Abstract
Endogenous and exogenous formaldehyde (FA) has been linked to cancer, neurotoxicity, and other pathophysiologic effects. Molecular and cellular mechanisms that underlie FA-induced damage are poorly understood. In this study, we investigated whether proteotoxicity is an important, unrecognized factor in cell injury caused by FA. We found that irrespective of their cell cycle phases, all FA-treated human cells rapidly accumulated large amounts of proteins with proteasome-targeting K48-linked polyubiquitin, which was comparable with levels of polyubiquitination in proteasome-inhibited MG132 controls. Both nuclear and cytoplasmic proteins were damaged and underwent K48-polyubiquitination. There were no significant changes in the nonproteolytic K63-polyubiquitination of soluble and insoluble cellular proteins. FA also rapidly induced nuclear accumulation and Ser326 phosphorylation of the main heat shock-responsive transcription factor HSF1, which was not a result of protein polyubiquitination. Consistent with the activation of the functional heat shock response, FA strongly elevated the expression of HSP70 genes. In contrast to the responsiveness of the cytoplasmic protein damage sensor HSF1, FA did not activate the unfolded protein response in either the endoplasmic reticulum or mitochondria. Inhibition of HSP90 chaperone activity increased the levels of K48-polyubiquitinated proteins and diminished cell viability after FA treatment. Overall, our results indicate that FA is a strong proteotoxic agent, which helps explain its diverse pathologic effects, including injury in nonproliferative tissues.
Exposure to formaldehyde (FA) is associated with a range of adverse health effects, including respiratory and other cancers,1, 2, 3 as well as other serious pathophysiologic manifestations.4, 5, 6 Metabolic formation of FA after methanol ingestion causes acute injury, including tissue failure and irreversible damage to the optical nerve, and long-term neurologic impairments, parkinsonism, and polyneuropathy.7, 8 Human and other cells also continuously produce FA endogenously as a by-product of several normal biochemical processes,5 including histone demethylation reactions in the nucleus.9 Recent studies using mouse genetic models with diminished FA detoxification have established a surprisingly high toxicity of endogenous FA, as evidenced by developmental defects in the embryos due to maternal aldehydes10 and multitissue degeneration and cancers in adult mice.11
FA is a well-known crosslinking agent that forms DNA-protein crosslinks (DPCs) due to its reactivity with NH2 groups of histone lysines and DNA bases.12, 13 DPCs have long been considered as the main genotoxic and cytotoxic lesions for FA, which led to the use of FA as a tool for understanding of DPC tolerance mechanisms using collections of mutant cells with different DNA repair defects.14, 15, 16, 17, 18 The results of these investigations were contradictory even for the same cell model, and one study has found that the sensitivity of specific mutants depended on the exposure protocol.14 Genotoxic signaling triggered by FA in human cells occurred only in the S phase,19, 20 which is consistent with the stalling of replication helicases by DNA-attached proteins.21 However, FA-induced activation of the transcription factor p5320 and the DNA damage-responsive kinase ataxia-telangiectasia mutated (ATM)22 showed bell-shaped dose dependence, suggesting that FA-treated cells may experience another major stress that becomes dominant at certain exposure conditions even in S-phase cells. The importance of the non-DPC damage is also supported by clinical manifestations of FA toxicity such as neurologic damage,6, 8 because neurons are nondividing cells, and the replication-associated mode of cytotoxicity is not applicable to them. FA-induced DPCs are repairable and unstable hydrolytically,23 suggesting that their transient presence is also not likely to exert long-lasting effects on neuronal cells through some replication-unrelated mechanisms. In contrast to DNA damage, adult neurons are sensitive to the presence of abnormal proteins, as evidenced by such protein conformation-associated pathologies as Parkinson, Huntington, and Alzheimer diseases.24
In this work, we examined a possibility that FA can act as a proteotoxic agent, inducing accumulation of abnormal proteins and DNA damage-independent cytotoxicity. We reasoned that FA conjugation with amino and SH groups can cause misfolding and aggregation of proteins, resulting in proteotoxic effects. The formation of abnormal proteins in cells is monitored by specialized protein quality controls that promote refolding and stabilization of misfolded proteins by molecular chaperones and eliminate severely damaged proteins via proteasomal and other degradation pathways.25, 26 We found that FA-treated human cells displayed clear evidence of proteotoxic stress, such as accumulation of polyubiquitinated proteins and the activation of the heat shock response. Inhibition of the heat shock protein (HSP)90 chaperone activity caused a loss of cell viability over a range of doses, including normally weakly cytotoxic or completely noncytotoxic doses. Thus, FA is a strong inducer of protein damage that requires engagement of protein quality controls for preserving cell viability.
Materials and Methods
Chemicals
MG132 was obtained from Selleckchem (Houston, TX). G5 was from Santa Cruz Biotechnology (Dallas, TX) and 17-demethoxy-17-[[2-(dimethylamino)ethyl]amino]geldanamycin hydrochloride (17-DMAG) and VER-155008 were purchased from Tocris Bioscience (Bristol, United Kingdom). FA solution (F8775), buffers, and salts were from Sigma-Aldrich (St. Louis, MO).
Cells and Treatments
H460 and IMR90 cells were purchased from the ATCC (Manassas, VA). Immortalized H460 cells were grown in 95% air/5% CO2 using 10% fetal bovine serum-supplemented RPMI-1640. IMR90 normal human lung fibroblasts were grown in Dulbecco's modified Eagle's medium containing 10% serum in the presence of 5% O2 and 5% CO2. Normal human bronchial epithelial (HBE) cells (Lonza, Walkersville, MD) were cultured in 95% air/5% CO2 using a vendor's recommended medium (CC-3170; Lonza). Cells were seeded for FA and other treatments at approximately 40% to 50% confluence and allowed to attach overnight. With the exception of a short time-course study involving 30- and 60-minute incubations, cells were treated with FA in complete growth media containing serum.
Western Blot Analysis
Attached cells were collected by scraping, and soluble cellular proteins were typically obtained by extraction with a cold lysis buffer [50 mmol/L Tris (pH 8.0), 250 mmol/L NaCl, 1% NP40, 0.1% SDS, 5 mmol/L EDTA, 2 mmol/L Na3VO4, 10 mmol/L Na2P2O7, 10 mmol/L NaF] supplemented with protease inhibitors and 20 mmol/L N-ethylmaleimide (referred to as 1% NP40 buffer). After incubation for 10 minutes on ice, lysates were spun down at 10,000 × g for 10 minutes, 4°C, and supernatants were collected and frozen at −80°C. For solubilization of the remaining insoluble cellular material, pellets were boiled for 10 minutes in a 2% SDS buffer (2% SDS, 50 mmol/L Tris-HCl pH 6.8, 10% glycerol, 20 mmol/L N-ethylmaleimide) supplemented with protease/phosphatase inhibitors (Thermo Scientific, Rockford, IL). Samples were cooled to room temperature, followed by centrifugation at 10,000 × g for 10 minutes to obtain clear supernatants. In the initial experiments assessing HSF1 status in FA-treated cells, a set of soluble lysates was also prepared using phosphate-buffered saline (PBS) containing 1% Triton X-100, 20 mmol/L N-ethylmaleimide, and protease/phosphatase inhibitors (referred to as 1% Triton buffer). Proteins were separated by SDS-PAGE on 8% gels and electrotransferred overnight onto ImmunoBlot polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). Primary antibodies were anti–K48-linked polyubiquitin (4289S; dilution 1:1000), anti–K63-linked polyubiquitin (5621S; dilution 1:1000), anti–phospho-T68-checkpoint kinase (CHK)2 (2661; dilution 1:1000), anti–phospho-S317-CHK1 (2344; dilution 1:1000), anti–phospho-S15-p53 (9284; dilution 1:1000), anti–glyceraldehyde-3-phosphate dehydrogenase (3683; dilution 1:2000), and anti-HSF1 (4356S; dilution 1:1000) from Cell Signaling Technology (Danvers, MA); anti-fibrillarin (ab5821; dilution 1:5000), anti–phospho-S1981-ATM (ab81292; dilution 1:1000), and anti–phospho-S326-HSF1 (ab76076; dilution 1:1000) from Abcam (Cambridge, MA); anti–phospho-S824-KAP1 (A300-767A; dilution 1:1000) from Bethyl Laboratories (Montgomery, TX); anti-tubulin (T6557; dilution 1:2000) from Sigma-Aldrich. Horseradish peroxidase-conjugated goat anti-mouse IgG (12-349; dilution 1:5000) and goat anti-rabbit IgG (7074; dilution 1:2000) were from Millipore (Billerica, MA) and Cell Signaling Technology, respectively.
Immunofluorescence
Cells were seeded on human fibronectin-coated coverslips and allowed to attach overnight before exposures to FA and other stressors. S-phase cells were labeled by the addition of 10 μmol/L 5-ethynyl-2′-deoxyuridine (EdU) for 1 hour before FA treatments. At the end of the incubation times, media were aspirated, and cells were rinsed with PBS. Two different methods were used for cell fixation: −20°C-chilled methanol for 10 minutes at 4°C or 3.7% paraformaldehyde for 10 minutes at 4°C (only for pS326-HSF1 staining). Both methanol- and paraformaldehyde-fixed cells were permeabilized with PBS-0.5% Triton X-100 for 15 minutes at room temperature. Coverslips were blocked with 2% fetal bovine serum for 30 minutes, followed by EdU staining using Click-iT EdU-Alexa Fluor 488 Imaging kit (Thermo Scientific). Primary antibodies for K48-linked polyubiquitin (05-61307; Millipore), K63-linked polyubiquitin (BML-PW0600; Enzo, Farmingdale, NY), phospho-S326-HSF1 (ab76076; Abcam), HSF1 (4356; Cell Signaling Technology), FK2 (BML-PW8810; Enzo), HSP72/73 (HSP01, Millipore) were all used at 1:200 dilution. The secondary antibodies, A11029 Alexa Fluor 488 goat anti-mouse and A11036 Alexa Fluor 568 goat anti-rabbit (both at 1:500 dilution), were from Life Technologies (Carlsbad, CA). All antibody dilutions were made in a PBS solution containing 1% bovine serum albumin and 0.5% Tween-20. Cells were incubated with primary antibodies for 2 hours at 37°C, washed three times with PBS, and then incubated with secondary antibodies for 30 minutes at 37°C. Coverslips were mounted on glass slides using a fluorescence mounting media with DAPI (H-1200; Vectashield, Burlingame, CA). Cells were viewed and imaged on a Nikon (Melville, NY) E-800 Eclipse fluorescent microscope at ×200 magnification. Confocal images were taken on a Zeiss (Thornwood, NY) LSM710 confocal microscope at ×630 magnification.
Quantitative RT-PCR
RNA was purified from cells using TRIzol (Thermo Scientific). Briefly, cells were collected by trypsinization, washed with PBS, and lysed in TRIzol (1 mL/1.2 × 106 cells) for 5 minutes at room temperature. Cell lysates were extracted with 200 μL of chloroform per sample, and the upper aqueous phase containing RNA was collected after centrifugation. RNA was then precipitated with 0.5 mL of isopropanol, washed with 75% ethanol, and resuspended in RNase-free water. Purity of RNA was assessed by a Nanodrop spectrophotometer. cDNA was synthesized using the RT First Strand kit (SABiosciences, Frederick, MD) following the manufacturer's instructions. PCR reactions were performed using the Human Heat Shock Proteins & Chaperones RT Profiler PCR Array (PAHS-076ZE; SABiosciences) in the ABI 7900HT Real-Time PCR System (Applied Biosystems, Carlsbad, CA). Four housekeeping genes B2M, HPRT1, RPL13A, and ACTB were used for normalization purposes. Differences in gene expression were calculated by the 2−ΔΔCt method. Individual quantitative RT-PCR reactions for DDIT3, ATF4, and PPP1R15A were performed using the ViiA7 system (Thermo Fischer). Three housekeeping genes B2M, TBP, and GAPDH were used for normalization. All individual sets of primers were purchased from SABiosciences (catalog no.: PPH00310A for DDIT3, PPH02016A for ATF4, PPH02081E for PPP1R15A, PPH001150F for GAPDH, PH01091G for TBP, and PPH01094E for B2M).
Cytotoxicity of FA Treatments
Measurements of the metabolic activity using the CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI) and scoring of the colony-forming ability were used for the assessment of cytotoxicity. In the clonogenic survival procedure, H460 cells were seeded onto 6-well plates at 40% to 50% confluence and grown overnight. On the next day, cells were treated with FA (three wells per dose), washed, trypsinized, and seeded at 200 cells/well in 6-well plates. Cells were allowed to form visible colonies over 7 to 8 days and then fixed with methanol and stained with the Giemsa solution (Sigma-Aldrich). For the CellTiter-Glo assay, cells were seeded into 96-well optical cell culture plates (2000 cells/well for H460 and 1000 cells/well for IMR90 and HBE cells), grown overnight, and then treated with FA (four wells per dose). The assay measurements were recorded at 72 hours after FA removal.
Statistical Analysis
Statistical differences between the groups were evaluated by two-tailed, unpaired t-test. Results in multiple comparisons were adjusted using the Bonferroni correction.
Results
K48-Polyubiquitination of Proteins in Response to FA
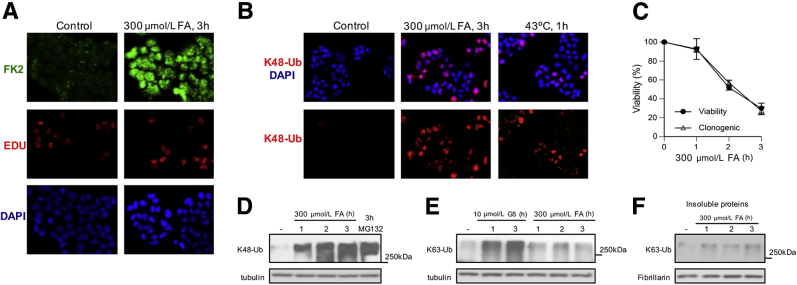
Cells target damaged proteins for proteasomal degradation by triggering their polyubiquitination. Therefore, if FA is a proteotoxic stressor, then FA-treated cells should show accumulation of polyubiquitinated proteins. In the first set of experiments, we used human H460 cells that we have previously characterized for various cytotoxic and DNA damage-related responses to FA.19, 20, 22 FA-induced signaling in H460 was similar to that in normal human cells. We found that a treatment of H460 cells with 300 μmol/L FA for 3 hours caused a massive formation of polyubiquitinated proteins as detected by immunostaining with a polyubiquitin-specific FK2 antibody (Figure 1A). This is a widely used antibody that recognizes a protein-conjugated polyubiquitin but not free ubiquitin. Our FA treatment corresponded to the conditions that inhibited p53 activation.20 Unlike DNA and chromatin damage-related stress signaling,19, 20, 22 accumulation of polyubiquitinated proteins was not restricted to S-phase cells (EdU-positive cells) and occurred in all cells (Figure 1A). Ubiquitin can form biologically very different polyubiquitin chains, depending which Lys position is used for linkages. The presence of Lys48(K48)-linked polyubiquitin chains leads to binding to proteasomes and triggers degradation of proteins.27 With the use of K48 linkage-specific antibodies, we found the formation of proteasome-targeted polyubiquitinated proteins in all FA-treated cells, which was more extensive than in cells subjected to a 43°C heat shock (Figure 1B). Although the used dose of FA clearly induced massive protein polyubiquitination, it was also quite toxic (25% to 30% viability) in two long-term viability assays that measured metabolic activity or the colony-formation ability (Figure 1C). However, the accumulation of K48-polyubiquitinated proteins was not limited to the highly toxic doses of FA, because a minimally cytotoxic 1-hour exposure to FA (90% to 95% viability) (Figure 1C) also caused a strong signal for the high molecular weight K48-polyubiquitin products in soluble cellular proteins (Figure 1D). The 3-hour long exposures to FA and the proteasome inhibitor MG132 produced comparable amounts of K48-polyubiquitinated proteins, demonstrating a high proteotoxic activity of FA. Proteins can also contain covalently attached K63-linked polyubiquitin chains that play a role in signaling processes, including DNA repair.27 We did not detect significant changes in the levels of K63-specific polyubiquitination in either soluble or insoluble cellular proteins (Figure 1, E and F). These negative results were not caused by technical factors, because simultaneously run samples of cells treated with a general deubiquitinase inhibitor G5 produced clear increases in K63-polyubiquitinated proteins.
Figure 1.
Formation of polyubiquitinated proteins in FA-treated H460 human lung cells. A: Representative images of cells stained for DNA (DAPI), S-phase cells (EdU), and polyubiquitinated proteins (FK2 antibody). Cells were treated with FA for 3 hours in complete medium. B: Immunostaining for K48-Ub in FA-treated (3 hours) and heat-shocked cells. C: Cell viability and colony formation after FA treatments. Viability was measured at 72 hours after FA exposure. D: Formation of K48-Ub by FA and MG132 (5 μmol/L). Tubulin is used as a loading control. E: Western blot analysis for K63-specific polyubiquitination of soluble proteins in cells treated with FA and the deubiquitinase inhibitor G5. F: Western blot analysis for K63-Ub in the insoluble cellular proteins. Fibrillarin was used as a loading control. Data are expressed as means ± SD. n = 2 independent experiments, each including three dishes (clonogenic survival) or four wells (viability assay) per dose (C). EdU, 5-ethynyl-2′-deoxyuridine; FA, formaldehyde; K48-Ub, K48-linked polyubiquitin; K63-Ub, K63-polyubiquitination.
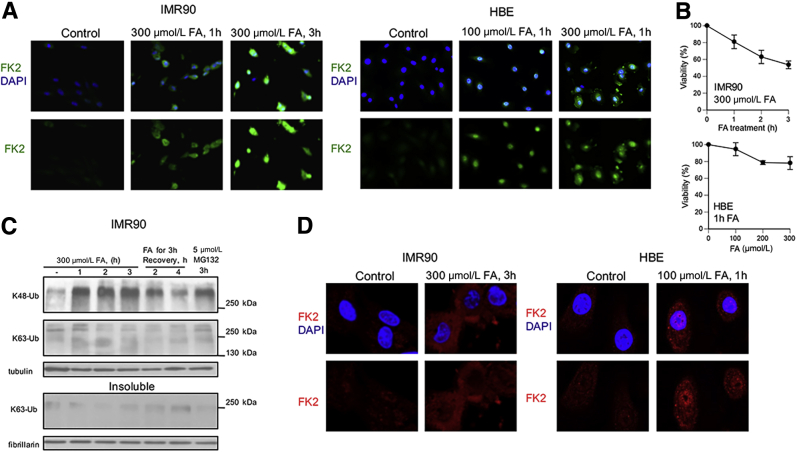
To examine proteotoxic responses in primary human cells, we analyzed FA-treated IMR90 and HBE cells by immunostaining with the polyubiquitin-binding FK2 antibody (Figure 2A). Both types of normal cells showed strong increases in the protein polyubiquitination already after brief 1-hour long treatments with FA, which caused only modest decreases in the long-term viability of these cells (Figure 2B). Western blot analysis with linkage-specific antibodies confirmed the accumulation of K48- but not K63-polyubiquitinated proteins in IMR90 cells at different times of FA exposure and recovery (Figure 2C). The lack of increases in K63-polyubiqitinated proteins in proteasome-inhibited cells (MG132 lane) is consistent with the nonproteolytic role of this type of ubiquitin linkages. Confocal imaging showed that most FA-induced protein polyubiquitination was cytoplasmic, but polyubiquitinated proteins were also detectable in the nucleus, especially in HBE cells (Figure 2D).
Figure 2.
Polyubiquitination of proteins in response to FA in primary human cells. A: Epifluorescence images of IMR90 and HBE cells stained for DNA (DAPI) and polyubiquitinated proteins (FK2 antibody). B: Viability of IMR90 cells treated with 300 μmol/L FA for 0 to 3 hours and HBE cells treated with 0 to 300 μmol/L FA for 1 hour. Cell viability was measured at 72 hours after FA removal. C: Western blots for linkage-specific polyubiquitination in the soluble (top panel) and insoluble (bottom panel) proteins. Tubulin and fibrillarin were used as loading controls. D: Confocal images of IMR90 and HBE cells immunostained for polyubiquitinated proteins with FK2 antibody. Data are expressed as means ± SD. n = 3 (IMR90) or 2 experiments (HBE), each including four wells per dose (B). FA, formaldehyde; HBE, human bronchial epithelial; K48-Ub, K48-linked polyubiquitin; K63-Ub, K63-polyubiquitin.
Heat Shock Response to FA
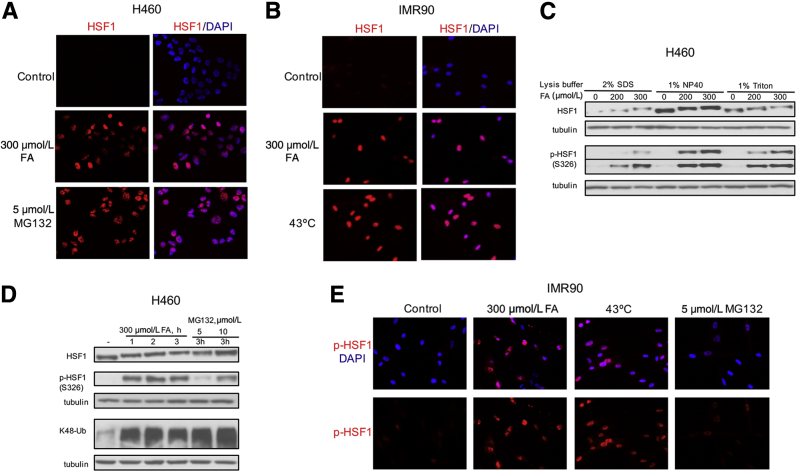
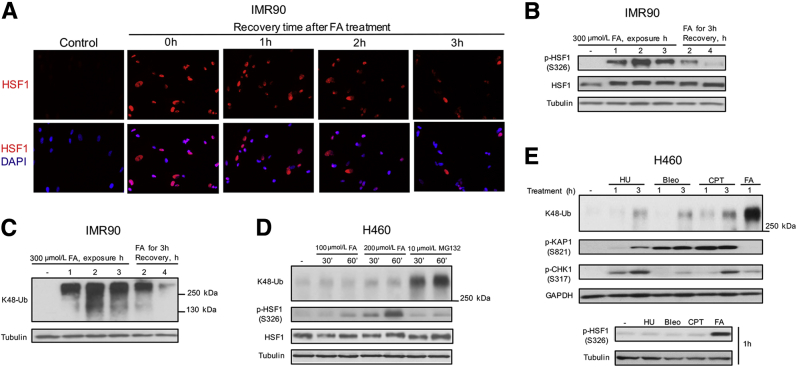
Accumulation of damaged or misfolded proteins triggers a heat shock response, which involves activation of the HSF1 transcription factor.28, 29 To test whether FA-induced proteotoxic stress induces the heat shock response, we examined classic markers of HSF1 activation such as its nuclear translocation and hyperphosphorylation. We found that FA-treated H460 and IMR90 cells showed large amounts of nuclear HSF1, which was comparable with responses induced by the proteasome inhibitor MG132 and the canonical HSF1 activator heat shock (Figure 3, A and B). The presence of hyperphosphorylated HSF1 is detected by its slower migration in SDS-PAGE gels. Heat shock-activated HSF1 forms nuclear complexes of different solubility.30 To examine the presence of phosphorylated HSF1 in FA-treated cells, we first tested cell lysates prepared with three different detergents. We found that our standard 1% NP40 lysis buffer used for extraction of soluble proteins for the determination of polyubiquitinated proteins gave the highest sensitivity for the detection of both total HSF1 and its Ser326-phosphorylated form (Figure 3C). The NP40 buffer has also been effective in extracting chromatin-bound DNA repair proteins31 and other stress-responsive transcription factors such as HIF1α and p53.32 Irrespectively of the lysis buffer, FA treatments clearly showed the hyperphosphorylation-associated upward shift in HSF1 mobility and extensive Ser326 phosphorylation. The presence of Ser326 phosphorylation has been associated with the acquisition of the transactivation activity by HSF1.33 The extent of HSF1 hyperphosphorylation by FA was comparable with that induced by the proteasome inhibitor MG132 at the same duration of exposure; however, FA was a more potent inducer of Ser326 phosphorylation than MG132 in H460 cells (Figure 3D). These differences in Ser326 phosphorylation were unrelated to the amounts of polyubiquitinated proteins, which were comparable for 5 μmol/L MG132 and all FA treatments. A stronger ability of FA to induce Ser326-HSF1 phosphorylation in comparison with MG132 was also observed in normal IMR90 cells by immunostaining, excluding cell- and assay-specific effects as causes of these differences (Figure 3E). The levels of Ser326 phosphorylation by FA were comparable with those by a 43°C treatment, pointing to a closer similarity of FA to heat shock than to proteasome inhibitors. A time-course study of cells treated with FA for 1 to 3 hours found a very good correlation between the amounts of K48-polyubiquitinated proteins and activation of HSF1 as assayed by its Ser326 phosphorylation, slower migration and nuclear HSF1 staining (Figure 4, A–C). Markers of activated HSF1 and protein polyubiquitination also showed similar kinetics of decay during recovery of cells after FA damage. These observations raised a question whether there is a cause-effect relationship between the accumulation of polyubiquitinated proteins and HSF1 activation. To explore a potential link between these two markers of proteotoxic stress, we compared responses after brief 30- and 60-minute treatments with FA and the proteasome inhibitor MG132 in serum-free medium, which permits comparable rates of diffusion of both chemicals into cells due to elimination of FA binding with serum proteins. We found that MG132 caused a rapid accumulation of polyubiquitinated proteins, which was not accompanied by significant changes in HSF1 mobility and Ser326 phosphorylation (Figure 4D). In contrast, FA induced very clear increases in Ser326 phosphorylation and slower HSF1 mobility without a detectable accumulation of polyubiquitinated proteins at these times. Thus, HSF1 activation is an early stress response to FA, which is triggered before the buildup of polyubiquitinated proteins.
Figure 3.
Nuclear accumulation and phosphorylation of HSF1 in FA-treated human cells. A and B: H460 (A) and IMR90 (B) cells were treated (FA and MG132 for 3 hours, 43°C for 1 hour) and immunostained for HSF1. C: Western blots for HSF1 and Ser326-phosphorylated HSF1 in protein extracts prepared from H460 cells using different lysis buffers. FA treatments were for 3 hours. Tubulin was used as a loading control. D: HSF1 phosphorylation and K48-Ub of soluble cellular proteins (1% NP40 extraction) after H460 treatments with FA and MG132. E: Representative images of IMR90 normal human cells immunostained for Ser326–p-HSF1. Cells were exposed to FA or MG132 for 3 hours, and 43°C for 1 hour. FA, formaldehyde; HSF1, heat shock factor 1; K48-Ub, K48-polyubiquitination; p-HSF1, phosphorylated HSF1.
Figure 4.
Time-dependent induction and recovery of proteotoxic responses. A: Epifluorescence images of IMR90 stained for HSF1 and DNA (DAPI). Cells were treated with 300 μmol/L FA for 3 hours in complete medium and fixed after 0 to 3 hours recovery. B and C: HSF1-Ser326 phosphorylation (B) and levels of K48-Ub during and after FA treatments of IMR90 cells in complete medium (C). D: Time-dependent activation of HSF1 by FA and MG132 in serum-free medium. E: Proteotoxic and genotoxic responses in H460 cells treated with 5 mmol/L HU, 10 μmol/L Bleo, 1 μmol/L CPT, or 300 μmol/L FA. Bleo, bleomycin; CPT, camptothecin; FA, formaldehyde; HSF1, heat shock factor 1; HU, hydroxyurea; K48-Ub, K48-polyubiquitinated proteins; p-HSF1, phosphorylated HSF1.
Heat shock responses have been induced by UV and ionization radiation,34, 35 which act as both DNA- and protein-damaging agents. Because FA also damages DNA and proteins, we sought to compare proteotoxic responses by FA and DNA-damaging chemicals with minimal or no protein damage. We selected a widely used replication stressor hydroxyurea, which inhibits synthesis of deoxyribonucleotides, the radiomimetic bleomycin, which causes strand breaks following intercalation into the DNA duplex, and the topoisomerase I inhibitor camptothecin, which traps topoisomerase I covalently attached to a DNA break. We found that none of the three DNA-damaging agents induced a detectable accumulation of K48-polyubiquitinated proteins after 1-hour long treatments despite their induction of higher levels of DNA damage in comparison with FA, as judged by a much stronger DNA damage-associated stress signaling reflecting phosphorylation of ATM (Ser824-KAP1) and ataxia telangiectasis and Rad3 related (ATR; Ser317-CHK1) targets (Figure 4E). In agreement with these results, only FA but not hydroxyurea, bleomycin, or camptothecin stimulated activating phosphorylation of HSF1 at Ser326 (Figure 4E). Longer 3-hour treatments with all three DNA-damaging agents produced small increases in K48-polyubiquitination (Figure 4E), likely reflecting the formation of DNA repair-related polyubiquitinated products.36, 37 Thus, both the extent and the timing of K48-polyubiquitination by FA are clearly consistent with its proteotoxic, not genotoxic properties.
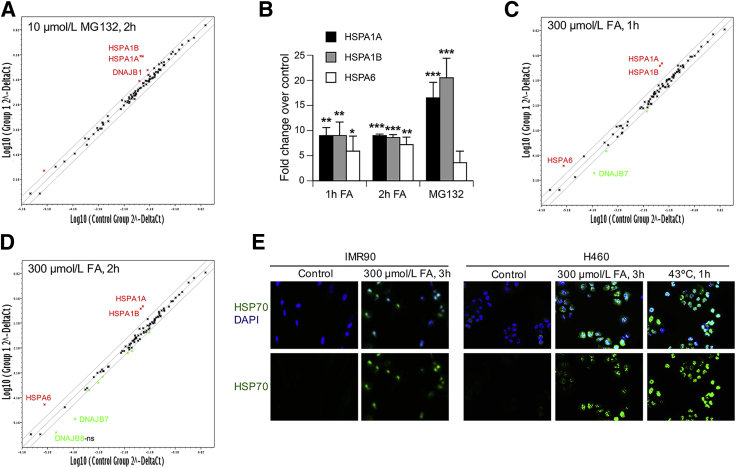
To assess the impact of FA on gene expression of proteins involved in protein folding and stability, we performed quantitative RT-PCR for 84 genes included in the Heat Shock Proteins & Chaperons PCR Array from SABiosciences. IMR90 normal human cells treated for 2 hours with 10 μmol/L MG132, a dose that causes a shift in HSF1 mobility and a strong nuclear accumulation of HSF1, were used as a positive control. MG132 caused large increases in gene expression of two inducible members of the HSP70 family, HSPA1A and HSPA1B (Figure 5, A and B). A more modest up-regulation was also observed for the HSP40 member DNAJB1 (3- ± 0.9-fold increase, P < 0.05). No other genes were significantly induced or repressed by MG132 above the twofold threshold (marked by the two-lane corridor). Cells treated with FA for 1 or 2 hours also strongly up-regulated expression of HSPA1A and HSPA1B genes (Figure 5, B–D). Unlike MG132, both FA treatments also induced expression of another HSP70 member, HSPA6, and repressed transcription of the HSP40 gene DNAJB7 (6.8- ± 2.6-fold decrease, P < 0.05, and 9.8- ± 4.2-fold decrease, P < 0.05, for 1 and 2 hours treatments, respectively). DNAJB7 is one of the HSP70 co-chaperones with a promiscuous client-binding specificity.38 A strong up-regulation of inducible HSP70 members by FA was also observed on the protein level in two types of cells, which was comparable with heat shock responses (Figure 5E). Supporting in vivo relevance of our gene expression findings, chronic inhalation exposure of rats to FA induced a testicular injury and elevated HSP70 expression in the seminiferous epithelium.39 Down-regulation of DNAJB7 expression by FA could result from its direct damage to specific transcription cofactors, making them nonfunctional. This suggestion is consistent with the fact that the protein-nondamaging stressor MG132 did not cause the same transcriptional repression.
Figure 5.
Gene expression of heat shock proteins and protein chaperones. The gene expression was determined by quantitative RT-PCR using the Human Heat Shock Proteins & Chaperones RT Profiler PCR Array (panels A–D). A: Scatter plot of relative expression for 84 heat shock/protein chaperones-related genes in control and MG132-treated IMR90 cells. Log-transformed values of the relative expression level of each gene (2−ΔCt) between MG132-treated (y axis) and control cells (x axis) are shown. The two-lane corridor indicates a twofold threshold in expression changes. Up-regulated and down-regulated genes by the treatment are indicated by red and green symbols, respectively. B: Expression of HSP70 heat shock genes in MG132-and FA-treated IMR90 cells. C and D: Relative expression for 84 heat shock/protein chaperones-related genes in 1 hour FA-treated (C) and 2 hours FA-treated (D) IMR90 cells. E: Epifluorescence images for protein expression of inducible HSP70 (HSP72/73) in cells treated with FA or subjected to heat shock. Data are expressed as means (A, C, and D) or means ± SD (B). n = 3 experiments (A–D). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 relative to untreated controls. FA, formaldehyde; HSP70, heat shock protein 70; ns, not significant; p-, phosphorylated.
Proteotoxic conditions frequently induce the unfolded proteins response (UPR), which is triggered by the accumulation of misfolded or unfolded proteins in the endoplasmic reticulum.40 Activation of specific UPR branches leads to the transcriptional up-regulation of a characteristic set of genes. None of the three canonical UPR target genes DNAJB6 (fold changes: 1.2 ± 0.1 for 1 hour and 0.8 ± 0.1 for 2 hours of FA), HSP90B1 (fold changes: 1.1 ± 0.1 for 1 hour and 0.8 ± 0.1 for 2 hours of FA), HSPA5 (fold changes: 1 ± 0.4 for 1 hour and 0.8 ± 0.2 for 2 hours of FA), three protein kinase RNA-like endoplasmic reticulum kinase pathway members DDIT3 (fold changes: 0.9 ± 0.2 for 1 hour and 0.9 ± 0.1 for 2 hours of FA), ATF4 (fold changes: 1 ± 0.1 for 1 hour and 1.1 ± 0.1 for 2 hours of FA), PPP1R15A (fold changes: 1 ± 0.2 for 1 hour and 1 ± 0.1 for 2 hours of FA), or the inositol-requiring enzyme 1 pathway member ATF6 (fold changes: 0.6 ± 0.2 for 1 hour and 0.5 ± 0.1 for 2 hours of FA) showed higher expression after FA treatments, indicating the absence of a significant endoplasmic reticulum stress. It is possible that UPR can be up-regulated at different doses or longer FA treatments. UPR activation by FA has been reported in A549 human lung cells; however, the observed increases in the UPR readouts were modest (approximately 1.5-fold) and coincided with the activation of apoptotic events.41 Similar to the endoplasmic reticulum UPR, gene expression of the components of the mitochondrial unfolded response (DNAJA3, HSPD1, HSPE1, HSPA9)42 was also not significantly altered by 1- and 2-hour long treatments of IMR90 with 300 μmol/L FA (0.8- to 1.3-fold changes range).
FA Toxicity in HSP90-Inhibited Cells
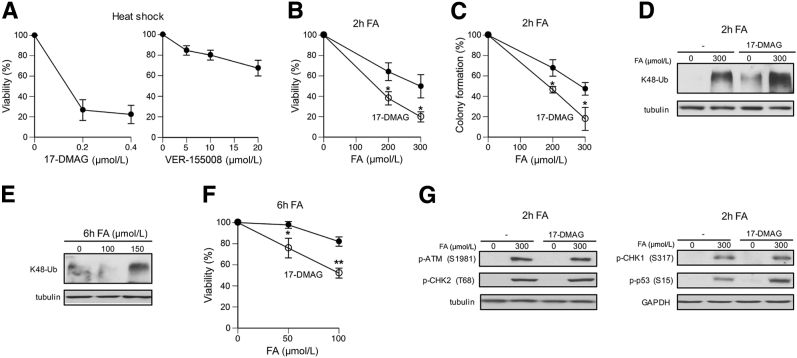
Heat shock proteins HSP70 and HSP90 are ATP-dependent molecular chaperones that promote folding and stabilization of client proteins in their active conformations.26 Inhibition of ATPase activity of HSP70/90 blocks their activity, which can be used for testing the importance of these major protein chaperones in cellular resistance to a particular stressor. To validate this approach, we examined cytotoxicity of heat shock in cells with transiently inhibited HSP90 and HSP70 (Figure 6A). We found that inhibition of HSP90 with a water-soluble analogue of the antibiotic geldanamycin, 17-DMAG, caused a strong reduction in cell viability, whereas inactivation of ATPase activity of HSP70 by VER-155008 had only a modest effect. Heat shock is a strong inducer of both HSP70 and HSP90,28 and the observed difference in protective effects of these proteins could be related to off-target effects of the used inhibitor of HSP70 or pleiotropic cellular effects of HSP70 inhibition. A selection of HSP70 inhibitors is currently limited, and VER-155008 is the most commonly used drug. In light of a high target selectivity by 17-DMAG and its potent effect on toxicity of heat shock, we used this inhibitor to assess the role of HSP90 in protection of cells against FA. We found that 2-hour long treatments of H460 cells with 200 and 300 μmol/L FA, doses that induced large amounts of K48-polyubiqitinated proteins (Figures 1 and 3), were significantly more cytotoxic to HSP90-inhibited cells, as measured by colony formation and cell viability assays (Figure 6, B and C). Consistent with its protein conformation-stabilizing role, inhibition of HSP90 activity increased the amount of proteins targeted for proteolysis in FA-treated cells (Figure 6D). Low FA concentrations, such as 100 μmol/L in H460 cells, did not cause detectable increases in the steady-state levels of K48-polyubiquitination (Figure 6E), raising a question whether cytoprotection by HSP90 is limited to high doses of FA. We found that inhibition of HSP90 activity also reduced cell viability after low FA doses that were either minimally or even noncytotoxic in H460 cells (Figure 6F). The inability of FA to up-regulate HSP90 expression (Figure 5, C and D) makes cells dependent on the constitutive levels of this chaperone and probably limits their potential to suppress adverse effects of FA-induced protein damage. In addition to its role in suppressing toxicity of FA-damaged proteins, HSP90 activity can potentially be important for DNA damage responses. We found that inhibition of HSP90 by 17-DMAG had no apparent effect on the activation of the apical DNA damage-responsive kinase ATM as evidenced by normal levels of its autophosphorylation and phosphorylation of its target, CHK2 kinase (Figure 6G). The main trigger of ATM activity in FA-treated cells is chromatin damage in S phase.22 FA-induced replication stress stimulates ATR kinase that phosphorylates CHK1 kinase and the transcription factor p53.20 HSP90-inhibited cells showed normal levels of phospho-CHK1 but elevated amounts of phospho-p53 (Figure 6G), pointing to a selectively enhanced ATR signaling.
Figure 6.
Role of HSP90 in FA resistance. Cell viability was measured at 72 hours after heat shock or FA treatments. A: Impact of HSP90 (17-DMAG) and HSP70 (VER-155008) inhibitors on viability of heat shocked H460 cells (43°C, 40 minutes). Inhibitors were present during heat shock and for the first 4 hours of recovery. B and C: Cell viability (B) and clonogenic survival (C) of H460 cells treated with FA for 2 hours; 17-DMAG (0.2 μmol/L) was present during FA treatments and for the first 4 hours of recovery. D: Higher levels of K48-polyubiquitinated proteins in FA-treated H460 cells in the presence of 17-DMAG. E: K48-polyubiquitination of proteins in H460 cells treated with FA for 6 hours. F: Viability of H460 cells treated with FA for 6 hours in the absence or presence of 0.2 μmol/L 17-DMAG. G: Effects of 17-DMAG on ATM-dependent (left panel) and ATR-dependent (right panel) signaling events in FA-treated cells. Data are expressed as means ± SD. n = 3. ∗P < 0.05, ∗∗P < 0.01. ATM, ataxia-telangiectasia mutated; ATR, ataxia telangiectasia and Rad3 related; CHK, checkpoint kinase; FA, formaldehyde; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSP90, heat shock protein 90; K48-Ub, K48-polyubiquitination; 17-DMAG, 17-demethoxy-17-[[2-(dimethylamino)ethyl]amino]geldanamycin hydrochloride.
Discussion
Protein Damage and FA Toxicity
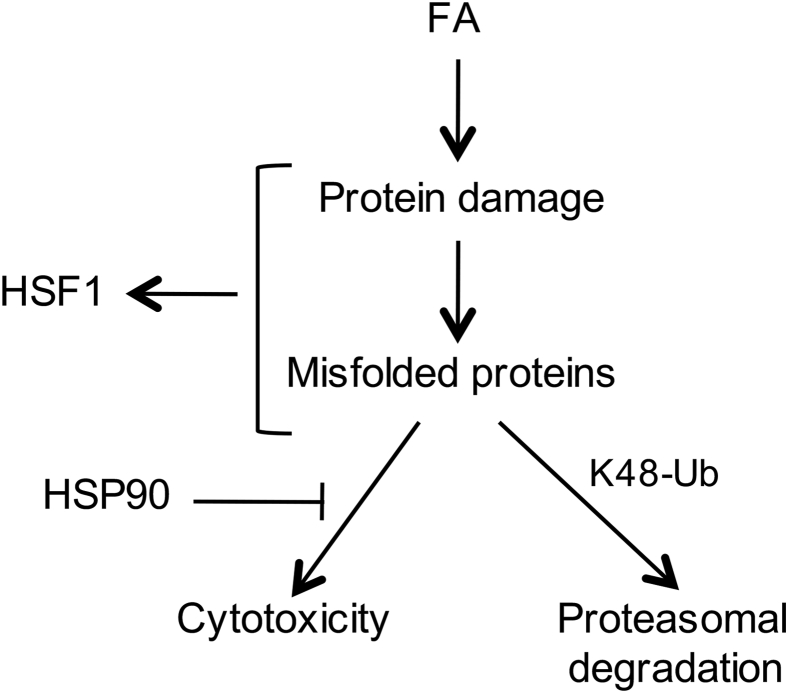
As summarized graphically (Figure 7), our results showed that FA is a potent proteotoxic chemical as evidenced by its induction of large amounts of K48-polyubiquitinated proteins, which are substrates for proteasomal degradation,27 and a rapid activation of the protein damage-sensitive transcription factor HSF1 in different types of human cells, including primary cells. The prominence of protein damage in FA-induced cellular injury was demonstrated by the detection of proteotoxic responses after brief, mildly cytotoxic exposures. A transient inhibition of HSP90 chaperone activity resulted in lower levels of cell viability, indicating the importance of the protein homeostatic mechanisms in FA resistance. FA has long been known and extensively used as a DNA-protein crosslinking agent. The formation of DPCs has generally been considered as the main cytotoxic form of cellular injury, and the kinetics of these genetic lesions has been used in human risk assessment.43 FA-induced and other forms of DPCs are potent blockers of DNA replication caused by stalling of ring-shaped replicative helicase complexes that are unable to slide through the sites of covalently attached proteins.20, 21 The genotoxic stress resulting from DPC-mediated replication blockage led to the stimulation of the damage-responsive kinase ATR that phosphorylated and activated the proapoptotic transcription factor p53 exclusively in the S-phase cells.20 Consequently, nonreplicating cells lack this DPC-dependent form of cytotoxicity. One of the main models for explanation of FA carcinogenicity is cytotoxicity-based regenerative proliferation.44, 45 In this mechanism, which is also used for risk assessment modeling, a critical parameter is the replicative state of cells, because only replicating cells were thought to experience cell death signaling caused by DPCs. Our results here showed that all FA-treated cells experience another major form of cytotoxic injury, namely, extensive protein damage and proteotoxic stress. Thus, induction of cell death by FA is not limited to replicating cells. If FA was only toxic to replicating cells, chronically exposed tissues should have shown degenerative pathology not regenerative proliferation, because FA would be killing only proliferating stem/progenitor cells but not differentiated cells. Direct exposures to FA or its production from ingested methanol are also associated with noncancer health effects, including neurotoxicity.6, 7, 8 Neurons are terminally differentiated cells that are known to be much more sensitive than other types of cells to a variety of proteotoxic conditions, such as the production of specific misfolded proteins,24 chronic exposure to low levels of cysteine-reactive metals,46, 47 or when cellular capacity for disposal of normally damaged/aged proteins is diminished.48 Proteotoxicity, therefore, fits well as a plausible cause of FA-associated neurotoxic effects. FA-damaged proteins can elicit cytotoxic effects via abnormal cellular biochemistry resulting from a direct inhibition of specific enzymes and structural defects in the multicomponent complexes. The formation of large amounts of damaged proteins can also potentially diminish the availability of the ubiquitin-proteasome system for removal of DNA-crosslinked proteins at high FA doses, which would indirectly enhance genotoxicity of FA. Proteolytic repair of FA-induced DPC has been shown to be a proteasome activity-dependent process.19, 23
Figure 7.
Summary of major findings on proteotoxicity of FA. FA conjugation to proteins results in their misfolding, which triggers activation of the protein damage-sensitive transcription factor HSF1. Accumulation of FA-damaged proteins causes cytotoxicity, which counteracted by the chaperone activity of HSP90. Misfolded proteins undergo K48-polyubiquitination and subsequent proteasomal degradation. FA, formaldehyde; HSF1, heat shock factor 1; HSP90 heat shock protein 90; K48-Ub, K48-polyubiquitination.
A loss of the Fanconi anemia proteins has been found to result in hypersensitivity of cells to toxic effects of FA in cultured cells and in vivo.11, 17, 18 The Fanconi pathway plays a major role in repair of interstrand DNA crosslinks, and its importance in FA resistance has also been assumed to be related to the involvement in repair of DNA damage. However, the absence of the hypersensitivity phenotype in the essential components of homologous recombination repair,18 which are all critical for resistance to DNA crosslinks, is inconsistent with the primary role of Fanconi anemia proteins in repair of FA-DNA damage. In addition to proteasomes, autophagy is another major cellular process for disposal of misfolded or otherwise damaged proteins.48 A recent discovery of the role of Fanconi pathway proteins in autophagy49 raises a possibility that FA hypersensitivity of cells lacking FANC genes could be related to the compromised removal of FA-damaged proteins via autophagy.
Proteotoxic Responses to FA
FA-treated cells showed accumulation of the proteasome-targeting K48-polyubiquitination of proteins and activation of HSF1, which is the main heat shock-responsive transcription factor in human and other mammalian cells.28, 29 Accumulation of K48-polyubiquitinated proteins in FA-treated cells reflected the presence of damaged proteins and not inhibition of deubiquitinases, as evidenced by the absence of changes in nonproteolytic K63-linked polyubiquitination. Inhibition of proteasomes also resulted in the buildup of K48-polyubiquitinated proteins, which preceded HSF1 activation, suggesting that HSF1 responded to the consequences of the inability of cells to digest ubiquitinated proteins. In the case of FA, HSF1 activation occurred earlier than the accumulation of polyubiquitinated proteins, indicating that HSF1 was responsive to the presence of protein damage and not the result of massive protein polyubiquitination. Although both proteasome inhibition and FA changed gel mobility of HSF1 indicative of its hyperphosphorylation,28, 30 FA caused a much stronger Ser326 phosphorylation, demonstrating another difference in HSF1 activation by proteasome inhibition and FA. Ser326 phosphorylation has been linked to a more efficient association of HSF1 with its coactivator death-domain associated protein.33 Heat shock-induced phosphorylation of Ser326 has been attributed to mammalian target of rapamycin complex 1.50 It remains to be determined whether the same kinase is responsible for this HSF1 modification in response to FA.
Activation of HSF1 involves several steps.28, 29 Under normal conditions, HSF1 resides as an inactive monomer in the cytoplasm where its activity is repressed via binding with the protein chaperones HSP90, HSP70, and HSP40. Accumulation of damaged/misfolded proteins during proteotoxic stress is thought to titrate away HSP90, allowing HSF1 to form homotrimers and move into the nucleus where it binds to promoters of heat shock-responsive genes. A full transactivation of HSF1 is associated with the dissociation of HSP70/HSP40 and hyperphosphorylation involving up to 12 amino acids. Other studies have also reported the importance of disulfide bridges in the formation of active HSF1 trimers, which is viewed as a direct sensing response for proteotoxic stressors.51, 52, 53 Both the classical chaperone displacement model and the intrinsic response mechanism can also be involved in the activation of HSF1 by FA. Accumulation of damaged proteins by FA exposure was evident from the massive presence of K48-polyubiquitinated proteins, whereas the heightened sensitivity of cells to FA cytotoxicity by HSP90 inhibition demonstrated the importance of this chaperone in stabilization of cellular proteins. As a crosslinking agent with a strong affinity for SH-groups, FA can also potentially cause crosslinking of HSF1 monomers via methylene bridging of their cysteines [monomer(A)-S-CH2-S-monomer(B)]. Testing the involvement of this crosslinking mechanism is likely to represent a significant technical challenge due to hydrolytic instability of S-adducted FA.
Footnotes
Supported by NIH R01 grant ES020689 (A.Z.).
Disclosures: None declared.
Current address of B.R., Department of Clinical Chemistry and Molecular Diagnostics, Poznan University of Medical Sciences, Poznan, Poland.
References
- 1.IARC (International Agency for Research on Cancer) Formaldehyde. IARC Monogr Eval Carcinog Risks Hum. 2006;88:39–325. [Google Scholar]
- 2.Hauptmann M., Stewart P.A., Lubin J.H., Beane Freeman L.E., Hornung R.W., Herrick R.F., Hoover R.N., Fraumeni J.F., Jr., Blair A., Hayes R.B. Mortality from lymphohematopoietic malignancies and brain cancer among embalmers exposed to formaldehyde. J Natl Cancer Inst. 2009;101:1696–1708. doi: 10.1093/jnci/djp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwilk E., Zhang L., Smith M.T., Smith A.H., Steinmaus C. Formaldehyde and leukemia: an updated meta-analysis and evaluation of bias. J Occup Environ Med. 2010;52:878–886. doi: 10.1097/JOM.0b013e3181ef7e31. [DOI] [PubMed] [Google Scholar]
- 4.Duong A., Steinmaus C., McHale C.M., Vaughan C.P., Zhang L. Reproductive and developmental toxicity of formaldehyde: a systematic review. Mutat Res. 2011;728:118–138. doi: 10.1016/j.mrrev.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Toxicology Program Final report on carcinogens background document for formaldehyde. Rep Carcinog Backgr Doc. 2010;(10-5981):i–512. [PubMed] [Google Scholar]
- 6.Pitten F.A., Kramer A., Herrmann K., Bremer J., Koch S. Formaldehyde neurotoxicity in animal experiments. Pathol Res Pract. 2000;196:193–198. doi: 10.1016/S0344-0338(00)80100-4. [DOI] [PubMed] [Google Scholar]
- 7.Paasma R., Hovda K.E., Jacobsen D. Methanol poisoning and long term sequelae - a six years follow-up after a large methanol outbreak. BMC Clin Pharmacol. 2009;9:5. doi: 10.1186/1472-6904-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy N.J., Sudini M., Lewis L.D. Delayed neurological sequelae from ethylene glycol, diethylene glycol and methanol poisonings. Clin Toxicol (Phila) 2010;48:967–973. doi: 10.3109/15563650.2010.532803. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Oberbeck N., Langevin F., King G., de Wind N., Crossan G.P., Patel K.J. Maternal aldehyde elimination during pregnancy preserves the fetal genome. Mol Cell. 2014;55:807–817. doi: 10.1016/j.molcel.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pontel L.B., Rosado I.V., Burgos-Barragan G., Garaycoechea J.I., Yu R., Arends M.J., Chandrasekaran G., Broecker V., Wei W., Liu L., Swenberg J.A., Crossan G.P., Patel K.J. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell. 1978;15:945–954. doi: 10.1016/0092-8674(78)90278-7. [DOI] [PubMed] [Google Scholar]
- 13.Solomon M.J., Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Graaf B., Clore A., McCullough A.K. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst) 2009;8:1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano T., Katafuchi A., Matsubara M., Terato H., Tsuboi T., Masuda T., Tatsumoto T., Pack S.P., Makino K., Croteau D.L., Van Houten B., Iijima K., Tauchi H., Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noda T., Takahashi A., Kondo N., Mori E., Okamoto N., Nakagawa Y., Ohnishi K., Zdzienicka M.Z., Thompson L.H., Helleday T., Asada H., Ohnishi T. Repair pathways independent of the Fanconi anemia nuclear core complex play a predominant role in mitigating formaldehyde-induced DNA damage. Biochem Biophys Res Commun. 2011;404:206–210. doi: 10.1016/j.bbrc.2010.11.094. [DOI] [PubMed] [Google Scholar]
- 17.Ridpath J.R., Nakamura A., Tano K., Luke A.M., Sonoda E., Arakawa H., Buerstedde J.M., Gillespie D.A., Sale J.E., Yamazoe M., Bishop D.K., Takata M., Takeda S., Watanabe M., Swenberg J.A., Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 18.Rosado I.V., Langevin F., Crossan G.P., Takata M., Patel K.J. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 19.Ortega-Atienza S., Green S.E., Zhitkovich A. Proteasome activity is important for replication recovery, CHK1 phosphorylation and prevention of G2 arrest after low-dose formaldehyde. Toxicol Appl Pharmacol. 2015;286:135–141. doi: 10.1016/j.taap.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong V.C., Cash H.L., Morse J.L., Lu S., Zhitkovich A. S-phase sensing of DNA-protein crosslinks triggers TopBP1-independent ATR activation and p53-mediated cell death by formaldehyde. Cell Cycle. 2012;11:2526–2537. doi: 10.4161/cc.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duxin J.P., Dewar J.M., Yardimci H., Walter J.C. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega-Atienza S., Wong V.C., DeLoughery Z., Luczak M.W., Zhitkovich A. ATM and KAT5 safeguard replicating chromatin against formaldehyde damage. Nucleic Acids Res. 2016;44:198–209. doi: 10.1093/nar/gkv957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quievryn G., Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 24.Morimoto R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg A.L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 26.Kriegenburg F., Ellgaard L., Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2012;279:532–542. doi: 10.1111/j.1742-4658.2011.08456.x. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda F., Crosetto N., Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Anckar J., Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 29.Home T., Jensen R.A., Rao R. Heat shock factor 1 in protein homeostasis and oncogenic signal integration. Cancer Res. 2015;75:907–912. doi: 10.1158/0008-5472.CAN-14-2905. [DOI] [PubMed] [Google Scholar]
- 30.Mathew A., Mathur S.K., Jolly C., Fox S.G., Kim S., Morimoto R.I. Stress-specific activation and repression of heat shock factors 1 and 2. Mol Cell Biol. 2001;21:7163–7171. doi: 10.1128/MCB.21.21.7163-7171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds M.F., Peterson-Roth E.C., Bespalov I.A., Johnston T., Gurel V.M., Menard H.L., Zhitkovich A. Rapid DNA double-strand breaks resulting from processing of Cr-DNA cross-links by both MutS dimers. Cancer Res. 2009;69:1071–1079. doi: 10.1158/0008-5472.CAN-08-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong V.C., Morse J.L., Zhitkovich A. p53 activation by Ni(II) is a HIF-1alpha independent response causing caspases 9/3-mediated apoptosis in human lung cells. Toxicol Appl Pharmacol. 2013;269:233–239. doi: 10.1016/j.taap.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boellmann F., Guettouche T., Guo Y., Fenna M., Mnayer L., Voellmy R. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc Natl Acad Sci U S A. 2004;101:4100–4105. doi: 10.1073/pnas.0304768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto H., Wang X., Ohnishi T. Binding between wild-type p53 and hsp72 accumulated after UV and gamma-ray irradiation. Cancer Lett. 1995;92:127–133. doi: 10.1016/0304-3835(95)03769-s. [DOI] [PubMed] [Google Scholar]
- 35.Ohnishi K., Matsumoto H., Takahashi A., Wang X., Ohnishi T. Heat shock transcription factor, HSF, is activated by ultraviolet irradiation. Photochem Photobiol. 1996;64:949–952. [PubMed] [Google Scholar]
- 36.Brinkmann K., Schell M., Hoppe T., Kashkar H. Regulation of the DNA damage response by ubiquitin conjugation. Front Genet. 2015;6:98. doi: 10.3389/fgene.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson S.P., Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 38.Kampinga H.H., Craig E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozen O.A., Akpolat N., Songur A., Kus I., Zararsiz I., Ozacmak V.H., Sarsilmaz M. Effect of formaldehyde inhalation on Hsp70 in seminiferous tubules of rat testes: an immunohistochemical study. Toxicol Ind Health. 2005;21:249–254. doi: 10.1191/0748233705th235oa. [DOI] [PubMed] [Google Scholar]
- 40.Wang M., Kaufman R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 41.Lim S.K., Choi H., Park M.J., Kim D.I., Kim J.C., Kim G.Y., Jeong S.Y., Rodionov R.N., Han H.J., Yoon K.C., Park S.H. The ER stress-mediated decrease in DDAH1 expression is involved in formaldehyde-induced apoptosis in lung epithelial cells. Food Chem Toxicol. 2013;62:763–769. doi: 10.1016/j.fct.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 42.Aldridge J.E., Horibe T., Hoogenraad N.J. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramaniam R.P., Chen C., Crump K.S., Devoney D., Fox J.F., Portier C.J., Schlosser P.M., Thompson C.M., White P. Uncertainties in biologically-based modeling of formaldehyde-induced respiratory cancer risk: identification of key issues. Risk Anal. 2008;28:907–923. doi: 10.1111/j.1539-6924.2008.01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conolly R.B., Kimbell J.S., Janszen D.B., Miller F.J. Dose response for formaldehyde-induced cytotoxicity in the human respiratory tract. Regul Toxicol Pharmacol. 2002;35:32–43. doi: 10.1006/rtph.2001.1515. [DOI] [PubMed] [Google Scholar]
- 45.Conolly R.B., Kimbell J.S., Janszen D., Schlosser P.M., Kalisak D., Preston J., Miller F.J. Human respiratory tract cancer risks of inhaled formaldehyde: dose-response predictions derived from biologically-motivated computational modeling of a combined rodent and human dataset. Toxicol Sci. 2004;82:279–296. doi: 10.1093/toxsci/kfh223. [DOI] [PubMed] [Google Scholar]
- 46.Aschner M., Onishchenko N., Ceccatelli S. Toxicology of alkylmercury compounds. Met Ions Life Sci. 2010;7:403–434. doi: 10.1039/BK9781847551771-00403. [DOI] [PubMed] [Google Scholar]
- 47.Mendez-Armenta M., Rios C. Cadmium neurotoxicity. Environ Toxicol Pharmacol. 2007;23:350–358. doi: 10.1016/j.etap.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Menzies F.M., Fleming A., Rubinsztein D.C. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 49.Sumpter R., Jr., Sirasanagandla S., Fernandez A.F., Wei Y., Dong X., Franco L., Zou Z., Marchal C., Lee M.Y., Clapp D.W., Hanenberg H., Levine B. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165:867–881. doi: 10.1016/j.cell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou S.D., Prince T., Gong J., Calderwood S.K. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One. 2012;7:e39679. doi: 10.1371/journal.pone.0039679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn S.G., Thiele D.J. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17:516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y., Zhang C., Chen J., Guo L., Li X., Li W., Yu Z., Deng J., Zhang P., Zhang K., Zhang L. Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol Biochem. 2013;64:92–98. doi: 10.1016/j.plaphy.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 53.Zhong M., Orosz A., Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]