Abstract
The bacterial spectrum in chronic rhinosinusitis (CRS) is clinically relevant. This study aimed to compare two sampling techniques and to characterise Staphylococcus aureus isolated from CRS patients. Bacterial specimens were collected from the nares and maxillary sinus in 42 CRS patients and from the nares in 57 healthy controls. Maxillary sinus sampling was performed in two ways in each patient: with a cotton-tipped aluminium swab through the enlarged sinus ostium, and with a protected brush. S. aureus was characterised by DNA-sequencing of the repeat region of the S. aureus protein A gene, spa typing. The protected brush technique was superior to the cotton-tipped aluminium swab in reducing contamination rate. However, the two sampling methods were consistent in terms of clinically relevant bacterial findings, and the easy-to-handle cotton-tipped swab can still be recommended when culturing the maxillary sinus. Patients showed a significantly higher presence of S. aureus in the nares compared with healthy controls, and healthy controls showed a significantly higher presence of coagulase-negative staphylococci in the nares compared with patients. The spa types were identical for the nares and maxillary sinus in all patients except one. The sampling techniques showed equivalent results, indicating a low risk of unnecessary antibiotic treatment when using the easy-to-handle cotton-tipped aluminium swab. The high rate of identical spa types of S. aureus isolated from the nares and maxillary sinus of CRS patients might indicate colonisation of the maxillary sinus from the nares.
Keywords: Staphylococcus aureus, Sinusitis, Nasal polyps, Sampling studies, Bacterial typing
Introduction
Chronic rhinosinusitis (CRS) affects more than 10 % of the European population [1]. The condition is characterised by nasal congestion, nasal discharge, headache, facial fullness, and changes in smell and taste lasting longer than 3 months, and is verified by nasal endoscopy and/or computerised tomography of the sinuses [2, 3]. The disease is probably multifactorial, and different factors have been suggested to affect its development, including environmental factors and host factors [4–7]. Reduced ventilation of the sinuses due to blockage of the ostiomeatal complex in the middle nasal meatus is thought to be one factor [6, 8]. Reduced oxygen pressure in the sinus and absorption of oxygen might promote bacterial growth [9]. Allergy and asthma are suggested to enhance mucosal swelling and cause obstruction of the ostium, thereby predisposing for CRS. Immune deficiencies, cell membrane sodium and chloride channel malfunction, and ciliary dysfunction may also be contributing factors [10]. Important environmental factors in development of CRS are probably microbial, especially fungi and bacteria contributing to chronic mucosal inflammation [7]. Bacterial biofilms are often found in sinuses in patients with CRS undergoing sinus surgery and are associated with more severe disease [11, 12]. Biofilms are complex structures composed of communities of microbs embedded within an extracellular matrix, predominantly polysaccharides. Bacterial involvement is well accepted in the pathogenesis of acute sinusitis, where Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are the most common bacterial findings [5, 13]. The role of bacteria in CRS is less clear, and findings of Staphylococcus aureus, coagulase-negative staphylococci (CoNS), and anaerobes seem to dominate to various extents in different studies [13–19]. However, a Brazilian study of 62 samples from maxillary sinuses of CRS patients found no anaerobes, and Pseudomonas aeruginosa was the most commonly found bacterium [20]. The variability in microbial presence in different studies might be a result of differences in culturing techniques, contamination of samples, patient selection, ethnic origin of the patients, and pre-treatment regimens. In addition, handling of samples can affect growth due to the high sensitivity of the anaerobes. S. aureus in maxillary sinus cultures has been reported in about 25 % of patients with CRS [21]. A meta-analysis supports the role of S. aureus in asthma and allergic rhinitis [22], and S. aureus has also been shown to have an association with inflammatory diseases, such as atopic dermatitis [23, 24]. S. aureus displays a wide range of virulence factors; among these, staphylococcal enterotoxins and toxic shock syndrome toxin-1 have been demonstrated to activate the immune system and affect proinflammatory cells by acting as superantigens. Some studies have suggested that chronic rhinosinusitis with nasal polyposis (CRSwNP) has a relationship with S. aureus infection and especially with staphylococcal enterotoxins as a modulator of the disease [6, 25–27]. The aim of this study was to investigate the bacterial spectrum patients with CRS, and especially the presence of S. aureus, and to characterise S. aureus isolated from the nares and maxillary sinus of CRS patients in comparison with samples from the nares in healthy controls. Another aim was to evaluate an optimised culturing technique for the maxillary sinus.
Materials and methods
Patients and controls
Forty-two patients with CRS were recruited at the Department of Otolaryngology, Örebro University Hospital, Sweden, from 2004 to 2010. Two ENT specialists (UT and SH) were responsible for the inclusion procedure. The diagnosis of CRS was based on history, clinical examination, and computed tomography scans according to the definitions and guidelines of the American Academy of Otolaryngology—Head and Neck Surgery [2]. A position paper on rhinosinusitis guidelines [28], prepared by the European Academy of Allergology and Clinical Immunology and approved by the European Rhinologic Society, [3] was published after the inclusion procedure for this study had started. However, the definition of this position paper matches that used in this study. Patients visiting the ENT outpatient clinic with CRS and an enlarged opening to the maxillary sinus due to previous surgery, and patients with CRS admitted for sinus surgery, were invited to participate. Nasal endoscopy was performed, and the presence or absence of nasal polyps was documented; thus, patients were identified as having either chronic rhinosinusitis without nasal polyposis (CRSsNP) or with nasal polyposis (CRSwNP). The inclusion procedure was not consecutive. Healthy volunteers were invited to participate in the study when visiting the Örebro travel consultation clinic. Fifty-seven controls were enrolled, and all of them did declare “no nasal polyps as an adult” and “no previous sinus surgery”. All participants were >18 years. There was 17/42 (40.5 %) male in the study group and 26/57 (45.6 %) male in the control group. The mean age was 52.5 years in the patient group and 50.0 years in the control group. Additional informed consent was obtained from all individual participants from whom identifying information is included in this study.
Samples
Specimens were collected from the nares and maxillary sinuses of the patients, and from the nares of the healthy controls. Sampling from the maxillary sinuses of CRS patients was performed with an endoscope, using a cotton-tipped aluminium swab (Copan, Brescia, Italy) passing the nasal cavity and placed into the sinus through the enlarged sinus ostium under visual control (Fig. 1). Care was taken to avoid contamination via contact with the nose wall. Samples were also taken from the maxillary sinuses of patients with a protected brush (Olympus, model no BC-202D-3010), which could be shielded with a cover when passing through the narrow space such as the nasal cavity. This brush is often used when collecting bronchial specimen (Fig. 2). The brush was used thought an enlarged sinus ostium. Nasal specimens were collected from patients and controls using a nasal swab touching the nares. The criteria for a concordance between the two different sampling methods were defined as equal bacterial findings regarding both type and number of bacterial species.
Fig. 1.
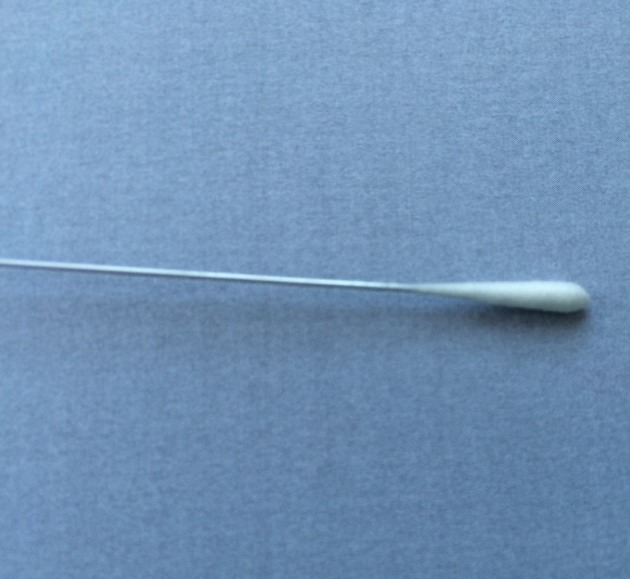
Cotton-tipped aluminium swab (Copan, Brescia, Italy)
Fig. 2.
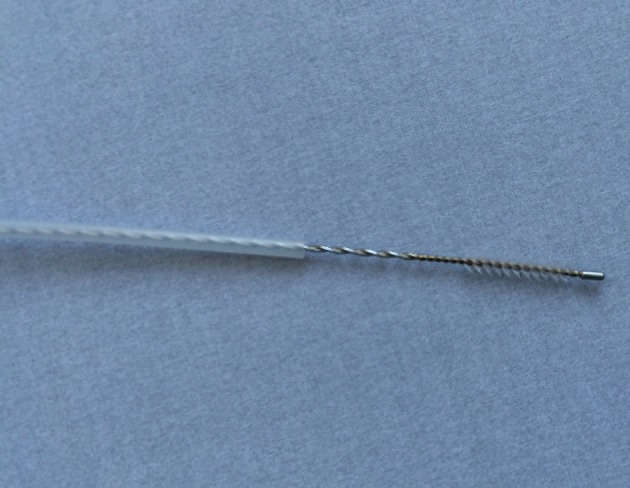
Protected brush (Olympus, model no BC-202D-3010)
Microbiological analysis
The culture and species verification of bacteria was performed in accordance with routine diagnostic procedures at the Department of Laboratory Medicine, Clinical Microbiology, Örebro University Hospital. Samples were cultured on blood agar medium [4.25 % Columbia II Agar (BBL, Becton–Dickinson, Baltimore, MD, USA), 0.3 % Agar No. 2 (Lab M Ltd., Bury, UK), and 5 % bovine blood] in an anaerobic atmosphere on FAA plates (4.6 % LAB 90 Fastidious Anaerobe Agar, LAB M, Lancashire, UK) supplemented with 5 % horse blood and incubated for 2 days at 37 °C. S. aureus isolates were dissolved in preservation medium [yeast extract (DIFCO Laboratories, Sparks, MD, USA) and horse serum added trypticase soy broth (BBL, Sparks, MD, USA)] and stored at −70 °C pending further spa typing that was performed as previously described [29].
Spa typing
Single locus DNA-sequencing of the repeat region of the Staphylococcus protein A gene (spa) for typing of S. aureus was performed as previously described [30].
Statistics
Fisher’s exact test was used to calculate p values reflecting the difference between groups containing categorical data, and the Chi-squared test was used for the same purpose with quantitative data. A p value <0.05 was considered statistically significant.
Results
Initial enrolment comprised 43 patients with CRS and 58 healthy controls. One patient was excluded due to having titanic dental implants reaching the maxillary sinus, and one of the controls was excluded due to an age below 18 years. The study group therefore consisted of 42 patients and 57 controls. Mean age was almost equal between the groups (51.5 and 50.0 years). The patient group was 46.0 % male, and the control group was 40.5 % male. Polyposis was present in 24 of the 42 (57 %) patients and none of the controls. All samples collected from the nares showed bacterial growth, though one sample in the patient group was missing. Fourteen different bacteria were identified. Table 1 shows the bacterial findings in patients and controls. The most common aerobic bacteria were CoNS, isolated from the maxillary sinus in 18/42 (43 %) patients. CoNS was found in nares in 17/42 (40 %) patients, and in 43/57 (75 %) controls (p = 0.0008). S. aureus was found in both nares and maxillary sinus in 15/42 (36 %) patients using the protected brush technique and in 18/42 (43 %) patients using the cotton-tipped aluminium swab technique (p = 0.66). S. aureus was isolated in the nares in 24/42 (57 %) patients and 16/57 (28 %) controls (p = 0.004). S. aureus findings in the maxillary sinus did not differ significantly between CRSwNP patients and CRSsNP patients (p = 0.35). Facultative anaerobic bacteria were found in five samples from nares of CRS patients and in two samples from controls. There was no significant difference between the patients and controls regarding the presence of facultative anaerobic bacteria in nares (p = 0.13). Maxillary sinus cultures using the protected brush technique showed two species present in 6/42 (14.3 %) patients, but none with three or more, while cultures using the conventional technique showed mixed flora in 14/42 (33 %) patients. At least one species of aerobic bacteria was cultured from 41/42 (98 %) patients, and anaerobic/facultative anaerobic bacteria was cultured from the maxillary sinus in 5/42 (12 %) patients. The nares showed mixed flora in 7/57 (12 %) controls and 14/42 (33 %) patients. Three samples (7 %) showed no growth, all collected with the brush technique from the maxillary sinus in CRS patients. In a comparison between the two sampling techniques, there was consistency in growth in 23/42 (56 %) patients. In 22/42 (52 %) of the brush samples, there was growth of only one species (Table 2). In one case, S. pneumoniae was found with the cotton-tipped aluminium swab and H. influenzae with the protected brush, and in another case, S. aureus was found with the cotton-tipped aluminium swab and H. influenzae with the protected brush. Furthermore, in two cases, α-streptococci and anaerobic Gram-positive cocci, respectively, were found with the protected brush but not with the cotton-tipped aluminium swab.
Table 1.
Frequency of bacterial findings in samples from patients with chronic rhinosinusitis with nasal polyposis (CRSwNP), chronic rhinosinusitis without nasal polyposis (CRSsNP), and healthy controls
| Agent identified | Maxillary sinus culture (CRS) | Nares culture (CRS) | Nares culture (controls) | ||||
|---|---|---|---|---|---|---|---|
| CRSwNP (n = 24) | CRSsNP (n = 18) | Total (n = 42) | CRSwNP* (n = 24) | CRSsNP (n = 18) | Total (n = 42) | Controls (n = 57) | |
| Aerobic bacteria | |||||||
| CoNS** | 9 (38 %) | 9 (50 %) | 18 (43 %) | 10 (42 %) | 7 (39 %) | 17 (40 %) | 43 (75 %) |
| Staphylococcus aureus | 8 (33 %) | 7 (39 %) | 15 (36 %) | 14 (58 %) | 10 (56 %) | 24 (57 %) | 16 (28 %) |
| Haemophilus influenzae | 2 (8 %) | 1 (5.6 %) | 3 (7.1 %) | 0 | 1 (2 %) | ||
| Diphtheroid rods | 2 (8 %) | 2 (4.7 %) | 2 (8 %) | 2 (11 %) | 4 (9.5 %) | 4 (7 %) | |
| Pseudomonas sp | 0 | 1 (5.6 %) | 1 (2.3 %) | 0 | |||
| Moraxella sp | 0 | 1 (4 %) | 1 (2.3 %) | 0 | |||
| α-haemolytic streptococci | 1 (4 %) | 1 (2.3 %) | 1 (4 %) | 1 (5.6 %) | 2 (4.7 %) | 1 (2 %) | |
| Staphylococcus lugdunensis | 1 (4 %) | 1 (2.3 %) | 0 | 0 | |||
| Micrococcus sp | 0 | 0 | 1 (2 %) | ||||
| Streptococcus pneumoniae | 1 (4 %) | 1 (2.3 %) | 2 (8 %) | 2 (4.7 %) | 0 | ||
| Enterobacteriaceae*** | 2 (8 %) | 2 (4.7 %) | 3 (12.5 %) | 1 (5.6 %) | 4 (9.5) | 1 (2 %) | |
| Escherichia coli | 0 | 1 (4 %) | 1 (2.3 %) | 1 (2 %) | |||
| Anaerobic | |||||||
| Propionibacterium acnes | 1 (4 %) | 1 (2.3 %) | 0 | 0 | |||
| Anaerobic Gram-positive cocci | 1 (4 %) | 1 (2.3 %) | 0 | 0 | |||
* One culture is missing
** Coagulase-negative staphylococci (CoNS)
*** Except Escherichia coli
Table 2.
Comparison of two different sampling techniques used in 42 patients with chronic rhinosinusitis (CRS)
| Case # | Maxillary sinus (Rayon-wire swab) | Maxillary sinus (Protected brush) | Concordance* | Assumed impact on the optimal choice of antibiotics treatment when based on findings with rayon wire swab |
|---|---|---|---|---|
| 1 | Streptococcus pneumoniae | Haemophilus influenzae | – | High |
| 2 |
Staphylococcus aureus
CoNS** |
Staphylococcus aureus | – | Low |
| 3 | CoNS Diphtheroid rods |
Diphtheroid rods | – | Low |
| 4 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 5 | CoNS | CoNS | + | Low |
| 6 |
Streptococcus
pneumoniae Moraxella catarrhalis |
Streptococcus pneumoniae | – | Low |
| 7 |
Staphylococcus aureus
Propionibacterium acnes |
Staphylococcus aureus | – | Low |
| 8 | CoNS | CoNS | + | Low |
| 9 | CoNS | CoNS | + | Low |
| 10 | Staphylococcus aureus |
Staphylococcus aureus
CoNS |
– | Low |
| 11 | Staphylococcus aureus | Haemophilus influenzae | – | Highs |
| 12 | Staphylococcus aureus |
Staphylococcus aureus
CoNS |
– | Low |
| 13 | CoNS | CoNS | + | Low |
| 14 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 15 | CoNS | CoNS | + | Low |
| 16 | CoNS | CoNS | + | Low |
| 17 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 18 | CoNS Haemophilus influenzae Klebsiella sp |
Haemophilus influenzae | – | Low |
| 19 | CoNS | CoNS | + | Low |
| 20 | Staphylococcus aureus | Negative | – | Low |
| 21 | CoNS Enterobacteriaceae |
Enterobacteriaceae | – | Low |
| 22 |
Staphylococcus aureus
Diphtheroid rods |
CoNS | – | Low |
| 23 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 24 | CoNS | CoNS | + | Low |
| 25 | Staphylococcus lugdunensis | Staphylococcus lugdunensis | + | Low |
| 26 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 27 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 28 | CoNS Propionibacterium acnes |
CoNS | – | Low |
| 29 | CoNS | CoNS | + | Low |
| 30 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 31 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 32 | CoNS Streptococcus pneumoniae |
CoNS | – | High |
| 33 | CoNS | CoNS | + | Low |
| 34 | CoNS | CoNS | + | Low |
| 35 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 36 | Staphylococcus aureus | Staphylococcus aureus | + | Low |
| 37 | Escherichia coli | Propionibacterium acnes | – | Highs |
| 38 | Staphylococcus aureus | Negative | – | Low |
| 39 | CoNS Propionibacterium acnes |
CoNS α-streptococci anaerobic Gram-positive cocci |
– | Low |
| 40 | CoNS Diphtheroid rods Propionibacterium acnes |
Negative | – | Low |
| 41 | CoNS Enterococcus sp |
CoNS | – | Low |
| 42 |
Staphylococcus aureus
Enterobacteriaceae |
Staphylococcus aureus
Enterobacteriaceae |
+ | Low |
Specimens obtained from the maxillary sinus
* Concordance meaning that the same type of bacteria was found with both culturing technique
** Coagulase-negative staphylococci (CoNS)
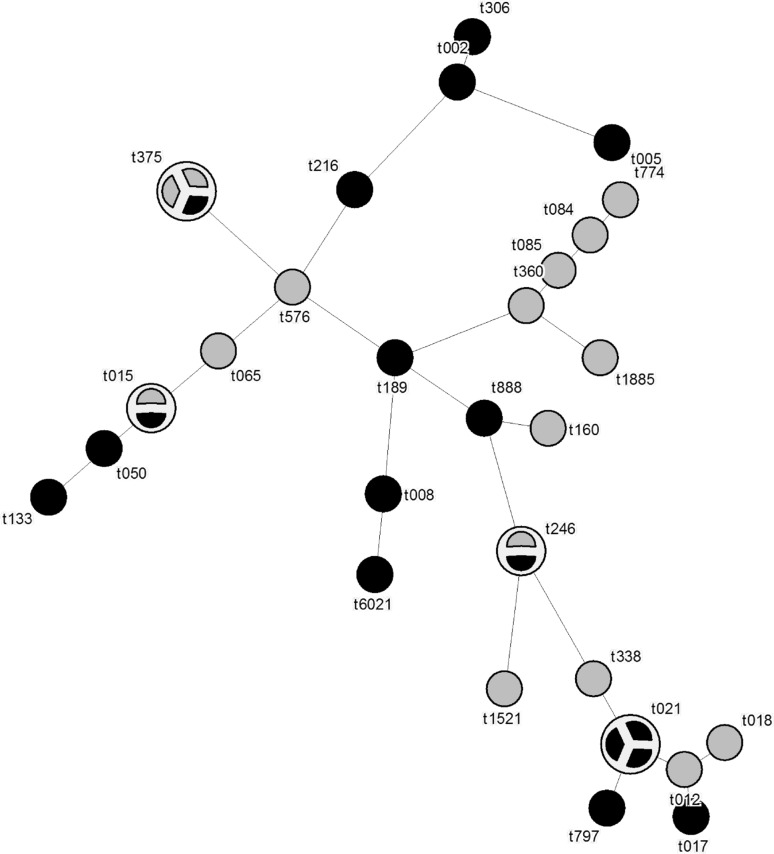
There was a wide variation in the distribution of spa types (Table 3; Fig. 3). We found identical spa types of S. aureus from the nares and from the maxillary sinus in 17/18 patients (94 %). The remaining patient with S. aureus isolates showed two unrelated spa types in the two areas: t084 and t189.
Table 3.
Distribution of spa types of S. aureus isolates from patients with chronic rhinosinusitis (CRS) in both maxillary sinus and nares
| Case # | S. aureus isolated in Maxillary sinus, CRS | S. aureus isolated in Nares, CRS |
|---|---|---|
| 1 | t246 | t246 |
| 2 | t050 | t050 |
| 3 | t008 | t008 |
| 4 | t021 | t021 |
| 5 | t021 | t021 |
| 6 | t015 | t015 |
| 7 | t021 | t021 |
| 8 | t375 | t375 |
| 9 | t216 | t216 |
| 10 | t017 | t017 |
| 11 | t133 | t133 |
| 12 | t002 | t002 |
| 13 | t084 | t189 |
| 14 | t61 | t6021 |
| 15 | t797 | t797 |
| 16 | t888 | t888 |
| 17 | t005 | t005 |
| 18 | t306 | t306 |
Fig. 3.
Distribution and genetic relationship of spa types of S. aureus isolates from the nares in controls (grey) and CRS patients (black). One circle represents one isolate
Discussion
The isolation rate of S. aureus in maxillary sinus cultures from patients with CRS has been reported to be about 25 % [21]. In this study, S. aureus was found in 15/42 (36 %) of the maxillary sinus cultures from CRS patients with both the optimised sampling technique (protected brush) and standard cotton-tipped aluminium swab. In another three patients, S. aureus were found using standard cotton-tipped aluminium swab but not with the brush. This indicates a contamination rate of 3/18 (17 %) for S. aureus. In 17/42 (40.5 %) samples, bacterial findings with standard cotton-tipped aluminium swab differ from findings using protected brush. However, CoNS in combination with other bacterial findings using cotton-tipped aluminium swab compromised most of these differences. The protected brush technique was superior to the standard cotton-tipped aluminium swab in reducing the contamination of CoNS and S. aureus when culturing maxillary sinus. However, the clinical relevance of using this optimised technique as the protected brush is questionable. In almost all patients cultured with the cotton-tipped aluminium swab, this culturing technique seemed to be adequate for collecting clinically relevant samples, and thus have a low impact on antibiotic treatment (Table 3). S. aureus is thought to persistently colonise the nares of about 20 % of the population (range 12–30 %), and approximately 30 % of the remainder is intermittent carriers (range 16–70 %) [31, 32]. Wertheim et al. regarded the nose as the major site of S. aureus carriage, and from here, the organism is thought to spread to other parts of the body [32]. S. aureus and CoNS were the predominant bacterial findings in the maxillary sinus of our patients with CRS. In addition, the number of patients with CRS with S. aureus in the nares was statistically significantly higher than the number of controls with S. aureus in the nares, while the number of patients with CoNS was statistically significantly lower compared to controls. The high S. aureus colonisation rate of the nares in patients with CRS may reflect its importance in the CRS disease. A hypothesis based on our findings might be that S. aureus counteracts the CoNS in the nares and promotes colonisation of S. aureus in the maxillary sinus through transport of bacteria across the mucus conjoining the nares with the sinus.
Furthermore, identical spa types in both the nares and maxillary sinus were found in all patients with findings of S. aureus in both locations (with one exception), which supports the theory that the nares can be the primary site from where the bacteria can spread and colonise the maxillary sinus. S. aureus has in certain circumstances the ability to produce enterotoxins acting as superantigens that bind to T cells and exaggerate disease severity and expression [25], and cause serious invasive diseases such as sepsis with or without infective endocarditis and necrotising pneumonia as well as mild diseases, such as superficial skin and soft tissue infections. S. aureus enterotoxins may play a role in the severity of CRS, especially CRSwNP [27, 33, 34]. The ability of superantigens to enhance inflammatory reactions [25] might have a connection to the predominance of S. aureus in CRS, again especially CRSwNP [35]. An increased immune response to S. aureus enterotoxins has been demonstrated in nasal polyposis tissue, resulting in more pronounced eosinophilic inflammation and higher local immunoglobulin E production against staphylococcal enterotoxins in patients affected by CRSwNP [26, 36]. Our study included 23 patients with CRSwNP and 19 with CRSsNP. We could not find any differences in bacterial findings in nares or maxillary sinus when comparing these groups, but the two groups were small and comparison is difficult. Brook et al., who studied the microbiology of the maxillary sinus in 48 patients with CRS, also found no difference between CRSwNP and CRSsNP [37] which recently also was shown by Brook et al. [38]. Another study showed no significant difference in the prevalence of genes encoding virulence factors between isolates from patients with CRS and healthy controls [39]. However, a large meta-analysis by Ou et al. based on 12 case–control studies with a total of 340 cases and 178 controls demonstrated a relationship between the presence of S. aureus superantigens and the persistence and severity of CRSwNP [27].
The high rate of S. aureus in the maxillary sinus in patients with CRS in our study strengthens the importance of S. aureus for the pathogenesis of the disease, but its role needs to be further investigated. Larger studies could hopefully lead to valuable knowledge about the role of S. aureus in CRS. Sweden is a low endemic area for methicillin-resistant S. aureus (MRSA), and there was only one MRSA isolate in our study; this is probably also due to a generally low prescription of antibiotics in Sweden, which leads to low incidence of resistant isolates. All participants were living in Sweden, but we have no information on ethnic origin, which may have been of interest. Some patients were included in conjunction with sinus surgery, and some had a previous history of sinus surgery and were included as outpatients at the otolaryngology clinic. However, a previous study indicates that endoscopic sinus surgery does not change the bacterial flora, though it does change the presence of bacterial biofilms in the sinus [40].
Conclusion
Contamination with CoNS was common using cotton-tipped aluminium swab technique. However, the protected brush technique did reduce the contamination rate but does not significantly improve the reliability of bacteriological diagnostics for practical clinical use. The risk of unnecessary antibiotic treatment is, therefore, thought to be low when using the easy-to-handle cotton-tipped aluminium swab. Furthermore, S. aureus was found in 36 % of the maxillary sinus samples from the CRS patients, and the number of patients with S. aureus in the nares was statistically significantly higher than the number of controls with S. aureus in the nares, while the number of patients with CoNS in the nares was statistically significantly lower compared to controls. These findings, together with a very high rate of identical spa types of S. aureus from patients with S. aureus in both nares and maxillary sinus, might indicate colonisation of the maxillary sinus from the nares and presumably a relationship between S. aureus and CRS.
Acknowledgments
This study was supported by Grants from the Research Committee of Örebro County Council. We thank Bengt Hellmark for performing the spa analysis. We also thank the nurses at the travel consultation clinic, Örebro, Sweden, who participated in sample collection.
Compliance with ethical standards
Conflict of interest
Bo Söderquist has been a consultant for Pfizer and Janssen-Cilag.
Ethical approval
All procedures performed in this study were in accordance with the ethical standards of the national research committee and with the 1964 Helsinki declaration. The study was approved by the regional ethical review board in Uppsala, Sweden.
References
- 1.Hastan D, Fokkens WJ, Bachert C, Newson RB, Bislimovska J, Bockelbrink A, Bousquet PJ, Brozek G, Bruno A, Dahlen SE, Forsberg B, Gunnbjornsdottir M, Kasper L, Kramer U, Kowalski ML, Lange B, Lundback B, Salagean E, Todo-Bom A, Tomassen P, Toskala E, van Drunen CM, Bousquet J, Zuberbier T, Jarvis D, Burney P. Chronic rhinosinusitis in Europe–an underestimated disease. A GA(2)LEN study. Allergy. 2011;66(9):1216–1223. doi: 10.1111/j.1398-9995.2011.02646.x. [DOI] [PubMed] [Google Scholar]
- 2.Benninger MS, Ferguson BJ, Hadley JA, Hamilos DL, Jacobs M, Kennedy DW, Lanza DC, Marple BF, Osguthorpe JD, Stankiewicz JA, Anon J, Denneny J, Emanuel I, Levine H. Adult chronic rhinosinusitis: definitions, diagnosis, epidemiology, and pathophysiology. Otolaryngol Head Neck Surg. 2003;129(3 Suppl):S1–S32. doi: 10.1016/S0194-5998(03)01397-4. [DOI] [PubMed] [Google Scholar]
- 3.Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, Georgalas C, Goossens H, Harvey R, Hellings P, Hopkins C, Jones N, Joos G, Kalogjera L, Kern B, Kowalski M, Price D, Riechelmann H, Schlosser R, Senior B, Thomas M, Toskala E, Voegels R, Wang de Y, Wormald PJ (2012) European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl (23): p 3 (preceding table of contents, 1–298) [PubMed]
- 4.Hamilos DL. Chronic sinusitis. J Allergy Clin Immunol. 2000;106(2):213–227. doi: 10.1067/mai.2000.109269. [DOI] [PubMed] [Google Scholar]
- 5.Meltzer EO, Hamilos DL, Hadley JA, Lanza DC, Marple BF, Nicklas RA, Bachert C, Baraniuk J, Baroody FM, Benninger MS, Brook I, Chowdhury BA, Druce HM, Durham S, Ferguson B, Gwaltney JM, Kaliner M, Kennedy DW, Lund V, Naclerio R, Pawankar R, Piccirillo JF, Rohane P, Simon R, Slavin RG, Togias A, Wald ER, Zinreich SJ, American Academy of Allergy A, Immunology, American Academy of Otolaryngic A, American Academy of O-H, Neck S, American College of Allergy A, Immunology, American Rhinologic S Rhinosinusitis: establishing definitions for clinical research and patient care. J Allergy Clin Immunol. 2004;114(6 Suppl):155–212. doi: 10.1016/j.jaci.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Cauwenberge P, Van Hoecke H, Bachert C. Pathogenesis of chronic rhinosinusitis. Curr Allergy Asthma Rep. 2006;6(6):487–494. doi: 10.1007/s11882-006-0026-3. [DOI] [PubMed] [Google Scholar]
- 7.Lam K, Schleimer R, Kern RC. The etiology and pathogenesis of chronic rhinosinusitis: a review of current hypotheses. Curr Allergy Asthma Rep. 2015;15(7):41. doi: 10.1007/s11882-015-0540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gwaltney JM,, Jr, Jones JG, Kennedy DW. Medical management of sinusitis: educational goals and management guidelines the International Conference on sinus Disease. Ann Otol Rhinol Laryngol Suppl. 1995;167:22–30. [PubMed] [Google Scholar]
- 9.Ragab A, Clement P, Vincken W. Objective assessment of lower airway involvement in chronic rhinosinusitis. Am J Rhinol. 2004;18(1):15–21. [PubMed] [Google Scholar]
- 10.Chaaban MR, Kejner A, Rowe SM, Woodworth BA. Cystic fibrosis chronic rhinosinusitis: a comprehensive review. Am J Rhinol Allergy. 2013;27(5):387–395. doi: 10.2500/ajra.2013.27.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foreman A, Jervis-Bardy J, Wormald PJ. Do biofilms contribute to the initiation and recalcitrance of chronic rhinosinusitis? Laryngoscope. 2011;121(5):1085–1091. doi: 10.1002/lary.21438. [DOI] [PubMed] [Google Scholar]
- 12.Hamilos DL. Host-microbial interactions in patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2014;133(3):640–653. doi: 10.1016/j.jaci.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Cauwenberge PB, Vander Mijnsbrugge AM, Ingels KJ. The microbiology of acute and chronic sinusitis and otitis media:a review. Eur Arch Otorhinolaryngol. 1993;250(Suppl 1):S3–S6. doi: 10.1007/BF02540108. [DOI] [PubMed] [Google Scholar]
- 14.Brook I. The role of anaerobic bacteria in sinusitis. Anaerobe. 2006;12(1):5–12. doi: 10.1016/j.anaerobe.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Brook I. Bacteriology of chronic sinusitis and acute exacerbation of chronic sinusitis. Arch Otolaryngol Head Neck Surg. 2006;132(10):1099–1101. doi: 10.1001/archotol.132.10.1099. [DOI] [PubMed] [Google Scholar]
- 16.Chin CW, Yeak CL, Wang DY. The microbiology and the efficacy of antibiotic-based medical treatment of chronic rhinosinusitis in Singapore. Rhinology. 2010;48(4):433–437. doi: 10.4193/Rhino09.071. [DOI] [PubMed] [Google Scholar]
- 17.Brook I. Microbiology and management of sinusitis. J Otolaryngol. 1996;25(4):249–256. [PubMed] [Google Scholar]
- 18.Damm M, Quante G, Jurk T, Sauer JA. Nasal colonization with Staphylococcus aureus is not associated with the severity of symptoms or the extent of the disease in chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2004;131(3):200–206. doi: 10.1016/j.otohns.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 19.Kostamo K, Richardson M, Virolainen-Julkunen A, Leivo I, Malmberg H, Ylikoski J, Toskala E. Microbiology of chronic hyperplastic sinusitis. Rhinology. 2004;42(4):213–218. [PubMed] [Google Scholar]
- 20.Mantovani K, Bisanha AA, Demarco RC, Tamashiro E, Martinez R, Anselmo-Lima WT. Maxillary sinuses microbiology from patients with chronic rhinosinusitis. Braz J Otorhinolaryngol. 2010;76(5):548–551. doi: 10.1590/S1808-86942010000500002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biel MA, Brown CA, Levinson RM, Garvis GE, Paisner HM, Sigel ME, Tedford TM. Evaluation of the microbiology of chronic maxillary sinusitis. Ann Otol Rhinol Laryngol. 1998;107(11 Pt 1):942–945. doi: 10.1177/000348949810701107. [DOI] [PubMed] [Google Scholar]
- 22.Pastacaldi C, Lewis P, Howarth P. Staphylococci and staphylococcal superantigens in asthma and rhinitis: a systematic review and meta-analysis. Allergy. 2011;66(4):549–555. doi: 10.1111/j.1398-9995.2010.02502.x. [DOI] [PubMed] [Google Scholar]
- 23.Breuer K, Kapp A, Werfel T. Bacterial infections and atopic dermatitis. Allergy. 2001;56(11):1034–1041. doi: 10.1034/j.1398-9995.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- 24.Jappe U. Superantigens and their association with dermatological inflammatory diseases: facts and hypotheses. Acta Derm Venereol. 2000;80(5):321–328. doi: 10.1080/000155500459231. [DOI] [PubMed] [Google Scholar]
- 25.Zhang N, Gevaert P, van Zele T, Perez-Novo C, Patou J, Holtappels G, van Cauwenberge P, Bachert C. An update on the impact of Staphylococcus aureus enterotoxins in chronic sinusitis with nasal polyposis. Rhinology. 2005;43(3):162–168. [PubMed] [Google Scholar]
- 26.Bachert C, van Zele T, Gevaert P, De Schrijver L, Van Cauwenberge P. Superantigens and nasal polyps. Curr Allergy Asthma Rep. 2003;3(6):523–531. doi: 10.1007/s11882-003-0065-y. [DOI] [PubMed] [Google Scholar]
- 27.Ou J, Wang J, Xu Y, Tao ZZ, Kong YG, Chen SM, Shi WD. Staphylococcus aureus superantigens are associated with chronic rhinosinusitis with nasal polyps: a meta-analysis. Eur Arch Otorhinolaryngol. 2014;271(10):2729–2736. doi: 10.1007/s00405-014-2955-0. [DOI] [PubMed] [Google Scholar]
- 28.Fokkens W, Lund V, Bachert C, Clement P, Helllings P, Holmstrom M, Jones N, Kalogjera L, Kennedy D, Kowalski M, Malmberg H, Mullol J, Passali D, Stammberger H, Stierna P. EAACI position paper on rhinosinusitis and nasal polyps executive summary. Allergy. 2005;60(5):583–601. doi: 10.1111/j.1398-9995.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- 29.Berglund C, Ito T, Ma XX, Ikeda M, Watanabe S, Soderquist B, Hiramatsu K. Genetic diversity of methicillin-resistant Staphylococcus aureus carrying type IV SCCmec in Orebro County and the western region of Sweden. J Antimicrob Chemother. 2009;63(1):32–41. doi: 10.1093/jac/dkn435. [DOI] [PubMed] [Google Scholar]
- 30.Berglund C, Soderquist B. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden-possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin Microbiol Infect. 2008;14(11):1048–1056. doi: 10.1111/j.1469-0691.2008.02090.x. [DOI] [PubMed] [Google Scholar]
- 31.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10(3):505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 33.Derycke L, Perez-Novo C, Van Crombruggen K, Corriveau MN, Bachert C. Staphylococcus aureus and chronic airway disease. World Allergy Organ J. 2010;3(8):223–228. doi: 10.1097/WOX.0b013e3181ecd8ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachert C, Zhang N. Chronic rhinosinusitis and asthma: novel understanding of the role of IgE ‘above atopy’. J Intern Med. 2012;272(2):133–143. doi: 10.1111/j.1365-2796.2012.02559.x. [DOI] [PubMed] [Google Scholar]
- 35.Seiberling KA, Grammer L, Kern RC. Chronic rhinosinusitis and superantigens. Otolaryngol Clin North Am. 2005;38(6):1215–1236. doi: 10.1016/j.otc.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Van Cauwenberge P, Van Zele T, Bachert C. Chronic rhinonsinusitis and nasal polyposis: the etiopathogenesis revealed? Verh K Acad Geneeskd Belg. 2008;70(5–6):305–322. [PubMed] [Google Scholar]
- 37.Brook I, Frazier EH. Bacteriology of chronic maxillary sinusitis associated with nasal polyposis. J Med Microbiol. 2005;54(Pt 6):595–597. doi: 10.1099/jmm.0.45767-0. [DOI] [PubMed] [Google Scholar]
- 38.Brook I. Microbiology of chronic rhinosinusitis. Eur J Clin Microbiol Infect Dis. 2016;35(7):1059–1068. doi: 10.1007/s10096-016-2640-x. [DOI] [PubMed] [Google Scholar]
- 39.Thunberg U, Hugosson S, Monecke S, Ehricht R, Soderquist B. Molecular characteristics of Staphylococcus aureus associated with chronic rhinosinusitis. APMIS. 2015;123(1):37–44. doi: 10.1111/apm.12299. [DOI] [PubMed] [Google Scholar]
- 40.Larson DA, Han JK. Microbiology of sinusitis: does allergy or endoscopic sinus surgery affect the microbiologic flora? Curr Opin Otolaryngol Head Neck Surg. 2011;19(3):199–203. doi: 10.1097/MOO.0b013e328344f67a. [DOI] [PubMed] [Google Scholar]