Abstract
The incidence of platelet bacterial contamination is approximately 1 per 2,000 units and has been acknowledged as the most frequent infectious risk from transfusion. In preliminary studies, the sterility of platelet concentrates (PCs) was tested with an automated bacterial blood culturing system and molecular genetic assays. Two real-time reverse transcriptase PCR (RT-PCR) assays performed in a LightCycler instrument were developed and compared regarding specificity and sensitivity by the use of different templates to detect the majority of the clinically important bacterial species in platelets. Primers and probes specific for the conserved regions of the eubacterial 23S rRNA gene or the groEL gene (encoding the 60-kDa heat shock protein Hsp60) were designed. During the development of the 23S rRNA RT-PCR, problems caused by the contamination of reagents with bacterial DNA were noted. Treatment with 8-methoxypsoralen and UV irradiation reduced the level of contaminating DNA. The sensitivity of the assays was greatly influenced by the enzyme system which was used. With rTth DNA polymerase in a one-enzyme system, we detected 500 CFU of Escherichia coli or Staphylococcus epidermidis/ml. With a two-enzyme system consisting of Moloney murine leukemia virus RT and Taq DNA polymerase, we detected 16 CFU/ml. With groEL mRNA as the target of RT-PCR under optimized conditions, we detected 125 CFU of E. coli/ml, and no problems with false-positive results caused by reagent contamination or a cross-reaction with human nucleic acids were found. Furthermore, the use of mRNA as an indicator of viability was demonstrated. Here we report the application of novel real-time RT-PCR assays for the detection of bacterial contamination of PCs that are appropriate for transfusion services.
The bacterial contamination of blood products remains a persistent but often ignored problem in transfusion medicine. The incidence of platelet bacterial contamination is approximately 1 per 2,000 to 3,000 units (13). In the United States, bacterial contamination is considered the second most common cause of death overall from transfusion, with mortality rates ranging from 1 in 20,000 to 1 in 85,000 donor exposures. Estimations of severe morbidity and mortality range from 100 to 150 transfused individuals each year (14). The risk of receiving bacterially contaminated platelets is estimated to be 50- to 250-fold higher than the combined risk of transfusion-related infections per unit associated with human immunodeficiency virus type 1, hepatitis B virus, and human T-cell leukemia virus types 1 and 2 (4). Regarding this, the American Association of Blood Banks (AABB) standards claim that blood banks or transfusion services shall have methods to limit and detect bacterial contamination in all platelet components and that these methods were to be implemented by 1 March 2004 (14). The screening of platelet concentrates (PCs) for bacterial contamination is already performed in some European countries. Sterility screening has been mandatory in Belgium since 1998, for example, and in The Netherlands it has been mandatory since November 2001 (25).
Platelets are stored aerobically for up to 5 days at 20 to 24°C, allowing a wide variety of bacteria to grow. For platelet transfusions, >30% of clinical complications are due to Staphylococcus species, particularly Staphylococcus epidermidis and S. aureus, but numerous other bacterial species may also be responsible (5, 31, 38). Conventional methods for the detection of these bacteria in blood components involve culturing and identification by morphological, biochemical, and immunological characteristics.
Different methods for bacterial detection have been investigated, including the direct detection of bacteria by microscopic analyses of blood smears, an indirect observation of bacterial metabolism by measurement of the platelet pH or glucose levels or by a change in the color of red blood cells, the detection of bacterial growth by pyrogen testing, flow cytometric methods, or the cessation of platelet swirling (for reviews, see references 9, 23, 38, and 39). Furthermore, an indirect method that monitored the percent oxygen (%O2) in the air above aliquots of PCs was evaluated (27). The detection of bacterial growth by culture methods is still the most sensitive method. A bacterial sterility test should be rapid, affordable, adequately sensitive, specific, and simple to perform (37). Only automated bacterial blood culturing systems meet many of the requirements of an ideal test because they detect a wide range of organisms at concentrations of only 1 to 10 CFU per ml (5). Recently, molecular genetic techniques based on bacterial genomic detection were developed, with the 16S rRNA gene as a target (7, 11, 29, 33). In real-time PCR, a sensitivity of about 30 CFU per ml was demonstrated (33).
Nevertheless, there are disadvantages with DNA- and rRNA-based methods. On the one hand, they do not distinguish between living and dead organisms (34). On the other hand, the implementation of a universal PCR can be hindered by problems such as nucleic acid contamination of reagents which may be derived from a bacterial source, such as Taq DNA polymerase or uracil-N-glycosylase (UNG) (8). Meier et al. (22) investigated a method to eliminate contaminating DNAs by the use of psoralen. Psoralens are known to intercalate into double-stranded nucleic acids and to form a covalent interstrand cross-link after photo activation with UV light. Therefore, the use of 8-methoxypsoralen (8-MOP) to extinguish the template activity of contaminating DNAs was suggested (16).
For our study, we chose a ubiquitous and highly conserved single-copy gene coding for a 60-kDa heat shock protein (Hsp60, also known as GroEL). Reverse transcriptase PCR (RT-PCR) is a potentially sensitive, specific, and rapid in vitro technique for the amplification of RNA molecules. RT-PCR might also allow the specific detection of viable cells, since RNA is generally less stable than DNA in dead cells. Recently, the development of real-time PCR technology has greatly facilitated PCR analysis, making possible the simultaneous amplification, detection, and quantitation of target nucleic acid molecules. Here we report the application of real-time groEL mRNA and 23S rRNA RT-PCRs for the detection of bacterial contamination of PCs.
MATERIALS AND METHODS
Blood collection.
Apheresis-derived single-donor platelets were obtained from the transfusion service OWL in Bad Oeynhausen, Germany, after standard processing from healthy blood donors with an MCS+ instrument (Haemonetics GmbH, Munich, Germany). After the preparation of PCs, 7 ml of platelet-rich plasma was transferred for sterility testing with a plasma extractor under aseptic conditions into a 150-ml transfer pack container from a quadruple transfer pack (Baxter Healthcare Co., Deerfield, Ill.) and stored at 20 to 24°C with agitation.
Microbiological sterility control.
The sterility of 451 clinical probes was tested by use of the BacT/Alert3D continuous monitoring system (bioMérieux, Durham, N.C.). Forty-milliliter aerobic culture bottles (BacT/Alert 3D; bioMérieux) were inoculated with 5-ml aliquots of the platelet samples and incubated at 37°C for up to 3 days in the automated culture system and for another 4 days in an incubator. After 7 days of incubation, a visual inspection of the bottles was performed, and then 1 ml of each culture was plated onto Columbia agar containing 5% sheep blood (bioMérieux). The culture plates were incubated at 37°C for another 48 h.
Bacterial strains and culture conditions.
The bacterial species listed in Table 1 were subcultured in Trypticase soy broth (TS; bioMérieux) at 37°C for 24 h under aerobic conditions. One hundred-microliter aliquots of serial dilutions of logarithmically growing cultures in TS were plated on TS agar for determinations of the bacterial titers.
TABLE 1.
Bacterial strains used for this study
| Species | Strain |
|---|---|
| Bacillus cereus | Isolate RV2 298 |
| Bacillus subtilis | ATCC 6633 |
| Citrobacter koseri | Isolate 013886/98 |
| Clostridium perfringens | Isolate 005398/98 |
| Enterobacter cancerogenes | Isolate 007367/97 |
| Enterobacter faecium | ATCC 6057 |
| Enterococcus faecalis | ATCC 29212 |
| Enterobacter sp. | Isolate 230703 |
| Escherichia coli | ATCC 35218 |
| Isolate 8338 | |
| Haemophilus influenzae | Isolate 009069/98 |
| Klebsiella oxycota | Isolate 005251/00 |
| Isolate 8232/96 | |
| Klebsiella pneumoniae | Isolate 04101 |
| Isolate RV1 04/01 | |
| Isolate 7724/99 | |
| Peptostreptococcus anaerobius | ATCC 27337 |
| Pseudomonas aeruginosa | ATCC 27853 |
| Isolate | |
| Pseudomonas fluorescens | Isolate 8393 |
| Isolate RV A020902 | |
| Pseudomonas putida | Isolate 790 |
| Isolate RV3 031/00 | |
| Serratia marcescens | Isolate 0004201/00 |
| Isolate 008330/98 | |
| Isolate 230703 | |
| Serratia rubidaea | Isolate 006745/99 |
| Serratia sp. | Isolate 230703 |
| Shigella flexneri | Isolate 070598 |
| ATCC 29903 | |
| ATCC 231298 | |
| Staphylococcus aureus | ATCC 29213 |
| Staphylococcus epidermidis | Isolate 015038/98 |
| Streptococcus mitis | Isolate 008492/98 |
| Streptococcus pneumoniae | ATCC 49619 |
| Yersinia enterocolitica | ATCC 9610 |
| Yersinia pseudotuberculosis | Isolate 110400 |
| Candida albicans | ATCC 10291 |
RNA isolation.
The isolation of total RNAs from overnight cultures and platelets was performed with a NucleoSpin RNA II kit (Macherey-Nagel GmbH, Düren, Germany). As a variation to the manufacturer's protocol, 1-ml aliquots of the bacterial or platelet samples were collected by centrifugation for 5 min at 20,000 × g. The supernatants were removed. The pellets were resuspended in 200 μl of AK1 (6.7% sucrose, 50 mM Tris-HCl [pH 8], 1 mM EDTA) containing 10 mg of lysozyme (Sigma-Aldrich, Taufkirchen, Germany) per ml, 100 μg of lysostaphin (Sigma-Aldrich) per ml, and 100 mg of 106-μm-diameter glass beads (Sigma-Aldrich). After incubation for 45 min at 37°C, cell disruption was achieved by use of the Ribolyser system (Hybaid GmbH, Heidelberg, Germany) for 5 min. Lysis buffer (350 μl) (Machery-Nagel GmbH) was added, and the samples were centrifuged for 10 min at 20,000 × g. The supernatants were removed and place into clean 1.5-ml reaction tubes. The following steps corresponded to the manufacturer's protocol. For elimination of the genomic DNA, a DNase I treatment was performed directly on the silica membrane.
RNA quantitation was carried out by a sensitive fluorescence-based solution assay for RNA that used the RiboGreen RNA quantitation reagent (Molecular Probes, Leiden, The Netherlands) as described by the manufacturer.
Real-time RT-PCR with rTth DNA polymerase (one-enzyme system).
DNase-treated RNA samples were analyzed by a one-step RT-PCR method using rTth DNA polymerase (Applied Biosystems, Darmstadt, Germany), which has both reverse transcriptase and DNA polymerase activities, to synthesize cDNAs from RNAs and to amplify the products in subsequent PCRs. RT-PCRs were carried out in a LightCycler instrument (Roche Diagnostics GmbH, Mannheim, Germany) in capillaries containing 15 μl of reaction mix and 5 μl of nucleic acid extract. The reaction mix consisted of 1× TaqMan EZ buffer, 5 mM manganese acetate, 500 ng of bovine serum albumin (Sigma-Aldrich) per μl, 500 nM (each) forward and reverse primers, a 300 μM concentration of each deoxynucleoside triphosphate, a 200 nM concentration of each TaqMan fluorescent probe, 0.1 U of rTth polymerase (Applied Biosystems) per μl, and 0.01 U of UNG (Applied Biosystems) per μl.
The cycling conditions for the 23S rRNA RT-PCR were 37°C for 5 min (UNG), 95°C for 5 min (UNG inactivation), and 50°C for 15 min (cDNA synthesis), followed by 30 cycles of denaturation at 95°C for 5 s, annealing at 57°C for 15 s, and extension at 72°C for 20 s. groEL RT-PCRs were performed using the following conditions: 37°C for 5 min, 95°C for 5 min, and 60°C for 15 min, followed by 45 cycles of 95°C for 5 s, 58°C for 15 s, and 72°C for 20 s.
Real-time RT-PCR with Moloney murine leukemia virus RT and DNA Taq polymerase (two-enzyme system).
A Superscript II one-step RT-PCR with a Platinum Taq kit (Invitrogen, Karlsruhe, Germany) was used as the basis for the reaction mixture. The RT-PCR mix consisted of 1× reaction buffer, 5 mM MgSO4, 500 ng of bovine serum albumin (Sigma-Aldrich) per μl, 600 nM (each) forward and reverse primers, a 250 nM concentration of each TaqMan fluorescent probe, and 0.6 μl of enzyme mix. The reactions were performed in a volume of 20 μl that included 5 μl of nucleic acid extract.
The cycling conditions were an initial single cycle at 48°C for 15 min to synthesize the cDNAs, followed by a single cycle at 95°C for 5 min to inactivate the reverse transcriptase and for denaturation. The cDNAs were amplified in PCRs of 30 to 45 cycles as described for the RT-PCR system using rTth polymerase.
Nucleic acid decontamination of master mixture.
The master mixture with the primers for the 23S rRNA region was prepared as mentioned above. 8-MOP (Sigma-Aldrich) was dissolved in dimethyl sulfoxide (Sigma-Aldrich). A concentration of 25 μg per ml was added to the master mix to a final concentration of 1% dimethyl sulfoxide in the PCR assay. The mixture was irradiated with a UV linker at 312 nm for 3, 5, or 15 min. The template was then added, and RT-PCR was performed in the LightCycler instrument (Roche Diagnostics).
Electrophoresis and imaging.
During evaluation studies, the PCR products were separated by agarose gel electrophoresis to look for nonspecific amplification products and false-negative results caused by imperfect probe binding. Therefore, aliquots (15 μl) of the real-time RT-PCR products were separated in 2% agarose gels. The amplification products were visualized with ethidium bromide staining under UV illumination. To determine the sizes of the products, we used the DNA molecular weight marker VIII (Roche Diagnostics).
Primer design for amplification of 23S bacterial rRNA and groEL mRNA.
Purified PCR products from the 23S rRNA gene (primers BLF1380 and BLR1686) (36) and the groEL gene (primers H297 and H280) (12) were sequenced by the BigDye dideoxy extension method (ABI Prism Dye Terminator cycle sequencing ready reaction kit, v. 2.0; Applied Biosystems). The sequencing products were analyzed with an ABI Prism 310 DNA sequencer (Applied Biosystems).
The nucleotide sequences were aligned with CLUSTAL X sequence alignment software (version 1.8). The degree of nucleotide sequence homology was checked with the BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST) by comparisons with sequences in the EMBL, GenBank, and DDBJ databases. The oligonucleotides (Table 2) were designed with Oligo 5.0 primer analysis software (National Biosciences, Plymouth, Minn.).
TABLE 2.
Oligonucleotide primers and TaqMan fluorescent probe sequences used for RT-PCRs
| Oligonucleotide | Target gene | Sequence (5′ to 3′) | Position of product (nt) | GenBank accession no. | Reference or source |
|---|---|---|---|---|---|
| BLF1380 | 23S rRNA | GGACAACAGGTTAATATTCC | 1380-1398 | AF053966 | 36 |
| BLR1686 | 23S rRNA | GGGGCCATTTTGCCGAGTTC | 1685-1666 | AF053966 | 36 |
| BLF-E | 23S rRNA | TTCCTGTACTTGGTGTTACTGCGA | 1395-1418 | AF053966 | This work |
| BLR1686-E | 23S rRNA | GGCACCATTTTGCCTAGTTC | 1685-1666 | AF053966 | This work |
| BL-1-TM | 23S rRNA | [6-FAM]ACCTGTGTCGGTTTGSGGTACGRTT[TAMRA] | 1620-1596 | AF053966 | This work |
| H279 | groEL | GAIIIIGCIGGIGAYGGIACIACIAC | 247-272 | X82212.2 | 12 |
| H280 | groEL | YKIYKITCICCRAAICCIGGIGCYTT | 855-830 | X82212.2 | 12 |
| HSP60-73F | groEL | TGAAACGYGGTATCGACAAA | 347-366 | X82212.2 | This work |
| HSP60-287R | groEL | CTGCATACCTTCAACMACGTCC | 583-562 | X82212.2 | This work |
| HSP60-217TM1 | groEL | [6-FAM]CCTTCTTTACCGACTTTITCCATCGCTT[TAMRA] | 519-492 | X82212.2 | This work |
| HSP60-217TM2 | groEL | [6-FAM]CCTTCTTTRCCMACTTTMTCCATCGCTT[TAMRA] | 519-492 | X82212.2 | This work |
Viability testing and inactivation of bacteria.
Log-phase cultures of Escherichia coli were diluted 1 to 100 in isotonic AK1 buffer in 1-ml aliquots. These samples were treated in different ways to kill the bacteria. Thermic inactivation was performed at 100°C for 5 min, 80°C for 10 min, and 60°C for 20 min. Other samples were exposed to 67% ethanol for 7 min, pelleted by centrifugation at 20,000 × g for 5 min, washed twice, and resuspended in 100 μl of TS broth. After this, the heat-inactivated cells were incubated for a further 0, 5, or 30 min or 1, 2, or 20 h at room temperature, and the ethanol-inactivated cells were incubated for 0, 5, 15, 45, and 70 min. To monitor the presence of viable cells, we plated 50-μl samples onto Columbia agar containing 5% sheep blood and incubated them at 37°C for 24 h just before RNA isolation.
Analytical sensitivities of 23S rRNA and mRNA real-time RT-PCRs.
To determine the detection limit of real-time RT-PCRs, we spiked a 1,350-μl sample of a pooled PC with 150 μl of a fresh overnight culture of E. coli ATCC 35218 (109 cells). The sample was diluted with a PC without further bacterial contamination down to 1 CFU per ml. 23S rRNA and groEL mRNA real-time RT-PCRs were performed as described above.
RESULTS
Study design.
During a study between 6 May and 15 August 2003, 451 platelet samples were tested for bacterial contamination in parallel by use of the automated BacT/Alert 3D system, with subsequent plating on TS agar. All specimens tested negative by the microbiological approach.
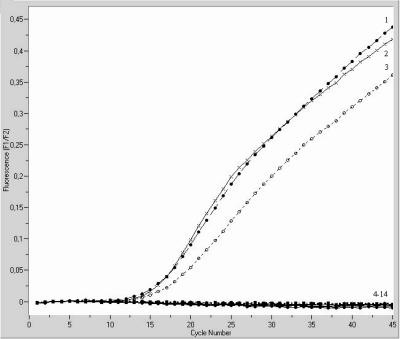
For this study, we developed a detection method that targets the abundant 23S rRNA and the groEL mRNA. The detection of rRNA should impart an increased sensitivity compared to assays based upon the detection of a single copy or even multiple copies of genomic sequences because each cell contains 1,000 to 10,000 copies of rRNA. In addition, an mRNA-based detection method should better reflect the number of viable organisms because mRNA is generally degraded within a few minutes after cell death. One-step real-time RT-PCRs based on LightCycler technology were successfully applied for the amplification of rRNA and mRNA from log-phase cells of bacterial species involved in PC contamination. The 23S rRNA primers amplify a 290-bp product, whereas the groEL primers amplify a 236-bp product. Figure 1 shows a representative RT-PCR screening for bacterial contamination of PCs by use of the groEL primer system. All PCs tested by rRNA- and mRNA-targeting RT-PCRs gave negative results, which corresponds to the results achieved by microbiological sterility testing.
FIG. 1.
Amplification plots for LightCycler groEL RT-PCR assay using the two-enzyme system for sterility testing of platelet concentrates. RNAs were isolated from an overnight culture of Shigella flexneri ATCC 29903 as a positive control and from different platelet samples. Sterile H2O was used instead of template RNA as a negative control. The amplification profiles of the positive controls are shown in plots 1, 2, and 3. Plots 4, 5, 6, 7, 8, 9, 10, and 11 represent the amplification profiles of the platelet samples, and plots 12, 13, and 14 are the profiles of the negative controls.
8-MOP and UV irradiation.
False-positive results for the no-template controls (water or platelets) with similar threshold cycle values (CT > 35) were given for all 23S rRNA RT-PCRs which were performed with a master mixture without 8-MOP and UV irradiation (data not shown). No false-positive signals were detected when the master mixture was treated with 8-MOP and UV irradiation. The CT values for the positive controls increased with the intensity of UV exposure, as shown in Table 3. Therefore, UV exposure times were optimized in the range of 3 to 5 min.
TABLE 3.
Effects of UV irradiation time and 8-MOP on CT values in 23S rRNA RT-PCR
| UV exposure time (min) | CT value in 23S RT-PCR
|
|
|---|---|---|
| Positive control (E. coli total RNA) | Negative control (water) | |
| 0 | 13.11 | >35 |
| 3 | 15.23 | >35 |
| 5 | 17.04 | >35 |
| 15 | 17.57 | >35 |
Specificity testing of 23S rRNA and groEL real-time RT-PCRs.
All of the bacterial strains listed in Table 1 were detected by 8-MOP-decontaminated PCR reagents in 23S rRNA real-time RT-PCRs (data not shown). The false-positive results of the no-template controls (water or platelets) could be reduced by the use of 8-MOP and UV irradiation.
In order to minimize nucleic acid contamination by recombinant enzymes and to test for a molecular genetic marker of viability, we developed a novel groEL mRNA real-time RT-PCR assay. Additionally, the yeast Candida albicans was also detected with the groEL RT-PCR assay.
The specificity of the RT-PCR assay seemed to be 100%, as shown by the testing of 451 sterile PC specimens. The no-template controls gave no false-positive results, as shown in Fig. 1.
Analytical sensitivity testing.
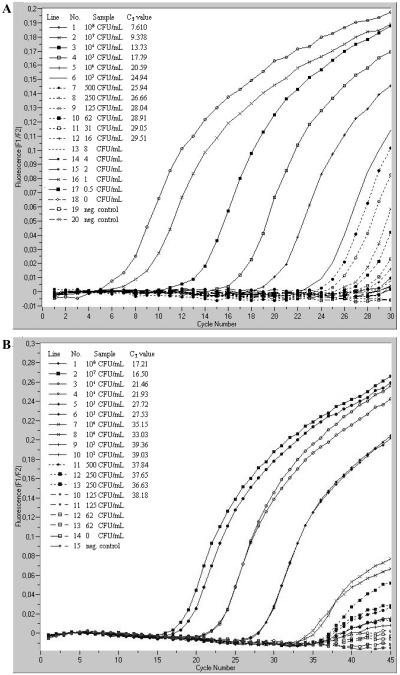
The TaqMan real-time 23S rRNA and groEL mRNA RT-PCRs were performed with E. coli strain ATCC 35218, using 20 μg (2 × 109 CFU per ml) to 0.02 pg of RNA per ml as a template. The concentrations of bacteria and RNA detected by each RT-PCR are shown in Fig. 2. In summary, 0.6 pg of RNA per ml (16 CFU per ml) was detected by the 23S rRNA RT-PCR, and 2.5 pg of RNA per ml (125 CFU per ml) was detected by the groEL RT-PCR using Moloney murine leukemia virus RT and DNA Taq polymerase. The detection limit of the 23S rRNA RT-PCR using rTth polymerase amounted to 2 pg per ml (500 CFU per ml), and that for groEL RT-PCR was 20 pg per ml (104 CFU per ml).
FIG. 2.
Determination of detection limits of RT-PCR assays targeting the 23S rRNA (A) and groEL mRNA (B). LightCycler RT-PCRs were performed with the two-enzyme system. A pooled platelet concentrate was spiked with a fresh overnight culture of E. coli ATCC 35218 (109 cells) and diluted with platelet concentrate without bacterial contamination down to 1 CFU per ml. Sterile H2O was used as a negative control.
Moreover, the TaqMan real-time 23S rRNA RT-PCR using rTth DNA polymerase was performed with S. epidermidis, using 9.2 μg (2 × 108 CFU per ml) to 0.009 pg of RNA per ml as a template. The detection limit was 4.5 pg of RNA per ml (500 CFU per ml).
Detection of groEL mRNA and 23S rRNA in heat- and ethanol-inactivated cells.
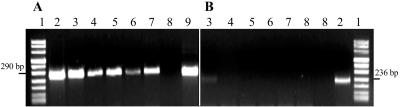
The correlation between the appearance of RT-PCR products and cell viability after ethanol or heat treatment was verified by time course experiments. E. coli strain ATCC 35218 was chosen so that we could compare the results to those from a previous study (34); the strain was killed by ethanol or heat treatment, and the RNA contents of cells stored at room temperature were monitored. Both groEL mRNA and 23S rRNA real-time RT-PCRs were performed with total RNAs. After the RT-PCRs, the amplification products were analyzed by agarose gel electrophoresis as well. Immediately after heating to 60, 80, and 100°C, groEL mRNA was undetectable (Fig. 3). In contrast, the mRNA was detected in untreated suspensions, an overnight culture, and a 1-to-100 dilution in TS broth, which were all used as positive controls. Moreover, 23S rRNA was detected by RT-PCR in all ethanol-treated and untreated cell suspensions. No viable cells were detected in any of the plated samples, except for the positive controls. In contrast to the mRNA, 23S rRNA was detected by RT-PCR in all heat-treated and untreated cell suspensions.
FIG. 3.
Time course of 23S rRNA and groEL mRNA stabilities in ethanol-treated E. coli cells. Gel electrophoresis of RT-PCR products of the 23S rRNA (A) and the groEL mRNA (B) of E. coli ATCC 35218 was performed after LightCycler RT-PCRs as described in Materials and Methods. E. coli cells (109 cells per ml, lane 9) were inactivated by incubation with 67% ethanol for 7 min at room temperature for 5 (lane 4), 15 (lane 5), 45 (lane 6), and 70 min (lane 7). Lane 3 contains the untreated 1-to-100 dilution of E. coli, lane 8 contains the no-template control, and lane 1 contains the DNA molecular weight marker.
DISCUSSION
The aim of our study was to develop a real-time RT-PCR method to detect bacterial contamination in platelets that is sensitive, specific for all transfusion-relevant bacterial species, and more suitable for routine sterility testing of PCs. The use of a rapid-cycle real-time format proved to be time- and labor-saving and minimized contamination risks related to the handling of PCR products.
Previous studies have used 16S ribosomal DNA as a target for nucleic acid amplification technology (NAT) screening (6, 7, 29, 33, 35). Analyses of the sequences of 23S ribosomal DNAs revealed more variation between species, so they might be useful for identification or for specific detection with specific fluorescent probes. As for the 16S rRNA, the high copy number of RNA molecules per cell (approximately 104 copies per cell) contributes to an elevated sensitivity when RNA-targeting methods such as nucleic acid sequence-based amplification or RT-PCR are used (7, 18, 21).
Primers and a universal fluorescent TaqMan probe were designed to amplify and detect a 290-bp product from the 23S rRNA. This allows the detection of diverse bacterial species without significant cross-reactions with human-derived nucleic acids. The first RT-PCR amplifications gave false-positive results for the no-template controls, similar to the results of other rRNA-based methods (8, 33). To exclude carryover contamination from previously amplified DNAs in which uracil was incorporated, we used the enzyme uracil-N-glycosylase, which cleaves the uracil base from the phosphodiester backbone of uracil-containing DNA. The enzyme has no effect on thymine-containing DNAs (20). Therefore, the additional contamination of the reagents by any kind of eubacterial DNA in the case of bacterial 16S and 23S rRNAs cannot be excluded by using UNG and still represents a serious problem. Taq polymerase has often been found to be contaminated with bacterial DNAs (3, 30) or rRNAs. To reduce the rate of false-positive results caused by the contaminating reagents, we used 8-MOP activated by UV irradiation for 3 to 5 min, which did not interfere with Taq DNA polymerase and which has already been used to successfully remove contaminating DNA from 16S rRNA PCR mixtures (15, 22). The CT values for the positive controls increased in proportion to the dose of UV irradiation and indicated a decrease in PCR efficiency, but with 8-MOP we could reduce the false-positive fluorescent signal.
The detection limit was 16 CFU per ml for the 23S rRNA RT-PCR and 125 CFU per ml for the groEL RT-PCR. However, the determination and comparison of detection limits for bacterial NAT assays are difficult because a standardization of methods and reference material are not yet available. Furthermore, the definition of bacterial titers by the numbers of CFU is problematic because huge amounts of non-cultivable, dead cells or free nucleic acids are present and can be amplified by DNA-based NAT. Therefore, the standardization of bacterial NAT, as is common for viral NAT (32), is strongly required for the detection of bacterial contamination of blood products as previously proposed by Montag (24).
A further consideration was to use a more unstable molecule that normally does not contaminate the reagents and therefore should not be detected. rRNA is more labile than DNA and is more susceptible to degradation caused by deleterious treatments (34), but it is not as labile as mRNA. mRNA is turned over rapidly in living bacterial cells, with most mRNA species having a half-life of only a few seconds (18). 16S and 23S rRNAs did not indicate the viability status of cells that were killed by heat or ethanol under in vitro conditions. Sheridan et al. (34) developed a method to examine the relationship between mRNA and viability: they exposed the cells to two different stress treatments (heat and ethanol) and assayed the mRNAs from three different genes (rpoH, groEL, and tufA). Because of its ubiquitous and highly conserved regions, we chose the groEL gene, as described previously (12, 19), and assayed the mRNA for this single-copy gene. No false-positive results caused by the contamination of reagents or by cross-reactions with human nucleic acids were detected for the no-template controls. The comparison of the two RT-PCR systems showed that the two-enzyme system was more sensitive because of its specific enzymes. Nevertheless, the advantage of rTth polymerase-based assays is the use of enzymatic carryover protection, although a slightly lower detection limit was determined for the RT-PCR system using rTth polymerase. Because of its short half-life, the presence of mRNA may be a good indicator of viability. Since there is no unique definition of the terms “life” and “alive,” we declared a cell viable when it was able to multiply under suitable conditions. One disadvantage of DNA-based methods is that they do not distinguish between living and dead cells (17). With our viability test, we have shown that groEL mRNA was undetectable immediately after cell inactivation, as previously shown for other primer systems (2, 28). The suitability of mRNAs as viability indicators needs to be proven before their use in a reliable technique for the sterility testing of PCs.
Due to the limited durability of PCs, a rapid diagnostic test for transfusion medicine-relevant bacteria is essential. Each bacterial contamination does not play an important role, because all species and even isolates of certain species are not able to grow within human plasma (25). Because of the low bacterial titers of about 10 to 100 bacteria per donation at the beginning, the sensitivity of the detection method plays a crucial role. As a result, false-negative results caused by sampling errors are a problem for both microbiological and molecular genetic methods. Microbiological detection using 10 to 20 ml of platelet concentrate for aerobic and anaerobic cultivation is susceptible to false-positive results (37) and requires a long incubation time, while NAT assays should be sensitive and fast enough for a routine contamination screening of PCs. Therefore, we suggest the screening of PCs during the second day after donation, for which a satellite bag of the PCs should be used. This pre-enrichment should enable the vast majority of contaminated PCs to be detected with current NAT methods, but it can be expected that rare, slow-growing bacteria may escape this detection scheme. If routine testing of PCs is accomplished, an extension of the storage of PCs will likely be demanded because platelet function or activation is not adversely affected over 7 days of storage (10) and because PCs transfused on day 6 or 7 yielded expected clinical responses (1, 26).
In the present study, we described a novel real-time RT-PCR for detecting bacterial mRNAs in PCs, which were suitable as an indicator of viability. The problem of contaminated reagents such as Taq polymerase was reduced by the use of 8-MOP activated by UV light. In ongoing studies, we will increase the sensitivity of the assay by using a larger nucleic acid input. Furthermore, the implementation of an internal positive control can detect false-negative results due to PCR inhibition. In conclusion, further studies with larger sample numbers of PCs should be conducted to demonstrate the applicability of routine contamination screening in transfusion services.
Acknowledgments
We thank Michael Schmidt for his critical reading of the manuscript and Sarah Kirkby for her linguistic advice.
REFERENCES
- 1.AuBuchon, J. P., L. K. Cooper, M. F. Leach, D. E. Zuaro, and J. D. Schwartzman. 2002. Experience with universal bacterial culturing to detect contamination of apheresis platelet units in a hospital transfusion service. Transfusion 42:855-861. [DOI] [PubMed] [Google Scholar]
- 2.Bej, A. K., W.-Y. Ng, S. Morgan, D. Jones, and M. H. Mahbubani. 1996. Detection of viable Vibrio cholerae by reverse-transcriptase polymerase chain reaction (RT-PCR). Mol. Biotechnol. 5:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Böttger, E. C. 1990. Frequent contamination of Taq polymerase with DNA. Clin. Chem. 36:1258. [PubMed] [Google Scholar]
- 4.Brecher, M. E. 2002. Methods to minimize transfusion related bacterial sepsis, p. 69-84. In F. Brown and R. Seitz (ed.), Advances in transfusion safety—2001. Developments in biologicals, vol. 108. Karger, Basel, Switzerland. [PubMed] [Google Scholar]
- 5.Brecher, M. E., N. Means, C. J. Jere, D. Heath, S. Rothenberg, and L. C. Stutzman. 2001. Evaluation of an automated culture system for detecting bacterial contamination of platelets: an analysis of 15 contaminating organisms. Transfusion 41:477-482. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, N. M., E. E. M. Jaeger, S. Choudhury, A. A. S. Dunlop, M. M. Matheson, P. Adamson, N. Okhravi, and S. Lightman. 2000. Detection of and discrimination between gram-positive and gram-negative bacteria in intraocular samples by using nested PCR. J. Clin. Microbiol. 38:1753-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaney, R., J. Rider, and D. Pamphilon. 1999. Direct detection of bacteria in cellular blood products using bacterial ribosomal RNA-directed probes coupled to electrochemiluminescence. Transfus. Med. 9:177-188. [DOI] [PubMed] [Google Scholar]
- 8.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depcik-Smith, N. D., S. N. Hay, and M. E. Brecher. 2001. Bacterial contamination of blood products: factors, options, and insights. J. Clin. Apheresis 16:192-201. [DOI] [PubMed] [Google Scholar]
- 10.Dumont, L. J., and T. VandenBroeke. 2003. Seven-day storage of apheresis platelets: report of an in vitro study. Transfusion 43:43-50. [DOI] [PubMed] [Google Scholar]
- 11.Feng, P., S. P. Keasler, and W. E. Hill. 1992. Direct identification of Yersinia enterocolitica in blood by polymerase chain reaction amplification. Transfusion 32:850-854. [DOI] [PubMed] [Google Scholar]
- 12.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Cow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative Staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodnough, L. T., A. Shander, and M. E. Brecher. 2003. Transfusion medicine: looking to the future. Lancet 361:161-169. [DOI] [PubMed] [Google Scholar]
- 14.Hillyer, C. D., C. D. Josephson, M. A. Blajchman, J. G. Vostal, J. S. Epstein, and J. L. Goodman. 2003. Bacterial contamination of blood components: risks, strategies, and regulation. Joint ASH and AABB educational session in transfusion medicine. Hematology 2003:575-589. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, M. S., L. A. Beck, and R. A. Skuce. 1994. Identification and elimination of DNA sequences in Taq DNA polymerase. J. Clin. Microbiol. 32:2007-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinno, Y., K. Yoshiura, and N. Niikawa. 1990. Use of psoralen as extinguisher of contaminating DNA in PCR. Nucleic Acids Res. 18:6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keer, J. T., and L. Birch. 2003. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53:175-183. [DOI] [PubMed] [Google Scholar]
- 19.Kwok, A. Y. C., S.-C. Su, R. P. Reynolds, S. J. Bay, Y. Av-Gay, N. J. Dovichi, and A. W. Chow. 1999. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int. J. Syst. Bacteriol. 49:1181-1192. [DOI] [PubMed] [Google Scholar]
- 20.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 21.McKillip, J. L., L.-A. Jaykus, and M. Drake. 1998. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier, A., D. H. Persing, M. Finken, and E. C. Böttger. 1993. Elimination of contaminating DNA within polymerase chain reaction reagents: implications for a general approach to detection of uncultured pathogens. J. Clin. Microbiol. 31:646-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell, K. M., and M. E. Brecher. 1999. Approaches to the detection of bacterial contamination in cellular blood products. Transfus. Med. Rev. 13:132-144. [DOI] [PubMed] [Google Scholar]
- 24.Montag, T. 3 July 2003. Bacterial standards suitable for validation of blood screening and pathogen reduction. Oral presentation at SoGAT XVI, Langen, Germany.
- 25.Montag, T., and D. de Korte. 17 September 2003. Screening-Methoden zur Erkennung bakterieller Kontaminationen in Blutkomponenten. Oral presentation at DGTI, Innsbruck, Austria.
- 26.Murphy, S. 2002. What's so bad about old platelets? Transfusion 42:809-811. [DOI] [PubMed] [Google Scholar]
- 27.Ortolano, G. A., L. F. Freundlich, S. Holme, R. L. Russell, M. A. Cortus, K. Wilkins, H. Nomura, C. Chong, R. Carmen, A. Capetandes, and B. Wenz. 2003. Detection of bacteria in WBC-reduced PLT concentrates using percent oxygen as a marker for bacteria growth. Transfusion 43:1276-1285. [DOI] [PubMed] [Google Scholar]
- 28.Patel, B. K. R., D. K. Banjerjee, and P. D. Butcher. 1993. Determination of Mycobacterium leprae viability by polymerase chain reaction amplification of 71-kDa heat shock protein mRNA. J. Infect. Dis. 168:799-800. [DOI] [PubMed] [Google Scholar]
- 29.Petershofen, E. K., R. Fislage, R. Faber, H. Schmidt, W. K. Roth, and E. Seifried. 2000. Detection of nucleic acid sequences from bacterial species with molecular genetic methods. Transfus. Sci. 23:21-27. [DOI] [PubMed] [Google Scholar]
- 30.Rand, V. H., and H. Houck. 1990. Taq polymerase contains bacterial DNA of unknown origin. Mol. Cell. Probes 4:445-450. [DOI] [PubMed] [Google Scholar]
- 31.Roth, V. R., M. J. Arduino, J. Nobiletti, C. S. Holt, L. A. Carson, C. F. Wolf, B. A. Lenes, P. M. Allison, and W. R. Jarvis. 2000. Transfusion-related sepsis due to Serratia liquefaciens in the United States. Transfusion 40:931-935. [DOI] [PubMed] [Google Scholar]
- 32.Saldanha, J. 1999. Standardization: a progress report. Biologicals 27:285-289. [DOI] [PubMed] [Google Scholar]
- 33.Sen, K. 2000. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J. Clin. Microbiol. 38:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheridan, G. E. C., C. I. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turenne, C. Y., E. Witwicki, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 2000. Rapid identification of bacteria from positive blood cultures by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 38:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Camp, G., S. Chapele, and R. De Watcher. 1993. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA genes with conserved primer sequences. Curr. Microbiol. 27:147-151. [DOI] [PubMed] [Google Scholar]
- 37.Wagner, S. J., and D. Robinette. 1998. Evaluation of an automated microbiologic blood culture device for detection of bacteria in platelet components. Transfusion 38:674-679. [DOI] [PubMed] [Google Scholar]
- 38.Wagner, S. J., L. I. Friedman, and R. Y. Dodd. 1994. Transfusion associated bacterial sepsis. Clin. Microbiol. Rev. 7:290-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yomtovian, R. 2002. Novel methods for detection of platelet bacterial contamination. Vox Sang. 38:129-131. [DOI] [PubMed] [Google Scholar]