardana C. L., Gayler K. K., Risinger J. R., Siefers N., Holt B. F., III. (2013). NUCLEAR FACTOR Y transcription factors have both opposing and additive roles in ABA-mediated seed germination. PLoS ONE 8:e59481 10.1371/journal.pone.0059481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumimoto R. W., Zhang Y., Siefers N., Holt B. F., III. (2010). NF-YC3 NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63 379–391. 10.1111/j.1365-313X.2010.04247.x [DOI] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18 157–166. 10.1016/j.tplants.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Lee H., Fischer R. L., Goldberg R. B., Harada J. J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 100 2152–2156. 10.1073/pnas.0437909100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-González M. A., Ibarra-Laclette E., Cruz-Ramirez A., Herrera-Estrella L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7:e48138 10.1371/journal.pone.0048138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zheng W., Zhu Y., Ye H., Tang B., Arendsee Z. W., et al. (2015). QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. U.S.A. 112 14734–14739. 10.1073/pnas.1514670112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. X., Oono Y., Zhu J., He X. J., Wu J. M., Iida K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20 2238–2251. 10.1105/tpc.108.059444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Hooft van Huijsduijnen R., Mantovani R., Benoist C., Mathis D. (1992a). Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J. Biol. Chem. 267 8984–8990. [PubMed] [Google Scholar]
- Li X. Y., Mantovani R., Hooft van Huijsduijnen R., Andre I., Benoist C., Mathis D. (1992b). Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res. 20 1087–1091. 10.1093/nar/20.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. J., Fang Y., Fu Y. R., Huang J. G., Wu C. A., Zheng C. C. (2013). NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis. PLoS ONE 8:e61289 10.1371/journal.pone.0061289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22 782–796. 10.1105/tpc.109.072173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Hu P., Huang M., Tang Y., Li Y., Li L., et al. (2016). The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 7: 12768. 10.1038/ncomms12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K. M., West M. A., Lo R., Kwong R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. 10.1016/S0092-8674(00)81463-4 [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239 15–27. 10.1016/S0378-1119(99)00368-6 [DOI] [PubMed] [Google Scholar]
- Meinke D. W., Franzmann L. H., Nickle T. C., Yeung E. C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 1049–1064. 10.1105/tpc.6.8.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Tan H., Hong S., Liang Y., Zuo J. (2013). Arabidopsis transcription factor genes NF-YA1 5 6 and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant 6 188–201. 10.1093/mp/sss061 [DOI] [PubMed] [Google Scholar]
- Myers Z. A., Kumimoto R. W., Siriwardana C. L., Gayler K. K., Risinger J. R., Pezzetta D., et al. (2016). NUCLEAR FACTOR Y, subunit C (NF-YC) transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 12:e1006333 10.1371/journal.pgen.1006333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M., Gnesutta N., Donati G., Gatta R., Forni C., Fossati A., et al. (2013). Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152 132–143. 10.1016/j.cell.2012.11.047 [DOI] [PubMed] [Google Scholar]
- Nardone V., Chaves-Sanjuan A., Nardini M. (2016). Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim. Biophys. Acta 10.1016/j.bbagrm.2016.09.006 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Nelson D. E., Repetti P. P., Adams T. R., Creelman R. A., Wu J., Warner D. C., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104 16450–16455. 10.1073/pnas.0707193104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D. A., Helfer A., Hamilton E. E., King J. J., Imaizumi T., Schultz T. F., et al. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475 398–402. 10.1038/nature10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya T., Maki S., Kawasaki Y., Sugino A. (2000). Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res. 28 3846–3852. 10.1093/nar/28.20.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield A. J., Yang P., Conway A. E., Cinghu S., Freudenberg J. M., Yellaboina S., et al. (2014). Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55 708–722. 10.1016/j.molcel.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B. D., Musialak-Lange M., Nuc P., May P., Buhtz A., Kehr J., et al. (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150 1541–1555. 10.1104/pp.109.139139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R. W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., et al. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24 4777–4792. 10.1105/tpc.112.105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier C., Cocchiarella F., Mantovani R., Moras D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278 1336–1345. 10.1074/jbc.M209635200 [DOI] [PubMed] [Google Scholar]
- Sato H., Mizoi J., Tanaka H., Maruyama K., Qin F., Osakabe Y., et al. (2014). Arabidopsis DPB3-1 a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 26 4954–4973. 10.1105/tpc.114.132928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Ye T., Zhong B., Liu X., Jin R., Chan Z. (2014). AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 203 554–567. 10.1111/nph.12812 [DOI] [PubMed] [Google Scholar]
- Siefers N., Dang K. K., Kumimoto R. W., Bynum W. E. T., Tayrose G., Holt B. F., III. (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149 625–641. 10.1104/pp.108.130591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardana C. L., Kumimoto R. W., Jones D. S., Holt B. F., III. (2014). Gene family analysis of the Arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol. Biol. Report. 32 971–986. 10.1007/s11105-014-0704-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Shim J. S., Kinmonth-Schultz H. A., Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66 441–464. 10.1146/annurev-arplant-043014-115555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C., Declerck M., Christ A., Blein T., Ma L., Lelandais-Briere C., et al. (2014). A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 202 1197–1211. 10.1111/nph.12735 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Wang H. H., McCarty D. R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143 902–911. 10.1104/pp.106.092320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Liu X., Liu X., Li Y., Wu K., Hou X. (2016). Arabidopsis NF-YCs mediate the light control of hypocotyl elongation via modulating histone acetylation. Mol. Plant 10.1016/j.molp.2016.11.007 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Tiwari S. B., Shen Y., Chang H. C., Hou Y., Harris A., Ma S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187 57–66. 10.1111/j.1469-8137.2010.03251.x [DOI] [PubMed] [Google Scholar]
- Warpeha K. M., Upadhyay S., Yeh J., Adamiak J., Hawkins S. I., Lapik Y. R., et al. (2007). The GCR1 GPA1 PRN1 NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 143 1590–1600. 10.1104/pp.106.089904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18 2971–2984. 10.1105/tpc.106.043299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., Yee K. M., Danao J., Zimmerman J. L., Fischer R. L., Goldberg R. B., et al. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 1731–1745. 10.2307/3869904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Li T., Xu P. B., Li L., Du S. S., Lian H. L., et al. (2016). DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 590 541–549. 10.1002/1873-3468.12076 [DOI] [PubMed] [Google Scholar]
- Xu J. J., Zhang X. F., Xue H. W. (2016). Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J. Exp. Bot. 67 6399–6411. 10.1093/jxb/erw409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58 843–856. 10.1111/j.1365-313X.2009.03817.x [DOI] [PubMed] [Google Scholar]
- Zambelli F., Pavesi G. (2016). Genome wide features, distribution and correlations of NF-Y binding sites. Biochim. Biophys. Acta 10.1016/j.bbagrm.2016.10.007 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Zemzoumi K., Frontini M., Bellorini M., Mantovani R. (1999). NF-Y histone fold alpha1 helices help impart CCAAT specificity. J. Mol. Biol. 286 327–337. 10.1006/jmbi.1998.2496 [DOI] [PubMed] [Google Scholar]
- Zhao M., Ding H., Zhu J. K., Zhang F., Li W. X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190 906–915. 10.1111/j.1469-8137.2011.03647.x [DOI] [PMC free article] [PubMed] [Google Scholar]