Abstract
Purpose
Cis-acting regulatory SNPs resulting in differential allelic expression (DAE) may, in part, explain the underlying phenotypic variation associated with many complex diseases. To investigate whether common variants associated with DAE were involved in breast cancer susceptibility among BRCA1 and BRCA2 mutation carriers, a list of 175 genes was developed based of their involvement in cancer-related pathways.
Methods
Using data from a genome-wide map of SNPs associated with allelic expression, we assessed the association of ~320 SNPs located in the vicinity of these genes with breast and ovarian cancer risks in 15,252 BRCA1 and 8211 BRCA2 mutation carriers ascertained from 54 studies participating in the Consortium of Investigators of Modifiers of BRCA1/2.
Results
We identified a region on 11q22.3 that is significantly associated with breast cancer risk in BRCA1 mutation carriers (most significant SNP rs228595 p = 7 × 10−6). This association was absent in BRCA2 carriers (p = 0.57). The 11q22.3 region notably encompasses genes such as ACAT1, NPAT, and ATM. Expression quantitative trait loci associations were observed in both normal breast and tumors across this region, namely for ACAT1, ATM, and other genes. In silico analysis revealed some overlap between top risk-associated SNPs and relevant biological features in mammary cell data, which suggests potential functional significance.
Conclusion
We identified 11q22.3 as a new modifier locus in BRCA1 carriers. Replication in larger studies using estrogen receptor (ER)-negative or triple-negative (i.e., ER-, progesterone receptor-, and HER2-negative) cases could therefore be helpful to confirm the association of this locus with breast cancer risk.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-016-4018-2) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Genetic modifiers, Differential allelic expression, Genetic susceptibility, Cis-regulatory variants, BRCA1 and BRCA2 mutation carriers
Introduction
Pathogenic mutations in the BRCA1 and BRCA2 genes substantially increase a woman’s lifetime risk of developing breast and ovarian cancers [1–4]. These risks vary significantly according to (a) age at disease diagnosis in carriers of identical mutations, (b) the cancer site in the individual who led to the family’s ascertainment, (c) the degree of family history of the disease [1, 4, 5], and (d) the type and location of BRCA1 and BRCA2 mutations [6]. These observations suggest that other factors, including lifestyle/hormonal factors [7] as well as other genetic factors, modify cancer risks in BRCA1 and BRCA2 mutation carriers. Direct evidence for such genetic modifiers of risk has been obtained through the association studies performed by the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA), which have shown that several common breast cancer susceptibility alleles identified through population-based genome-wide association studies (GWASs) are also associated with breast cancer risk among BRCA1 and BRCA2 mutation carriers [8–10].
Global analysis of GWAS data has shown that the vast majority of common variants associated with susceptibility to cancer lie within genomic non-coding regions and are predicted to account for cancer risk through regulation of gene expression [11, 12]. A recent expression quantitative trait loci (cis-eQTL) analysis for mRNA expression in 149 known cancer risk loci performed in five tumor types (breast, colon, kidney, lung, and prostate) has shown that approximately 30 % of such risk loci were significantly associated with eQTLs present in at least one gene within 500 kb [13]. These results suggest that additional cancer susceptibility loci may be identified through studying genetic variants that affect the regulation of gene expression. In the present study, we selected genes of interest for their known involvement in cancer etiology, identified 320 genetic variants in the vicinity of these genes with evidence of differential allelic expression (DAE), and then investigated the associations of these variants with breast and ovarian cancer risks among BRCA1 and BRCA2 mutation carriers. These included variants in genes involved in DNA repair (homologous recombination and DNA interstrand crosslink repair), interaction with and/or modulation of BRCA1 and BRCA2 cellular functions, cell cycle control, centrosome amplification and interaction with AURKA, apoptosis, ubiquitination, as well as known tumor suppressors, mitotic kinases, and other kinases, sex steroid action, and mammographic density.
Materials and methods
Subjects
All study participants were female carriers of a deleterious germline mutation in either BRCA1 or BRCA2 and aged 18 years or older [14]. Fifty-four collaborating CIMBA studies contributed a total of 23,463 samples (15,252 BRCA1 mutation carriers and 8211 BRCA2 mutation carriers) to this study, including 12,127 with breast cancer (7797 BRCA1 and 4330 BRCA2 carriers) and 3093 with ovarian cancer (2462 BRCA1 and 631 BRCA2 carriers). The number of samples included from each study is provided in Online Resource 1. The recruitment strategies, clinical, demographic, and phenotypic data collected from each participant have been previously reported [14].
Ethics statement
BRCA1 and BRCA2 mutation carriers were recruited through the CIMBA initiative, following approval of the corresponding protocol by the Institutional Review Board or Ethics Committee at each participating center (Online Resource 2); written informed consent was obtained from all study participants [8, 9].
SNP selection and differential allelic expression
SNP selection was performed by first identifying a list of 175 genes of interest involved in cancer-related pathways and/or mechanisms. The list of genes was established by analyzing published results and by using available public databases such as the Kyoto encyclopedia of genes and genomes (http://www.genome.jp/kegg/). Next, DAE SNPs located within these gene regions were identified using previously reported data on allelic expression cis-associations, derived using (1) the lllumina Human1M-duo BeadChip for lymphoblastoid cell lines from Caucasians (CEU population) (n = 53) [15], the Illumina Human 1M Omni-quad for primary skin fibroblasts derived from Caucasian donors (n = 62) [13, 16], and the Illumina Infinium II assay with Human 1.2M Duo custom BeadChip v1 for human primary monocytes (n = 188) [17]. Briefly, 1000 Genomes project data were used as a reference set (release 1000G Phase I v3) for the imputation of genotypes from HapMap individuals. Genotypes were inferred using algorithms implemented in IMPUTE2 [18]. The unrelated fibroblast panel consisted of 31 parent–offspring trios, in which the genotypes of offspring were used to permit accurate phasing. Mapping of each allelic expression trait was carried out by first normalizing allelic expression ratios at each SNP using a polynomial method [19] and then calculating average phased allelic expression scores across annotated transcripts, followed by correlation of these scores to local (transcript ± 500 kb) SNP genotypes in fibroblasts as described earlier [16]. A total of 355 genetic variants were selected on the basis of evidence of association with DAE in the selected 175 genes (see Online Resource 3 for a complete list of SNPs and genes). Following the selection process, SNPs were submitted for design and inclusion on a custom-made Illumina Infinium array (iCOGS) as previously described [8, 9]. Following probe design and post-genotyping quality control, 316 and 317 SNPs were available for association analysis in BRCA1 and BRCA2 mutation carriers, respectively. Genotyping and quality control procedures have been described in detail elsewhere [8, 9].
Statistical analysis
Associations between genotypes and breast and ovarian cancer risks were evaluated within a survival analysis framework, using a one degree-of-freedom score test statistic based on modeling the retrospective likelihood of the observed genotypes conditional on the disease phenotypes [20, 21]. To estimate the magnitude of the associations [hazard ratios (HRs)], we maximized the retrospective likelihood, which was parameterized in terms of the per-allele HR. All analyses were stratified by country of residence and using calendar year and cohort-specific incidence rates of breast and ovarian cancers for mutation carriers. Given 320 tests, the cutoff value for significance after a Bonferroni adjustment for multiple testing was p < 1.5 × 10−4.
The associations between the genotypes and tumor subtypes were evaluated using an extension of the retrospective likelihood approach that models the association with two or more subtypes simultaneously [22].
Imputation was performed separately for BRCA1 and BRCA2 mutation carriers to estimate genotypes for other common variants across a ±50-kb region centered around the 12 most strongly associated SNPs (following the NCBI Build 37 assembly), using the March 2012 release of the 1000 Genomes Project as the reference panel and the IMPUTE v.2.2 software [18]. In all analyses, only SNPs with an imputation accuracy coefficient r 2 >0.30 were considered [8, 9].
Functional annotation
Publicly available genomic data were used to annotate the SNPs most strongly associated with breast cancer risk at locus 11q22.3. The following regulatory features were obtained for breast cell types from ENCODE and NIH Roadmap Epigenomics data through the UCSC Genome Browser: DNase I hypersensitivity sites, chromatin hidden Markov modeling (ChromHMM) states, and histone modifications of epigenetic markers, more specifically commonly used marks associated with enhancers (H3K4Me1 and H3K27Ac) and promoters (H3K4Me3 and H3K9Ac). To identify putative target genes, we examined potential functional chromatin interactions between distal and proximal regulatory transcription factor-binding sites and the promoters at the risk loci, using the chromatin interaction analysis by paired end tag (ChiA-PET) and genome conformation capture (Hi-C, 3C, and 5C) datasets downloaded from GEO and from 4D-genome [23]. Maps of active mammary super-enhancer regions in human mammary epithelial cells (HMECs) were obtained from Hnisz et al. [24]. Enhancer–promoter specific interactions were predicted from the integrated method for predicting enhancer targets (IM-PETs) [25]. RNA-Seq data from ENCODE was used to evaluate the expression of exons across the 11q22.3 locus in MCF7 and HMEC cell lines. For MCF7 and HMEC, alignment files from 19 and 4 expression datasets, respectively, were downloaded from ENCODE using a rest API wrapper (ENCODExplorer R package) [26] in the bam format and processed using metagene R packages [27] to normalize in Reads per Millions aligned and to convert into coverages.
eQTL analyses
The influence of germline genetic variations on gene expression was assessed using a linear regression model, as implemented in the R library eMAP (http://www.bios.unc.edu/~weisun/software.htm). An additive effect was inferred by modeling subjects’ copy number of the rare allele, i.e., 0, 1, or 2 for a given genotype. Only relationships in cis (defined as those for which the SNP is located at <1 Mb upstream or downstream from the center of the transcript) were investigated. The eQTL analyses were performed on both normal and tumor breast tissues (see Online Resource 4 for the list and description of datasets, as well as the sources of genotype and expression data). For all sample sets, the genotyping data were processed as follows: SNPs with call rates <0.95 or minor allele frequencies, MAFs (<0.05) were excluded, as were SNPs out of Hardy–Weinberg equilibrium with P < 10−13. All samples with a call rate <80 % were excluded. Identity by state was computed using the R GenABEL package [28], and samples from closely related individuals whose identity by state was lower than 0.95 were removed. The SNP and sample filtration criteria were applied iteratively until all samples and SNPs met the set thresholds.
Results
From the 175 genes selected for their involvement in cancer-related pathways and/or mechanisms, we identified a set of 355 genetic variants showing evidence of association with DAE (see Online Resource 3 for the complete list of genes and SNPs). Of those, 39 and 38 SNPs were excluded because of low Illumina design scores, low call rates, and/or evidence of deviation from Hardy–Weinberg equilibrium (P value <10−7), for BRCA1 and BRCA2 analyses, respectively. A total of 316 and 317 SNPs (representing 227 independent SNPs with a pairwise r 2 <0.1) were successfully genotyped in 15,252 BRCA1 and 8211 BRCA2 mutation carriers, respectively. Association results for breast and ovarian cancer risks for all SNPs are presented in Online Resource 5.
Breast cancer association analysis
Evidence of association with breast cancer risk (at p < 10−2) was observed for nine SNPs in BRCA1 mutation carriers and three SNPs in BRCA2 mutation carriers (Table 1). The strongest association with breast cancer risk among BRCA1 carriers was observed for rs6589007, located at 11q22.3 in intron 15 of the NPAT gene (p = 4.6 × 10−3) at approximately 54 kb upstream of the ATM gene. Similar associations were observed for two other highly correlated variants (r 2 >0.8) on chromosome 11, namely rs183459 (p = 5.7 × 10−3) also located within NPAT and rs228592 (p = 5.5 × 10−3) located in intron 11 of ATM. No association was observed between SNPs at this locus and breast cancer risk for BRCA2 carriers (Online Resource 5).
Table 1.
Associations with breast cancer risk in BRCA1 and BRCA2 mutation carriers for SNPs observed at p < 10−2
| Locations | Positions | SNPs | Nearest genes | Unaffected (number) | Affected (number) | Unaffected (MAF) | Affected (MAF) | HR* (95 % CI) | p values |
|---|---|---|---|---|---|---|---|---|---|
| BRCA1 mutation carriers | |||||||||
| 1q42.13 | 227,308,416 | rs11806633 | CDC42BPA | 7455 | 7797 | 0.07 | 0.06 | 1.128 (1.039–1.225) | 4.8 × 10−3 |
| 2p23.2 | 28,319,320 | rs6721310 | BRE | 7454 | 7793 | 0.33 | 0.33 | 1.064 (1.018–1.111) | 5.4 × 10−3 |
| 2q11.2 | 100,019,496 | rs2305354 | REV1 | 7451 | 7796 | 0.44 | 0.45 | 1.057 (1.015–1.100) | 7.1 × 10−3 |
| 4p15.33 | 14,858,341 | rs1389999 | CEBP | 7454 | 7795 | 0.35 | 0.35 | 0.940 (0.901–0.982) | 5.3 × 10−3 |
| 5q14.1 | 79,901,952 | rs425463 | DHFR, MSH3 | 7430 | 7755 | 0.33 | 0.35 | 1.058 (1.013–1.105) | 9.5 × 10−3 |
| 11q22.3 | 108,040,104 | rs6589007 | NPAT, ACAT1, ATM | 7451 | 7797 | 0.41 | 0.42 | 1.062 (1.019–1.107) | 4.6 × 10−3 |
| 11q22.3 | 108,089,197 | rs183459 | NPAT, ATM | 7447 | 7789 | 0.40 | 0.41 | 1.061 (1.018–1.105) | 5.7 × 10−3 |
| 11q22.3 | 108,123,189 | rs228592 | ATM | 7449 | 7792 | 0.42 | 0.41 | 1.061 (1.018–1.106) | 5.5 × 10−3 |
| 12p13.33 | 986,004 | rs7967755 | WNK1, RAD52 | 7454 | 7797 | 0.16 | 0.152 | 0.927 (0.876–0.980) | 7.5 × 10−3 |
| BRCA2 mutation carriers | |||||||||
| 6p22.1 | 28,231,243 | rs9468322 | NKAPL | 3880 | 4329 | 0.04 | 0.05 | 1.235 (1.080–1.412) | 4.2 × 10−3 |
| 8q11.21 | 48,708,742 | rs6982040 | PRKDC | 3876 | 4327 | 0.006 | 0.002 | 0.497 (0.292–0.843) | 2.7 × 10−3 |
| 16p13.3 | 1,371,154 | rs2268049 | UBE2I | 3880 | 4325 | 0.14 | 0.16 | 1.116 (1.031–1.207) | 4.5 × 10−3 |
CI confidence interval, HR hazard ratio, MAF minor allele frequency, SNP single-nucleotide polymorphism
* Hazard ratio per allele (one degree of freedom) estimated from the retrospective likelihood analysis
The strongest evidence of association with breast cancer risk in BRCA2 mutation carriers was observed for rs6982040, located at 8q11.21 in intron 74 of the PRKDC gene (p = 2.7 × 10−3). However, this variant had a very low frequency in affected and unaffected individuals (MAF values of 0.002 and 0.006, respectively). No association was observed for this locus in BRCA1 carriers (Online Resource 5).
Of the nine SNPs associated with breast cancer risk in BRCA1 mutation carriers, three were primarily associated with estrogen receptor (ER)-negative breast cancer: rs11806633 at 1q42.13 in the CDC42BPA gene (p = 9.0 × 10−3), rs6721310 at 2p23.2 in the BRE gene (p = 3.0 × 10−3), and rs2305354 at 2q11.2 in the REV1 gene (p = 1.0 × 10−3), although the differences between ER-positive and ER-negative disease associations were not statistically significant (Table 2). Of the three BRCA2-associated loci, only rs9468322 at 6p22.1 was associated with ER-positive disease (p = 5.0 × 10−4), although the differences in HRs between ER-positive and ER-negative tumors were not statistically significant (Table 2).
Table 2.
Associations with breast cancer risk by tumor subtype in BRCA1 and BRCA2 mutation carriers
| Locations | Positions | SNPs | ER-positive | ER-negative | ER-diff | ||
|---|---|---|---|---|---|---|---|
| HR (95 % CI) | p values | HR (95 % CI) | p values | p-diff | |||
| BRCA1 mutation carriers | |||||||
| 1q42.13 | 227,308,416 | rs11806633 | 1.10 (0.90–1.33) | 0.35 | 1.14 (1.03–1.25) | 9.0 × 10−3 | 0.73 |
| 2p23.2 | 28,319,320 | rs6721310 | 1.00 (0.88–1.09) | 0.96 | 1.08 (1.04–1.15) | 3.0 × 10−3 | 0.20 |
| 2q11.2 | 100,019,496 | rs2305354 | 0.98 (0.91–1.10) | 0.71 | 1.09 (1.03–1.13) | 1.0 × 10−3 | 0.09 |
| 4p15.33 | 14,858,341 | rs1389999 | 0.94 (0.85–1.04) | 0.20 | 0.94 (0.89–0.99) | 2.0 × 10−2 | 0.95 |
| 5q14.1 | 79,901,952 | rs425463 | 1.04 (0.94–1.15) | 0.48 | 1.07 (1.01–1.12) | 1.6 × 10−2 | 0.67 |
| 11q22.3 | 108,040,104 | rs6589007 | 1.08 (0.99–1.19) | 9.8 × 10−2 | 1.06 (1.01–1.11) | 2.0 × 10−2 | 0.66 |
| 11q22.3 | 108,089,197 | rs183459 | 1.08 (0.99–1.19) | 9.3 × 10−2 | 1.05 (1.00–1.11) | 3.7 × 10−2 | 0.62 |
| 11q22.3 | 108,123,189 | rs228592 | 1.08 (0.96–1.19) | 9.7 × 10−2 | 1.06 (1.00–1.11) | 3.4 × 10−2 | 0.64 |
| 12p13.33 | 986,004 | rs7967755 | 0.96 (0.84–1.09) | 0.54 | 0.92 (0.86–0.98) | 1.0 × 10−2 | 0.56 |
| BRCA2 mutation carriers | |||||||
| 6p22.1 | 28,231,243 | rs9468322 | 1.30 (1.12–1.51) | 5.0 × 10−4 | 1.00 (0.72–1.40) | 0.99 | 0.17 |
| 8q11.21 | 48,708,742 | rs6982040 | N/A | N/A | N/A | N/A | N/A |
| 16p13.3 | 1,371,154 | rs2268049 | 1.10 (1.01–1.21) | 4.0 × 10−2 | 1.17 (0.98–1.39) | 8.0 × 10−2 | 0.56 |
CI confidence interval, HR hazard ratio, SNP single-nucleotide polymorphism, N/A not available
* Hazard ratio per allele (one degree of freedom) estimated from the retrospective likelihood analysis
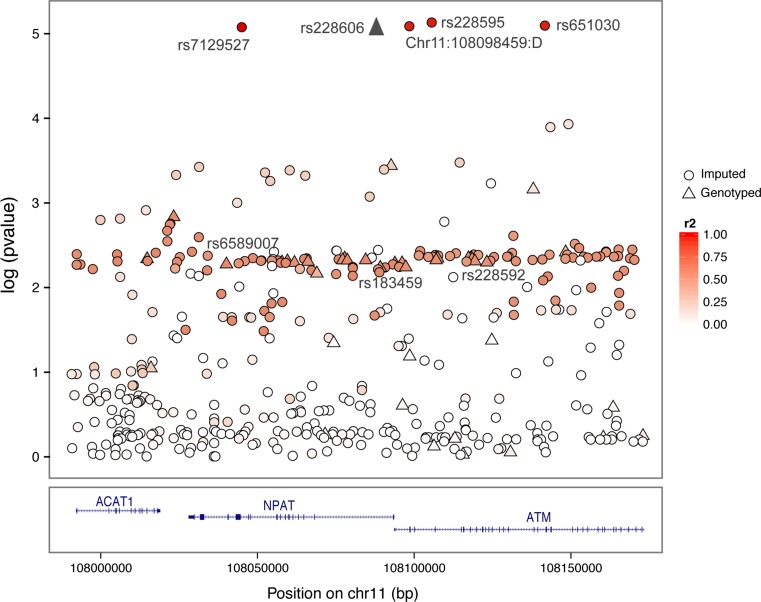
Although evidence of association with breast cancer risk was observed for the above-described loci in BRCA1 and BRCA2 mutation carriers, none of these associations reached significance after a Bonferroni adjustment for multiple testing. Imputation using the 1000 Genomes data (encompassing ± 50 kb centered on each of the 12 associated variants, Online Resource 6) identified several SNPs with significant associations in BRCA1 mutation carriers at the 11q22.3 locus (with SNP rs228595 as the most significant, p = 7.38 × 10−6), and which were partly correlated with the genotyped SNPs (r 2 <0.4, Fig. 1). After imputation, we also found associations (albeit not statistically significant after multiple testing adjustments), between one imputed SNP at locus 12p13 (rs2255390, p = 5.0 × 10−4) and breast cancer risk for BRCA1 carriers, and two SNPs and breast cancer risk for BRCA2 carriers, namely 6p22 (chr6:28226644:I, p = 9.0 × 10−4) and 8q11 (rs189286892, p = 2.0 × 10−4).
Fig. 1.
Manhattan plot depicting the strength of association between breast cancer risk in BRCA1 mutation carriers and all imputed and genotyped SNPs located across the 11q22.3 locus bound by hg19 coordinates chr11:107990104_108173189. Directly genotyped SNPs are represented as triangles and imputed SNPs (r 2 > 0.3, MAF > 0.02) are represented as circles. The linkage disequilibrium (r 2) for the most strongly associated genotyped SNP with each SNP was computed based on subjects of European ancestry that were included in the 1000 Genome Mar 2012 EUR release. Pairwise r 2 values are plotted using a red scale, where white and red means r 2 = 0 and 1, respectively. SNPs are plotted according to their chromosomal position: physical locations are based on the GRCh37/hg19 map. SNP rs228606 was genotyped in the iCOGS array but was not included in our original hypothesis of association with DAE. Gene annotation is based on the NCBI RefSeq gene descriptors from the UCSC genome browser
Ovarian cancer association analyses
Evidence of association with ovarian cancer risk (p < 10−2) was observed for six SNPs in BRCA1 mutation carriers and three SNPs in BRCA2 mutation carriers (Table 3). The strongest association with ovarian cancer risk in BRCA1 carriers was observed for rs12025623 located at 1p36.12 (p = 7 × 10−3) in an intron of the ALPL gene. Another correlated variant (r 2 >0.7) on chromosome 1 was also genotyped, namely rs1767429 (p = 9 × 10−3), which was also located within ALPL. The strongest evidence of association with ovarian cancer risk in BRCA2 mutation carriers was observed for rs2233025 (p = 5 × 10−3), located at 1p32.22 within the MAD2L2 gene. None of these associations remained statistically significant after multiple testing adjustments. Imputed genotypes of SNPs in a region encompassing ± 50 kb centered on each of the nine associated variants did not identify stronger associations.
Table 3.
Associations with ovarian cancer risk in BRCA1 and BRCA2 mutation carriers for SNPs observed at p < 10−2
| Locations | Positions | SNPs | Nearest genes | Unaffected (number) | Affected (number) | Unaffected (MAF) | HR* (95 % CI) | p values |
|---|---|---|---|---|---|---|---|---|
| BRCA1 mutation carriers | ||||||||
| 1p36.12 | 21,889,340 | rs1767429 | ALPL, RAP1GAP | 12,765 | 2460 | 0.42 | 1.092 (1.024–1.164) | 9 × 10−3 |
| 1p36.12 | 21,892,479 | rs12025623 | ALPL, RAP1GAP | 12,789 | 2460 | 0.36 | 1.098 (1.027–1.173) | 7 × 10−3 |
| 6p21.32 | 32,913,246 | rs1480380 | BRD2, HLA-DMB, HLA-DMA | 12,790 | 2462 | 0.07 | 1.178 (1.041–1.333) | 9 × 10−3 |
| 10p12.1 | 27,434,716 | rs788209 | ANKRD26, YME1L1, MASTL, ACBD5 | 12,754 | 2455 | 0.15 | 0.879 (0.804–0.961) | 5 × 10−3 |
| 17p13.1 | 8,071,592 | rs3027247 | MIR3676, C17orf59, AURKB, C17orf44, C17orf68, PFAS | 12,786 | 2461 | 0.29 | 0.905 (0.844–0.970) | 5 × 10−3 |
| 17q22 | 53,032,425 | rs17817865 | MIR4315-1, TOM1L1, COX11, STXBP4 | 12,790 | 2462 | 0.27 | 0.905 (0.842–0.971) | 8 × 10−3 |
| BRCA2 mutation carriers | ||||||||
| 1p32.22 | 11,735,652 | rs2233025 | MAD2L2, FBXO6 | 7574 | 631 | 0.18 | 0.777 (0.657–0.919) | 5 × 10−3 |
| 9p13.3 | 35,055,669 | rs595429 | VCP, FANCG, c9orf131 | 7579 | 631 | 0.46 | 0.856 (0.758–0.964) | 6 × 10−3 |
| 17q25.3 | 76,219,783 | rs2239680 | DHX29, SKIV2L2 | 7579 | 630 | 0.28 | 0.828 (0.722–0.948) | 7 × 10−3 |
CI confidence interval, HR hazard ratio, MAF minor allele frequency, SNP single-nucleotide polymorphism
* Hazard ratio per allele (one degree of freedom) estimated from the retrospective likelihood analysis
eQTL analysis in breast tissue
To identify the genes influenced via the observed associations with breast cancer at locus 11q22.3, eQTL analysis was performed using gene expression data from tumor and normal breast tissues (for detailed descriptions of datasets, refer to Online Resource 4), and all genotyped as well as imputed SNPs within a 1-Mb region on either side of the most significant genotyped SNP. eQTL associations were observed in both normal and tumor breast tissues in this region, although none of those were correlated with our most significant risk SNPs (Online Resource 7). The strongest eQTL associations were observed in the breast cancer tissue dataset BC241 for the SLC35F2 gene (rs181187590, p = 1.4 × 10−5, r 2 = 0.08, i.e., 8 % of the variation in SLC35F2 expression was attributable to this SNP). Other eQTLs observed in this dataset included ELMOD1 (rs181187590, p = 1.3 × 10−4, r 2 = 0.06), EXPH5 (rs181187590, p = 3 × 10−4, r 2 = 0.054), and ATM (rs4987915, p = 3.7 × 10−4, r 2 = 0.05). In The Cancer Genome Atlas (TCGA) BC765 breast cancer dataset, the strongest associations with gene expression were observed for the non-coding RNA lLOC643923 (rs183293362, p = 2.3 × 10−4, r 2 = 0.02), ATM (rs4987924, p = 8.3 × 10−4, r 2 = 0.015), and KDELC2 (rs4753834, p = 8.6 × 10−4, r 2 = 0.015) loci. The eQTL analysis performed for the TCGA normal breast tissue dataset (NB93) showed an association between SNP chr11:108075271:D and ACAT1 gene expression level (p = 6.5 × 10−3, r 2 = 0.08). No association was observed in the normal breast tissue dataset NB116.
Functional annotation
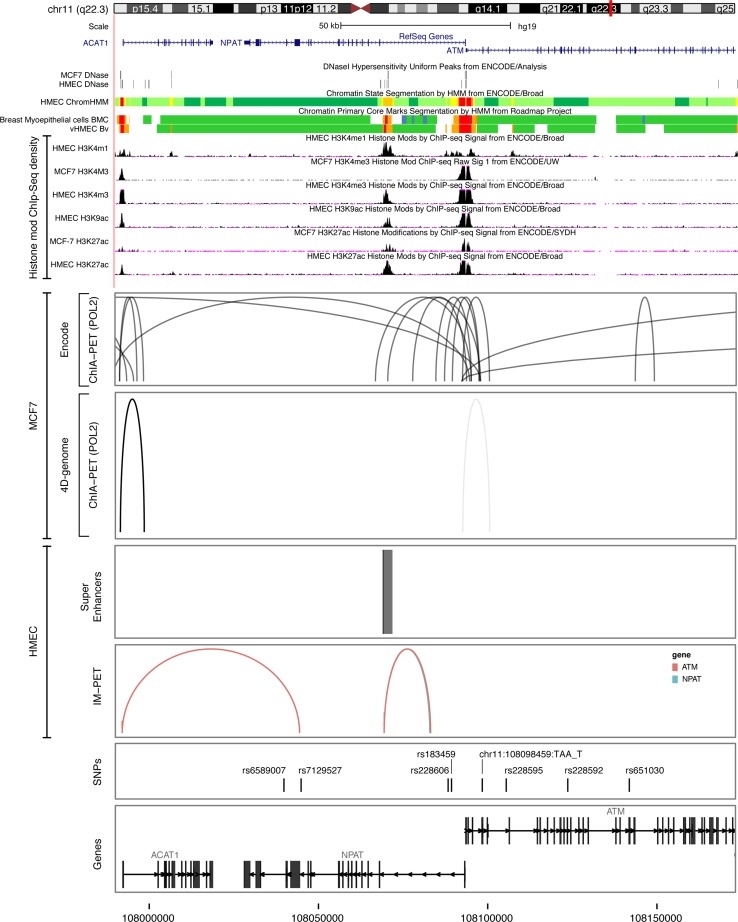
In order to assess the potential functional role of the most significant risk SNPs in the 11q22.3 region, ENCODE chromatin biological features were evaluated in available breast cells, namely HMECs, breast myoepithelial cells, and MCF7 breast cancer cells. We observed some overlap between features of interest and candidate SNPs within the 11q22.3 region (Fig. 2). The most interesting variant was rs228606, which overlapped a monomethylated H3K4 mark in HMECs. Analysis of data from the Roadmap Epigenomics project also showed overlap with a monomethylated H3K4 mark and with an acetylated H3K9 mark in primary breast myoepithelial cells. From ChiA-PET data, chromosomal interactions were found in the NPAT and ATM genes in MCF7 cells, located mainly in the vicinity of the promoter regions of these genes, which encompassed a strongly associated imputed SNP at this locus, namely chr11:108098459_TAA_T. Lastly, although super-enhancers and predicted enhancer–promoter interactions mapped to the 11q22.3 locus in HMECs, none overlapped with our top candidate SNPs (Fig. 2).
Fig. 2.
Functional annotation of the 11q22.3 locus. Upper panel functional annotations using data from the ENCODE and NIH Roadmap Epigenomics projects. From top to bottom, epigenetic signals evaluated included DNase clusters in MCF7 cells and HMECs, chromatin state segmentation by hidden Markov model (ChromHMM) in HMECs, breast myoepithelial cells, and variant human mammary epithelial cells (vHMECs), where red represents an active promoter region, orange a strong enhancer, and yellow a poised enhancer (the detailed color scheme of chromatin states is described in the UCSC browser), and histone modifications in MCF7 and HMEC cell lines. All tracks were generated by the UCSC genome browser (hg 19 release). Lower panel long-range chromatin interactions: from top to bottom, ChiA-PET interactions for RNA polymerase II in MCF-7 cells identified through ENCODE and 4D-genome. The ChiA-PET raw data available from the GEO database under the following accession (GSE33664, GSE39495) were processed with the GenomicRanges package. Maps of mammary cell super-enhancer locations as defined in Hnisz et al. [24] are shown in HMECs. Predicted enhancer–promoter determined interactions in HMECs, as defined by the integrated method for predicting enhancer targets (IM-PET), are shown. The annotation was obtained through the Bioconductor annotation package TxDb.Hsapiens.UCSC.hg19.knownGene. The tracks have been generated using ggplot2 and ggbio library in R
Discussion
DAE is a common phenomenon in human genes, which represents a new approach to identifying cis-acting mechanisms of gene regulation. It offers a new avenue for the study of GWAS variants significantly associated with various diseases/traits. Indeed, the majority of GWAS hits localize outside known protein-coding regions [11, 12], suggesting a regulatory role for these variants. In the present study, we have assessed the association between 320 SNPs associated with DAE and breast/ovarian cancer risk among BRCA1 and BRCA2 mutation carriers. Using this approach, we found evidence of association for a region at 11q22.3, with breast cancer risk in BRCA1 mutation carriers. Analysis of imputed SNPs across a 185-kb region (±50 kb from the center of each of the three genotyped SNPs at this locus) revealed a set of five strongly correlated SNPs that were significantly associated with breast cancer risk. This region contains several genes including ACAT1, NPAT, and ATM. ACAT1 (acetyl-CoA acetyltransferase 1) encodes a mitochondrial enzyme that catalyzes the reversible formation of acetoacetyl-CoA from two molecules of acetyl-CoA. Defects in this gene are associated with ketothiolase deficiency, an inborn error of isoleucine catabolism [29]. NPAT (nuclear protein, co-activator of histone transcription) is required for progression through the G1 and S phases of the cell cycle, for S phase entry [30], and for the activation of the transcription of histones H2A, H2B, H3, and H4 [31]. NPAT germline mutations have been associated with Hodgkin lymphoma [32]. Finally, ATM (ataxia telangiectasia mutated) encodes an important cell cycle checkpoint kinase that is required for cellular response to DNA damage and for genome stability. Mutations in this gene are associated with ataxia telangiectasia, an autosomal recessive disorder [33]. ATM is also an intermediate-risk breast cancer susceptibility gene, with rare heterozygous variants being associated with increased risk of developing the disease [34]. Although several studies have assessed the role of the most common ATM variants in breast cancer susceptibility, the results obtained are inconsistent [35]. A recent study had identified an association between an ATM haplotype and breast cancer risk in BRCA1 mutation carriers with a false discovery rate-adjusted p value of 0.029 for overall association of the haplotype [36]. Four of the five SNPs making up the haplotype were almost perfectly correlated (r 2 >0.9) with the three originally genotyped SNPs of the present study. These SNPs were, however, only moderately correlated (r 2 >0.4) with the most significant risk SNPs (p = 10−6), identified later through imputation.
Although eQTL analysis has identified cis-eQTL associations between several variants and ACAT1, ATM as well as other neighboring genes in both breast carcinoma and normal breast tissues, none of these associations involved the most significantly associated risk SNPs. Furthermore, the correlation between eQTLs and the most significant risk SNPs was weak. The lack of consistency between the eQTL results and the allelic imbalance data originally used for SNP selection in the design of the present study can probably be explained by the differences between the cell types used in these analyses. The list of allelic imbalance-associated SNPs was selected from studies performed in lymphoblastoid cell lines [15], primary skin fibroblasts [13, 16], and primary monocytes [17], while eQTLs were analyzed in breast carcinoma and normal breast tissue. This tissue heterogeneity in the data sources used represents one of the limitations of this study, although no such data were available in mammary cells when this study was originally designed.
The identification of a region at 11q22.3 that is associated specifically with breast cancer risk in BRCA1 mutation carriers may explain why association studies performed using breast cancer cases from the general population have so far yielded conflicting results with regard to common variants at this locus. The majority of tumors arising in BRCA1 carriers show either low or absent ER expression, while the majority of BRCA2-associated tumors are ER positive, as in most sporadic cancers arising in the general population. Large-scale studies using only ER-negative or triple-negative (i.e., ER-, progesterone receptor-, and HER2-negative) cases could therefore be helpful to confirm the association of this locus with breast cancer risk.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Silje Nord was financed by a Carrier Grant from the Norwegian Regional Health authorities (Grant Number 2014061). BCFR-AU: Maggie Angelakos, Judi Maskiell, Gillian Dite, Helen Tsimiklis. BCFR-NY: we wish to thank members and participants in the New York site of the Breast Cancer Family Registry for their contributions to the study. BCFR-ON: we wish to thank the members and participants in the Ontario Familial Breast Cancer Registry for their contributions to the study. BFBOCC-LT thank Vilius Rudaitis and Laimonas Griškevičius. BFBOCC-LV thank Drs. Janis Eglitis, Anna Krilova, and Aivars Stengrevics. BMBSA wish to thank the families who contribute to the BMBSA study. BRICOH: we wish to thank Yuan Chun Ding and Linda Steele for their work in participant enrollment and biospecimen and data management. CBCS: we thank Bent Ejlertsen and Anne-Marie Gerdes for the recruitment and genetic counseling of participants. CNIO: we thank Alicia Barroso, Rosario Alonso, and Guillermo Pita for their assistance. CONSIT TEAM: Daniela Zaffaroni of the Fondazione IRCCS Istituto Nazionale Tumori (INT), Milan, Italy; Monica Barile and Irene Feroce of the Istituto Europeo di Oncologia, Milan; Maria Grazia Tibiletti of the Ospedale di Circolo-Università dell’Insubria, Varese, Italy; Liliana Varesco of the IRCCS AOU San Martino: IST Istituto Nazionale per la Ricerca sul Cancro, Genoa, Italy; Alessandra Viel of the CRO Aviano National Cancer Institute, Aviano, Italy; Gabriele Capone of the University of Florence, Florence, Italy; Laura Ottini and Giuseppe Giannini of the “Sapienza” University, Rome, Italy; Antonella Savarese and Aline Martayan of the Istituto Nazionale Tumori Regina Elena, Rome, Italy; Stefania Tommasi and Brunella Pilato of the Istituto Nazionale Tumori “Giovanni Paolo II,” Bari, Italy; and the personnel of the Cogentech Cancer Genetic Test Laboratory, Milan, Italy. CORE: the CIMBA data management and analysis was funded through Cancer Research: UK Grant C12292/A11174. ACA is a Senior Cancer Research: UK Research Fellow. We wish to thank Sue Healey for her enormous contribution to CIMBA, in particular taking on the task of mutation classification with Olga Sinilnikova. EMBRACE: RE was supported by NIHR support to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. FCCC: we thank Ms. JoEllen Weaver and Dr. Betsy Bove for their technical support. GC-HBOC: we would like to thank LIFE: Leipzig Research Centre for Civilization Diseases (Markus Loeffler, Joachim Thiery, Matthias Nüchter, Ronny Baber). Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) Study: National Cancer Genetics Network «UNICANCER Genetic Group», France. We wish to pay a tribute to Olga M. Sinilnikova, who with Dominique Stoppa-Lyonnet initiated and coordinated GEMO until she sadly passed away on the 30th June 2014, and to thank all the GEMO Collaborating Groups for their contribution to this study. GEMO Collaborating Centers are as follows: Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Hospices Civils de Lyon: Centre Léon Bérard, and Equipe «Génétique du cancer du sein», Centre de Recherche en Cancérologie de Lyon: Olga Sinilnikova†, Sylvie Mazoyer, Francesca Damiola, Laure Barjhoux, Carole Verny-Pierre, Mélanie Léone, Nadia Boutry-Kryza, Alain Calender, Sophie Giraud; and Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Bruno Buecher, Claude Houdayer, Etienne Rouleau, Lisa Golmard, Agnès Collet, Virginie Moncoutier, Muriel Belotti, Antoine de Pauw, Camille Elan, Catherine Nogues, Emmanuelle Fourme, Anne-Marie Birot. Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Olivier Caron, Marine Guillaud-Bataille. Centre Jean Perrin, Clermont–Ferrand: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona, Sandrine Handallou. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet, Dominique Vaur, Laurent Castera. Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, Audrey Remenieras, François Eisinger. CHU Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier, Pascal Pujol. Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin†, Claude Adenis. Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker. Institut Bergonié, Bordeaux: Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Nicolas Sevenet, Michel Longy. Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel. CHU Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung, Magalie Peysselon. CHU Dijon: Fanny Coron, Laurence Faivre. CHU St-Etienne: Fabienne Prieur, Marine Lebrun, Caroline Kientz. Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice: Marc Frénay. CHU Limoges: Laurence Vénat-Bouvet. CHU Nantes: Capucine Delnatte. CHU Bretonneau, Tours: Isabelle Mortemousque. Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier, Mathilde Warcoin. CHU Vandoeuvre-les-Nancy: Johanna Sokolowska, Myriam Bronner. CHU Besançon: Marie-Agnès Collonge-Rame, Alexandre Damette. Creighton University, Omaha, USA: Henry T. Lynch, Carrie L. Snyder. G-FAST: we wish to thank the technical support of Ilse Coene en Brecht Crombez. HCSC: we acknowledge the technical assistance of Alicia Tosar and Paula Diaque. HEBCS would like to thank Drs. Sofia Khan, Taru A. Muranen, Carl Blomqvist and RNs Irja Erkkilä and Virpi Palola for their help with the HEBCS data and samples. The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) consists of the following Collaborating Centers: Coordinating Center: Netherlands Cancer Institute, Amsterdam, NL: M.A. Rookus, F.B.L. Hogervorst, F.E. van Leeuwen, S. Verhoef, M.K. Schmidt, N.S. Russell, D.J. Jenner; Erasmus Medical Center, Rotterdam, NL: J.M. Collée, A.M.W. van den Ouweland, M.J. Hooning, C. Seynaeve, C.H.M. van Deurzen, I.M. Obdeijn; Leiden University Medical Center, NL: C.J. van Asperen, J.T. Wijnen, R.A.E.M. Tollenaar, P. Devilee, T.C.T.E.F. van Cronenburg; Radboud University Nijmegen Medical Center, NL: C.M. Kets, A.R. Mensenkamp; University Medical Center Utrecht, NL: M.G.E.M. Ausems, R.B. van der Luijt, C.C. van der Pol; Amsterdam Medical Center, NL: C.M. Aalfs, T.A.M. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz, H.E.J. Meijers-Heijboer; University Hospital Maastricht, NL: E.B. Gómez-Garcia, M.J. Blok; University Medical Center Groningen, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits, G.H. de Bock; The Netherlands Foundation for the Detection of Hereditary Tumours, Leiden, NL: H.F. Vasen; The Netherlands Comprehensive Cancer Organization (IKNL): S. Siesling, J. Verloop; The Dutch Pathology Registry (PALGA): L.I.H. Overbeek. The HEBON study was supported by the Dutch Cancer Society Grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the Netherlands Organization of Scientific Research Grant NWO 91109024, the Pink Ribbon Grants 110005 and 2014-187.WO76, the BBMRI Grant NWO 184.021.007/CP46, and the Transcan Grant JTC 2012 Cancer 12-054. HEBON thanks the registration teams of IKNL and PALGA for their help in part of the data collection. HRBCP: we wish to thank Hong Kong Sanatorium and Hospital for their continued support. HUNBOCS: we wish to thank the Hungarian Breast and Ovarian Cancer Study Group Members (Janos Papp, Tibor Vaszko, Aniko Bozsik, Judit Franko, Maria Balogh, Gabriella Domokos, Judit Ferenczi, Department of Molecular Genetics, National Institute of Oncology, Budapest, Hungary) and the clinicians and patients for their contributions to this study. HVH: we wish to thank the Oncogenetics Group (VHIO) and the High Risk and Cancer Prevention Unit of the University Hospital Vall d’Hebron. Thanks to the Cellex Foundation for providing research facilities and equipment. ICO: we wish to thank the ICO Hereditary Cancer Program Team led by Dr. Gabriel Capella. INHERIT: we would like to thank Dr. Martine Dumont for sample management and skillful assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. J.S. and P.S. were part of the QC and Genotyping Coordinating Group of iCOGS (BCAC and CIMBA). IPOBCS: we wish to thank Drs. Ana Peixoto, Catarina Santos, and Pedro Pinto for their skillful contribution to the study. KCONFAB: we wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study [which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)] for their contributions to this resource, and the many families who contribute to kConFab. Modifier Study of Quantitative Effects on Disease (MODSQUAD) thank ModSQuaD members Csilla Szabo (National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA); Lenka Foretova and Eva Machackova (Department of Cancer Epidemiology and Genetics, Masaryk Memorial Cancer Institute and MF MU, Brno, Czech Republic); and Michal Zikan, Petr Pohlreich, and Zdenek Kleibl (Oncogynecologic Center and Department of Biochemistry and Experimental Oncology, First Faculty of Medicine, Charles University, Prague, Czech Republic). MSKCC: Anne Lincoln, Lauren Jacobs. NICCC: we wish to thank the NICCC National Familial Cancer Consultation Service Team led by Sara Dishon, the Lab Team led by Dr. Flavio Lejbkowicz, and the Research Field Operations Team led by Dr. Mila Pinchev. NRG Oncology: we thank the investigators of the Australia New Zealand NRG Oncology Group. OCGN: we wish to thank the members and participants in the Ontario Cancer Genetics Network for their contributions to the study. OSUCCG: Leigha Senter, Kevin Sweet, Caroline Craven, Julia Cooper, and Michelle O’Conor were instrumental in accrual of study participants, ascertainment of medical records, and database management. SEABASS: we would like to thank Yip Cheng Har, Nur Aishah Mohd Taib, Phuah Sze Yee, Norhashimah Hassan, and all the research nurses, research assistants, and doctors involved in the MyBrCa Study for their assistance in patient recruitment, data collection, and sample preparation. In addition, we thank Philip Iau, Sng Jen-Hwei, and Sharifah Nor Akmal for contributing samples from the Singapore Breast Cancer Study and the HUKM-HKL Study. The Malaysian Breast Cancer Genetic Study was funded by Research Grants from the Malaysian Ministry of Science, Technology and Innovation, Ministry of Higher Education (UM.C/HIR/MOHE/06), and charitable funding from Cancer Research Initiatives Foundation. SMC Team wishes to acknowledge the assistance of the Meirav Comprehensive Breast Cancer Center Team at the Sheba Medical Center for their assistance in this study. SWE-BRCA: Swedish scientists participating as SWE-BRCA Collaborators are from Lund University and University Hospital: Åke Borg, Håkan Olsson, Helena Jernström, Karin Henriksson, Katja Harbst, Maria Soller, Ulf Kristoffersson; from Gothenburg Sahlgrenska University Hospital: Anna Öfverholm, Margareta Nordling, Per Karlsson, Zakaria Einbeigi; from Stockholm and Karolinska University Hospital: Anna von Wachenfeldt, Annelie Liljegren, Annika Lindblom, Brita Arver, Gisela Barbany Bustinza, Johanna Rantala; from Umeå University Hospital: Beatrice Melin, Christina Edwinsdotter Ardnor, Monica Emanuelsson; from Uppsala University: Hans Ehrencrona, Maritta Hellström Pigg, Richard Rosenquist; and from Linköping University Hospital: Marie Stenmark-Askmalm, Sigrun Liedgren. UCHICAGO: we wish to thank Cecilia Zvocec, Qun Niu, physicians, genetic counselors, research nurses and staff of the Cancer Risk Clinic for their contributions to this resource, and the many families who contribute to our program. UCLA: we thank Joyce Seldon MSGC and Lorna Kwan, MPH, for assembling the data for this study. UCSF: we would like to thank Dr. Robert Nussbaum and the following genetic counselors for participant recruitment: Beth Crawford, Kate Loranger, Julie Mak, Nicola Stewart, Robin Lee, Amie Blanco, and Peggy Conrad. And thanks to Ms. Salina Chan for her data management. UKFOCR: we thank Susan Ramus, Carole Pye, Patricia Harrington, and Eva Wozniak for their contributions toward the UKFOCR. VFCTG: Geoffrey Lindeman, Marion Harris, Martin Delatycki of the Victorian Familial Cancer Trials Group. We thank Sarah Sawyer and Rebecca Driessen for assembling the data and Ella Thompson for performing all DNA amplification.
Funding
Funding for the iCOGS infrastructure came from the European Community’s Seventh Framework Programme under Grant Agreement No. 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-cancer GWAS Initiative (1U19 CA148537, 1U19 CA148065, and 1U19 CA148112: the GAME-ON Initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. BCFR: this work was supported by Grant UM1 CA164920 from the National Cancer Institute. BFBOCC was supported by Lithuania (BFBOCC-LT): Research Council of Lithuania Grant SEN-18/2015. BIDMC was supported by the Breast Cancer Research Foundation. BRCA-gene mutations and breast cancer in South African women (BMBSA) was supported by Grants from the Cancer Association of South Africa (CANSA) to Elizabeth J. van Rensburg. BRICOH: SLN was partially supported by the Morris and Horowitz Families Endowed Professorship. CNIO: this work was partially supported by Spanish Association Against Cancer (AECC08), RTICC 06/0020/1060, FISPI08/1120, Mutua Madrileña Foundation (FMMA), and SAF2010-20493. COH-CCGCRN: patients were recruited for this study from the City of Hope Clinical Cancer Genomics Community Research Network, supported in part by Award Number RC4CA153828 (PI: J. Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. CONSIT Team: Associazione Italiana Ricerca sul Cancro (AIRC) to P. Peterlongo (IG 2012 Id.12821) and P. Radice (IG 2014 Id.15547). Funds from Italian citizens who allocated the 5 × 1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional Strategic Projects ‘5x1000’) to S. Manoukian. FiorGen Foundation for Pharmacogenomics to L. Papi. CORE: the CIMBA Data Management and Data Analysis were supported by Cancer Research: UK Grants C12292/A20861, C12292/A11174. ACA is a Cancer Research-UK Senior Cancer Research Fellow. GCT is an NHMRC Senior Principal Research Fellow. J. Lecarpentier has been financially supported by the Fondation ARC (FONDATION ARC, 9 rue Guy Môquet 94803 Villejuif: France), Grant Number SAE20131200623. This work was supported by the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” Program: Grant No. CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade: Grant No. PSR-SIIRI-701. The PERSPECTIVE Project was supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the Ministère de l’Économie, Innovation et Exportation du Québec through Genome Québec, and The Quebec Breast Cancer Foundation. This work was also supported by the Ministère de l’Économie, Innovation et Exportation du Québec: Grant No. PSR-SIIRI-701. DEMOKRITOS: this research has been co-financed by the European Union (European Social Fund: ESF) and Greek National Funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF): Research Funding Program of the General Secretariat for Research and Technology: SYN11_10_19 NBCA. Investing in Knowledge Society through the European Social Fund. The DKFZ Study was supported by the DKFZ. EMBRACE was supported by Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo were supported by an NIHR Grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust were supported by an NIHR Grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles and Elizabeth Bancroft were supported by Cancer Research UK Grant C5047/A8385. Ros Eeles was also supported by NIHR support to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. FCCC: the authors acknowledge the support from The University of Kansas Cancer Center (P30 CA168524) and the Kansas Bioscience Authority Eminent Scholar Program. A.K.G. was funded by 5U01CA113916, R01CA140323, and by the Chancellors Distinguished Chair in Biomedical Sciences Professorship. The German Consortium of Hereditary Breast and Ovarian Cancer (GC-HBOC) was supported by the German Cancer Aid (Grant No. 110837, Rita K. Schmutzler). This work was supported by the European Regional Development Fund and Free State of Saxony, Germany (LIFE: Leipzig Research Centre for Civilization Diseases, Project Numbers 713-241202, 713-241202, 14505/2470, 14575/2470). GEMO: the study was supported by the Ligue Nationale Contre le Cancer; the Association “Le cancer du sein, parlons-en!” Award; the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” Program and the French National Institute of Cancer (INCa). GEORGETOWN: CI received support from the Non-therapeutic Subject Registry Shared Resource at Georgetown University (NIH/NCI Grant P30-CA051008), the Fisher Center for Hereditary Cancer and Clinical Genomics Research, and Swing Fore the Cure. G-FAST: Bruce Poppe is a Senior Clinical Investigator of FWO. Mattias Van Heetvelde obtained funding from IWT. HCSC was supported by a Grant RD12/0036/0006 and 15/00059 from ISCIII (Spain), partially supported by European Regional Development FEDER Funds. The HEBCS was financially supported by the Helsinki University Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society and the Sigrid Juselius Foundation. HEBON Study was supported by the Dutch Cancer Society Grants NKI1998-1854, NKI2004-3088, and NKI2007-3756, the Netherlands Organization of Scientific Research Grant NWO 91109024, the Pink Ribbon Grants 110005 and 2014-187.WO76, the BBMRI Grant NWO 184.021.007/CP46, and the Transcan Grant JTC 2012 Cancer 12-054. HEBON thanks the Registration Teams of Dutch Cancer Registry (IKNL; S. Siesling, J. Verloop) and the Dutch Pathology database (PALGA; L. Overbeek) for their help in part of the data collection. HRBCP was supported by the Hong Kong Sanatorium and Hospital, Dr. Ellen Li Charitable Foundation, The Kerry Group Kuok Foundation, National Institute of Health 1R 03CA130065, and North California Cancer Center. Hungarian Breast and Ovarian Cancer Study (HUNBOCS) was supported by Hungarian Research Grants KTIA-OTKA CK-80745 and OTKA K-112228. ICO: Contract Grant Sponsor: Asociación Española Contra el Cáncer, Spanish Health Research Fund; Carlos III Health Institute; Catalan Health Institute and Autonomous Government of Catalonia. Contract Grant Numbers: ISCIIIRETIC RD06/0020/1051, RD12/0036/008, PI10/01422, PI10/00748, PI13/00285, PIE13/00022, 2009SGR290, and 2014SGR364. The IHCC was supported by Grant PBZ_KBN_122/P05/2004. The ILUH Group was supported by the Icelandic Association “Walking for Breast Cancer Research” and by the Landspitali University Hospital Research Fund. INHERIT: this work was supported by the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” Program: Grant No. CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade: Grant No. PSR-SIIRI-701. The PERSPECTIVE Project was supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research (Grant GPH-129344), the Ministère de l’Économie, Science et Innovation du Québec through Genome Québec, and the Quebec Breast Cancer Foundation. IOVHBOCS was supported by Ministero della Salute and “5x1000” Istituto Oncologico Veneto Grant. IPOBCS: this study was in part supported by Liga Portuguesa Contra o Cancro. kConFab was supported by a Grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania, and South Australia, and the Cancer Foundation of Western Australia. KOHBRA was supported by a Grant from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (1020350). MAYO was supported by NIH Grants CA116167, CA192393, and CA176785, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), and a Grant from the Breast Cancer Research Foundation. MCGILL: Jewish General Hospital Weekend to End Breast Cancer, Quebec Ministry of Economic Development, Innovation and Export Trade. MODSQUAD was supported by MH CZ: DRO (MMCI, 00209805) and by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101) to LF, and by Charles University in Prague Project UNCE204024 (MZ). MSKCC was supported by Grants from the Breast Cancer Research Foundation, the Robert and Kate Niehaus Clinical Cancer Genetics Initiative, and the Andrew Sabin Research Fund. NAROD: 1R01 CA149429-01. NCI: the research of Drs. MH Greene and PL Mai was supported by the Intramural Research Program of the US National Cancer Institute, NIH, and by Support Services Contracts NO2-CP-11019-50 and N02-CP-65504 with Westat, Inc., Rockville, MD. NICCC was supported by Clalit Health Services in Israel. Some of its activities were supported by the Israel Cancer Association and the Breast Cancer Research Foundation (BCRF), NY. NNPIO: this work has been supported by the Russian Federation for Basic Research (Grants 15-04-01744 and 16-54-00055). NRG Oncology: this study was supported by NRG Oncology Operations Grant Number U10 CA180868 as well as NRG SDMC Grant U10 CA180822, Gynecologic Oncology Group (GOG) Administrative Office and the GOG Tissue Bank (CA 27469), and the GOG Statistical and Data Center (CA 37517). Drs. Greene, Mai, and Savage were supported by Funding from the Intramural Research Program, NCI. OSUCCG was supported by the Ohio State University Comprehensive Cancer Center. PBCS: this work was supported by the Italian Association of Cancer Research (AIRC) [IG 2013 N.14477] and Tuscany Institute for Tumors (ITT) Grant 2014–2015–2016. SEABASS: Ministry of Science, Technology and Innovation, Ministry of Higher Education (UM.C/HlR/MOHE/06), and Cancer Research Initiatives Foundation. The SMC Team was in part sponsored by a Grant from the Israeli Cancer Association to the Israeli Consortium of Hereditary Breast Cancer. SWE-BRCA Collaborators were supported by the Swedish Cancer Society. UCHICAGO was supported by NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA125183), R01 CA142996, 1U01CA161032, and by the Ralph and Marion Falk Medical Research Trust, the Entertainment Industry Fund National Women’s Cancer Research Alliance, and the Breast Cancer Research Foundation. OIO is an ACS Clinical Research Professor. UCLA: Jonsson Comprehensive Cancer Center Foundation; Breast Cancer Research Foundation. UCSF: UCSF Cancer Risk Program and Helen Diller Family Comprehensive Cancer Center. UKFOCR was supported by a Project Grant from CRUK to Paul Pharoah. UPENN: National Institutes of Health (NIH) (R01-CA102776 and R01-CA083855; Breast Cancer Research Foundation; Susan G. Komen Foundation for the Cure, Basser Research Center for BRCA. VFCTG: Victorian Cancer Agency, Cancer Australia, National Breast Cancer Foundation. WCP: Dr. Karlan was funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Study participants were recruited through the CIMBA Initiative, following the approval of the corresponding protocol by the Institutional Review Board or Ethics Committee at each participating center. Written informed consent was obtained from all study participants.
Footnotes
The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the Collaborating Centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Rita Katharina Schmutzler: On behalf of the German Consortium of Hereditary Breast and Ovarian Cancer (GC-HBOC).
References
- 1.Begg CB, Haile RW, Borg A, Malone KE, Concannon P, Thomas DC, et al. Variation of breast cancer risk among BRCA1/2 carriers. JAMA. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105:812–822. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simchoni S, Friedman E, Kaufman B, Gershoni-Baruch R, Orr-Urtreger A, Kedar-Barnes I, et al. Familial clustering of site-specific cancer risks associated with BRCA1 and BRCA2 mutations in the Ashkenazi Jewish population. Proc Natl Acad Sci USA. 2006;103:3770–3774. doi: 10.1073/pnas.0511301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313:1347–1361. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebel TM, Domchek SM, Rebbeck TR. Modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: systematic review and meta-analysis. J Natl Cancer Inst. 2014;106:dju091. doi: 10.1093/jnci/dju091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudet MM, Kuchenbaecker KB, Vijai J, Klein RJ, Kirchhoff T, McGuffog L, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9:e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes DR, Antoniou AC. Unravelling modifiers of breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers: update on genetic modifiers. J Intern Med. 2012;271:331–343. doi: 10.1111/j.1365-2796.2011.02502.x. [DOI] [PubMed] [Google Scholar]
- 11.Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Stram A, Chen C, Kar S, Gayther S, Pharoah P, et al. Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum Mol Genet. 2014;23:5294–5302. doi: 10.1093/hmg/ddu228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104–107. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 16.Adoue V, Schiavi A, Light N, Almlöf JC, Lundmark P, Ge B, et al. Allelic expression mapping across cellular lineages to establish impact of non-coding SNPs. Mol Syst Biol. 2014;10:754. doi: 10.15252/msb.20145114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almlöf JC, Lundmark P, Lundmark A, Ge B, Maouche S, Göring HH, et al. Powerful identification of cis-regulatory SNPs in human primary monocytes using allele-specific gene expression. PLoS ONE. 2012;7:e52260. doi: 10.1371/journal.pone.0052260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, Lam KC, et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet. 2011;7:e1001279. doi: 10.1371/journal.pgen.1001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes DR, Lee A, EMBRACE Investigators. kConFab Investigators. Easton DF, Antoniou AC. Evaluation of association methods for analysing modifiers of disease risk in carriers of high-risk mutations. Genet Epidemiol. 2012;36:274–291. doi: 10.1002/gepi.21620. [DOI] [PubMed] [Google Scholar]
- 21.Antoniou AC, Sinilnikova OM, Simard J, Léoné M, Dumont M, Neuhausen SL, et al. RAD51 135G/C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulligan AM, Couch FJ, Barrowdale D, Domchek SM, Eccles D, Nevanlinna H, et al. Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res. 2011;13:R110. doi: 10.1186/bcr3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng L, He B, Tan K. 4DGenome: a comprehensive database for chromatin interactions. Bioinformatics. 2015;31:2560–2564. doi: 10.1093/bioinformatics/btv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He B, Chen C, Teng L, Tan K. Global view of enhancer–promoter interactome in human cells. Proc Natl Acad Sci USA. 2014;111:E2191–E2199. doi: 10.1073/pnas.1320308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beauparlant CJ, Lemacon A, Droit A (2015) ENCODExplorer: a compilation of ENCODE metadata. R package version 1.4.3
- 27.Beauparlant CJ, Lamaze F, Deschenes A, Samb R, Lemaçon A, Belleau P, Bilodeau S, Droit A. Metagene profiles analyses reveal regulatory element’s factor-specific recruitment patterns. PLoS Comput Biol. 2016;12:e1004751. doi: 10.1371/journal.pcbi.1004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 29.Fukao T, Yamaguchi S, Kano M, Orii T, Fujiki Y, Osumi T, et al. Molecular cloning and sequence of the complementary DNA encoding human mitochondrial acetoacetyl-coenzyme A thiolase and study of the variant enzymes in cultured fibroblasts from patients with 3-ketothiolase deficiency. J Clin Investig. 1990;86:2086–2092. doi: 10.1172/JCI114946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. doi: 10.1101/gad.827700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saarinen S, Aavikko M, Aittomäki K, Launonen V, Lehtonen R, Franssila K, et al. Exome sequencing reveals germline NPAT mutation as a candidate risk factor for Hodgkin lymphoma. Blood. 2011;118:493–498. doi: 10.1182/blood-2011-03-341560. [DOI] [PubMed] [Google Scholar]
- 33.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 34.Tavtigian SV, Oefner PJ, Babikyan D, Hartmann A, Healey S, Le Calvez-Kelm F, et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet. 1009;85:427–446. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milne RL. Variants in the ATM gene and breast cancer susceptibility. Genome Med. 2009;1:12. doi: 10.1186/gm12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebbeck TR, Mitra N, Domchek SM, Wan F, Friebel TM, Tran TV, et al. Modification of BRCA1-associated breast and ovarian cancer risk by BRCA1-interacting genes. Cancer Res. 2011;71:5792–5805. doi: 10.1158/0008-5472.CAN-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.