Abstract
Isolates of Candida dubliniensis may be misidentified as Candida albicans in microbiological laboratories if only the germ tube and/or the chlamydospore test is used for identification to the species level. In this study, we have evaluated the efficacy of tobacco agar for the differentiation of C. dubliniensis from C. albicans. On this medium at 28°C, all 30 C. dubliniensis isolates produced yellowish-brown colonies with hyphal fringes and abundant chlamydospores, whereas 54 C. albicans isolates formed smooth, white-to-cream-colored colonies with no chlamydospore production. This medium provides a simple tool for presumptive differentiation of C. dubliniensis from C. albicans.
Candida dubliniensis is a newly described species that was first isolated from oral cavities of human immunodeficiency virus (HIV)-infected individuals (19). Although initially found to be principally associated with oral carriage and infection, recent reports indicate that this species is capable of causing a variety of clinical conditions, including candidemia, both in HIV-infected and in non-HIV-infected individuals (7, 9, 10, 14, 20). Since C. dubliniensis, like Candida albicans, forms germ tubes and chlamydospores, it may be misidentified in routine diagnostic laboratories (12). While a number of useful tests based on phenotypic characteristics, such as discrimination based on colony color on CHROMagar Candida medium, growth at 42 to 45°C, and assimilation profiles, have been developed (8, 10, 15, 16), they are not completely reliable (10). PCR-based molecular methods offer the most reliable alternative for definite discrimination of C. dubliniensis, but these tests are labor-intensive and may not be readily available (2, 5, 6).
In 1999, Staib and Morschhauser (18) reported that on Staib's Niger seed (Guizotia abyssinica)-creatinine agar, C. dubliniensis isolates formed rough colonies and abundant chlamydospores but that C. albicans isolates grew as smooth colonies without chlamydospores. Those authors concluded that under appropriate growth conditions, the formation of rough colonies and chlamydospores can be used as species-specific markers for the identification of C. dubliniensis. Subsequently, Al-Mosaid et al. (3) evaluated the efficacy of Staib agar and caffeic acid-ferric citrate agar for discrimination between C. dubliniensis and C. albicans and confirmed the usefulness of only Staib agar for this purpose. Those authors inferred that differentiation between these two species was best achieved on the basis of colony morphology (97.7% efficacy). In 2003, three additional publications confirmed the usefulness of Staib agar (11) and sunflower (Helianthus annuus) seed agar (1, 4) for this purpose. On the basis of the formation of hyphal fringes alone, Al-Mosaid et al. (4) were able to differentiate with 100% accuracy 128 C. dubliniensis isolates on sunflower seed agar. Niger seed agar and sunflower seed agar media were initially developed to differentiate Cryptococcus neoformans from other yeasts, as it forms brown colonies on these media (13, 17). Taking a clue from a very recent publication by Tendolkar et al. (21) showing that C. neoformans also develops brown colonies on tobacco agar, we thought it would be interesting to test this medium for the differentiation of C. dubliniensis and C. albicans. The results of the study are presented in this communication.
Twenty-eight local clinical isolates and two reference strains (CD 36 and CBS 7987) of C. dubliniensis were studied in comparison with 4 reference strains and 50 randomly selected clinical isolates of C. albicans. The identities of the test isolates were established by using the germ tube test, the chlamydospore test on cornmeal-Tween 80 agar, and the Vitek 2 ID-YST system (BioMerieux, Marcy l'Etiole, France). The identities of C. dubliniensis and C. albicans isolates were further confirmed by seminested PCR amplification of their rRNA genes with species-specific primers corresponding to unique sequences within the internally transcribed spacer 2 regions of C. dubliniensis and C. albicans and/or by direct sequencing of the internally transcribed spacer 2 regions (2, 5). In addition, five clinical isolates each of Candida tropicalis, Candida glabrata, Candida parapsilosis, and Candida krusei and two reference strains of Cryptococcus neoformans, ATCC 90112 (serotype A) and CBS 1930 (serotype B), were also tested for colony characteristics on tobacco agar. The method used for the preparation of tobacco agar was the same as that described by Tendolkar et al. (21), except that we used cigarette tobacco instead of tobacco leaves. Briefly, 50 g of tobacco from commercially available cigarette brands (Marlboro; tar, 8 mg; nicotine, 0.6 mg; Philip Morris Products SA, Richmond, Va.) was mixed with 1 liter of distilled water. The mixture was boiled for 30 min and then filtered through several layers of gauze. To this filtrate, 20 g of agar was added, and the volume was made up to 1 liter. The pH of the medium at this point was 5.4. It was autoclaved at 121°C for 15 min. Twenty milliliters of medium was poured into each petri plate (90-mm diameter). All the test isolates were freshly subcultured on Sabouraud dextrose agar (Difco), and tobacco agar plates were streaked with a small amount of inoculum from the isolated colonies. The culture plates were incubated at 28°C and observed daily up to 96 h for colony characteristics, such as surface topography (rough or smooth), formation of hyphal fringes at the periphery, and color. Colonies were also observed directly at low-power (×10) and high-power (×40) magnifications for the formation of hyphal fringes and chlamydospores. In addition, tobacco agar prepared from two other commercial brands of cigarettes (Wills Navy Cut [ITC Ltd., Calcutta, India] and Charminar [VST Industries Ltd., Hyderabad, India]) was also used for comparison.
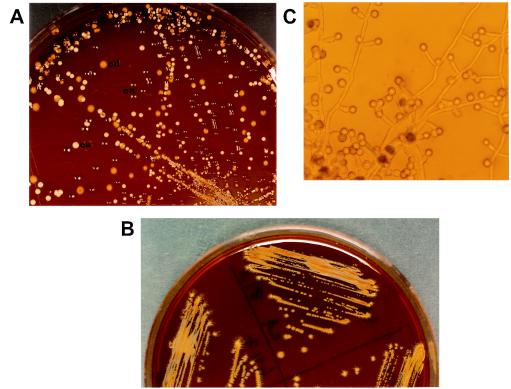
On tobacco agar, all 30 isolates of C. dubliniensis produced rough, yellowish-brown colonies with peripheral hyphal fringes after incubation for 48 to 72 h at 28°C (Fig. 1A and B). In addition, all the isolates formed abundant chlamydospores on the peripheral hyphal fringes after 24 to 48 h of incubation at 28°C (Fig. 1C). In contrast, on this medium, all C. albicans isolates showed smooth, white-to-cream-colored colonies without hyphal fringes or chlamydospores even after extended incubation for up to 10 days. These observations were reproducible when experiments were repeated on three different occasions. Like C. albicans, none of the isolates of C. tropicalis, C. glabrata, C. parapsilosis, or C. krusei formed yellowish-brown colonies or chlamydospores on tobacco agar and thus were indistinguishable from each other. However, microscopic examination of C. krusei, C. tropicalis, and C. parapsilosis colonies after 48 h of incubation showed peripheral hyphal fringes in five, three, and one of the isolates, respectively. No peripheral hyphal structures were seen in C. glabrata colonies. Two reference strains of C. neoformans produced dark-brown colonies on this medium when they were examined individually and in mixed cultures with C. dubliniensis and C. albicans (Fig. 1A). Similar observations were made when tobacco agar prepared from other commercial brands (Wills Navy Cut and Charminar cigarettes) were used. Although all initial experiments were performed on freshly prepared media, no appreciable difference in results was obtained with media stored for 7 days. Likewise, no difference in phenotypic characteristics was observed when tobacco agar plates were incubated at 30°C. However, when plates were incubated at 37°C, all 30 C. dubliniensis isolates formed rough and fringed colonies, with considerable reduction in the intensity of yellowish-brown pigmentation. Chlamydospores were formed by only two of the isolates, and their numbers were slight. None of the isolates of C. albicans formed fringed colonies or chlamydospores at this temperature. Our results suggest that all germ tube- and/or chlamydospore-positive yeast isolates may be tested on tobacco agar to differentiate between C. dubliniensis and C. albicans.
FIG. 1.
(A) Tobacco agar showing mixed growth of C. dubliniensis, C. albicans, and Cryptococcus neoformans at 28°C. Note the yellowish-brown colonies of C. dubliniensis with hyphal fringes (cd), the white-to-cream-colored colonies with no hyphal fringes of C. albicans (ca), and the brown colonies (white spots are due to light reflection) of C. neoformans (cn) after 72 h of incubation. (B) Characteristic yellowish-brown, rough colonies with peripheral hyphal fringes of three additional isolates of C. dubliniensis on tobacco agar. (C) Chlamydospores of C. dubliniensis on tobacco agar after 48 h of incubation (magnification, ×280).
In conclusion, based on phenotypic characteristics, i.e., the development of yellowish-brown rough colonies, the formation of hyphal fringes around colonies, and the production of abundant chlamydospores, tobacco agar offers an additional simple means for differentiating C. dubliniensis from C. albicans with 100% accuracy.
Acknowledgments
This study was supported by Kuwait University research grant MPI 118.
We thank Daad Farhat for her excellent technical assistance.
REFERENCES
- 1.Adou-Bryn, K., C. Douchet, A. Ferrer, L. Grimaud, R. Robert, and D. Richard-Lenoble. 2003. Morphologic features of Candida dubliniensis on a modified Pal's medium: primary study. J. Mycol. Med. 13:99-103. [Google Scholar]
- 2.Ahmad, S., Z. Khan, A. S. Mustafa, and Z. U. Khan. 2002. Seminested PCR in the diagnosis of candidemia: comparison with culture, antigen detection, and biochemical methods for identification. J. Clin. Microbiol. 40:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Mosaid, A., D. Sullivan, I. F. Salkin, D. Shanley, and D. C. Coleman. 2001. Differentiation of Candida dubliniensis from Candida albicans on Staib agar and caffeic acid-citrate agar. J. Clin. Microbiol. 39:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Mosaid, A., D. J. Sullivan, and D. C. Coleman. 2003. Differentiation of Candida dubliniensis from Candida albicans on Pal's agar. J. Clin. Microbiol. 41:4787-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellepola, A. N., S. F. Hurst, C. M. Elie, and C. J. Morrison. 2003. Rapid and unequivocal differentiation of Candida dubliniensis from other Candida species using species-specific DNA probes: comparison with phenotypic identification methods. Oral Microbiol. Immunol. 18:379-388. [DOI] [PubMed] [Google Scholar]
- 7.Fotedar, R., and S. S. Al-Hedaithy. 2003. Candida dubliniensis at a university hospital in Saudi Arabia. J. Clin. Microbiol. 41:1907-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gales, A. C., M. A. Pfaller, A. K. Houston, S. Joly, D. J. Sullivan, D. C. Coleman, and D. R. Soll. 1999. Identification of Candida dubliniensis based on temperature and utilization of xylose and alpha-methyl-d-glucoside as determined with the API 20C AUX and Vitek YBC systems. J. Clin. Microbiol. 37:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb, G. S., A. P. Limaye, Y. C. Chen, and W. C. Van Voorhis. 2001. Candida dubliniensis fungemia in a solid organ transplant patient: case report and review of the literature. Med. Mycol. 39:483-485. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez, J., P. Morales, M. A. Gonzalez, and G. Quindos. 2002. Candida dubliniensis, a new fungal pathogen. J. Basic Microbiol. 42:207-227. [DOI] [PubMed] [Google Scholar]
- 11.Lees, E., and R. C. Barton. 2003. The use of niger seed agar to screen for Candida dubliniensis in the clinical microbiology laboratory. Diagn. Microbiol. Infect. Dis. 46:13-17. [DOI] [PubMed] [Google Scholar]
- 12.Odds, F. C., L. Van Nuffel, and G. Dams. 1998. Prevalence of Candida dubliniensis isolates in a yeast stock collection. J. Clin. Microbiol. 36:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal, M., and B. S. Mehrotra. 1982. Studies on the efficacy of sunflower seed agar for the isolation and identification of Cryptococcus neoformans Arogya. J. Health Sci. 8:74-79. [Google Scholar]
- 14.Polacheck, I., J. Strahilevitz, D. Sullivan, S. Donnelly, I. F. Salkin, and D. C. Coleman. 2000. Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel. J. Clin. Microbiol. 38:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancak, B., J. H. Rex, V. Paetznick, E. Chen, and J. Rodriguez. 2003. Evaluation of a method for identification of Candida dubliniensis bloodstream isolates. J. Clin. Microbiol. 41:489-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoofs, A., F. C. Odds, R. Colebunders, M. Ieven, and H. Goossens. 1997. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur. J. Clin. Microbiol. Infect. Dis. 16:296-300. [DOI] [PubMed] [Google Scholar]
- 17.Staib, F. 1962. Cryptococcus neoformans und Guizotia abyssinica (Syn G. oleifera) Farbreaktion für Cr. neoformans. J. Hyg. 148:466-475. [Google Scholar]
- 18.Staib, P., and J. Morschhauser. 1999. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses 42:521-524. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennet, and D. C. Coleman. 1995. Candida dubliniensis sp. nov., phenotypic and molecular characterization of a novel species associated with oral candidiasis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan, D., K. Haynes, J. Bille, P. Boerlin, L. Rodero, S. Lloyd, M. Henman, and D. Coleman. 1997. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 35:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tendolkar, U., S. Taniwala, S. Jog, and M. Mathur. 2003. Use of a new medium—tobacco agar, for pigment production of Cryptococcus neoformans. Indian J. Med. Microbiol. 21:277-279. [PubMed] [Google Scholar]